Abstract
Protein hydration is important to protein structure and function. Molecular distribution functions have been an invaluable tool to study protein hydration. Proximal radial distribution functions (pRDFs) have been postulated as being transferable across proteins based on evidence collected from two proteins [V. A. Makarov, B. K. Andrews, and B. M. Pettitt, Biopolymers 45(7), 469 (1998)]. Here we selected nine proteins with different sizes as well as different secondary topologies. We show that pRDFs are universal for proteins with compact structures. We further compare these pRDFs with those calculated from polyglycines that have no defined structures to consider the extent of the validity of this approach.
Proteins most often exist and perform their functions in an aqueous environment.1 Much knowledge about hydration has been obtained from studying molecular pair distribution functions both experimentally and theoretically. These functions2 can be cumbersome to evaluate. Orientationally averaged or radial distribution functions are less useful analyzing solutions of macromolecules with low symmetry and considerable structural anisotropy.3
In view of these difficulties, a distribution function based on the proximity criteria, called proximal radial distribution function (pRDF), was proposed.4, 5 The resultant distribution function can be defined as perpendicular to the surface of the molecule of interest6, 7, 8 which has been used to elucidate the hydration for small organic molecules,4 proteins,9, 10 and DNAs.8, 11 The pRDF can be used to rapidly build an approximate solvent distribution which can be used for experimental and theoretical analysis.12
It has been hypothesized that the pRDF is a universal descriptor of solvation due to the fact that interactions between proteins and solvent are largely local9, 10, 13 once correlations with the rest of the solute are treated as a condition of the distribution. Makarov et al. obtained pRDFs from molecular dynamics simulations of two dissimilar proteins using the same force field and found that they are nearly identical including comparison with the experimentally determined functions providing further evidence in support of this hypothesis.9 More extensive studies of various proteins with different sizes and∕or different secondary topologies are thus needed to provide more evidence to support the hypothesis.
We carried out solvated molecular dynamics simulations on 9 proteins with different chain lengths from 16 amino acids to 274 amino acids with protein data bank identifiers, 1A0M, 2JOF, 1SHG, 4PTI, 4AZU, 2MGK, 1PSD, 1QAE, and 1A8Q. Structures include α, β, α∕β, even a de novo designed protein (2JOF). The CHARMM force field14 was chosen and CHARMM (Ref. 15) (version c34b1) was used. The proteins were solvated in a rectangular box of TIP3P water16 such that no protein surface is closer than 10 Å to any side of the box. Sodium or chloride ions were added to neutralize the system. Lennard-Jones interactions were truncated at 10 Å with a switching function applied starting at 8 Å. After 200 steps of minimization, each system was equilibrated for 400 ps. The production run was continued to 2 ns using the NPT ensemble at 300 K and 1 atm pressure.17 Electrostatics were calculated using the particle mesh Ewald method.18SHAKE19 was used to constraint all bonds involving hydrogen atoms. The leapfrog algorithm was utilized with a time step of 2 fs. The trajectory coordinates were saved every 0.1 ps.
A pRDF for each protein was calculated in the same way as by Makarov et al.9 A 0.5 Å grid spacing was used for collection and normalization as it proved sufficiently accurate previously.9 It is possible to group a certain set of solute atoms on which pRDF is calculated together based on chemical similarities or force field atom types.8, 9, 10 Previous results showed that simple grouping based on chemical similarities is sufficient.8, 9 In this study, we followed the minimally useful convention to group the solute atoms according to three kinds of atom types: oxygen, nitrogen, and carbon.
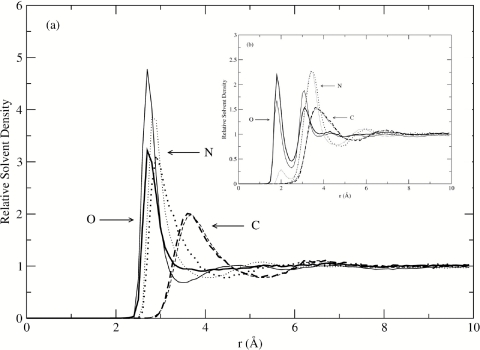
The pRDFs for water oxygen atoms are presented in Fig. 1a and pRDFs for water hydrogen atoms are presented in Fig. 1b. We find that the pRDFs of all nine proteins are in good agreement with each other. Despite a wide range of conformational flexibility, different chain lengths, and protein structural classifications, the observation that pRDFs are nearly constant among all the globular proteins investigated here agrees with the initial hypothesis.
Figure 1.
(a) Comparison of pRDF of water oxygen atoms relative to the oxygen, nitrogen, and carbon atoms of proteins with various sizes (1A0M: conotxin, black; 1A8Q: bromoperoxidase, red; 1QAE: Serratia endonuclease, green; 1SHG: SH3, blue; 1SPD: SOD, yellow; 2JOF: W-cage, brown; 2MGK: myoglobin, violet; 4AZU: azurin, cyan; 4PTI: BPTI, magenta); solid line: oxygen atoms; dotted line: nitrogen atoms; dashed line: carbon atoms. (b) The same as above for water hydrogen atoms.
We observed that although the first peak positions for all the proteins investigated here are in excellent agreement with each other, there are some variations for the water–oxygen peak heights to the oxygen and nitrogen atoms of the proteins. The standard deviations for the first peak in pRDFs of water oxygen atoms relative to protein oxygen atoms, nitrogen atoms, and carbon atoms are 0.27, 0.28, and 0.02, respectively, and 0.21 and 0.06 for the two peaks in pRDFs of water hydrogen relative to protein oxygen atoms, 0.06 and 0.02 to protein nitrogen and carbon atoms, respectively. The variations are due to both the differences of chemical nature in the selected solute atom groups (C, N, O) and the local protein geometric range and, thus, the local environment of the atoms. Most of the exposed carbon atoms of the proteins pRDF are nonpolar and so contain the common features of hydrophobic hydration,20 matching well both for peak positions and for peak heights. On the other hand, O and N atoms on the protein surfaces are able to form hydrogen bonds with water molecules. Considering variations for the number of hydrogen bonds, the variations for the pRDF heights are relatively slight.
We expect pathologies where the apparent universality may not hold for some systems. One notable case is that of short polypeptides without a well-formed, solvent-excluded hydrophobic core. To demonstrate this point, simulations were performed with Gly15, which has been observed not to form well-defined secondary structures in simulations.21 The pRDFs are plotted in Fig. 2. The peak positions still agree well, not surprisingly, because they are primarily determined by the short-ranged repulsive interactions, while the peak height of water oxygen atoms and hydrogen atoms relative to the O and N atoms on the globular proteins is much higher than that on polyglycine. Note the peak position and the height of water relative to the C atoms on both the globular proteins and the polyglycine are almost identical.
Figure 2.
(a) Comparison of pRDF for water oxygen atoms relative to the oxygen, nitrogen, and carbon atoms of myoglobin (thin line) and polyglycine (thick line); solid line: oxygen atoms; dotted line: nitrogen atoms; dashed line: carbon atoms. (b) The same as in (a) with water hydrogen atoms.
The universality of the pRDF of proteins has applications in protein solvation modeling because approximate reconstruction of the solvation shells is computationally trivial.9, 22 Uses in crystallographic refinement and other experimental structural methods have been around for some time.7, 9 Simulations, despite their successes in investigating biomolecular structure and dynamics,3, 23 demand relatively extensive computation in many data refinement schemes. This work allows the possibility to evaluate structure and thermodynamics including solvation quickly with reasonable accuracy,7, 8, 9 which is essential for instance to virtual screening of databases in rational drug design.24
Acknowledgments
The authors thank G. C. Lynch, K.-Y. Wong, H. Kokubo, and C. Y. Hu for many valuable discussions. We thank NIH (GM-037657) and the R. A. Welch Foundation (E-1024) for partial financial support. The computations were performed in part using the NSF metacenter facilities and the Molecular Science Computing Facility sponsored by DOE’s Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory, operated by Battelle.
REFERENCES
- Prabhu N. and Sharp K., Chem. Rev. 106(5), 1616 (2006). 10.1021/cr040437f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. P. and McDonald I. R., Theory of Simple Liquids, 3rd ed. (Academic, Amsterdam, the Netherlands, 2006). [Google Scholar]
- Brooks C. L., Karplus M., and Pettitt B. M., Proteins: A Theoretical Perspective of Dynamics, Structure and Thermodynamics (Wiley, New York, 1990). [Google Scholar]
- Mehrotra P. K. and Beveridge D. L., J. Am. Chem. Soc. 102(13), 4287 (1980). 10.1021/ja00533a001 [DOI] [Google Scholar]
- Mezei M. and Beveridge D. L., Methods Enzymol. 127, 21 (1986). 10.1016/0076-6879(86)27005-6 [DOI] [PubMed] [Google Scholar]
- Lounnas V. and Pettitt B. M., Proteins 18(2), 148 (1994). 10.1002/prot.340180207 [DOI] [PubMed] [Google Scholar]
- Lounnas V., Pettitt B. M., and Phillips G. N., Biophys. J. 66(3 Pt 1), 601 (1994). 10.1016/S0006-3495(94)80835-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki W. R. and Pettitt B. M., Biopolymers 41(1), 107 (1997). [DOI] [PubMed] [Google Scholar]
- Makarov V. A., Andrews B. K., and Pettitt B. M., Biopolymers 45(7), 469 (1998). [DOI] [PubMed] [Google Scholar]
- Jha A. K., Colubri A., Zaman M. H., Koide S., Sosnick T. R., and Freed K. F., Biochemistry 44(28), 9691 (2005). 10.1021/bi0474822 [DOI] [PubMed] [Google Scholar]
- Feig M. and Pettitt B. M., Biopolymers 48(4), 199 (1998). [DOI] [PubMed] [Google Scholar]
- G. N.PhillipsJr. and Pettitt B. M., Protein Sci. 4(2), 149 (1995). 10.1002/pro.5560040202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig M. and Pettitt B. M., Structure 6(11), 1351 (1998). 10.1016/S0969-2126(98)00135-X [DOI] [PubMed] [Google Scholar]
- MacKerell A. D., Bashford D., Bellott M., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T. K., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiorkiewicz-Kuczera J., Yin D., and Karplus M., J. Phys. Chem. B 102(18), 3586 (1998). 10.1021/jp973084f [DOI] [PubMed] [Google Scholar]
- Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., and Karplus M., J. Comput. Chem. 4(2), 187 (1983). 10.1002/jcc.540040211 [DOI] [Google Scholar]
- Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., and Klein M. L., J. Chem. Phys. 79(2), 926 (1983). 10.1063/1.445869 [DOI] [Google Scholar]
- Hoover W. G., Phys. Rev. A 31(3), 1695 (1985); 10.1103/PhysRevA.31.1695 [DOI] [PubMed] [Google Scholar]; Andersen H. C., J. Chem. Phys. 72(4), 2384 (1980). 10.1063/1.439486 [DOI] [Google Scholar]
- Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., and Pedersen L. G., J. Chem. Phys. 103(19), 8577 (1995). 10.1063/1.470117 [DOI] [Google Scholar]
- Ryckaert J. P., Ciccotti G., and Berendsen H. J. C., J. Comput. Phys. 23(3), 327 (1977). 10.1016/0021-9991(77)90098-5 [DOI] [Google Scholar]
- Choudhury N. and Pettitt B. M., J. Am. Chem. Soc. 127(10), 3556 (2005). 10.1021/ja0441817 [DOI] [PubMed] [Google Scholar]
- Hu C. Y., Kokubo H., Lynch G. C., Bolen D. W., and Pettitt B. M., Protein Sci. 19(5), 1011 (2010). 10.1002/pro.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen J. J., Makowski L., Sosnick T. R., and Freed K. F., Biophys. J. 99(5), 1611 (2010). 10.1016/j.bpj.2010.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M. and McCammon J. A., Nat. Struct. Biol. 9(9), 646 (2002). 10.1038/nsb0902-646 [DOI] [PubMed] [Google Scholar]
- Klebe G., Drug Discovery Today 11(13–14), 580 (2006); 10.1016/j.drudis.2006.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]; Shoichet B. K., Nature 432 (7019), 862 (2004). 10.1038/nature03197 [DOI] [PMC free article] [PubMed] [Google Scholar]