Abstract
Epithelial cells lining the adult colon do not normally express gastrin-releasing peptide (GRP) or its receptor (GRPR). In contrast, GRP/GRPR can be aberrantly expressed in colon cancer where they are associated with improved patient survival rates. However, the mechanism of action whereby these proteins mediate their beneficial effects is not known. Heterochromatin protein 1 is an epigenetic modifier of gene transcription for which three different isoforms exist in humans: HP1Hsα, HP1Hsβ, and HP1Hsγ. In breast cancer and melanoma, respectively, HP1Hsα and HP1Hsβ have been shown to modulate the aggressiveness of tumor cells in vivo. In contrast, the role of HP1 in colon cancer has not been elucidated, and a mechanism of regulating the expression of any HP1 isoform in any context has not yet been identified. In this article we demonstrate that abrogating GRP/GRPR signaling specifically down-regulates HP1Hsβ expression and that inhibiting GRPR signaling, or ablating HP1Hsβ expression, increases colon cancer cell invasiveness in vitro. These findings identify for the first time a signaling pathway regulating heterochromatin protein expression and suggest a mechanism whereby aberrantly expressed GRPR might alter the outcome of patients with colorectal cancer.
Epithelial cells lining the colon express gastrin-releasing peptide (GRP) and its receptor (GRPR) only during gut development where they contribute, albeit modestly, to organogenesis.1 In contrast, both proteins can be aberrantly expressed in the colon after malignant transformation; under these circumstances, GRP/GRPR expression is associated with increased patient survival rates and decreased lymph node metastases.2 Yet the mechanism whereby GRP/GRPR mediates its beneficial effects remains to be determined. In this article, we present evidence that GRPR signaling attenuates colon cancer invasiveness, a critical step in metastasis, by altering the expression of one of the members of the heterochromatin protein 1 (HP1) family.
In mammals, three different HP1 isoforms have been identified; in humans they are HP1Hsα, HP1Hsβ, and HP1Hsγ.3,4 Although located primarily within heterochromatin, HP1 proteins are also present in euchromatin, where they act to repress gene transcription by interacting with methylated histones.5 Most studies of HP1 have focused primarily on its role in gene silencing in the context of normal development,3,6 but recent studies suggest that the degree to which various isoforms are expressed in cancer can modify tumor cell behavior in ways that improve patient survival rates.7 Consequently, modulating HP1 isoform expression has been proposed as a potential therapeutic approach for various types of cancer.8
However, no study to date has identified a mechanism whereby any HP1 isoform is expressed or activated in cancer. We have shown that inhibiting GRP/GRPR signaling in human colon cancer cell lines specifically down-regulates HP1Hsβ expression, and that down-regulating HP1Hsβ expression increases the invasiveness of human colon cancer cell lines. These data identify a hitherto unappreciated mechanism whereby aberrantly expressed GRP/GRPR might improve the outcome of patients with colon cancer and identify a mechanism for regulating HP1Hsβ expression that might have therapeutic implications for the treatment of colon cancer.
Materials and Methods
Materials
Anhydrous ethanol (100%, 95%) and xylene were purchased from Pharmco (Brookfield, CT). Wash buffer, target retrieval solution, protein block serum, antibody diluent, LSAB2 System-HRP, liquid DAB substrate, chromogen system, EnVision+ HRP (DAB)-Rabbit System, and automated hematoxylin were obtained from DAKO (Carpentaria, CA). Auto/Iodine, Redusol, and PVDF membranes came from Fisher Scientific (Pittsburgh, PA). Chamber slides came from Nunc (Roskilde, Denmark), and QCM collagen wells came from Corning (Lowell, MA). Mammalian protease inhibitor cocktail and GRPR-specific antagonist RC-30959,10 were purchased from Sigma (St. Louis, MO). Matrigel came from Becton-Dickenson (Franklin Lakes, NJ). Anti-HP1Hsα is a rabbit polyclonal antibody directed to residues 1 and 100 of the human protein. Anti-HP1Hsβ is a rabbit polyclonal antibody directed between residues 150 and the C terminus of the human, mouse, and marsupial protein. Anti-HP1Hsγ is a rabbit polyclonal antibody directed between residues 1 and 100 of the human protein. Antibodies were purchased from AbCAM (Cambridge, MA). All cells were all obtained from ATCC (Rockville, MD) and maintained as recommended. Caco-2 and LS-174T cells are human colon epithelial cancer cells. siRNA targeted to the relevant mRNA was obtained from Ambion (Austin, TX) and used as directed.
RNA Detection by RT-PCR
Total RNA was isolated from the relevant cells using RNA-Stat 60 according to the manufacturer's specifications. Chloroform was added to solutions before they were centrifuged at 12,000×g for 15 minutes at 4°C. The upper aqueous layer was retrieved and mixed with isopropranol and subsequently centrifuged at 12,000×g for 10 minutes at 4°C. The pellet was washed with 75% ethanol, centrifuged at 7500×g for 5 minutes at 4°C, air-dried, and resuspended in water. mRNA was isolated using Qiagen (Valencia, CA) and Invitrogen (Carlsbad, CA) kits according to manufacturers' instructions. To amplify human HP1Hsα we used forward primer 5′-ATCGCTCGGGGCTTTGAGAGAG-3′ and reverse primer 5′-TTCCGCATCCTCAGGATATGCAT-3′. Human HP1Hsβ was amplified using forward primer 5′-AAGCTGGCGGGCACTATGGG-3′ and reverse primer 5′-GGGGCTGGTACTCAGGAGCG-3′. To amplify human HP1Hsγ we used forward primer 5′-GAGGCAGAGCCTGAGAATTTGTC-3′ and reverse primer 5′-TTTGCTGTCATCAGATTCACTGTC-3′. Actin was amplified using forward primer 5′-ATGGAAGAAGAGATCGC-3′ and reverse primer 5′-GGATGCCACGCTTGCTC-3′. PCR amplification was carried out for 35 cycles at 94°C for 30 seconds, 62°C for 30 seconds, and 72°C for 30 seconds.
Western Blot Analysis
Cells were grown to at least 60% confluence. Thereafter, cell monolayers were rinsed in PBS and lysed in ice-cold RIPA (50 mmol/L HEPES, pH 7.4; 150 mmol/L NaCl; 1% Triton X-10; 0.1% SDS; 0.5% sodium deoxycholate; 1 mmol/L sodium orthovanadate; 5 mmol/L EDTA; 5 mmol/L sodium fluoride) containing a 1:20 dilution of mammalian protease inhibitor cocktail (Sigma). In all instances, 200 μg of total cell protein were loaded into a preparative well and electrophoresed across a 12% polyacrylamide gel under denaturing and reducing conditions. Membranes were incubated with primary antibody for 2 hours at the following concentrations: actin 1:50, HP1Hsα 1:250, HP1Hsβ 1:750, and HP1Hsγ 1:500. This was followed by two 10-minute washes with TBST with immunoreactive bands that were visualized using HRP-conjugated goat anti-rabbit IgG and the ECL Plus detection system.
Immunohistochemistry
Confocal Microscopy
Cultured cells were seeded on polycarbonate membrane transwell inserts at various densities for 24 (preconfluent cells), 48 (preconfluent cells), or 168 hours (postconfluent cells). All cells were fixed in PBS containing 3.7% formaldehyde, washed with PBS for 5 minutes, and permeabilized for 2 minutes using 0.2% Triton-X100. Membranes were blocked with normal goat serum (5%) for 30 minutes at room temperature. Membranes were then incubated with primary antibody solution (all antibodies 1 mg/ml) at 1:100 overnight at 4°C in a humidity chamber. Membranes were then rinsed with PBS for 15 minutes and incubated with Alexa 488 goat anti-rat IgG for 60 minutes. In all instances, images were obtained using a Zeiss LSM 510 confocal microscope (New York, NY). Images were obtained at 1-μm increments using 40× plan neoflux objectives attached to a Zeiss LSM 510 confocal microscope.
Immunoperoxidase
A modified indirect immunoperoxidase technique was performed on 4-μm-thick paraffin-embedded sections that were hydrated in graded alcohols and rinsed in a running water bath. Slides were incubated for 5 minutes in a 3% hydrogen peroxide solution to quench endogenous peroxidase activity. Mercuric pigments were removed by incubating in auto/iodine for 1 minute, followed by two incubations in Redusol for 2 minutes each. Hydration was completed by rinsing three times in DAKO 1X wash buffer for 3 minutes. Antigen retrieval was accomplished by submerging tissue sections in 1X DAKO target retrieval solution for 30 minutes at 100°C in a steamer. Slides were allowed to cool to room temperature and rinsed in 1X DAKO wash buffer. Sections were blocked for 15 minutes with protein block serum and then incubated with antibodies to GRP (1:125); GRPR (1:750); or HP1Hsβ (1:750) overnight at 4°C. After rinsing with wash buffer, slides were incubated with Streptavidin conjugated to horseradish peroxidase (LSAB2 kit from DAKO) for 10 minutes. Slides were then rinsed and incubated with a liquid DAB substrate-chromogen system for 5 minutes to identify bound antibody. After a final rinse in 1X wash buffer, all slides were counterstained with DAKO automated hematoxylin for 2 minutes, dehydrated in alcohol, and mounted with a coverslip using Permount. Control tissues were processed simultaneously with the treated slides, with the exception that primary antibody was not applied.
Quantitative Immunohistochemistry
Chromogen abundance was quantified by determining the cumulative signal strength of the digital image file of the relevant region or cell of interest as previously described.11–15 For all specimens, control tissues were processed identically and at the same time, except that primary antibody was not applied. Control tissues were within 5 μm of the tested tissue and treated with secondary antibody, DAB, and counterstained with hematoxylin for precisely the same amount of time as the experimental tissue. All differences between the experimental tissue and the control tissue were ultimately related to DAB identification of the relevant protein. The amount of chromogen per pixel was determined by subtracting the mathematical “energy” (EM), or chromogen quantity of the control slide (ie, not exposed to primary antibody) from that in the homologous region of the experimental slide (ie, exposed to primary antibody). EM is expressed in the arbitrary values of energy units per pixels (eu/pix). Maximal and half-maximal Em values were determined after curve-fitting the data by least-squares nonlinear regression.
Cell Invasion
The number of cells invading was assessed using a modified Boyden chamber assay using QCM collagen wells (Corning) as previously described.16,17 Briefly, collagen-coated membranes with 8-μm diameter pores were plated with 5 × 105 cells in 500 μL. Those exposed to the GRPR-specific antagonist RC-3095 were exposed to peptide hormone for 24 hours before plating on collagen-coated membranes. In contrast, those exposed to siRNA were harvested and plated on the membranes 48 hours after transfection. In all instances, cell migration was assessed 5 hours after plating, as recommended by the manufacturer.
Results
GRP/GRPR Signaling Specifically Regulates HP1Hsβ Expression
We previously showed that GRP/GRPR signaling in colon cancer cell lines altered the protein level of an unspecified member of the HP1 family.18 To determine the specific HP1 isoform involved, we cultured GRP/GRPR-expressing preconfluent Caco-2 cells (Figure 1A) or LS-174T cells (data not shown) in the absence or presence of the GRPR-specific antagonist RC-30959,10 for 24 hours. RC-3095 is a potent GRPR-specific competitive antagonist, meaning that it binds with high affinity to this receptor without initiating signaling.9,10 The expression of HP1Hsα, HP1Hsβ, and HP1Hsγ were examined at the mRNA level by RT-PCR, and at the protein level by Western blotting and immunofluorescence. Whereas preconfluent cells expressed all three mRNA HP1 isoforms, exposure to GRPR antagonist specifically eliminated HP1Hsβ transcripts without affecting HP1Hsα or HP1Hsγ (Figure 1A).
Figure 1.
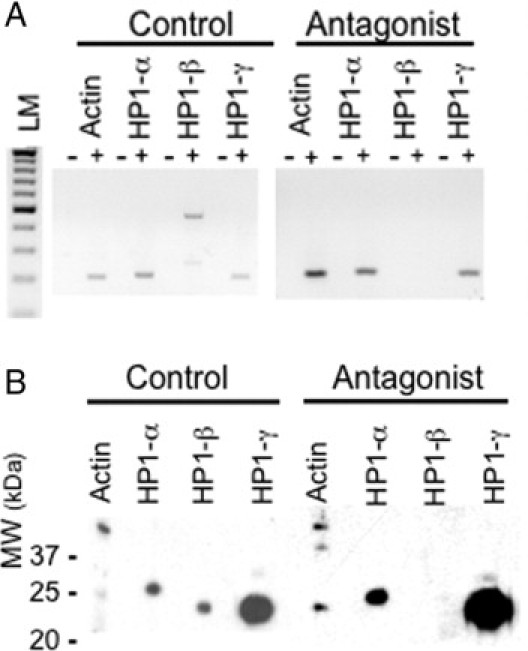
HP1 mRNA expression (A) and protein (B) expression as a function of GRP/GRPR signaling. Preconfluent Caco-2 cells expressing GRP/GRPR were incubated in the absence (control) or the presence of the specific GRPR antagonist RC-3095 (10 μmol/L, antagonist) for 24 hours. A: RNA was isolated and RT-PCR performed using gene-specific primers as described in Materials and Methods; LM, 100 bp lane marker. B: Total proteins were isolated, resolved by SDS-PAGE, transferred to nitrocellulose, and probed with the indicated antibody using the Mini-Protean II Multi-Screen, allowing for identical amounts of protein to be probed simultaneously with different antibodies. Symbols in A identify samples that were (+) or were not (−) exposed to reverse transcriptase.
To confirm that these changes were also observed at the level of protein expression, we performed Western blot analysis using specific antibodies. Proteins were isolated from preconfluent GRP/GRPR-expressing Caco-2 (Figure 1B) and LS-174T (data not shown) in the presence or absence of the GRPR-specific antagonist RC-3095. As previously described,15 cells were exposed to antagonist for 24 hours and the proteins extracted and evaluated by Western blot. Using specific HP1 antibodies, we determined that GRPR antagonist eliminated expression of HP1Hsβ without altering the presence of HP1Hsα or HP1Hsγ (Figure 1B).
Immunohistochemical assays further confirmed the dependence of HP1Hsβ expression on GRP/GRPR signaling. We cultured GRP/GRPR-expressing preconfluent Caco-2 cells (Figure 2) or LS-174T cells (data not shown) in the absence (Figure 2, A–C) or presence (Figure 2, D–F) of the GRPR-specific antagonist RC-3095 for 24 hours. In the absence of antagonist, all HP1 isoforms were detected in the nuclei of preconfluent GRP/GRPR-expressing cells (Figure 2, A–C). In contrast, exposure to the GRPR-specific antagonist RC-3095 for 24 hours specifically eliminated the expression of HP1Hsβ in preconfluent GRP/GRPR-expressing Caco-2 cells (Figure 2E) without altering the expression or location of HP1Hsα (Figure 2D) and HP1Hsγ (Figure 2F). These data show that GRP/GRPR signaling is essential for the transcription of HP1Hsβ, but not HP1Hsα or HP1Hsγ, in human colorectal cancer cells.
Figure 2.
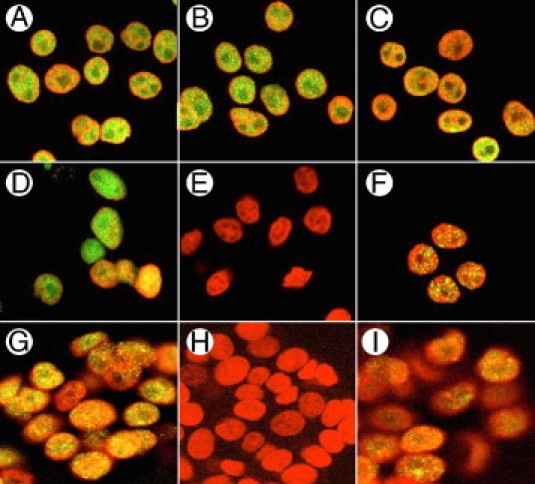
A–C: HP1 expression in Caco-2 cell nuclei in the absence of the GRPR-specific antagonist; D–F: HP1 expression in Caco-2 cell nuclei in the presence of the GRPR-specific antagonist RC-3095; G–I: siRNA directed to the GRPR. As described in Materials and Methods, cells were treated with 10 μmol/L RC-3095 every 12 hours for 24 hours, or to GRPR siRNA for 48 hours, and then exposed to antibodies for HP1Hsα (A, D, G); HP1Hsβ (B, E, H); or HP1Hsγ (C, F, I). They were then counterstained with DAPI (red) to identify nuclei.
We assessed HP1Hsβ expression in human colorectal cancer cells and determined whether this expression correlated with the amount of GRP/GRPR present in the same tissues. Using an approach identical to that which we have described previously,15,19 we randomly selected 10 human colorectal cancers from our gastrointestinal tissue bank and evaluated consecutive 4-μm-thick sections for GRP, GRPR, and HP1Hsβ expression. The tumors contained 25 separate and distinct well-differentiated regions, 16 moderately differentiated regions, and 20 poorly differentiated regions. For each region of distinct differentiation stages, between approximately 50 and 2000 cells were evaluated. Similar to results we have shown previously,2,15,19,20 GRP/GRPR were not detected in nonmalignant cells lining the colon but were present in colorectal cancer as a function of tumor cell differentiation (data not available). The greatest amount of GRP/GRPR was found in well-differentiated colon cancer cells; 340 ± 25 eu/pixel were detected in the non-nuclear regions of the cell. Similarly, the greatest amount of HP1Hsβ immunostaining was present in the nuclei of well-differentiated colon cancer cells (Figure 3B), with 171 ± 19 eu/pixel present. In contrast to what was observed for GRP/GRPR, HP1Hsβ immunostaining was also detectable in the nuclei of normal colonic cells (Figure 3A), but at levels of approximately 80% of that detectable in well-differentiated cancer cells (Figure 3D), whereas significantly less was detected in the nuclei of moderately differentiated cancer cells, and none was detected in poorly differentiated (Figure 3C) tumor cells. Excepting that which was present in normal colonic tissues, the amount of HP1Hsβ immunostaining strongly correlated with the amount of GRP/GRPR expressed in colon cancer cells (r2 = 0.975) (Figure 3D). However, our observation that HP1Hsβ was also expressed in the nuclei of normal colonic epithelial cells (Figure 3A), which do not express GRP/GRPR, also indicates that this peptide hormone and its cognate receptor do not regulate expression exclusively.
Figure 3.
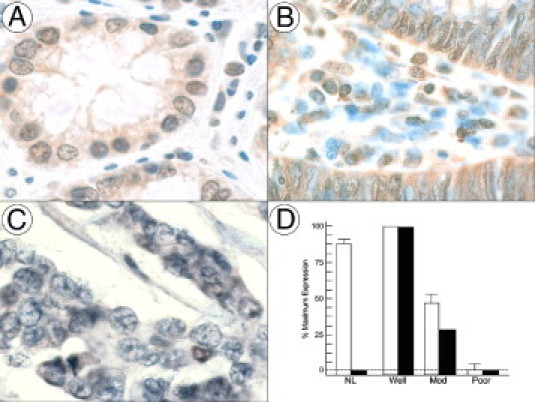
A: HP1Hsβ expression in the nuclei of normal adult human colon; B: HP1Hsβ expression in well-differentiated colon cancers; C: HP1Hsβ expression in poorly differentiated colon cancers. D: Quantification of the amount of HP1Hsβ (open bars) and GRP/GRPR (solid bars) present in normal tissues (NL) and in colon cancers of indicated differentiation (well, well differentiated; mod, moderately differentiated; poor, poorly differentiated). Tissues were processed as described in Materials and Methods, and images were acquired at ×400. Brown, HP1Hsβ; blue, nuclei.
Time-Course of HP1Hsβ Expression
To determine the time-course of HP1Hsβ down-regulation after blocking GRPR signaling, we exposed cells to the GRPR-specific antagonist RC-3095 at a concentration of 10 μmol/L (Figure 4). This concentration was determined to be the minimum dose required to eliminate ICAM1 expression (data not shown), the expression of which is GRPR-dependent.15 Whereas HP1Hsβ RNA was readily detectable by RT-PCR 20 hours after exposure to an antagonist, it was not detectable 4 hours later (data not shown). In contrast, whereas roughly 75% of HP1Hsβ protein remained present 16 hours after initial exposure to antagonist, only about 40% remained present 4 hours later (Figure 4A). After 24 hours of exposure to antagonist, HP1Hsβ protein could no longer be detected (Figure 4C).
Figure 4.
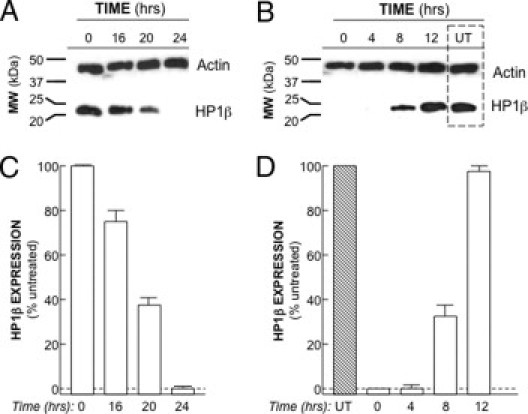
Time-course of HP1Hsβ protein expression. A: Caco-2 cells were exposed to the GRPR-specific antagonist RC-3095 (10 μmol/L every 12 hours) for the indicated times, followed by protein extraction and Western analysis using antibodies to the indicated proteins. B: Cells were exposed to GRPR antagonist for 24 hours, washed, and then cultured in antagonist-free media for the indicated times, followed by protein extraction and Western analysis using antibodies to the indicated proteins. UT, untreated (ie, never exposed to antagonist). C: The amount of HP1Hsβ at the times shown in A, and expressed as a percentage of that present in the absence of antagonist. The data are presented as means ± SE for a minimum of three separate experiments. D: The amount of HP1Hsβ at the indicated times, as shown in B, and expressed as a percentage of that present in UT cells. The data are presented as means ± SE for a minimum of three separate experiments.
To examine the time-course of HP1Hsβ up-regulation, cells were exposed to GRPR antagonist RC-3075 for 24 hours, then washed ×2 in PBS, followed by exposure to standard media for the indicated time points. For the first 4 hours after removal of antagonist HP1Hsβ, RNA (data not shown) and protein (Figure 4, B and D) could not be detected. HP1Hsβ RNA was first detected 6 hours after removal of the antagonist (data not shown), and 2 hours later the amount of HP1Hsβ protein was at levels that were approximately 35% of those noted in untreated cells (ie, those not exposed to antagonist; Figure 4B). Twelve hours after removal of antagonist HP1Hsβ, protein levels were similar to those observed in untreated cells (Figure 4, B and D).
Elimination of HP1Hsβ Using siRNA
To assess the contribution of HP1Hsβ to the alteration of colon cancer cell behavior, we eliminated its expression using siRNA in both Caco-2 (Figure 5) and LS-174T (data not shown) cells. We first assessed the specificity of HP1Hsβ siRNA. Transfecting cells with 45 μg siRNA for 48 hours resulted in a complete elimination of HP1Hsβ expression at both the RNA (Figure 5A) and protein (Figure 5B) levels. The expression of HP1Hsα and HP1Hsγ was not affected. We next determined that the duration of HP1Hsβ protein expression was eliminated by using siRNA. After 24, 48, and 72 hours of exposure to HP1Hsβ siRNA, the protein level of HP1Hsβ in treated cells was 8%, 0%, and 1% of that in untreated cells, respectively (Figure 5C). These data show that siRNA for HP1Hsβ specifically decreases this protein's expression for up to 72 hours post-transfection and completely eliminates expression 48 hours after transfection.
Figure 5.
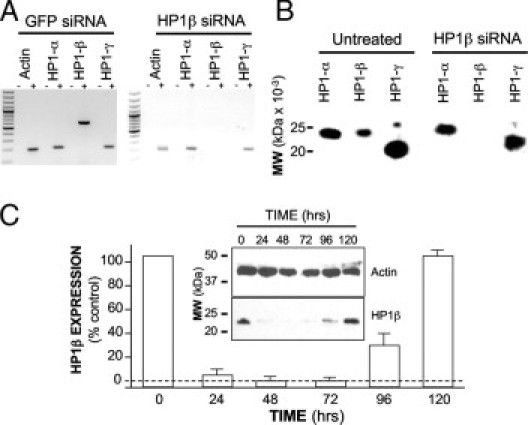
HP1Hsβ mRNA and protein expression are specifically eliminated by siRNA. Caco-2 cells were treated with HP1Hsβ siRNA, as described in Materials and Methods; 48 hours later the expression of all HP1 family members was assessed at the RNA level by RT-PCR (A) and at the protein level by Western analysis (B). C: The time span of decreased HP1Hsβ protein expression was determined by performing Western analysis at the indicated time points after siRNA exposure. +, samples that were exposed to reverse transcriptase; −, samples that were not exposed to reverse transcriptase. SiRNA to Green Fluorescent Protein (GFP) in A was used as a negative control. Data are means ± SE for a minimum of three separate experiments.
Effect of HP1Hsβ on Cell Invasion
Recent studies of breast cancer cell lines revealed an inverse correlation between HP1Hsα expression and tumor cell aggressiveness21 and showed that modulating the expression of this protein directly affects invasiveness.22 We therefore investigated whether GRPR-mediated alterations in HP1Hsβ expression might likewise alter colon cancer cell motility and present as increased invasiveness. To do this, we specifically determined the number of cells that migrated into collagen-coated membranes in a modified Boyden chamber.
The modified Boyden chamber assay was used as previously described.16,17 When 5 × 105 GRP/GRPR-expressing Caco-2 cells/500 μL were plated on collagen-coated membranes, 15.0 to 2.4 cells per high-powered field were able to traverse the 8-μm pores (n = 32) (Figure 6, A and C). In contrast, exposure to the GRPR antagonist RC-3095 increased the number of cells traversing the membrane by nearly 2.6- to 0.6-fold (Figure 6, B and C) (P < 0.05, paired t-test). Exposing cells to GRPR RNA interference likewise increased the number of cells invading by 2.5- to 0.4-fold, whereas those knocked down for HP1Hsβ increased the number invading by 1.9- to 0.3-fold (Figure 6C) (P < 0.05, respectfully). Similar results were observed with LS-174T cells. At baseline, 29.1 ± 4.6 LS-174T cells per high-powered field traversed the membrane (n = 24) (Figure 6C). Similar to what was noted for Caco-2 cells, exposure to the GRPR antagonist RC-3095 increased the number of cells traversing the membrane by 1.5- to 0.3-fold (P < 0.05) (Figure 6C). Exposing LS-174T cells to GRPR RNA interferences likewise increased the number of cells invading by 1.5- to 0.2-fold (P < 0.05) whereas those exposed to RNA interference for HP1 invaded 2.0- to 0.2-fold (P < 0.05) (Figure 6C).
Figure 6.
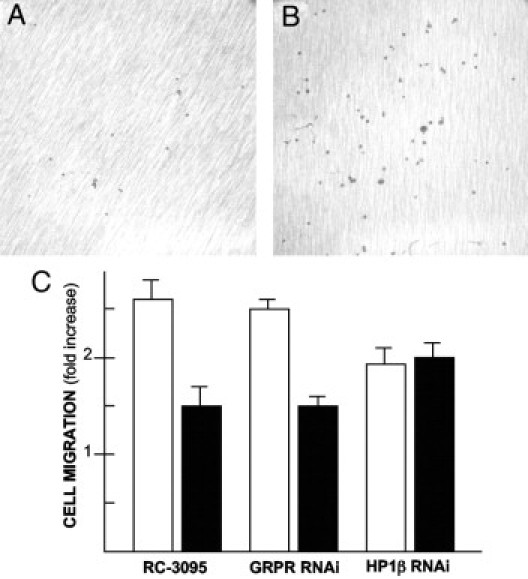
Cell invasion through collagen-coated membranes. A: GRP/GRPR-expressing Caco-2 cells in the absence of any additional treatment 5 hours after plating on the membrane's other side. B: Caco-2 cells after treatment with the GRPR-specific antagonist RC-3095 (10 μmol/L) every 12 hours for 24 hours before plating on membranes, and evaluating them 5 hours thereafter. C: Data for Caco-2 cells (closed bars) and LS-174T cells (open bars) in the absence of treatment or after exposure to GRPR siRNA (GRPR-RNAi), GRPR antagonist (RC-3095), or HP1Hsβ siRNA (HP1β RNAi). Data are expressed as cells per high-powered field (100), with three separate such fields averaged per experiment. Data are means ± SE for a minimum of three separate experiments. All conditions were significantly different from those of control cells processed in parallel (P < 0.05, paired t-test).
Discussion
HP1 consists of a family of evolutionarily conserved proteins that act to epigenetically alter gene expression. As might be expected given their name, HP1s are fundamental units of heterochromatin, but members of this family are also found in euchromatin, where they primarily repress gene transcription by interacting with methylated histones.5 HP1 members are encoded by chromobox (CBX) genes, with the gene for HP1Hsβ encoded by CBX1 located on chromosome 17.23 In normal development, various HP1 isoforms have been suggested to be important in adipocyte differentiation,8 blood cell maturation,24 and neuronal development.25 However, little is known about the regulation of CBX gene expression, particularly in the context of cancer. This is important, given that it is increasingly evident that decreased expression of various HP1 isoforms results in increased aggressiveness or invasiveness of a variety of or invasiveness of a variety of malignancies.7
In vitro studies of HP1 expression in cancer have suggested that this protein protects against tumor cell aggressiveness and metastasis.21,22,26,27 This has been most convincingly shown in breast cancer cell lines. In elegant work, breast cancer cell lines that were most highly invasive and metastatic were found to express little to no HP1Hsα, whereas the converse was true for poorly invasive tumor cells.21 When these cells were studied in an in vitro invasion model, increasing HP1Hsα expression in cells that normally express little of this protein showed decreased invasiveness, whereas decreasing the expression of this protein by RNAi in cells otherwise replete with HP1Hsα increased their invasiveness.22 Consequently, it has been suggested28 that HP1Hsα may be a member of a recently described, albeit small, class of proteins known as metastasis suppressors.29,30 Because HP1Hsβ has also been shown to attenuate metastasis of melanoma cells,31 it may be, given the data presented here, that all HP1 isoforms act as metastasis suppressors. However, the mechanisms whereby the expression of HP1 proteins are regulated in cancer are not well understood.
In this article, we show for the first time that antagonizing a specific ligand, GRP, or eliminating the expression of this ligand's cognate receptor, reduces the expression of HP1Hsβ. GRP is a 27 amino acid protein32 that is the mammalian homologue of bombesin, originally isolated from the skin of the frog Bombina bombina.33 GRP binds to a specific 7 transmembrane-spanning G protein-coupled receptor that in the gut is normally expressed only during development.1 After malignant transformation, however, GRP and GRPR can be aberrantly expressed in a variety of cancers, including those affecting the colon. Because activation induces the proliferation of a wide variety of cancer cell lines,34 GRP has been commonly considered to be a growth factor whose function should be attenuated or inhibited when aberrantly expressed in cancer.
Yet GRP is a modest mitogen in cancer, with micromolar concentrations of agonist increasing proliferation only 1.8 ± 0.2-fold.35 The weak effect of GRP on tumor cell growth is supported by clinical studies indicating that GRP/GRPR coexpression does not adversely affect the outcomes of patients with cancers of the stomach,36 colon,20 or lung.37 Furthermore, we recently showed that individuals with stage II and III colon cancer whose tumors express GRPR have significantly longer disease-free survival rates than do those whose colon cancers do not express this protein.2 The extant clinical data do not support the theory that GRP acts as a physiologically relevant growth factor in cancer.
A mechanism whereby GRP might improve patient outcome is suggested by the results of our recent in vitro studies. These investigations were focused by an earlier proteomics approach that identified proteins whose expression appeared to be regulated by GRP signaling.18 Briefly, we studied GRPR-expressing human colon cancer cells in the presence and absence of GRPR-specific antagonist. Proteins were resolved by two-dimensional gel electrophoresis and identified by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectroscopy. In addition to an unspecified member of the HP1 family—identified in the current study—we also determined that intracellular adhesion protein 1 and heat shock protein 72 (Hsp72) are up-regulated subsequent to GRP signaling.18 We have shown that intracellular adhesion protein 1 mediates GRP-induced colon cancer cell attachment to the extracellular matrix,15 and that Hsp72 promotes GRP-induced natural killer cell binding and tumor cytolysis.38 Based on the data presented in this paper, we add decreased tumor invasiveness to enhanced tumor cell attachment to the extracellular matrix and tumor cytolysis as mechanisms possibly mediating the improved outcome of patients with colorectal cancer whose tumors express GRP/GRPR.2
Acknowledgments
The authors gratefully appreciate the statistical assistance of Dr. Jae Kim (University of Illinois at Chicago) and the assistance of Dr. Kristina Matkowskyj in histopathology (Northwestern University, Evanston, IL).
Footnotes
Supported by a Veterans Administration Merit Review award (R.V.B.).
References
- 1.Carroll R.E., Matkowskyj K.A., Saunthararajah Y., Sekosan M., Battey J.F., Benya R.V. Contribution of gastrin-releasing peptide and its receptor to villus development in the murine and human gastrointestinal tract. Mech Dev. 2002;113:121–130. doi: 10.1016/s0925-4773(02)00032-1. [DOI] [PubMed] [Google Scholar]
- 2.Rivera C.A., Ahlberg N.C., Taglia L., Kumar M., Blunier A., Benya R.V. Expression of GRP and its receptor is associated with improved survival in patients with colon cancer. Clin Exp Metastasis. 2009;26:663–671. doi: 10.1007/s10585-009-9265-8. [DOI] [PubMed] [Google Scholar]
- 3.Eissenberg J.C., Elgin S.C. The HP1 protein family: getting a grip on chromatin. Curr Opin Genet Dev. 2000;10:204–210. doi: 10.1016/s0959-437x(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 4.Paro R., Hogness D.S. The polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci USA. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grewal S.I., Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 6.Dillon N. Heterochromatin structure and function. Biol Cell. 2004;96:631–637. doi: 10.1016/j.biolcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Dialynas G.K., Vitalini M.W., Wallrath L.L. Linking heterochromatin protein 1 (HP1) to cancer progression. Mutat Res. 2008;647:13–20. doi: 10.1016/j.mrfmmm.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takanashi M., Oikawa K., Fujita K., Kudo M., Kinoshita M., Kuroda M. Heterochromatin protein 1gamma epigenetically regulates cell differentiation and exhibits potential as a therapeutic target for various types of cancers. Am J Pathol. 2009;174:309–316. doi: 10.2353/ajpath.2009.080148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin Y., Ertl T., Cai R.Z., Halmos G., Schally A.V. Inhibitory effect of bombesin receptor antagonist RC-3095 on the growth of human pancreatic cancer cells in vivo and in vitro. Cancer Res. 1994;54:1035–1041. [PubMed] [Google Scholar]
- 10.Roesler R., Kopschina M.I., Rosa R.M., Henriques J.A., Souza D.O., Schwartsmann G. RC-3095, a bombesin/gastrin-releasing peptide receptor antagonist, impairs aversive but not recognition memory in rats. Eur J Pharmacol. 2004;486:35–41. doi: 10.1016/j.ejphar.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Matkowskyj K.A., Schonfeld D., Benya R.V. Quantitative immunohistochemistry by measuring cumulative signal strength using commercially available software Photoshop and Matlab. J Histochem Cytochem. 2000;48:303–311. doi: 10.1177/002215540004800216. [DOI] [PubMed] [Google Scholar]
- 12.Matkowskyj K.A., Cox R., Jensen R.T., Benya R.V. Quantitative immunohistochemistry by measuring cumulative signal strength accurately measures protein concentration. J Histochem Cytochem. 2003;51:205–214. doi: 10.1177/002215540305100209. [DOI] [PubMed] [Google Scholar]
- 13.Matkowskyj K.A., Cox R., Benya R.V. Quantitative immunohistochemistry by determining the norm of the image file. In: Hayat M.A., editor. Academic Press; Burlington, MA: 2006. pp. 279–284. [Google Scholar]
- 14.Matkowskyj K., Glover S., Benya R. Quantitative immunohistochemistry: an algorithm measuring cumulative signal strength and receptor number. Microsc Anal. 2004;18:5–6. [Google Scholar]
- 15.Taglia L., Matusiak D., Matkowskyj K.A., Benya R.V. Gastrin-releasing peptide mediates its morphogenic properties in human colon cancer by up-regulating intracellular adhesion protein-1 (ICAM-1) via focal adhesion kinase. Am J Physiol. 2007;292:G182–G190. doi: 10.1152/ajpgi.00201.2006. [DOI] [PubMed] [Google Scholar]
- 16.Itoh F., Yamamoto H., Hinoda Y., Imai K. Enhanced secretion and activation of matrilysin during malignant conversion of human colorectal epithelium and its relationship with invasive potential of colon cancer cells. Cancer. 1996;77:1717–1721. doi: 10.1002/(SICI)1097-0142(19960415)77:8<1717::AID-CNCR45>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 17.Zhu X.L., Liang L., Ding Y.Q. Overexpression of FMNL2 is closely related to metastasis of colorectal cancer. Int J Colorectal Dis. 2008;23:1041–1047. doi: 10.1007/s00384-008-0520-2. [DOI] [PubMed] [Google Scholar]
- 18.Ruginis T.A., Taglia L., Matusiak D., Bao-Shiang L., Benya R.V. Consequence of gastrin-releasing peptide receptor activation in a human colon cancer cell line: a proteomic approach. J Proteome Res. 2006;5:1460–1468. doi: 10.1021/pr060005g. [DOI] [PubMed] [Google Scholar]
- 19.Matkowskyj K.A., Keller K., Glover S., Kornberg L., Tran-Son-Tay R., Benya R.V. Expression of GRP and its receptor in well-differentiated colon cancer cells correlates with the presence of focal adhesion kinase phosphorylated at tyrosines 397 and 407. J Histochem Cytochem. 2003;51:1041–1048. doi: 10.1177/002215540305100807. [DOI] [PubMed] [Google Scholar]
- 20.Carroll R.E., Matkowskyj K.A., Chakrabarti S., McDonald T.J., Benya R.V. Aberrant expression of gastrin-releasing peptide and its receptor by well differentiated colon cancers in humans. Am J Physiol. 1999;276:G655–G665. doi: 10.1152/ajpgi.1999.276.3.G655. [DOI] [PubMed] [Google Scholar]
- 21.Kirschmann D.A., Lininger R.A., Gardner L.M., Seftor E.A., Odero V.A., Ainsztein A.M., Earnshaw W.C., Wallrath L.L., Hendrix M.J. Down-regulation of HP1Hsalpha expression is associated with the metastatic phenotype in breast cancer. Cancer Res. 2000;60:3359–3363. [PubMed] [Google Scholar]
- 22.Norwood L.E., Moss T.J., Margaryan N.V., Cook S.L., Wright L., Seftor E.A., Hendrix M.J., Kirschmann D.A., Wallrath L.L. A requirement for dimerization of HP1Hsalpha in suppression of breast cancer invasion. J Biol Chem. 2006;281:18668–18676. doi: 10.1074/jbc.M512454200. [DOI] [PubMed] [Google Scholar]
- 23.Lomberk G., Wallrath L., Urrutia R. The heterochromatin protein 1 family. Genome Biol. 2006;7:228. doi: 10.1186/gb-2006-7-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olins D.E., Olins A.L. Granulocyte heterochromatin: defining the epigenome. BMC Cell Biol. 2005;6:39. doi: 10.1186/1471-2121-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panteleeva I., Boutillier S., See V., Spiller D.G., Rouaux C., Almouzni G., Bailly D., Maison C., Lai H.C., Loeffler J.P., Boutillier A.L. HP1alpha guides neuronal fate by timing E2F-targeted genes silencing during terminal differentiation. EMBO J. 2007;26:3616–3628. doi: 10.1038/sj.emboj.7601789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norwood L.E., Grade S.K., Cryderman D.E., Hines K.A., Furiasse N., Toro R., Li Y., Dhasarathy A., Kladde M.P., Hendrix M.J., Kirschmann D.A., Wallrath L.L. Conserved properties of HP1(Hsalpha) Gene. 2004;336:37–46. doi: 10.1016/j.gene.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Pomeroy S.L., Tamayo P., Gaasenbeek M., Sturla L.M., Angelo M., McLaughlin M.E., Kim J.Y., Goumnerova L.C., Black P.M., Lau C., Allen J.C., Zagzag D., Olson J.M., Curran T., Wetmore C., Biegel J.A., Poggio T., Mukherjee S., Rifkin R., Califano A., Stolovitzky G., Louis D.N., Mesirov J.P., Lander E.S., Golub T.R. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 28.Moss T.J., Wallrath L.L. Connections between epigenetic gene silencing and human disease. Mutat Res. 2007;618:163–174. doi: 10.1016/j.mrfmmm.2006.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steeg P.S., Ouatas T., Halverson D., Palmieri D., Salerno M. Metastasis suppressor genes: basic biology and potential clinical use. Clin Breast Cancer. 2003;4:51–62. doi: 10.3816/cbc.2003.n.012. [DOI] [PubMed] [Google Scholar]
- 30.Berger J.C., Vander Griend D.J., Robinson V.L., Hickson J.A., Rinker-Schaeffer C.W. Metastasis suppressor genes: from gene identification to protein function and regulation. Cancer Biol Ther. 2005;4:805–812. doi: 10.4161/cbt.4.8.1865. [DOI] [PubMed] [Google Scholar]
- 31.Nishimura K., Hirokawa Y.S., Mizutani H., Shiraishi T. Reduced heterochromatin protein 1-beta (HP1beta) expression is correlated with increased invasive activity in human melanoma cells. Anticancer Res. 2006;26:4349–4356. [PubMed] [Google Scholar]
- 32.Erspamer V. Discovery, isolation and characterization of bombesin-like peptides. Ann NY Acad Sci. 1988;547:3–9. doi: 10.1111/j.1749-6632.1988.tb23870.x. [DOI] [PubMed] [Google Scholar]
- 33.Erspamer V., Erspamer G.F., Inslevini M., Negri L. Occurrence of bombesin and alytensin in extracts of the skin of three European discoglossid frogs and pharmacological actions of bombesin on extravascular smooth muscle. Br J Pharmacol. 1972;45:333–348. doi: 10.1111/j.1476-5381.1972.tb08087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen R.T., Battey J.F., Spindel E.R., Benya R.V. International Union of Pharmacology: LXVIII Mammalian bombesin receptors: nomenclature, distribution, pharmacology, signaling, and functions in normal and disease states. Pharmacol Rev. 2008;60:1–42. doi: 10.1124/pr.107.07108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen J.G., Carroll R.E., Benya R.V. The case for gastrin-releasing peptide acting as a morphogen when it and its receptor are aberrantly expressed in cancer. Peptides. 2001;22:689–699. doi: 10.1016/s0196-9781(01)00380-1. [DOI] [PubMed] [Google Scholar]
- 36.Carroll R.E., Carroll R., Benya R.V. Characterization of gastrin-releasing peptide receptors aberrantly expressed by non-antral gastric adenocarcinomas. Peptides. 1999;20:229–237. doi: 10.1016/s0196-9781(98)00164-8. [DOI] [PubMed] [Google Scholar]
- 37.Toi-Scott M., Jones C.L., Kane M.A. Clinical correlates of bombesin-like peptide receptor subtype expression in human lung cancer cells. Lung Cancer. 1996;15:341–354. doi: 10.1016/0169-5002(95)00597-8. [DOI] [PubMed] [Google Scholar]
- 38.Taglia L., Matusiak D., Benya R.V. GRP-induced up-regulation of Hsp72 promotes CD16+/94+ natural killer cell binding to colon cancer cells causing tumor cell cytolysis. Clin Exp Metastasis. 2008;25:451–463. doi: 10.1007/s10585-008-9151-9. [DOI] [PubMed] [Google Scholar]


