Abstract
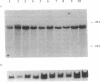
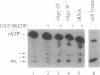
Helicases are essential enzymes for life because DNA replication, DNA repair, recombination, transcription, RNA splicing and translation all involve more than one helicase to unwind DNA or RNA. We have discovered, cloned and partially characterized a novel human helicase gene, SKI2W. The human SKI2W is located between the RD and RP1 genes in the class III region of the major histocompatibility complex (MHC) on chromosome 6, a genomic region associated with many malignant, genetic and autoimmune diseases. Derived amino acid sequence of human SKI2W showed an open reading frame for 1246 residues. It contains consensus sequences for structural motifs of an RNA helicase with a DEVH box. It has a leucine zipper motif that may be important for protein dimerization, and an RGD motif close to the N-terminus that might serve as a ligand for integrin or cell adhesion molecules. SKI2W shares a striking and extensive similarity to the yeast Ski2p that is involved in the inhibition of translation of poly(A) negative [poly(A)-] RNA, and plays an important role in antiviral activities. Human SKI2W fusion protein expressed in insect cells using a baculovirus vector has ATPase activity. The human SKI2W protein and the yeast Ski2p share extensive sequence similarities to another putative human protein KIAA0052, suggesting the presence of a new gene family that may be involved in translational regulation of cellular and viral RNA.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bairoch A., Bucher P. PROSITE: recent developments. Nucleic Acids Res. 1994 Sep;22(17):3583–3589. [PMC free article] [PubMed] [Google Scholar]
- Bayer P., Kraft M., Ejchart A., Westendorp M., Frank R., Rösch P. Structural studies of HIV-1 Tat protein. J Mol Biol. 1995 Apr 7;247(4):529–535. doi: 10.1006/jmbi.1995.0158. [DOI] [PubMed] [Google Scholar]
- Bork P., Koonin E. V. An expanding family of helicases within the 'DEAD/H' superfamily. Nucleic Acids Res. 1993 Feb 11;21(3):751–752. doi: 10.1093/nar/21.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers T. K., Moldow C. F., Bloomfield C. D., Yunis E. J. Familial Hodgkin's disease and the major histocompatibility complex. Vox Sang. 1977;33(5):273–277. doi: 10.1111/j.1423-0410.1977.tb04474.x. [DOI] [PubMed] [Google Scholar]
- Bristow J., Tee M. K., Gitelman S. E., Mellon S. H., Miller W. L. Tenascin-X: a novel extracellular matrix protein encoded by the human XB gene overlapping P450c21B. J Cell Biol. 1993 Jul;122(1):265–278. doi: 10.1083/jcb.122.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Macon K. J., Volanakis J. E. cDNA cloning and characterization of the protein encoded by RD, a gene located in the class III region of the human major histocompatibility complex. Biochem J. 1993 Sep 1;294(Pt 2):589–593. doi: 10.1042/bj2940589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin C. M., Rich S. S., Dehner L. P. Familial aggregation of nasopharyngeal carcinoma and other malignancies. A clinicopathologic description. Cancer. 1991 Sep 15;68(6):1323–1328. doi: 10.1002/1097-0142(19910915)68:6<1323::aid-cncr2820680623>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Company M., Arenas J., Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991 Feb 7;349(6309):487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- Dangel A. W., Mendoza A. R., Baker B. J., Daniel C. M., Carroll M. C., Wu L. C., Yu C. Y. The dichotomous size variation of human complement C4 genes is mediated by a novel family of endogenous retroviruses, which also establishes species-specific genomic patterns among Old World primates. Immunogenetics. 1994;40(6):425–436. doi: 10.1007/BF00177825. [DOI] [PubMed] [Google Scholar]
- DeMars R., Spies T. New genes in the MHC that encode proteins for antigen processing. Trends Cell Biol. 1992 Mar;2(3):81–86. doi: 10.1016/0962-8924(92)90077-z. [DOI] [PubMed] [Google Scholar]
- Demant P., Oomen L. C., Oudshoorn-Snoek M. Genetics of tumor susceptibility in the mouse: MHC and non-MHC genes. Adv Cancer Res. 1989;53:117–179. doi: 10.1016/s0065-230x(08)60281-x. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace F. V. RNA helicases: modulators of RNA structure. Trends Cell Biol. 1994 Aug;4(8):271–274. doi: 10.1016/0962-8924(94)90210-0. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Donchenko A. P., Blinov V. M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989 Jun 26;17(12):4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V. Endonuclease (R) subunits of type-I and type-III restriction-modification enzymes contain a helicase-like domain. FEBS Lett. 1991 Oct 21;291(2):277–281. doi: 10.1016/0014-5793(91)81301-n. [DOI] [PubMed] [Google Scholar]
- Hill A., Ploegh H. Getting the inside out: the transporter associated with antigen processing (TAP) and the presentation of viral antigen. Proc Natl Acad Sci U S A. 1995 Jan 17;92(2):341–343. doi: 10.1073/pnas.92.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. E., Henderson S. T., Petes T. D., Prakash S., Bankmann M., Prakash L. Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol Cell Biol. 1992 Sep;12(9):3807–3818. doi: 10.1128/mcb.12.9.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalidi I., Masset M., Gony J., Marcelli A., Jeannet M., Irlé C., Bauters F., Tchernia G., Turpin F., Gisselbrecht C. MHC related genetic susceptibility to Hodgkin's disease. Nouv Rev Fr Hematol. 1989;31(2):149–152. [PubMed] [Google Scholar]
- Kiledjian M., Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992 Jul;11(7):2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Smith J., Claude A., Lin R. J. The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J. 1992 Jun;11(6):2319–2326. doi: 10.1002/j.1460-2075.1992.tb05291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V. Similarities in RNA helicases. Nature. 1991 Jul 25;352(6333):290–290. doi: 10.1038/352290c0. [DOI] [PubMed] [Google Scholar]
- Kühn K., Eble J. The structural bases of integrin-ligand interactions. Trends Cell Biol. 1994 Jul;4(7):256–261. doi: 10.1016/0962-8924(94)90124-4. [DOI] [PubMed] [Google Scholar]
- Lévi-Strauss M., Carroll M. C., Steinmetz M., Meo T. A previously undetected MHC gene with an unusual periodic structure. Science. 1988 Apr 8;240(4849):201–204. doi: 10.1126/science.3353717. [DOI] [PubMed] [Google Scholar]
- Malo D., Skamene E. Genetic control of host resistance to infection. Trends Genet. 1994 Oct;10(10):365–371. doi: 10.1016/0168-9525(94)90133-3. [DOI] [PubMed] [Google Scholar]
- Masison D. C., Blanc A., Ribas J. C., Carroll K., Sonenberg N., Wickner R. B. Decoying the cap- mRNA degradation system by a double-stranded RNA virus and poly(A)- mRNA surveillance by a yeast antiviral system. Mol Cell Biol. 1995 May;15(5):2763–2771. doi: 10.1128/mcb.15.5.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melhem M. F., Kunz H. W., Gill T. J., 3rd A major histocompatibility complex-linked locus in the rat critically influences resistance to diethylnitrosamine carcinogenesis. Proc Natl Acad Sci U S A. 1993 Mar 1;90(5):1967–1971. doi: 10.1073/pnas.90.5.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S., Pelham H. R. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987 Mar 13;48(5):899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- Nomura N., Miyajima N., Sazuka T., Tanaka A., Kawarabayasi Y., Sato S., Nagase T., Seki N., Ishikawa K., Tabata S. Prediction of the coding sequences of unidentified human genes. I. The coding sequences of 40 new genes (KIAA0001-KIAA0040) deduced by analysis of randomly sampled cDNA clones from human immature myeloid cell line KG-1. DNA Res. 1994;1(1):27–35. doi: 10.1093/dnares/1.1.27. [DOI] [PubMed] [Google Scholar]
- Oomen L. C., van der Valk M. A., Hart A. A., Demant P., Emmelot P. Influence of mouse major histocompatibility complex (H-2) on N-ethyl-N-nitrosourea-induced tumor formation in various organs. Cancer Res. 1988 Dec 1;48(23):6634–6641. [PubMed] [Google Scholar]
- Patel S. S., Rosenberg A. H., Studier F. W., Johnson K. A. Large scale purification and biochemical characterization of T7 primase/helicase proteins. Evidence for homodimer and heterodimer formation. J Biol Chem. 1992 Jul 25;267(21):15013–15021. [PubMed] [Google Scholar]
- Pause A., Méthot N., Svitkin Y., Merrick W. C., Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994 Mar 1;13(5):1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perozzi G., Prakash S. RAD7 gene of Saccharomyces cerevisiae: transcripts, nucleotide sequence analysis, and functional relationship between the RAD7 and RAD23 gene products. Mol Cell Biol. 1986 May;6(5):1497–1507. doi: 10.1128/mcb.6.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumpton M., McGarvey M., Beggs J. D. A dominant negative mutation in the conserved RNA helicase motif 'SAT' causes splicing factor PRP2 to stall in spliceosomes. EMBO J. 1994 Feb 15;13(4):879–887. doi: 10.1002/j.1460-2075.1994.tb06331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer F. M., Palermos J., Zhu Z. B., Barger B. O., Cooper M. D., Volanakis J. E. Individuals with IgA deficiency and common variable immunodeficiency share polymorphisms of major histocompatibility complex class III genes. Proc Natl Acad Sci U S A. 1989 Oct;86(20):8015–8019. doi: 10.1073/pnas.86.20.8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., Guthrie C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 1992 Dec;11(13):5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby C. P., Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993 Apr 2;260(5104):53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- Speiser P. W., White P. C. Structure of the human RD gene: a highly conserved gene in the class III region of the major histocompatibility complex. DNA. 1989 Dec;8(10):745–751. doi: 10.1089/dna.1989.8.745. [DOI] [PubMed] [Google Scholar]
- Strauss E. J., Guthrie C. PRP28, a 'DEAD-box' protein, is required for the first step of mRNA splicing in vitro. Nucleic Acids Res. 1994 Aug 11;22(15):3187–3193. doi: 10.1093/nar/22.15.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surowy C. S., Hoganson G., Gosink J., Strunk K., Spritz R. A. The human RD protein is closely related to nuclear RNA-binding proteins and has been highly conserved. Gene. 1990 Jun 15;90(2):299–302. doi: 10.1016/0378-1119(90)90194-v. [DOI] [PubMed] [Google Scholar]
- Torres A., Martinez F., Gómez P., Gómez C., Garcia J. M., Nuñez-Roldan A. Simultaneous Hodgkin's disease in three siblings with identical HLA-genotype. Cancer. 1980 Aug 15;46(4):838–843. doi: 10.1002/1097-0142(19800815)46:4<838::aid-cncr2820460433>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Troelstra C., Jaspers N. G. Recombination and repair. Ku starts at the end. Curr Biol. 1994 Dec 1;4(12):1149–1151. doi: 10.1016/s0960-9822(00)00260-8. [DOI] [PubMed] [Google Scholar]
- Trowsdale J., Ragoussis J., Campbell R. D. Map of the human MHC. Immunol Today. 1991 Dec;12(12):443–446. doi: 10.1016/0167-5699(91)90017-n. [DOI] [PubMed] [Google Scholar]
- Tso J. Y., Sun X. H., Kao T. H., Reece K. S., Wu R. Isolation and characterization of rat and human glyceraldehyde-3-phosphate dehydrogenase cDNAs: genomic complexity and molecular evolution of the gene. Nucleic Acids Res. 1985 Apr 11;13(7):2485–2502. doi: 10.1093/nar/13.7.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widner W. R., Wickner R. B. Evidence that the SKI antiviral system of Saccharomyces cerevisiae acts by blocking expression of viral mRNA. Mol Cell Biol. 1993 Jul;13(7):4331–4341. doi: 10.1128/mcb.13.7.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. C., Morley B. J., Campbell R. D. Cell-specific expression of the human complement protein factor B gene: evidence for the role of two distinct 5'-flanking elements. Cell. 1987 Jan 30;48(2):331–342. doi: 10.1016/0092-8674(87)90436-3. [DOI] [PubMed] [Google Scholar]