Abstract
The excision repair cross completing group 1 gene product (ERCC1) and the regulatory subunit of ribonucleotide reductase (RRM1) have been reported as being prognostic of outcome and predictive of therapeutic efficacy in patients with non–small cell lung cancer. Routinely processed surgical specimens from 784 patients from the International Adjuvant Lung Trial were arrayed as tissue microarrays. In situ protein levels were scored with an automated, quantitative analysis system, dichotomized into high and low marker categories, and analyzed for associations with patients' characteristics, survival, and benefit from adjuvant chemotherapy. Scores for both markers were significantly associated with contributing center (P < 0.001) and skewed, with the bulk of scores being low. High scores were more frequent in women for ERCC1 and RRM1 and in older patients and those with adenocarcinoma for RRM1. Low ERCC1 scores indicated significant benefit from adjuvant chemotherapy [hazard ratio (HR) = 0.73 for chemotherapy versus control, P = 0.02]. Although all other survival associations were not statistically significant, low RRM1 scores trended to indicate benefit from adjuvant chemotherapy (HR = 0.84, P = 0.25), and ERCC1 scores were marginally prognostic of survival (HR = 0.77 for high versus low scores, P = 0.10). We conclude that contributing center and specimen quality substantially affect the levels of both markers. Future trials should incorporate the collection and processing of tumor specimens prospectively on standardized protocols to better reveal the impact of biomarkers on clinically relevant outcomes.
Tumor progression and resistance to therapeutic interventions are major obstacles to improving the outcome for patients with non–small cell lung cancer (NSCLC). Although notable progress has been made in the treatment of NSCLC, it is still associated with a poor prognosis for most patients. Modern techniques have facilitated the identification of genes and their products that may play a role in disease progression and patient response to therapy. Among these are the excision repair cross completing group 1 gene product (ERCC1) and the regulatory subunit of ribonucleotide reductase (RRM1).
ERCC1 is a component of the 5′ endonuclease of the nucleotide excision repair complex, and it is crucial for the repair of DNA damage caused by interstrand and intrastrand cross-links that prevent both replication and transcription.1 Previous work in patient-derived specimens has shown that elevated levels of ERCC1 are associated with improved outcomes in patients treated with surgical resection alone.2,3 It is presumed that this improvement in survival is a result of increased DNA damage repair capacity, and evidence of a reduced lung cancer risk in individuals with increased levels for nucleotide excision repair genes exists.4,5 However, in patients who require cytotoxic chemotherapy, in particular with agents that induce DNA adducts and cross-links, high levels of ERCC1 are associated with reduced efficacy presumably through the increased efficiency of repair of platinum-induced DNA damage.1,3,6
RRM1 functions as the regulatory subunit of ribonucleotide reductase and controls substrate specificity and the on/off function of ribonucleotide reductase, whereas the catalytic subunit (RRM2) converts nucleoside diphosphates to the corresponding deoxynucleotides.7 Results from multiple independent laboratories and clinical studies have shown that RRM1 is the dominant determinant of efficacy for the chemotherapeutic agent gemcitabine, a nucleoside analog.6,8–13 In addition, RRM1 suppresses carcinogen-induced lung tumorigenesis in experimental animal models, which appears to be a result of increased DNA damage repair capacity in the presence of elevated RRM1.14 It also suppresses tumor progression as shown by decreased cellular migration and invasion in vitro and suppression of metastasis formation in mouse models, resulting in increased animal survival.15,16 In patients with NSCLC and pancreatic cancer who had a complete surgical resection and no adjuvant therapy, survival was longer for those with high levels of expression compared with those with low levels of expression.17–19
Although little controversy exists regarding the association between ERCC1 and RRM1 with platinum and gemcitabine efficacy, there are recent reports of ERCC1 and RRM1 either not being significantly associated or potentially being inversely associated with the survival of NSCLC patients after a complete surgical resection.20,21 Knowing the prognostic impact of both genes on the survival of patients not undergoing chemotherapy with a platinum agent and/or gemcitabine is important for the design and interpretation of clinical trials that incorporate both genes into a treatment decision algorithm, in particular if this treatment is given in a curative attempt in completely resected patients. Two clinical studies in patients with advanced NSCLC have already used this approach and reported a favorable disease response when treatment is selected based on gene expression.22,23 Finally, most clinico-correlative investigations involving both genes have reported a highly significant coexpression between ERCC1 and RRM1.6,9,11,18,20,24
We investigated tumor specimens from the International Adjuvant Lung Trial (IALT25) for ERCC1 and RRM1 in situ protein expression using accurate quantitative analysis (AQUA), a recently developed automated immunofluorescence-based technology. One goal was to compare results obtained for ERCC1 by AQUA on tissue microarrays (TMAs) with those previously reported and obtained by standard manual immunohistochemical analysis (IHC) on full-section specimens. Another goal was to corroborate the role of RRM1 alone and combined with ERCC1 on survival of patients treated with surgery alone and those who had received adjuvant chemotherapy.
Materials and Methods
The IALT-Biospecimen Dataset
The IALT-biospecimen collection is the largest existing repository of surgically resected NSCLC specimens with prospectively collected clinical outcomes data on a phase III randomized clinical trial.3,25 Approval was obtained from the local institutional review boards, according to the legal regulations in each participating country. The collection consists of paraffin blocks from 784 patients with stage I to III (stage I, 270; stage II, 180; stage III, 334) cancer who had been randomly assigned to observation (N = 382) or adjuvant chemotherapy with cis-platin plus etoposide (N = 218), cis-platin plus vinorelbine (N = 122), cis-platin plus vindesine (N = 25), or cis-platin plus vinblastine (N = 37). The specimens were from 28 centers in 14 different countries in Europe and South America that contributed 1042 patients to the IALT. Initially, blocks from a total of 867 patients were collected. Blocks from 43 patients could not be used because they contained insufficient material. A central pathologic review was performed on full sections stained with H&E, and the quality of each specimen was rated as poor, average, or good. Specimens with poor quality (N = 40) were removed from the collection. In addition, information on pleural, vascular, and lymphatic invasion and lymphoid infiltration was collected. The results on the level of expression of ERCC1 by standard IHC on full-section specimens using an H-scoring scale from 0 to 3 [staining intensity (0, 1, 2, 3) × proportion score (0, 0.1, 0.5, 1.0)] have been previously reported.3
Description of the TMAs
Specimen cores from patients with average or good rating (N = 784) were arrayed as triplicate spots of 0.6-mm diameter in a total of 13 TMA blocks. A pathology review of the selected cores was performed (E.B.) to verify the presence of tumor tissue and tissue quality (classified as present or absent and average or good). From two patients two separate blocks were arrayed for a total of six spots (782 patients with three tumor spots and two patients with six tumor spots). All specimens from each patient were on the same slide (including the two patients who had two blocks), and patients from the 28 centers were clustered on slides (ie, they were not randomly distributed in the 13 TMA blocks).
Clinical and Pathologic Variables Collected
The following variables were prospectively collected on all patients: disease stage by TNM classification (AJCC sixth edition, 2002), tumor histologic findings, sex, age, performance status, and type of surgery. In addition, data on whether patients received adjuvant radiation and the random assignment to observation or adjuvant chemotherapy, including the type of chemotherapy, were known. The outcomes data used for this analysis were those originally reported with a median follow-up time of 56 months.3,25
Automated Quantitative in Situ Protein Analysis
Immunofluorescence combined with AQUA was used to assess in situ expression of the target molecules.26 Microtome sections of 4-μm thickness were microwaved in 10 mmol/L Tris-EDTA, pH 9.0 (for ERCC1 staining), or 10 mmol/L Tris-HCl, pH 12.0 (for RRM1 staining), for 10 minutes, cooled to room temperature, and rinsed with PBS. Endogenous peroxidase was inactivated with 3% hydrogen peroxide for 20 minutes. Slides were then incubated overnight at 4°C in appropriately diluted primary antiserum (R1AS-6, 1:800, pH 12.0, for RRM1) or antibody (clone 8F1, 1:300, pH 9.0, for ERCC1; cat D8903, lot 9475; Sigma-Aldrich, St. Louis, MO) as described.18 Cells of epithelial origin, consisting mostly of malignant cells, were identified by cytokeratin staining. The primary antibody was then visualized with different fluorochrome-labeled secondary antibodies (Envision labeled polymer horseradish peroxidase antirabbit or antimouse for RRM1 or ERCC1 detection; Alexa 555 goat antimouse or goat antirabbit for cytokeratin detection). Fluorescence of the target signals was amplified with Cy5-tyramide. 4′,6-diamidine-2′-phenylindole, which strongly binds to DNA minor groove, was added to the coverslip mounting solution and used for identification of nuclei. The final TMA slides were scanned with SpotGrabber, and image data were analyzed with AQUA (PM-2000, HistoRx, New Haven, CT). Software version 1.2 was used with an exposure time of 30 milliseconds. The maximal range of the AQUA scores with this software version is 0 to 255.
Statistical Analysis
The analysis was performed at two different levels. One was solely of the AQUA scores taking the characteristics of the TMA into account, and the other was to compare the individual patients' average scores with clinical outcomes parameters. The primary intent was to test for the prognostic value of RRM1, ERCC1, and both combined on patient survival in the group who did not receive chemotherapy and for interaction between these markers and benefit from adjuvant chemotherapy using the raw ERCC1 and RRM1 values with dichotomization of patients into high and low marker level categories using the median values as threshold. Provisions for preplanned optimal cut point analyses and normalization of marker values had not been made a priori.
AQUA scores were analyzed for skewness, kurtosis, and other summary statistics, and they were compared by slides, centers, and specimen quality. The relationships between marker categories and histologic findings, tumor size, lymph node involvement, and tumor stage were analyzed using logistic models stratified by center. The agreement between IHC H-scores3 and AQUA scores was studied by Spearman's correlation coefficient on the continuous values and by κ coefficient using the high and low expression categories.
A Cox model that included the clinical prognostic factors isolated in the main analysis of the IALT together with the treatment was used for the prognostic analysis, and it was adjusted for the factors used in the randomization process (including contributing center and specimen quality). All other factors statistically related to the marker in the multivariate logistic model were added to the Cox model. The predictive value of the marker was studied by testing the interaction between the marker category (high or low) and the attributed treatment (chemotherapy or observation) in the same Cox model. Survival curves by marker levels and treatment groups were generated using the Kaplan-Meier method.
All reported P values were two-sided. All analyses were performed using SAS software, version 8.2 (SAS Institute Inc., Cary, NC).
Results
ERCC1 and RRM1 Expression Characteristics
The expression of both proteins was predominantly nuclear as previously described3,18 and displayed a fine granular pattern for ERCC1 and a coarse granular pattern for RRM1.
ERCC1 values were obtained on 763 of 784 patients. Unavailability of values was a result of absence of tissue in the assigned location. Replicate core values (2× in 77, 3× in 672 and 6× in 1) were averaged. A histopathologic review of H&E-stained tissue cores confirmed the presence of NSCLC in 747 cases, which were used for further analyses and specimen quality (classified as average or good). Expression levels ranged from 2.2 to 149.1, with a median of 9.0, a mean of 13.2, and an SD of 12.8. They were highly skewed to the right (skewness, 4.3; kurtosis, 29.1) (ie, high scores were much farther away from the median than low scores).
RRM1 values were obtained on 752 patients, and values on patients with replicates were averaged (2× in 112, 3× in 610 and 6× in 1). Presence of NSCLC in tissue cores was confirmed in 738 cases. Levels ranged from 0.2 to 91.8, with a median of 9.7, a mean of 15.1, and an SD of 15.0. The RRM1 levels were skewed toward the right (skewness, 2.3; kurtosis, 6.0).
ERCC1 values in patients with three replicates were highly correlated (Spearman's r = 0.78 to 0.84; P < 0.0001), and the coefficients of variation were below 1.4%. Likewise, replicate RRM1 values were highly correlated (r = 0.76 to 0.80; P < 0.0001), and the coefficients of variation were below 1.6%. ERCC1 and RRM1 expression levels were modestly correlated (N = 730, r = 0.21; P < 0.001) (Figure 1).
Figure 1.
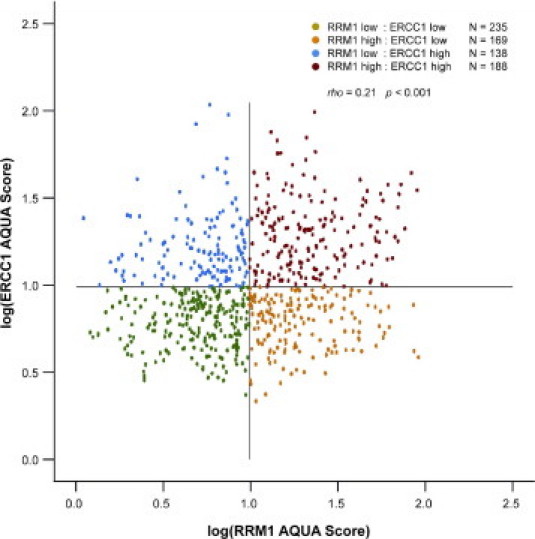
Distribution of ERCC1 and RRM1 in situ protein expression scores. Lines indicate scores of 10. ERCC1 and RRM1 expression levels were modestly correlated (N = 730, correlation between log expressions: r = 0.22, 95% confidence interval, 0.15-0.29).
Specimen origin by center and specimen quality contributed significantly to the differences seen for both ERCC1 and RRM1 values (Table 1), a result that had previously been observed for ERCC1 using full-section specimens and standard immunohistochemical analysis.3 Center accounted for 11% of the overall variation in ERCC1 AQUA levels (P < 0.0001), 10% in ERCC1 IHC levels (P < 0.0001), and 33% in the variation of RRM1 levels (P < 0.001).
Table 1.
ERCC1 and RRM1 Expression Categories by Patient Demographics and Disease Characteristics Stratified by Contributing Center
|
P value (adjusted by center) |
||
|---|---|---|
| Characteristics | ERCC1 | RRM1 |
| Sex | ||
| Male | 0.04 | 0.04 |
| Female | (F > M) | (F > M) |
| Age, y | ||
| <55 | ||
| 55–64 | 0.50 | 0.04 |
| >64 | (older > young) | |
| WHO-PS | ||
| 0 | ||
| 1 | 0.79 | 0.06 |
| 2 | (0 > 1 > 2) | |
| Stage | ||
| I | ||
| II | 0.53 | 0.75 |
| III | ||
| Tumor by TNM | ||
| 1 | ||
| 2 | 0.93 | 0.82 |
| 3 and 4 | ||
| Nodes by TNM | ||
| 0 | ||
| 1 | 0.65 | 0.82 |
| 2 | ||
| Histopathologic findings | ||
| Adenocarcinoma | ||
| Squamous cell carcinoma | 0.70 | 0.02 |
| Other | (adenocarcinoma > squamous cell carcinoma) | |
| Pleural invasion | ||
| Absent | 0.68 | 0.46 |
| Present | ||
| Vascular invasion | ||
| Absent | 0.06 | 0.13 |
| Present | (absent > present) | |
| Lymphatic invasion | ||
| Absent | 0.55 | 0.40 |
| Present | ||
| Lymphoid infiltration | ||
| Not intense | 0.66 | 0.38 |
| Intense | ||
| Type of surgery | ||
| Lobe/segmentectomy | 0.72 | 0.18 |
| Pneumonectomy | ||
| Quality of specimen | ||
| Good | 0.007 | 0.78 |
| Average (poor specimens excluded) | (average > good) | |
WHO-PS, World Health Organization Performance Scale; F, female; M, male.
ERCC1 and RRM1 Levels by Patient Demographics and Disease Characteristics
According to the prespecified analysis plan, ERCC1 expression was dichotomized into high versus low using 10 or greater versus less than 10 (high in 331 patients, low in 416). The threshold of 10, rather than the sample median of 9, was used because this resulted in a proportional cohort split similar to the one previously used using IHC-based H-scores (H-score of 1.5 to 3.0 in 335 and 0.0 to 1.0 in 426 patients). For RRM1 dichotomization, the sample median was used as threshold (10 or greater versus less than 10; high in 363 patients, low in 375).
We observed that high ERCC1 and RRM1 levels were more frequent in women compared with men (P = 0.04). High levels of RRM1 were also more frequent in older patients (P = 0.04) and in those with adenocarcinoma (P = 0.02). When all variables were assessed in a logistic model stratified by center, only age (P = 0.05) was retained to explain high RRM1 levels. All other parameters appeared equally distributed between patients in the high and low marker categories (Table 1).
ERCC1 Expression by Standard IHC and AQUA
Data for both the previously reported ERCC1 H-scores that had been generated using standard IHC on full-section specimens3 and the AQUA scores generated on TMAs were available on 727 patients. We observed a small correlation between these values (correlation between log-expressions: r = 0.22, 95% confidence interval, 0.15 to 0.29), with the upper limit being largely below 1 (test of r = 1: P < 0.0001) (Figure 2). Furthermore, when the ERCC1 IHC categories and AQUA categories were compared (156 IHC high and AQUA high, 164 IHC high and AQUA low, 166 IHC low and AQUA high, 241 IHC low and AQUA low), the agreement between both methods was very low (κ coefficient = 0.08; 95% confidence interval, 0.01 to 0.15).
Figure 2.
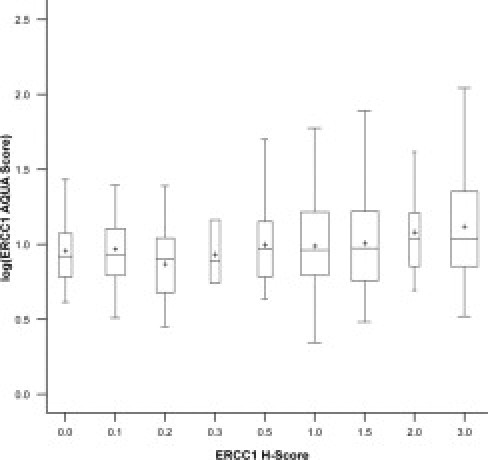
Distribution of ERCC1 AQUA scores by H-score categories. The box width is proportional to the number of patients in each category, the cross is the mean, and the horizontal bar is the median value. The correlation coefficient between log AQUA scores and raw H-score was 0.27 (95% confidence interval, 0.20 to 0.33).
The Prognostic and Predictive Impact of the High and Low ERCC1 Expression Categories
The differences in overall survival (OS) and disease-free survival (DFS) in a multivariate Cox model that included the variables sex, age, performance status, stage, histopathologic findings, type of surgery, lymphoid infiltration, and specimen quality with stratification by center (hereafter referred to as the Cox model) between patients in the high and low ERCC1 expression categories in those treated with adjuvant chemotherapy and those observed are summarized in Table 2 (Figure 3, A and B). The results showed similar trends as those previously reported with IHC on whole tumor sections. Patients with high ERCC1 expression had a better survival than those with low ERCC1 expression in the control group (HR = 0.77, P = 0.10, for OS; HR = 0.80, P = 0.12, for DFS), and only patients with low ERCC1 expression benefited from adjuvant chemotherapy (HR = 0.73, P = 0.02, for OS; HR = 0.76, P = 0.04, for DFS). However, the test for interaction fell short of statistical significance (P = 0.12 for OS; P = 0.24 for DFS).
Table 2.
Survival for Patients with High and Low ERCC1 and RRM1 Expression in the Observation and Adjuvant Chemotherapy Arms
| ERCC1 (N = 747) |
RRM1 (N = 738) |
|||||
|---|---|---|---|---|---|---|
| Low (<10) (N = 416) | High (≥10) (N = 331) | HR for high/low groups (95% CI) | Low (<10) (N = 375) | High (≥10) (N = 363) | HR for high/low groups (95% CI) | |
| OS | ||||||
| Control group | 0.77 (0.56–1.05) (P = 0.10) | 1.06 (0.77–1.46) (P = 0.74) | ||||
| No. of deaths/No. of patients | 108/195 | 85/170 | 90/179 | 98/180 | ||
| Median survival, months | 44 | 50 | 48 | 45 | ||
| 2-Year survival rate⁎ | 0.65 (0.58–0.71) | 0.72 (0.65–0.78) | 0.68 (0.61–0.75) | 0.69 (0.62–0.76) | ||
| 5-Year survival rate | 0.39 (0.32–0.47) | 0.43 (0.35–0.52) | 0.46 (0.38–0.54 | 0.38 (0.29–0.46) | ||
| Chemotherapy group | 1.07 (0.79–1.45) (P = 0.67) | 1.15 (0.84–1.57) (P = 0.38) | ||||
| No. of deaths/No. of patients | 108/221 | 88/161 | 96/196 | 101/183 | ||
| Median survival, months | 51 | 52 | 55 | 48 | ||
| 2-Year survival rate | 0.75 (0.68–0.80) | 0.76 (0.69–0.82) | 0.73 (0.67–0.79) | 0.76 (0.70–0.82) | ||
| 5-Year survival rate | 0.46 (0.39–0.53) | 0.40 (0.32–0.49) | 0.47 (0.39–0.55) | 0.39 (0.31–0.47) | ||
| HR for chemotherapy/control groups (95% CI) | 0.73 (0.55–0.96)(P = 0.02) | 1.01 (0.74–1.38)(P = 0.94) | 0.84 (0.62–1.13)(P = 0.25) | 0.91 (0.69–1.22)(P = 0.87) | ||
| Interaction P value (ERCCI or RRMI × treatment) | 0.12 | 0.69 | ||||
| DFS | ||||||
| Control group | 0.80 (0.60–1.08) (P = 0.12) | 1.08 (0.80–1.46) (P = 0.61) | ||||
| No. of events/No. of patients | 122/195 | 98/170 | 109/179 | 107/180 | ||
| Median survival, months | 33 | 39 | 34 | 37 | ||
| 2-Year survival rate | 0.57 (0.50–0.64) | 0.66 (0.58–0.72) | 0.60 (0.52–0.67) | 0.62 (0.55–0.69) | ||
| 5-Year survival rate | 0.33 (0.28–0.45) | 0.36 (0.28–0.45) | 0.35 (0.28–0.43) | 0.35 (0.28–0.43) | ||
| Chemotherapy group | 1.01 (0.76–1.34) (P = 0.97) | 1.09 (0.81–1.46) (P = 0.59) | ||||
| No. of events/No. of patients | 122/221 | 95/161 | 105/196 | 112/183 | ||
| Median survival, months | 43 | 45 | 50 | 40 | ||
| 2-Year survival rate | 0.65 (0.59–0.71) | 0.66 (0.58–0.73) | 0.65 (0.58–0.71) | 0.65 (0.58–0.72) | ||
| 5-Year survival rate | 0.41 (0.35–0.49) | 0.36 (0.28–0.45) | 0.44 (0.36–0.52) | 0.35 (0.28–0.43) | ||
| HR for chemotherapy/control groups (95% CI) | 0.76 (0.59–0.99)(P = 0.04) | 0.97 (0.72–1.30)(P = 0.82) | 0.87 (0.66–1.15)(P = 0.32) | 0.87 (0.66–1.15)(P = 0.32) | ||
| P value (ERCCI or RRMI × treatment) | 0.24 | 0.99 | ||||
Survival rate data are shown as mean (range).
Figure 3.
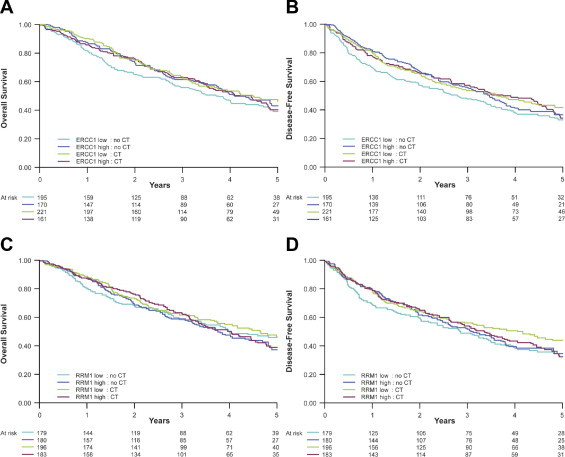
Overall (A and C) and disease-free (B and D) survival by high and low ERCC1 (A and B) and RRM1 (C and D) expression categories and treatment arm.
The Prognostic and Predictive Impact of the High and Low RRM1 Expression Categories
The survival differences in the Cox model between patients in the high and low RRM1 expression categories in those treated with adjuvant chemotherapy and those observed are summarized in Table 2 (Figure 3, C and D). There was no statistically significant impact of RRM1 expression on OS or DFS (interaction P = 0.69). However, patients with low RRM1 expression who received adjuvant chemotherapy had a numerically longer survival than those observed (HR = 0.84, P = 0.25, for OS; HR = 0.87, P = 0.32, for DFS).
The Prognostic and Predictive Impact of the Combined ERCC1 and RRM1 Expression Categories
Patients were categorized into four groups according to high and low ERCC1 and RRM1 expression levels. There were 188 patients with high levels for both, 138 with high ERCC1 and low RRM1 expression, 169 with low ERCC1 and high RRM1 expression, and 235 with low levels for both (Figure 1). The 25 samples with either ERCC1 or RRM1 missing were discarded for this analysis. In the patients observed after surgical resection (N = 357), there was no appreciable impact of the four marker categories on OS in the Cox model. The HRs for death were 0.83 (95% CI, 0.52 to 1.34; P = 0.45) for the high ERCC1 and low RRM1 expression group, 1.03 (95% CI, 0.66 to 1.62; P = 0.89) for the low ERCC1 and high RRM1 expression group, and 0.89 (95% CI, 0.56 to 1.42; P = 0.63) for the high ERCC1 and high RRM1 expression groups using the low ERCC1 and low RRM1 expression group as reference (test for equality of HR, P = 0.80; Kaplan-Meier plots in Figure 4).
Figure 4.
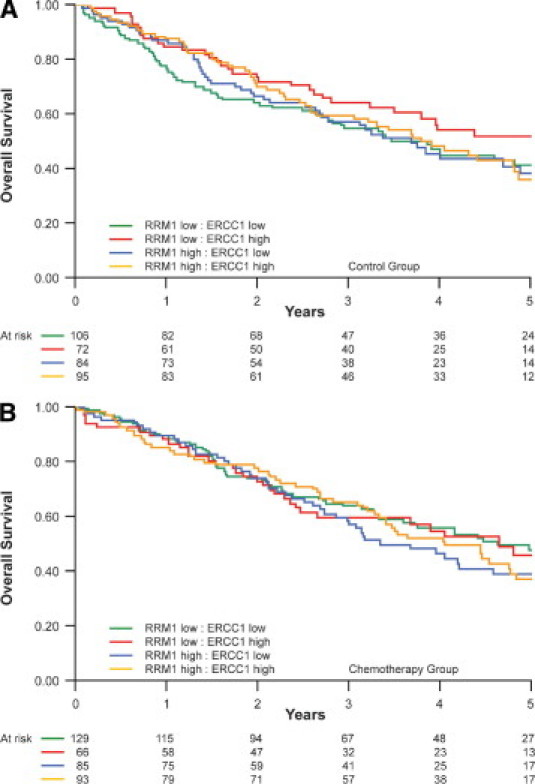
Overall survival by high and low ERCC1 and RRM1 expression categories and treatment arm (A, control group; B, chemotherapy group).
Finally, we assessed the impact of adjuvant chemotherapy in each marker group using the Cox model (Table 3). Although improvement of survival was best for patients with low ERCC1 and RRM1 expression in the adjuvant chemotherapy group, this difference was not statistically significant (P = 0.08). Overall, the four marker categories were not predictive of benefit from adjuvant chemotherapy (test for equality of HR, P = 0.60).
Table 3.
OS by ERCC1 and RRM1 Categories in the Observation and Adjuvant Chemotherapy Arms (P = 0.60, Test for Equality of the Four HRs)
| Chemotherapy group | Control group | HR for death (95% CI) (P value) | |
|---|---|---|---|
| ERCC1 <10/RRM1 <10 group (n = 235) | 0.72 (0.50–1.04) (P = 0.08) | ||
| No. of deaths/No. of patients | 62/129 | 57/106 | |
| Median survival, month | 56 | 42 | |
| 2-Year survival rate, mean (range) | 0.74 (0.65–0.80) | 0.64 (0.55–0.73) | |
| 5-Year survival rate, mean (range) | 0.47 (0.38–0.57) | 0.41 (0.32–0.52) | |
| ERCC1 <10/RRM1 <10 (n = 138) | 1.04 (0.63–1.72) (P = 0.89) | ||
| No. of deaths/No. of patients | 33/66 | 33/72 | |
| Median survival, month | 56 | 71 | |
| 2-Year survival rate | 0.74 (0.63–0.83) | 0.73 (0.62–0.82) | |
| 5-Year survival rate | 0.46 (0.32–0.60) | 0.52 (0.39–0.64) | |
| ERCC1 <10/RRM1 ≥10 (n = 169) | 0.88 (0.58–1.35) (P = 0.57) | ||
| No. of events/No. of patients | 46/85 | 47/84 | |
| Median survival, month | 40 | 44 | |
| 2-Year survival rate, mean (range) | 0.74 (0.63–0.82) | 0.66 (0.56–0.76) | |
| 5-Year survival rate, mean (range) | 0.39 (0.28–0.51) | 0.38 (0.27–0.51) | |
| ERCC1 ≥10/RRM1 ≥10 (n = 188) | 0.99 (0.66–1.47) (P = 0.95) | ||
| No. of events/No. of patients | 54/93 | 51/95 | |
| Median survival, month | 49 | 46 | |
| 2-Year survival rate, mean (range) | 0.77 (0.68–0.85) | 0.70 (0.60–0.78) | |
| 5-Year survival rate, mean (range) | 0.37 (0.26–0.48) | 0.36 (0.25–0.48) |
Discussion
We had previously reported that patients in the high ERCC1 expression category had a significantly longer survival (median OS, 55 months) than patients in the low ERCC1 category (median OS, 42 months; HR = 0.66; P = 0.009) for those who did not receive adjuvant chemotherapy as determined by an IHC-based H-scoring system on full-specimen sections.3 In addition, we had found that ERCC1 was also predictive of adjuvant chemotherapy efficacy because only patients in the low ERCC1 category benefited from this intervention (median OS, 56 versus 42 months for adjuvant therapy versus observation; HR = 0.65; P = 0.002).
The data presented here using an automated, objective scoring system (AQUA) on TMAs are similar, although less convincing. The only statistically significant difference observed was the effect of adjuvant chemotherapy on survival in patients with low ERCC1 expression (HR = 0.73); this effect was not observed in patients with high ERCC1 expression. In the observation group, patients in the high ERCC1 expression category lived longer than those in the low expression category (HR = 0.77); this difference, however, was not statistically significant. As will be discussed, the differences between the two studies may be related to different sampling procedures (whole tumor sections versus cores in TMAs) and different analysis systems (traditional IHC versus fluorescence-based automated scoring).
We had also previously reported that patients in the high RRM1 expression category had a significantly longer survival than patients in the low RRM1 category for those with completely resected stage I disease who did not receive adjuvant chemotherapy, as determined by the AQUA system in a different dataset using TMAs.18 Our results from the dataset presented do not support a differential survival between patients in the high and low RRM1 expression categories who did not receive adjuvant chemotherapy (HR = 1.06). We also did not observe a restriction of the survival benefit to patients in the high ERCC1 and high RRM1 expression category as previously reported, although a statistically nonsignificant trend was found (HR = 0.89). It is possible that this discrepancy may be explained by the fact that the earlier reported dataset only included patients with stage I disease from a single center with 45% in stage IA, whereas the current dataset includes patients from multiple centers with stage I to III, which is further discussed below.
Does this mean that ERCC1 and RRM1 are not prognostic of survival in patients who do not receive adjuvant chemotherapy after a complete surgical resection and that ERCC1 is a predictive marker for benefit from adjuvant platinum-based chemotherapy, whereas RRM1 is unlikely to be predictive for nongemcitabine, platinum-based, adjuvant chemotherapy?
To discuss these questions, it is important to recall that only the clinical data were collected prospectively for this specimen set. The collection of pathologic specimens was an afterthought and not mandatory for trial participation. The handling, processing, and fixative to be used were not a priori specified by the protocol, and they are likely to have affected the AQUA expression results of ERCC1 and RRM1. The handling of specimens includes the ischemia time (ie, the time elapsed from clamping of the blood vessels to resection and fixation), which may affect marker expression levels more than the fixation method. Given the global nature of IALT, which is a strength from the clinical outcomes perspective, it is also a weakness from the biomarker perspective because we were unable to control for these factors and in particular technical surgical and pathologic issues.
Our analysis of marker expression by contributing center and specimen quality showed that both contributed significantly to the observed variability. In addition, the marker scores in this dataset are overall low (ERCC1: median, 9.0; range, 2.2 to 149.1; mean, 13.2; RRM1: median, 9.7; range, 0.2 to 91.8; mean, 15.1), which suggests a loss of protein expression and consequently a loss of the full spectrum of ERCC1 and RRM1 levels and falsely low values in a substantial proportion of specimens. In fact, using the same technology and reagents on TMAs generated from single centers in the United States, Japan, and Italy, we had previously reported median ERCC1 values of 65.9 (range, 1.9 to 178.7; mean, 66.3) for resected lung cancers,18 34.7 (range, 5.2 to 131.3; mean, 45.7) for advanced lung cancers,24 59.2 (range, 19.5 to 110.4; mean, 61.0) for resected pancreatic cancers,19 and 56.6 (range, 18.5 to 102.6; mean, 57.4) for metastatic breast cancers.27 The corresponding median RRM1 values were 40.5 (range, 8.3 to 96.2; mean, 43.2), 34.1 (range, 5.3 to 105.6; mean, 38.4), 52.2 (range, 29.3 to 107.3; mean, 54.6), and 64.5 (range, 12.3 to 138.7; mean, 61.9). Also, the lower correlation between ERCC1 and RRM1 AQUA scores in this dataset (r = 0.21) compared with those previously reported (r = 0.26 and 0.36)18 in conjunction with the observed ranges of scores suggests that the RRM1 expression levels may have been affected more severely than the ERCC1 levels.
The observed small but significant correlation (r = 0.19, P < 0.0001) between ERCC1 H-scores and AQUA scores is remarkable given the technical limitations. When specimens are categorized into high and low groups, there is only slight agreement in specimen classification (κ = 0.08). An experienced pathologist, when scoring a full-section specimen for ERCC1 expression, may take morphologic features that affect specimen quality and preservation into account in a “supervised” process that may be elusive to a fully automated, “unsupervised,” quantitative, fluorescence-based scoring system such as the one used in our study (ie, in manual scoring it is easy to focus on areas that appear best suited for data acquisition). We assessed each tissue core for quality using an H&E-stained section of the arrays. Although this approach may reduce the impact of the “unsupervised” scoring process, it does not completely eliminate this element because the precise tissue core section used for AQUA analysis is different from the H&E quality assessed section. This may explain why the impact of ERCC1 expression categories on prognosis in the control group and the predictive benefit of adjuvant chemotherapy in the low ERCC1 category are attenuated with the AQUA-scoring compared to the H-scoring system. However, in specimens that have been collected under a standardized collection protocol, as is the case for most single-center collections, the AQUA scoring system has been shown to outperform H-scoring.26,28 An alternate explanation may be that tumoral ERCC1 expression is heterogeneous and thus not fully appreciated in the cores used for TMA construction. However, it is remarkable that ERCC1 and RRM1 scores among replicate cores were highly correlated (r > 0.76), suggesting that both markers are relatively homogenously expressed in tumor specimens.
Finally, McCabe et al29 showed that the concentration of antibodies used for visualization and quantitative analysis of antigens may lead to contradictory results. This has been shown for targets with a nonlinear relationship between marker levels and clinical outcome (ie, outcome may be poor for very low and very high levels but good for intermediate levels or vice versa). Therefore, if the antibody concentration used for staining does not cover the full range of target expression, an arbitrarily chosen cut point for cohort dichotomization, such as the sample median, may reveal a poor outcome for patients with low expression in one experiment yet a good outcome in another experiment. Although evidence for such a phenomenon for the markers examined here is lacking, the fact that our expression spectrum is skewed toward low values suggests that substantial cross-contamination in the low expression range with truly high expressers exists, which would severely attenuate our ability to appreciate the full range of the prognostic and predictive potential of ERCC1 and RRM1.
What can be concluded from the results presented and those referenced is that multicenter clinical trials that seek to investigate the impact of biomarkers on patient outcome and the efficacy of a chosen therapy require a cross-center, standardized operating procedure for acquisition, processing, and handling of biospecimens that is optimized for the biomarkers under investigation. Although a perfect synchronization of all aspects may be difficult to achieve, at the very minimum, all specimens must be fixed using the same methods. For construction of TMAs from paraffin-embedded specimens that require multiple blocks, we recommend that specimens be distributed randomly using a preassigned grid location and that specimen replicates be distributed in different blocks. We further recommend validation of the tumor material and the assignment of a quality score to each core. Depending on the intended biomarkers to be studied, appropriate and identical control samples should be included in each of the TMA blocks.
In summary, using an automated quantitative in situ protein analysis system, we confirmed that benefit from adjuvant, platinum-based chemotherapy is only seen in patients with low levels of ERCC1 expression. Although there was a similar trend for RRM1 protein expression, this was not statistically significant. We conclude that further corroboration of these markers requires a prospective study design that incorporates a standardized procedure for collection and processing of biospecimens and clinical data. Upcoming clinical trials integrating ERCC1 and RRM1 scoring on manually and automatically processed whole tumor sections are under way. In addition, interinstitutional controls are performed with identical batches of antibodies to optimize and validate the best standardized protocol.
Footnotes
Supported by grant R01-CA129343 from the National Cancer Institute, an unrestricted research grant from Eli Lilly, and by grants from Programme Hospitalier de Recherche Clinique 2005, Cancéropôle Rhône-Alpes.
References
- 1.Reed E. ERCC1 and clinical resistance to platinum-based therapy. Clin Cancer Res. 2005;11:6100–6102. doi: 10.1158/1078-0432.CCR-05-1083. [DOI] [PubMed] [Google Scholar]
- 2.Simon G.R., Sharma S., Cantor A., Smith P., Bepler G. ERCC1 expression is a predictor of survival in resected patients with non-small cell lung cancer. Chest. 2005;127:978–983. doi: 10.1378/chest.127.3.978. [DOI] [PubMed] [Google Scholar]
- 3.Olaussen K.A., Dunant A., Fouret P., Brambilla E., Andre F., Haddad V., Taranchon E., Filipits M., Pirker R., Popper H.H., Stahel R., Sabatier L., Pignon J- P., Tursz T., Le Chavalier T., Soria J.-C. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 4.Wei Q., Cheng L., Hong W.K., Spitz M.R. Reduced DNA repair capacity in lung cancer patients. Cancer Res. 1996;56:4103–4107. [PubMed] [Google Scholar]
- 5.Cheng L., Spitz M.R., Hong W.K., Wei Q. Reduced expression levels of nucleotide excision repair genes in lung cancer: a case-control analysis. Carcinogenesis. 2000;8:1527–1530. [PubMed] [Google Scholar]
- 6.Bepler G., Kusmartseva I., Sharma S., Gautam A., Cantor A., Sharma A., Simon G. RRM1-modulated in vitro and in vivo efficacy of gemcitabine and platinum in non-small cell lung cancer. J Clin Oncol. 2006;24:4731–4737. doi: 10.1200/JCO.2006.06.1101. [DOI] [PubMed] [Google Scholar]
- 7.Stubbe J. Ribonucleotide reductases in the twenty-first century. Proc Natl Acad Sci U S A. 1998;95:2723–2724. doi: 10.1073/pnas.95.6.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson J.D., Ma L., Flagella M., Geeganage S., Gelbert L.M., Slapak C.A. An increase in the expression of ribonucleotide reductase large subunit 1 is associated with gemcitabine resistance in non-small cell lung cancer cell lines. Cancer Res. 2004;64:3761–3766. doi: 10.1158/0008-5472.CAN-03-3363. [DOI] [PubMed] [Google Scholar]
- 9.Rosell R., Danenberg K.D., Alberola V., Bepler G., Sanchez J.J., Camps C., Provencio M., Isla D., Taron M., Diz P., Artal A. Ribonucleotide reductase messenger RNA expression and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res. 2004;10:1318–1325. doi: 10.1158/1078-0432.ccr-03-0156. [DOI] [PubMed] [Google Scholar]
- 10.Bergman A., Eijk P., van Haperen V., Smid K., Veerman G., Hubeek I., van den Ijssel P., Ylstra B., Peters G. In vivo induction of resistance to gemcitabine results in increased expression of ribonucleotide reductase subunit M1 as a major determinant. Cancer Res. 2005;65:9510–9516. doi: 10.1158/0008-5472.CAN-05-0989. [DOI] [PubMed] [Google Scholar]
- 11.Ceppi P., Volante M., Novello S., Rapa I., Danenberg K.D., Danenberg P.V., Cambieri A., Selvaggi G., Saviozzi S., Calogero R., Papotti M., Scagliotti G.V. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol. 2006;17:1818–1825. doi: 10.1093/annonc/mdl300. [DOI] [PubMed] [Google Scholar]
- 12.Nakahira S., Nakamori S., Tsujie M., Takahashi Y., Okami J., Yoshioka S., Yamasaki M., Marubashi S., Takemasa I., Miyamoto A., Takeda Y., Nagano H., Dono K., Umeshita K., Sakon M., Monden M. Involvement of ribonucleotide reductase M1 subunit overexpression in gemcitabine resistance of human pancreatic cancer. Int J Cancer. 2007;120:1355–1363. doi: 10.1002/ijc.22390. [DOI] [PubMed] [Google Scholar]
- 13.Kim S.O., Jeong J.Y., Kim M.R., Cho H.J., Ju J.Y., Kwon Y.S., Oh I.J., Kim K.S., Kim Y.I., Lim S.C., Kim Y.C. Efficacy of gemcitabine in patients with non-small cell lung cancer according to promoter polymorphisms in the ribonucleotide reductase M1 gene. Clin Cancer Res. 2008;14:3083–3088. doi: 10.1158/1078-0432.CCR-07-4591. [DOI] [PubMed] [Google Scholar]
- 14.Gautam A., Bepler G. Suppression of lung tumor formation by the regulatory subunit of ribonucleotide reductase. Cancer Res. 2006;66:6497–6502. doi: 10.1158/0008-5472.CAN-05-4462. [DOI] [PubMed] [Google Scholar]
- 15.Fan H., Huang A., Villegas C., Wright J.A. The R1 component of mammalian ribonucleotide reductase has malignancy-suppressing activity as demonstrated by gene transfer experiments. Proc Natl Acad Sci U S A. 1997;94:13181–13186. doi: 10.1073/pnas.94.24.13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gautam A., Li Z.R., Bepler G. RRM1-induced metastasis suppression through PTEN-regulated pathways. Oncogene. 2003;22:2135–2142. doi: 10.1038/sj.onc.1206232. [DOI] [PubMed] [Google Scholar]
- 17.Bepler G., Sharma S., Cantor A., Gautam A., Haura E., Simon G., Sharma A., Sommers E., Robinson L. RRM1 and PTEN as prognostic parameters for overall and disease-free survival in patients with non-small-cell lung cancer. J Clin Oncol. 2004;22:1878–1885. doi: 10.1200/JCO.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Zheng Z., Chen T., Li X., Haura E., Sharma A., Bepler G. The DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med. 2007;356:800–808. doi: 10.1056/NEJMoa065411. [DOI] [PubMed] [Google Scholar]
- 19.Akita H., Zheng Z., Takeda Y., Chiwan K., Kittaka N., Kobayashi S., Marubashi S., Takemasa I., Nagano H., Dono K., Nkamori S., Monden M., Mori M., Doki Y., Bepler G. Significance of RRM1 and ERCC1 expression in resectable pancreatic adenocarcinoma. Oncogene. 2009;28:2903–2909. doi: 10.1038/onc.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosell R., Skrzypski M., Jassem E., Taron M., Bartolucci R., Sanchez J.J., Mendez P., Chaib I., Perez-Roca L., Szymanowska A., Rzyman W., Puma F., Kobierska-Gulida G., Farabi R., Jassem J. BRCA1: a novel prognostic factor in resected non-small-cell lung cancer. PLoS One. 2007;2:e1129. doi: 10.1371/journal.pone.0001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saviozzi S., Ceppi P., Novello S., Ghio P., Lo Iacono M., Borasia P., Cambieri A., Volante M., Papotti M., Calogero R.A., Scagliotti G.V. Non-small cell lung cancer exhibits transcript overexpression of genes associated with homologous recombination and DNA replication pathways. Cancer Res. 2009;69:3390–3396. doi: 10.1158/0008-5472.CAN-08-2981. [DOI] [PubMed] [Google Scholar]
- 22.Simon G., Sharma A., Li X., Hazelton T., Walsh F., Williams C., Chiappori A., Haura E., Tanvetyanon T., Antonia S., Cantor A., Bepler G. Feasibility and efficacy of molecular analysis-directed individualized therapy in advanced non-small-cell lung cancer. J Clin Oncol. 2007;25:2741–2746. doi: 10.1200/JCO.2006.08.2099. [DOI] [PubMed] [Google Scholar]
- 23.Cobo M., Isla D., Massuti B., Montes A., Sanchez J.M., Provencio M., Vinolas N., Paz-Ares L., Lopez-Vivanco G., Munoz M.A., Felip E., Alberola V., Camps C., Domine M., Sanchez J.J., Sanchez-Ronco M., Danenberg K., Taron M., Gandara D., Rosell R. Customizing cisplatin based on quantitative excision repair cross-complementing 1 mRNA expression: a phase III trial in non-small-cell lung cancer. J Clin Oncol. 2007;25:2747–2754. doi: 10.1200/JCO.2006.09.7915. [DOI] [PubMed] [Google Scholar]
- 24.Reynolds C., Obasaju C., Schell M.J., Li X., Zheng Z., Boulware D., Caton J.R., DeMarco L.C., O'Rourke M.A., Shaw Wright G., Boehm K.A., Asmar L., Bromund J., Peng G., Monberg M.J., Bepler G. Randomized phase III trial of gemcitabine-based chemotherapy with in situ RRM1 and ERCC1 protein levels for response prediction in non-small-cell lung cancer. J Clin Oncol. 2009;27:5808–5815. doi: 10.1200/JCO.2009.21.9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The International Adjuvant Lung Cancer Trial Collaborative Group Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med. 2004;350:351–360. doi: 10.1056/NEJMoa031644. [DOI] [PubMed] [Google Scholar]
- 26.Camp R.L., Chung G.G., Rimm D.L. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 27.Metro G., Zheng Z., Fabi A., Schell M., Antoniani B., Mottolese M., Monteiro A.N., Vici P., Lara Rivera S., Boulware D., Cognetti F., Bepler G. In situ protein expression of RRM1, ERCC1, and BRCA1 in metastatic breast cancer patients treated with gemcitabine-based chemotherapy. Cancer Invest. 2010;28:172–180. doi: 10.3109/07357900903095722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gustavson M.D., Bourke-Martin B., Reilly D., Cregger M., Williams C., Mayotte J., Zerkowski M., Tedeschi G., Pinard R., Christiansen J. Standardization of HER2 immunohistochemistry in breast cancer by automated quantitative analysis. Arch Pathol Lab Med. 2009;133:60–66. doi: 10.5858/133.9.1413. [DOI] [PubMed] [Google Scholar]
- 29.McCabe A., Dolled-Filhart M., Camp R.L., Rimm D.L. Automated quantitative analysis (AQUA) of in situ protein expression, antibody concentration, and prognosis. J Natl Cancer Inst. 2005;97:1808–1815. doi: 10.1093/jnci/dji427. [DOI] [PubMed] [Google Scholar]


