Abstract
Peptides have been proposed to function in intracellular signaling within the cytosol. Although cytosolic peptides are considered to be highly unstable, a large number of peptides have been detected in mouse brain and other biological samples. In the present study, we evaluated the peptidome of three diverse cell lines: SH-SY5Y, MCF7, and HEK293 cells. A comparison of the peptidomes revealed considerable overlap in the identity of the peptides found in each cell line. The majority of the observed peptides are not derived from the most abundant or least stable proteins in the cell, and approximately half of the cellular peptides correspond to the N- or C- termini of the precursor proteins. Cleavage site analysis revealed a preference for hydrophobic residues in the P1 position. Quantitative peptidomic analysis indicated that the levels of most cellular peptides are not altered in response to elevated intracellular calcium, suggesting that calpain is not responsible for their production. The similarity of the peptidomes of the three cell lines and the lack of correlation with the predicted cellular degradome implies the selective formation or retention of these peptides, consistent with the hypothesis that they are functional in the cells.
Keywords: Peptides, peptidomics, hemopressin, HEK293, SH-SY5Y, MCF7
INTRODUCTION
Most proteomic studies utilize digestive enzymes such as trypsin to generate peptides that are subsequently analyzed by mass spectrometry.1 However, it is possible to detect peptides in extracts of tissues and cell lines without prior enzymatic digestion of the sample.2–5 These endogenous peptides are referred to as the peptidome. Initially, peptidomic analyses were conducted as a method to study neuropeptides and peptide hormones; these are signaling molecules that function in a variety of physiological processes.6 In addition to the commonly known neuropeptides, previous peptidomic studies have also detected fragments of intracellular proteins.7 These peptides do not appear to be postmortem breakdown fragments of the proteins, based on their presence in extracts from tissues that were heat-inactivated to eliminate protease activity and extracted with mild conditions that do not cleave peptide bonds.8–13
The proteasomal system does not directly convert proteins into amino acids, but instead breaks proteins into peptides of ~4–25 amino acids.14–16 Thus, peptides are expected to be present within cells as part of the normal turnover process of proteins. Some of these proteasome products are presented on the cell surface in complex with major histocompatibility complex class I molecules but the vast majority of the proteasome products are thought to be rapidly degraded by exopeptidases, with half-lives of several seconds, suggesting that peptides do not accumulate to detectable levels within the cell.17–19 However, only a small number of peptides were tested in experiments measuring turnover and it is possible that a subset of peptides are considerably more stable.
Recent studies have found large numbers of cellular peptides, raising the possibility that they may be involved in biological functions.7 Some of the peptides derived from cytosolic proteins have been found to be secreted 20 and to interact with specific G-protein coupled receptors.21–24 Although the mechanism of secretion is not known, it is presumably distinct from that of classical neuropeptides which are initially translated in the ER, routed through the Golgi, and processed into peptides within secretory granules which are subsequently released from the cell upon stimulation.25 Peptides produced in the cytoplasm, released from the cell by unconventional mechanisms, and which stimulate extracellular receptors have been recently termed ‘non-classical’ neuropeptides, by analogy with the non-classical neurotransmitters such as nitric oxide and lipid endocannabinoids.26 One likely example of a non-classical neuropeptide is RVD-hemopressin, an alpha hemoglobin-derived mouse-brain peptide which binds to CB1 cannabinoid receptors.21, 27
Another possibility is that the cellular protein-derived peptides may function in intracellular signaling.28 Some cytosolic peptides, such as those cleaved from Sterol Regulatory Element Binding Protein and Notch, translocate to the nucleus and function as transcription elements.29, 30 Small peptides that are directly produced from translation of small RNAs have recently been shown to function as transcription factors that affect epidermal differentiation in Drosophila.31 In addition, cytosolic peptides have been proposed to function in protein-protein interactions,32 and some evidence for such a role has been provided.28 Recently, peptides derived from mitochondrial proteins have been shown to contribute to the unfolded protein response in mitochondrial stress.33 Taken together, these studies suggest that the cellular peptidome plays a complex regulatory role in inter- and intracellular signaling.
To better understand the cellular peptidome, it is important to characterize the enzymatic pathway of peptide production. A recent analysis of the mouse brain peptidome found that approximately 50% of the cytosolic and mitochondrial protein-derived peptides represented the N- or C-termini of the precursor proteins.7 This raised the possibility that these peptides were produced by selective proteolysis, and not simply the result of proteasomal protein degradation because the latter would have been expected to produce a large number of internal protein fragments. In the present study, we examined the peptidome of two human cell lines, MCF7, a breast cancer line, and SH-SY5Y, a neuroblastoma. These were selected because they were reported to express alpha hemoglobin mRNA,34, 35 and we were interested in seeing if the hemopressins and other hemoglobin-derived peptides were produced. A quantitative peptidomics approach was used to examine the peptidome of these two cell lines under basal conditions, and under conditions with elevated intracellular calcium levels in order to activate calpains, a family of intracellular proteases known to cleave proteins at limited sites.36 In addition to studying these two cell lines, we compared the peptidome of these cells to that of HEK293 cells, a human embryonic kidney line, and to mouse brain. Only a minority of peptides detected in the MCF7 and SH-SY5Y cells were unique to each cell line, with the vast majority of peptides common to multiple cell lines and/or mouse brain. Only a small subset of the peptides detected in this study were derived from proteins that were highly abundant or highly unstable, and the majority were from moderately abundant and stable proteins. Approximately 50% of the identified peptides represented the N- or C-termini of the precursor protein, consistent with previous studies on mouse brain. However, levels of most peptides did not change upon elevation of intracellular Ca++, indicating that enzymes such as calpain are not involved with the production of these peptides.
MATERIALS AND METHODS
Cells
MCF7 and SH-SY5Y cells were grown to 100% confluence in 15 cm cell culture plates in high glucose Dulbecco’s Modified Eagle’s Medium (D-MEM, Invitrogen 11995), supplemented with 10% fetal bovine serum and 1% pen/strep antibiotic. Three plates were used per group. At the start of the experiment, media were removed from all plates and cells were washed twice with Dulbecco’s phosphate-buffered saline (DPBS, Invitrogen 14040). DPBS (10 ml) with or without 1 µM A23187 was then added to each plate of cells. The plates were incubated at 37°C for 30 min. After the incubation, DPBS was removed, new DPBS was added, and cells were scraped and collected into 50 ml tubes. The tubes were centrifuged at 1,500 rpm for 5 min. The supernatant was removed and cells were resuspended in another 10 ml DPBS, transferred to a new 15 ml tube, and centrifuged for 5 min. The supernatant was removed and the pellet resuspended in 1 ml hot water (80°C). The samples were incubated in an 80°C water bath for 20 min. The cell lysate was then transferred to a 2 ml microfuge tube and centrifuged at 4°C, 13,000 rpm and stored at −80°C overnight. The following day the samples were thawed and centrifuged again at 4°C, 13,000 rpm and the supernatant collected for peptide extraction. HEK293 cells were grown and collected as previously described.37
Quantitative peptidomics
Samples were prepared for mass spectrometry analysis as previously described for mouse brain tissues.10, 12, 38 Briefly, for peptide extraction, samples were concentrated to 550 µl and centrifuged at 4°C, 13,000 rpm for 15 min. The samples were placed on ice for 10 min to cool down and then 55 µl of ice cold 0.1 N HCl was added (for a final concentration of 10 mM HCl), vortex mixed and incubated on ice for 15 min. The samples were centrifuged at 4°C, 13,000 rpm for 40 min and the supernatant was transferred to a new low retention tube and 1/3 final volume of 0.4 M phosphate buffer (pH 9.5) was added. The samples were then stored at −80°C until labeling.
Quantitative peptidomics was performed using the differential isotopic method and trimethylammonium butyrate (TMAB) activated with N-hydroxysuccinimide (NHS), as described 39–41. For isotopic labeling, 22.4 mg of D0-, D3-, D9-, or D12-TMAB-NHS was dissolved in 56 µl DMSO. Briefly, 8 µl of TMAB-NHS solutions were added separately to each group of cells and incubated for 10 min at 23–25°C. Then 1.0 M NaOH was added to adjust the pH to 9.5 and the sample extract solution was incubated another 10 min at 23–25°C. These two steps were repeated six more times to ensure that all peptides were completely labeled. The mixture was then incubated at 23–25°C for two hours and then 44.8 µl of 2.5 M glycine was added to quench the remaining TMAB-NHS reagents. After labeling, all of the labeled samples were pooled and filtered through an Amicon Ultracel 10 kDa centrifugal filter device (Millipore) to remove proteins larger than 10 kDa. To remove any labels from Tyr residues in the peptides, the filtrate was adjusted to pH 9.0 with 1.0 M NaOH, 10 µl of 2.0 M hydroxylamine in DMSO was added, and the sample was incubated for 10 min. This was repeated two more times, for a total added volume of 30 µl hydroxylamine. The peptides were desalted with a PepClean™ C-18 spin column (Pierce) and eluted out of the column with 160 µl of 70% acetonitrile and 0.5% trifluoroacetic acid in water. The samples were frozen, evaporated in a vacuum centrifuge, and stored at −70°C until analysis. Mass spectrometry was performed on a Synapt G1 quadrupole time-of-flight mass spectrometer (Waters Co., EUA). The peptide mixture was desalted on line for 15 min using a Symmetry C18 trapping column (5 µm particles, 180 µm inner diameter × 20mm, Waters). The mixture of trapped peptides was then separated by elution with a water/acetonitrile, 0.1% formic acid gradient through a BEH 130-c18 column (1.7 µm particles, 100 µm inner diameter × 100 mm, Waters). Data were acquired in data-dependent mode, and peptides were automatically selected and dissociated in MS/MS by 10–30eV collisions with argon. Typical LC and electrospray ionization conditions were a flow rate of 600 nl/min, and capillary voltage of 3.5 Kv, block temperature of 100°C, and cone voltage of 100 V as previously described37.
Data Analysis
Data was analyzed using the Mass Lynx program, V4.0. Peptides were identified by MS/MS sequencing by the Mascot program followed by manual verification (described below). For Mascot searches, raw MS/MS data was converted to a peak list using Mascot Distiller, without smoothing or de-isotoping of peaks, using standard default parameters that included the determination of charge state (1+ to 5+). The peaklist was searched against the entire NCBInr database (version 20100702) with the taxonomy Homo sapiens selected (232,404 sequences), peptide and MS/MS tolerance of 0.1 Da, no cleavage enzymes specified, and variable modifications of GIST-Quat (K), GIST-Quat (N-term), GIST-Quat:2H(3) (K), GIST-Quat: 2H(3) (N-term), GIST-Quat:2H(9) (K), GIST-Quat:2H(9) (N-term), Acetyl (Protein N-term), Met-oxide, phospho-Ser/Thr, C-terminal amidation, and N-terminal pyroGlu. Manual verification of the Mascot results considered the following criteria: The observed mass had to be close to the theoretical mass (less than 50 parts per million difference between the masses); peptides had to have the correct charge state (as determined by the number of charged groups) and the number of tags had to be the same as the number of free amines (N-terminus and Lys residues); the Mascot result had to represent the top score of all possible hits, with at least 5 b- and/ or y-ions identified; and >80% of the major fragment ions observed had to match predicted fragments. In addition, for peptides containing 2 or more free amines, all of the TMAB tags incorporated into the peptide had to be the same isotopic form for the particular ion examined by Mascot, and this form had to match that observed from the MS data. For example, if a peptide incorporated three TMAB tags due to an N-terminal amine and two Lys residues, all three of the TMAB labels had to be either the D0, D3, or D9 form of TMAB (the D12 form is not an option in the Mascot program and so it wasn’t considered). This criterion greatly reduced the number of false positives because the Mascot program did not consider that the variable TMAB modifications needed to be consistent within a peptide. Mascot summary reports for peptides that represent a single fragment of a protein are included in the Supplemental Information.
To calculate the relative levels of peptides, the average height of each peak set in a spectrum was measured. For this, the monoisotopic peak and the first 13C-containing peaks were considered. In cases where there was not complete separation of the D0 and D3 peaks, or the D9 and D12 peaks, the isotopic distribution for the peptide was calculated and used to subtract the background for the multiple 13C-containing peaks. The ratio of the A23187-treated samples relative to the average for the untreated control cells was calculated and the average and standard deviation calculated. The variation between the individual untreated control plates versus the average control value was also calculated and used for the scatter plot analysis.
In addition to the replicates of the A23187-treated and untreated samples within each cell line, the entire analysis was performed on two separate cell lines with the expectation that peptides that were consistently altered in both cell lines by the elevation of intracellular Ca++ would be further evaluated by statistical testing. Unfortunately, even though many peptides were detected in common between the two cell lines, none of the peptides met the criterion of showing a consistent change in both cell lines upon elevation of Ca++ levels.
Data analysis for interactome
Data were analyzed through the use of Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com) software version 8.7. A data set containing gi numbers of the precursor protein of each peptide identified in the present study was uploaded into the application. Each identifier was mapped to its corresponding object in Ingenuity's Knowledge Base. Networks of network-eligible molecules were algorithmically generated based on their connectivity. The functional analysis identified the biological functions and/or diseases that were most significant to the data set and to the network. The network molecules associated with biological functions and/or diseases in Ingenuity’s Knowledge Base were considered for the analysis. Right-tailed Fisher’s exact test was used to calculate a p-value determining the probability that each biological function and/or disease assigned to that network is due to chance alone. A network is a graphical representation of the molecular relationships between molecules. Molecules are represented as nodes, and the biological relationship between two nodes is represented as a line. All relationships are supported by at least one reference from the literature, from a textbook, or from canonical information stored in the Ingenuity Pathways Knowledge Base. Human, mouse, and rat orthologs of a gene are stored as separate objects in the Ingenuity Pathways Knowledge Base, but are represented as a single node in the network. Nodes are displayed using various shapes that represent the functional class of the gene product. Filled nodes (grey) represent the proteins identified in the data set and unfilled nodes represent proteins that are part of the network but which were not identified in the present study. Lines indicate the nature of the relationship between the nodes (e.g., solid lines indicate direct interactions, dashed lines represent indirect interactions).
RESULTS
SH-SY5Y and MCF7 cells were used in the present study because of recent reports that these cell lines expressed alpha hemoglobin mRNA.34, 35 Utilizing quantitative real-time PCR, we were able to confirm the presence of alpha hemoglobin mRNA in both the SH-SY5Y and the MCF7cell lines (data not shown). Alpha hemoglobin mRNA was detected at an average of 28 cycles in both cell lines, which is well above the background detection limit of 35–40 cycles. A low level of α hemoglobin protein was found in the SH-SY5Y cell line upon Western blot analysis (data not shown). Therefore, we examined the peptidome of these two cell lines and detected hundreds of peptides in both cell lines, of which we were able to identify 110 peptides in MCF7 and 102 in SH-SY5Y cells. All of the peptides that were found in the SH-SY5Y and the MCF7 cells are listed in Supplemental Tables S1 and S2. This table includes charge state, number of tags and mass of the identified and non-identified peptides, along with GI number and name of the precursor protein for those peptides that were identified, Furthermore, the ratio of abundance of the peptide in A23187 treated cells versus that of controls can be found in these tables along with the standard error between replicates (Supplemental Tables S1 and S2). No hemoglobin-derived peptides were found in our analysis.
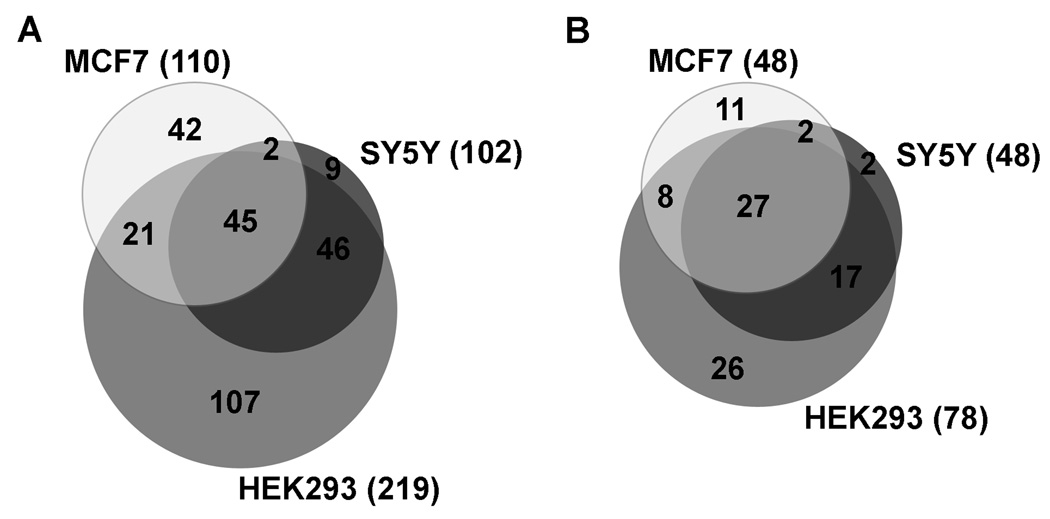
Analysis of the peptides found in each cell line revealed considerable overlap in the peptidome of these cells with each other and with data for HEK293 cells that had been previously obtained (37 and unpublished). Of the 102 peptides that were found in SH-SY5Y cells, only 9 were unique to these cells alone. Ninety-one of these peptides had been previously found in HEK293 cells and 47 were also detected in MCF7 cells. In the MCF7 cells, 68 of the 110 peptides found in these cells had been also found in at least one other cell line. Altogether, 45 of these peptides were common to all three cell lines (Figure 1A). Most of the peptides found in the SH-SY5Y and/or MCF7 cells are fragments of larger proteins, and only 2 are small proteins that passed through the 10 kDa filters. The total 272 distinct peptides found in any of the three cell lines represent 93 distinct proteins (Supplemental Table S3). The identity of all of these peptides can be found in Supplemental Table S3, along with what precursor protein they come from and in which of the three cell lines each peptide was found. Peptides from 27 of these protein precursors are found in all three of the cell lines, and 54 protein precursors are common to at least any two of the cell lines (Figure 1B). Peptides from 26 precursors were found only in the HEK293 cells, but this is likely due to the fact that more experiments have been performed in our laboratory on these cells than on the MCF7 and SH-SY5Y cells. Only 2 proteins were unique to the SH-SY5Y cells and 11 to the MCF7 cells.
Figure 1.
Analysis of the overlap among the peptidome of SH-SY5Y, MCF7, and HEK293 cells. A: Comparison of the peptides found in each of the cell lines. For this analysis, peptides were considered present in multiple cell lines only if the identical peptide was present in the cell lines, and not merely a related peptide from the same protein precursor. The total number of peptides identified in each cell line are indicated in parentheses. B: Comparison of the precursor proteins of the identified peptides found in each cell line. For this analysis, protein precursors were considered to be present in multiple cell lines even if different peptides from the same precursor were identified in each of the cells. The total number of precursor proteins identified in each cell line are indicated in parentheses.
Previous studies on peptides naturally occurring in mouse brain identified 427 peptides that were derived from 112 different cytosolic/mitochondrial proteins.7 Of the 93 distinct protein precursors of the peptides found in the three cell lines, 28 of these proteins were also found to be precursors of peptides found in the mouse brain. At the peptide level, 47 of the 272 different peptides found in any of the three cell lines were also found in brain as the exact homolog (i.e. 100% sequence conservation). Allowing for species differences in the sequence between the human cell lines and mouse brain, the number of similar peptides would be higher.
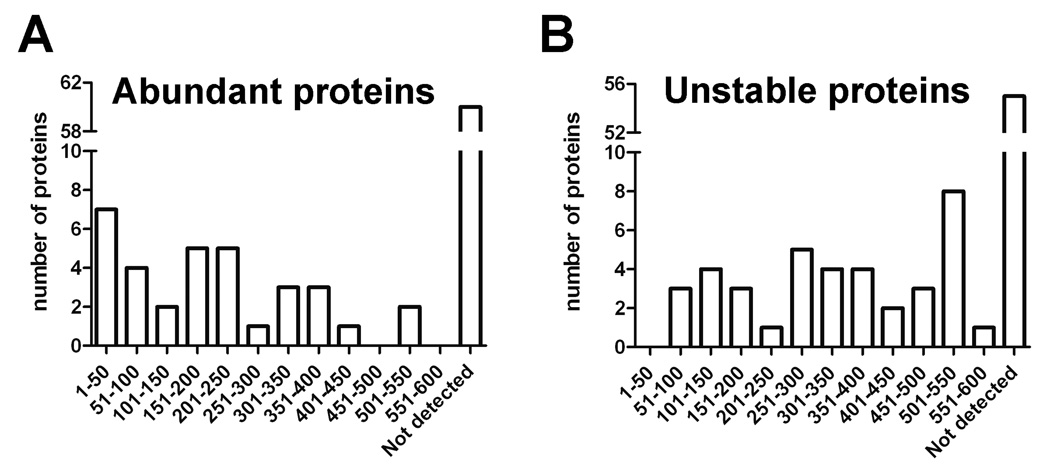
The finding that many of the same peptides are present in multiple human cell lines of diverse origin and also found in mouse brain raised the question as to why these peptides are so highly represented in the peptidome. We first addressed the possibility that these peptides are coming from the major cellular proteins in the cell. Schirle et al conducted a study in which they evaluated the most abundant proteins in HEK293 cells along with some other cell lines42. Only 33 of the peptide precursors found in our study were among the >600 proteins identified in HEK293 cells by Schirle et al. Of these 33 proteins, only 7 were reported to be among the 50 most abundant proteins (Figure 2A). The majority of the precursors of the peptides detected in the present study were not found in the previous proteomics study, either because the proteins were not highly abundant or for technical reasons. Overall, there is no correlation between the peptides found in the present study and the abundance of the proteins from which these peptides are derived (Figure 2A). We further considered the possibility that the protein precursors of the peptides detected in the present study may represent the most unstable proteins in the cell, which would be expected to have high turnover rates. Doherty et al. determined degradation rates for proteins found in HeLa cells.43 From their list of 573 proteins, we matched 39 of the 93 precursor proteins found in our study. None of these 39 proteins are among the top 50 most unstable proteins, and only 3 are among the 100 most unstable proteins (Figure 2B). As with abundance, there is no correlation between protein stability and the detection of a peptide in our peptidomic analyses (Figure 2B).
Figure 2.
The majority of peptides found in the human cell lines do not come from the most abundant cellular proteins (A) or the least stable proteins (B). A: Protein precursors of cellular peptides identified in the present study in SH-SY5Y, MCF7, and HEK293 cells were compared to those previously reported by Schirle et al in HEK293 cells and other cell lines42. The number of proteins found in both studies is plotted on the y-axis, with the results divided into groups based on the relative abundance values from Schirle et al, with 1–50 being the most abundant. Proteins identified in the present study but not detected in the previous proteomic studies are listed as “not detected.” B: Protein precursors of cellular peptides identified in the present study were compared to those previously reported by Doherty et al. in a study examining the degradation rates of HeLa cell proteins43. Of the 93 precursor proteins found in the present study, 38 of these were included in the 573 proteins examined by Doherty et al. The number of proteins in both studies is plotted on the y-axis, with results divided into groups based on the stability; 1–50 represents the least stable group of proteins. Not detected indicates precursor proteins of peptides found in the present study that were not detected by Doherty et al.
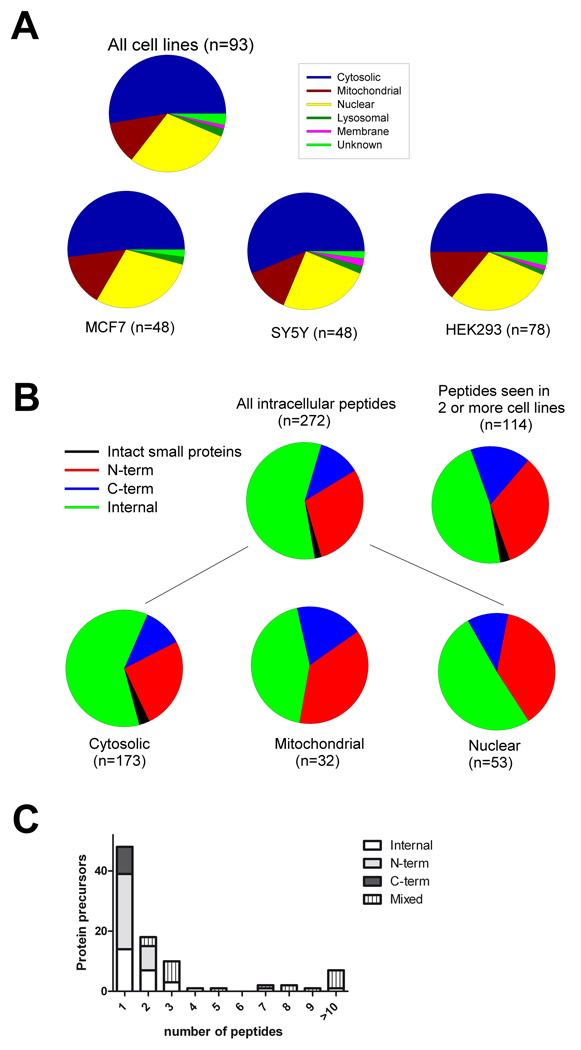
To further examine the characteristics of the peptides and their precursor proteins, we compared the intracellular localization of the precursor protein. Approximately 50% of the 272 peptides found in the present study arise from cytosolic proteins, and a similar fraction is found when the MCF7 and SH-SY5Y peptidomes are considered separately (Figure 3A). Nuclear proteins represent 25–30% of the protein precursors of the peptides found in the present study, and mitochondrial proteins represent 10–15% (Figure 3A). Previously, it was noted that approximately 50% of the non-secretory pathway peptides in mouse brain corresponded to the N- or C-terminus of the cytosolic, mitochondrial, or nuclear protein precursor.7 A similar analysis for the peptidome from the human cell lines revealed that of the 272 peptides found in any of the cell lines, approximately 45% are N- or C-terminal peptides (Figure 3B). Approximately 50% of the peptides derived from nuclear precursors are N- or C-termini, from the mitochondrial precursors slightly more than 50% are N- or C-termini, and of the cytosolic precursors approximately 40–45% are N- or C-termini (Figure 3B). Furthermore, if the analysis is limited to peptides that were seen in 2 or more of the cell lines, the percentage of N- and C-terminal peptides is 50% (Figure 3B). The finding that approximately half of the detected peptides are from the N- or C-termini of the protein fits with a scenario of an endoprotease cleaving proteins at a single site. To evaluate this further, we analyzed how many peptides are typically derived from each protein precursor. Approximately half of all 93 peptide precursors identified in the present study give rise to a single detectable peptide, and only 7 proteins are precursors to 10 or more peptides (Figure 3C).
Figure 3.
Analysis of the intracellular location of the protein precursors identified in the present study, and the relative position of the peptide within the protein. A: The top panel shows the intracellular location of the protein precursors of peptides found in any of the three cell lines. The bottom panels illustrate the intracellular location of the proteins broken down by each individual cell line. B: The relative positions of the identified peptides within the precursor proteins are indicated. The top left pie graph shows the results for all peptides, and the top right graph shows the results only for peptides seen in at least two of the cell lines analyzed. The bottom graphs show the analysis for all peptides broken down by intracellular location of the precursor proteins. C: Analysis of the number of peptides identified from each protein precursor. The y-axis indicates the number of protein precursors and the x-axis indicates the number of peptides from each protein. These peptides were also classified by their location within the protein, either internal fragments, N- or C- termini, or a combination of the above (“mixed”).
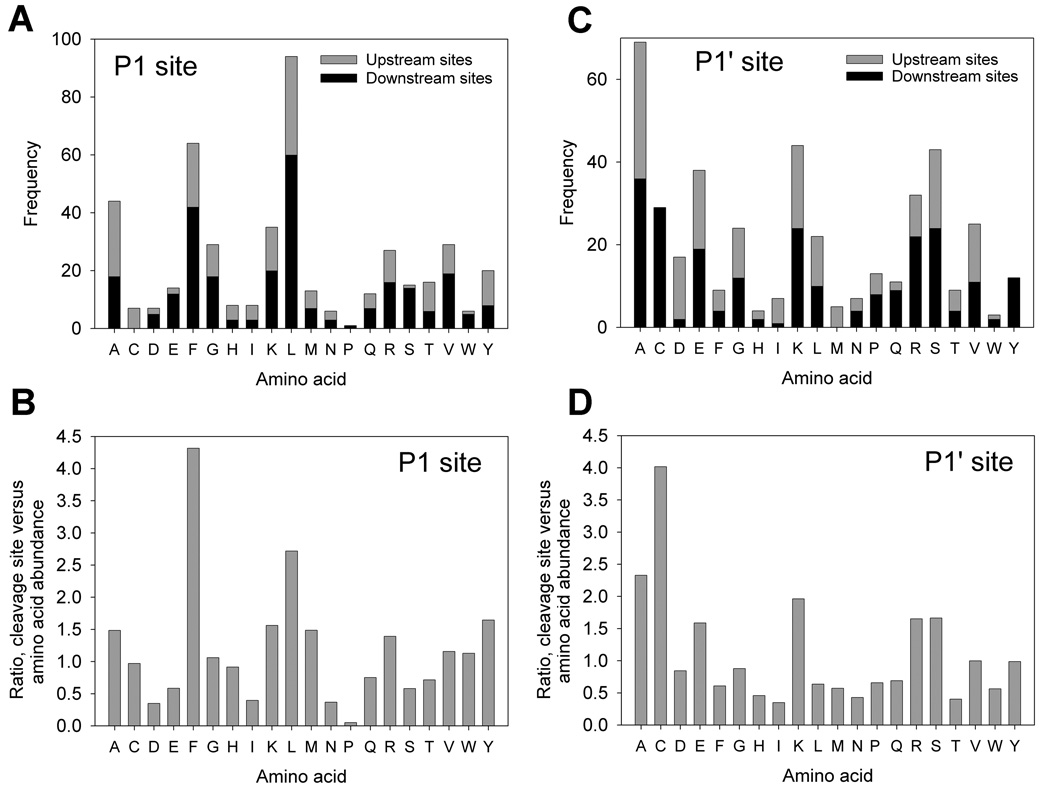
The frequency of various amino acids within the cleavage site was analyzed in order to provide insights as to the enzymes responsible for the cleavages. The most frequently seen amino acids at the P1 location are leucine, phenylalanine, and alanine (Figure 4A). When adjusted to the relative abundance of each amino acid within all human proteins, leucine, phenylalanine, and alanine are still very prominent at the P1 position of the peptides (Figure 4B). In addition, some preference for other hydrophobic residues (such as methionine, tyrosine, tryptophan, and valine) and basic (lysine and arginine) are observed (Figure 4B). Thus, the preferred residues in the P1 position of the cleavage sites are large hydrophobic amino acids. A similar analysis of the residues in the P1’ site of cleavage, based on the observed peptides, shows an abundance of alanine, glutamate, lysine, and serine (Figure 4C). When adjusted to the amino acid abundance in human proteins, these four amino acids are still among the most frequent in the P1’ position, in addition to cysteine and arginine (Figure 4D).
Figure 4.
Analysis of the cleavage site required to produce the peptides found in the three human cell lines. The cleavage site is defined as P1- P1’, with cleavage occurring between the two sites. Peptides located on the N-terminus of the protein require only a downstream cleavage (removal of the N-terminal initiator Met was not considered for the present analysis), and peptides located on the C-terminus of the protein require only an upstream cleavage. Internal peptides require both upstream and downstream cleavages. A: The number of peptides requiring cleavages is shown on the y-axis for each P1 amino acid. B: The P1 cleavage site analysis in panel A was adjusted to the relative abundance of each amino acid within human proteins, and plotted as a ratio such that 1.0 indicates the amino acid to be present in the cleavage sites at a level equal to the overall amino acid composition. Ratios above 1.0 indicate residues more frequently found in the cleavage site than elsewhere in the peptide. C: As in Panel A, except for the P1’ position. D: As in Panel B, except for the P1’ position.
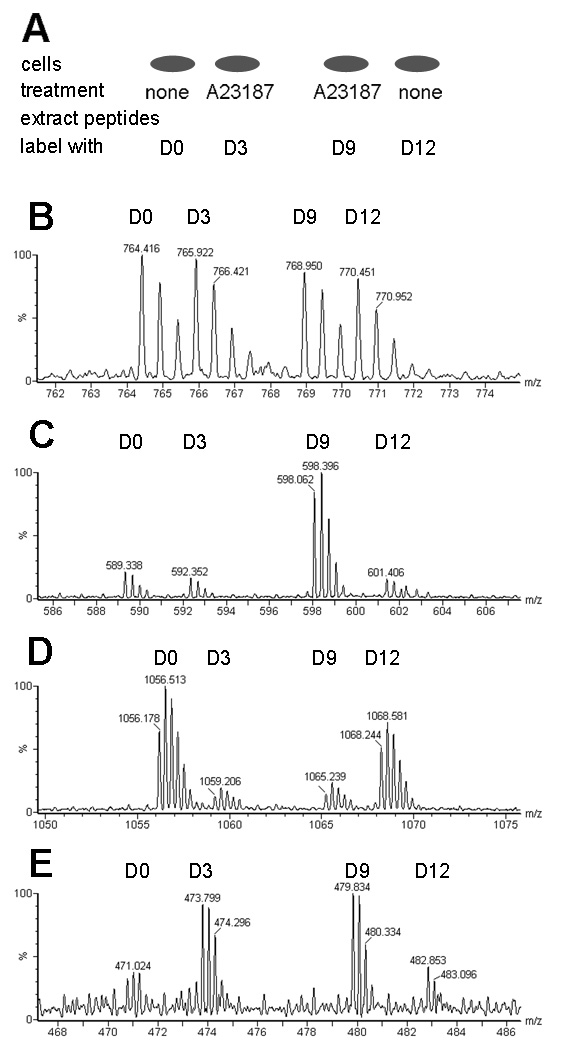
Based on the above data, calpain seems to be a likely candidate for the production of the peptides found in the cell lines. Calpains are proteases that cleave at specific sites within proteins, often at hydrophobic residues,44 and typically perform limited cleavages rather than protein degradation.45 Calpain activity has been reported to be elevated by treatment of SH-SY5Y cells with 1 µM A23187 for 30 min;46 a similar increase was observed with our SH-SY5Y cells using an assay for calpain activity (Supplemental Figure S1). To examine the effect of calpain activation on peptide levels, a quantitative peptidomics technique was used. Two groups of cells from each cell line were treated with 1 µM A23187 for 30 min to raise intracellular calcium levels and increase calpain activity, and another two groups received no treatment. The isotopic labeling strategy for the quantitative peptidomics technique is illustrated in Figure 5A, and representative data are shown in Figures 5B–E. Quantitative analysis was performed on all peptides detected by MS, including those identified by MS/MS sequencing as well as the unidentified peptides (Supplemental Figure S2, Table S1, and Table S2).
Figure 5.
Quantitative peptidomics approach to examine the effect of the calcium ionophore A23187 on levels of peptides in SH-SY5Y and MCF7 cells. A: Labeling scheme for experiments. Cells were either treated with A23187 in DPBS or incubated in DPBS without treatment (“none”). Peptides were extracted from each group and labeled with TMAB tags; the D0 and D12 tags were used with untreated cells and the D3 and D9 tags were used with A23187-treated cells. B: A representative spectrum showing a peptide from SH-SY5Y cells whose levels are not altered by the addition of A23187. The x-axis is mass/charge (m/z) and the y axis is relative abundance of peptide (%). This peptide was subsequently identified from M/S/MS analysis as Ac-MDTSRVQPIKLA (derived from 40S ribosomal protein S28) with 1 TMAB group (on the K) and 1 proton (on the R), for a charge state of 2+ (the TMAB group contributes a positive charge). C: An example of a peptide whose levels are variable among replicates. This unidentified peptide found in SH-SY5Y cells is present at the same relative abundance in the two untreated control groups and in one group of cells treated with A23187 whereas the other treated group has higher levels of this peptide. D: An example of a peptide that is decreased in response to treatment with A23187 in MCF7 cells. The peptide was subsequently identified by MS/MS as Ac-MEQVNELKEKGNKALSVGNIDDALQ (from stress-induced phosphoprotein 1), with 3 TMAB groups and no protons, for a charge of 3+. E: An example of an unidentified peptide that is increased in the groups that were treated with A23187 in MCF7 cells.
When the untreated control cells were compared to each other, the relative levels of peptides was close to the expected ratio of 1.0, and most controls fell within the range of 0.5 to 1.5 (Supplemental Figure S2, Table S1, and Table S2). The majority of peptides detected in both cell lines were not dramatically affected by the A23187 treatment (Figure 5B, Figure S2, Table S1, and Table S2). A few peptides did show an increase (Figure 5E) or a decrease (Figure 5D) with treatment, although most of these changes were not consistent in both groups of A23187-treated cells (Figure 5C). A total of 5 peptides were elevated with a ratio of 2 or higher over controls (Figure S2, Table S1, and Table S2). Of these 5 peptides, 2 are unidentified peptides found in SH-SY5Y cells and the increase was not consistent among the two treatment groups (Figure 5C illustrates the spectrum from one of these peptides). The other 3 peptides increased in MCF7 cells, with consistent increases in both groups of A23187-treated cells, but none of these peptides were identified. Additionally, 28 other peptides (10 of these identified) had a ratio of treated versus controls cells that were above 1.5, but it is not clear if these are consistent increases in response to the treatment. For example, the peptide SAMTEEAAVAIKAMAK derived from Eukaryotic translation initiation factor 5A, was found to be increased 59% (ratio = 1.59±0.09) in SH-SY5Y cells, but the same peptide was found in MCF7 cells at a ratio of 0.81±0.02. This suggests that the increase of this peptide in SH-SY5Y cells is not a universal change associated with the elevation of calcium in all cell lines. Another example is VVRHQLLKT, a peptide derived from Cytochrome c oxidase subunit 7c which is detected in multiple charge states. The peptide with a 4+ charge seen in SH-SY5Y cells is increased with a ratio of 1.50±0.01, but the 3+ charged peptide is not increased substantially, with a ratio of 1.16± 0.05. Furthermore, this same peptide is not increased in MCF7 cells, with values of 0.89±0.08 for the 4+ charge state and 0.94±0.07 for the 3+ charge state. There were also 26 peptides (12 of these identified) that were less abundant in treated cells are compared to controls (ratio of 0.5 or lower). The decreases were only seen in MCF7 cells (Supplemental Table S2). Of the 12 identified peptides, 9 were not found in SH-SY5Y cells and the other 3 were found in SH-SY5Y cell but the levels of these peptides were not changed in the SH-SY5Y cell line (Supplemental Table S1).
To evaluate whether the diverse peptide precursor proteins of the observed cellular peptidome shared any functional similarity, we examined whether these proteins are known to interact with one another. Data were analyzed through the use of Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). This analysis was performed for the protein precursors of peptides found in each individual cell line (data not shown) as well as for those proteins precursors found in multiple cell lines, which represent the more abundant and broadly-expressed peptidome (Supplemental Figure S3 and Table S4). Of the 54 proteins identified as peptide precursors in two or more cell lines, 50 were found to form an interactome, with many of these proteins involved in cell cycle, development, post-translational modifications, protein synthesis, and cell death. The network analysis of the precursor proteins is listed in Supplemental Table S4. These proteins have also been implicated to be involved with cancer, hepatic system disease, and other genetic, neurological, and skeletal/muscular disorders.
DISCUSSION
A major finding of the present study is that the peptidome of three diverse human cell lines, MCF7 breast cancer cells, SH-SY5Y neuroblastoma cells, and HEK293 kidney cells, are remarkably similar. Furthermore, there is a large overlap in peptides and precursor proteins that are found in these three cell lines and in mouse brain. Only a small fraction of the identified peptides represent the most abundant or least stable proteins in the cells, based on proteomic studies from other groups.42, 43 Thus, the cellular peptidome is not merely the degradome of the cellular proteins, but appears to represent a distinct pool of peptides.
We initially used the SH-SY5Y and MCF7 cell lines because they were reported to express alpha hemoglobin mRNA,34, 35 and we were interested in characterizing the enzymes that produce the hemopressins and other hemoglobin-derived peptides. We confirmed the expression of low levels of alpha hemoglobin mRNA and protein in SH-SY5Y cells. In brain that was extensively perfused to remove blood, Western blot analysis detected a strong signal for alpha hemoglobin protein and quantitative real-time PCR detected alpha hemoglobin mRNA after an average of 23–24 cycles.27 This contrasts with the present study, which found alpha hemoglobin mRNA after ~28 PCR cycles in the SH-SY5Y and MCF7 cell lines. While this is still considerably above the background threshold for detection of mRNA, it is an order of magnitude lower than the expression in mouse brain and this presumably explains the low signal for alpha hemoglobin protein and undetectable hemoglobin-derived peptides. As a side point, we also evaluated alpha hemoglobin mRNA levels in SH-SY5Y cells that were differentiated with either retinoic acid and/or nerve growth factor. There was no difference in the levels of alpha hemoglobin mRNA with either of the differentiation conditions (data not shown).
A central question concerns the enzymatic pathway(s) responsible for production of the cellular peptidome. In theory, a likely candidate is the proteasome, a large multi-subunit complex that degrades proteins into peptides.16 One of the major proteolytic activities of the proteasome is the chymotrypsin-like subunit (the β5 subunit), which cleaves proteins after hydrophobic residues. Another subunit (the β2 subunit) has trypsin-like activity and cleaves proteins after basic residues.16, 47 Both of these cleavages are highly represented in the observed cellular peptidome (Figure 4C). Furthermore, the peptides identified in the present study range in size from ~578 to ~3256 Da, with an average of approximately 1374 Daltons; these masses are within the range of proteasome products.15 However, proteasomal degradation would be expected to produce a larger number of internal fragments than terminal fragments. Although internal fragments of some proteins were found to be highly represented in the cellular peptidome (discussed below), the majority of proteins gave rise to only a single detectable peptide which was most often the N- or C-terminus (Figure 3C). Thus, it is unlikely that the proteasome is responsible for the formation of the majority of the observed peptides, although direct studies testing this possibility are necessary.
There were seven proteins which were precursors to ten or more peptides, including many internal peptides. These proteins are triosephosphate isomerase 1, splicing factor arginine/serine-rich 1, heat shock 10kDa protein 1 (chaperonin 10), peptidylprolyl isomerase A, nucleophosmin, heat shock 20kDa protein 1, and keratin 8. The first five of these proteins were previously detected in HEK293 and other cell lines: triosephosphate isomerase 1 is the 13th most abundant, peptidylprolyl isomerase is the 21st most abundant, and heat shock 10kDa protein 1 is the 70th most abundant.42 Splicing factor arginine/serine-rich 1 is lower on the list of abundant proteins, at number 289, and is the 165th most unstable protein while nucleophosmin is number 396 in abundance and 514th-most unstable. 42,43 Peptides derived from heat shock 20kDa protein 1 and keratin 8 were found only in the MCF7 cells and are not on the list of most abundant proteins found in HEK cells.42 For the abundant cellular proteins that give rise to many internal peptides detected in the peptidome, it is possible that the proteasome is involved in their production and the observed fragments therefore represent normal protein turnover intermediates. However, because the majority of the peptides found in the cellular peptidomes represent only a limited region of the protein, typically the N- or C-termini, it is likely that these peptides are preferentially cleaved. The calpains were considered to be likely candidates based on their broad distribution in many cell types, their preference for hydrophobic residues (which fits the observed cleavage sites), and their known preference for limited cleavages of proteins rather than a degradative role.36 However, elevating intracellular Ca++ with the calcium ionophore A23187 does not substantially alter the levels of the majority of the observed peptides, suggesting that the calpains are not involved with the production of most of the observed peptides. Further research is needed to identify the proteases that generate the observed peptides. Interestingly, proteomics studies using specific N-terminal labeling strategies found that only a minority of the proteins in Jurkat cells had N-termini that were the proteins’ N-terminal initiator Met or the penultimate residue after removal of the initiator Met.48 Some of the observed N-termini were due to a known cleavage site due to transit peptide, signal peptide, or propeptide removal, but the majority of observed N-termini were not the predicted N-termini, raising the possibility that selective proteolysis is a common cellular process.
An alternative possibility to explain the large representation of N- and C-termini, or other single fragments from proteins is that the observed peptides may be more stable than others. In this model, the proteins would be broken down by the usual degradation processes (such as the proteasome) and then while most of the products are degraded, some specific peptides are resistant to further degradation. The major pathway for degradation of the peptides formed by the proteasome is thought to involve aminopeptidases, although endopeptidases and carboxypeptidases may also participate.17, 49, 50 Of the 272 peptides that were found in any of the three cell lines studied in the present analysis, 69 are acetylated and would be therefore resistant to aminopeptidase action. While this can contribute to the selective detection of these peptides, and not other fragments from these proteins, the other 203 peptides are not acetylated and would therefore not be resistant to aminopeptidase activity. It is possible that the peptides are stabilized by interactions with cellular proteins or other molecules. Evidence for stabilization of cytosolic peptides has been provided from studies examining antigenic peptides, and appears to involve binding to a heat-shock protein.51
Although the majority of peptide precursor proteins are common to multiple cell lines, a small number of proteins were found in only the MCF7 or SH-SY5Y cell lines, and not also in HEK293 cells (which have been subjected to numerous studies in our laboratory and have a larger dataset than the other cell lines). The two proteins found only in SH-SY5Y cells are heat shock 70kDa protein 1A/1B and LSM5 homolog (U6 small nuclear RNA associated). The eleven proteins found only in MCF7 cells are 14-3-3 protein zeta/delta, proteasome activator subunit 1, cathepsin D, heat shock 27kDA protein 1, beta-galactosidase precursor (lactase), LEM domain-containing protein 2, nuclear casein kinase and cyclin-dependent kinase substrate 1, fructosamine-3-kinase-related protein, and keratins 8, 18, and 19. The majority of these proteins are presumably not unique to just one of the cell lines, and several of these (14-3-3 protein, cathepsin D, and proteasome activator subunit 1) are known to be broadly expressed in various cell types. The three proteins that do stand out are the keratins. While keratins are generally regarded as proteins of contamination in mass spectrometry,52 keratins 8, 18, and 19 have been reported to be present in MCF7 cells.53, 54 Therefore, our finding of peptides derived from keratin 8, 18, and 19 in the MCF7 cells presumably reflects the proteins’ presence in these cells; if contaminants, we should also have detected other keratins not known to be expressed in the MCF7 cells.
The finding that the cellular peptidome reflects a subset of proteins, and within most of these proteins, a subset of the potential fragments implies that these peptides are not merely protein degradation products but are selectively produced and/or selectively retained. Either of these possibilities implies a function for the peptides. An emerging concept is that peptides may function in regulating a large number of protein-protein, protein-lipid, and protein-nucleic acid interations.32 Some evidence for this hypothesis has been recently obtained in a variety of systems including C.elegans,33 Drosophila,31 and mammalian cells.28 Furthermore, many studies have introduced synthetic peptides into cells to influence protein-protein interactions, and low nanomolar concentrations of peptides are often sufficient to affect an array of processes.55–57
In summary, our findings illustrate that the majority of cellular peptides found in three distinct cell lines are similar in identity. We have shown that these peptides are made by selective cleavages that result in a large number of N- and C-terminal peptides which are not coming from the most abundant or unstable proteins. Furthermore, the abundance of the peptides is not increased in response to elevated intracellular Ca2+ levels. Taken together, it is likely that the cellular peptidome plays a complex regulatory role in a variety of intracellular processes, and further studies are needed to identify the precise functions of each peptide as well as the enzymes involved in the formation and degradation of the peptidome.
Supplementary Material
ACKNOWLEDGMENTS
This work was primarily supported by National Institutes of Health grant DA-04494 (L.D.F.). Mass spectrometry was supported by São Paulo State Research Foundation (FAPESP; grant 04/14846 – Rede Proteoma SP), Financiadora de Estudos e Projetos (FINEP grant A-03/134 – Rede Proteoma SP) and Brazilian National Research Council (CNPq; grant 559698/2009-7 - Rede GENOPROT. ESF and LMC are supported by CNPq research fellowships).
REFERENCES
- 1.Wysocki VH, Resing KA, Zhang Q, Cheng G. Methods. 2005;35:211–222. doi: 10.1016/j.ymeth.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 2.Fricker LD, Lim J, Pan H, Che FY. Mass Spectrom Rev. 2006;25:327–344. doi: 10.1002/mas.20079. [DOI] [PubMed] [Google Scholar]
- 3.Svensson M, Skold K, Nilsson A, Falth M, Svenningsson P, Andren PE. Biochem Soc Trans. 2007;35:588–593. doi: 10.1042/BST0350588. [DOI] [PubMed] [Google Scholar]
- 4.Bora A, Annangudi SP, Millet LJ, Rubakhin SS, Forbes AJ, Kelleher NL, Gillette MU, Sweedler JV. J Proteome Res. 2008;7:4992–5003. doi: 10.1021/pr800394e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cape SS, Dowell JA, Li L. Methods Mol Biol. 2009;492:381–393. doi: 10.1007/978-1-59745-493-3_23. [DOI] [PubMed] [Google Scholar]
- 6.Strand FL. Prog Drug Res. 2003;61:1–37. doi: 10.1007/978-3-0348-8049-7_1. [DOI] [PubMed] [Google Scholar]
- 7.Fricker LD. Mol Biosyst. 2010;6:1355–1365. doi: 10.1039/c003317k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skold K, Svensson M, Norrman M, Sjogren B, Svenningsson P, Andren PE. Proteomics. 2007;7:4445–4456. doi: 10.1002/pmic.200700142. [DOI] [PubMed] [Google Scholar]
- 9.Svensson M, Skold K, Svenningsson P, Andren PE. J Proteome Res. 2003;2:213–219. doi: 10.1021/pr020010u. [DOI] [PubMed] [Google Scholar]
- 10.Che FY, Zhang X, Berezniuk I, Callaway M, Lim J, Fricker LD. J Proteome Res. 2007;6:4667–4676. doi: 10.1021/pr060690r. [DOI] [PubMed] [Google Scholar]
- 11.Sturm RM, Dowell JA, Li L. Methods Mol Biol. 615:217–226. doi: 10.1007/978-1-60761-535-4_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Che FY, Lim J, Pan H, Biswas R, Fricker LD. Mol Cell Proteomics. 2005;4:1391–1405. doi: 10.1074/mcp.T500010-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Svensson M, Boren M, Skold K, Falth M, Sjogren B, Andersson M, Svenningsson P, Andren PE. J Proteome Res. 2009;8:974–981. doi: 10.1021/pr8006446. [DOI] [PubMed] [Google Scholar]
- 14.Goldberg AL. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 15.Wenzel T, Eckerskorn C, Lottspeich F, Baumeister W. FEBS Lett. 1994;349:205–209. doi: 10.1016/0014-5793(94)00665-2. [DOI] [PubMed] [Google Scholar]
- 16.Voges D, Zwickl P, Baumeister W. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 17.Rock KL, York IA, Goldberg AL. Nat Immunol. 2004;5:670–677. doi: 10.1038/ni1089. [DOI] [PubMed] [Google Scholar]
- 18.Kloetzel PM. Biochim Biophys Acta. 2004;1695:225–233. doi: 10.1016/j.bbamcr.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Reits E, Griekspoor A, Neijssen J, Groothuis T, Jalink K, van Veelen P, Janssen H, Calafat J, Drijfhout JW, Neefjes J. Immunity. 2003;18:97–108. doi: 10.1016/s1074-7613(02)00511-3. [DOI] [PubMed] [Google Scholar]
- 20.Annangudi SP, Luszpak AE, Kim SH, Ren S, Hatcher NG, Weiler IJ, Thornley KT, Kile BM, Wightman RM, Greenough WT, Sweedler JV. ACS Chem Neurosci. 2010;1:306–314. doi: 10.1021/cn900036x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomes I, Grushko JS, Golebiewska U, Hoogendoorn S, Gupta A, Heimann AS, Ferro ES, Scarlata S, Fricker LD, Devi LA. Faseb J. 2009;23:3020–3029. doi: 10.1096/fj.09-132142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heimann AS, Gomes I, Dale CS, Pagano RL, Gupta A, de Souza LL, Luchessi AD, Castro LM, Giorgi R, Rioli V, Ferro ES, Devi LA. Proc Natl Acad Sci U S A. 2007;104:20588–20593. doi: 10.1073/pnas.0706980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomes I, Dale CS, Casten K, Geigner MA, Gozzo FC, Ferro ES, Heimann AS, Devi LA. Aaps J. 2010 doi: 10.1208/s12248-010-9217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nyberg F, Sanderson K, Glamsta EL. Biopolymers. 1997;43:147–156. doi: 10.1002/(SICI)1097-0282(1997)43:2<147::AID-BIP8>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 25.Nickel W, Rabouille C. Nat Rev Mol Cell Biol. 2009;10:148–155. doi: 10.1038/nrm2617. [DOI] [PubMed] [Google Scholar]
- 26.Gelman JS, Fricker LD. Aaps J. 2010;12:279–289. doi: 10.1208/s12248-010-9186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gelman JS, Sironi J, Castro LM, Ferro ES, Fricker LD. J Neurochem. 2010;113:871–880. doi: 10.1111/j.1471-4159.2010.06653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cunha FM, Berti DA, Ferreira ZS, Klitzke CF, Markus RP, Ferro ES. J Biol Chem. 2008;283:24448–24459. doi: 10.1074/jbc.M801252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kopan R, Schroeter EH, Weintraub H, Nye JS. Proc Natl Acad Sci U S A. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown MS, Goldstein JL. Proc Natl Acad Sci U S A. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, Kageyama Y. Science. 329:336–339. doi: 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- 32.Ferro ES, Hyslop S, Camargo AC. J Neurochem. 2004;91:769–777. doi: 10.1111/j.1471-4159.2004.02757.x. [DOI] [PubMed] [Google Scholar]
- 33.Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. Mol Cell. 2010;37:529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medvedeva VP, Richter F, Chesselet MF. Sfn Abstract. 2009;630.9 [Google Scholar]
- 35.Shankavaram UT, Varma S, Kane D, Sunshine M, Chary KK, Reinhold WC, Pommier Y, Weinstein JN. BMC Genomics. 2009;10:277. doi: 10.1186/1471-2164-10-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Croall DE, Ersfeld K. Genome Biol. 2007;8:218. doi: 10.1186/gb-2007-8-6-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berti DA, Morano C, Russo LC, Castro LM, Cunha FM, Zhang X, Sironi J, Klitzke CF, Ferro ES, Fricker LD. J Biol Chem. 2009;284:14105–14116. doi: 10.1074/jbc.M807916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, Che FY, Berezniuk I, Sonmez K, Toll L, Fricker LD. J Neurochem. 2008;107:1596–1613. doi: 10.1111/j.1471-4159.2008.05722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang R, Sioma CS, Thompson RA, Xiong L, Regnier FE. Anal Chem. 2002;74:3662–3669. doi: 10.1021/ac025614w. [DOI] [PubMed] [Google Scholar]
- 40.Morano C, Zhang X, Fricker LD. Anal Chem. 2008;80:9298–9309. doi: 10.1021/ac801654h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gelman JS, Wardman J, Bhat VB, Gozzo FC, Fricker LD. Methods Mol Biol. doi: 10.1007/978-1-61779-458-2_31. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schirle M, Heurtier MA, Kuster B. Mol Cell Proteomics. 2003;2:1297–1305. doi: 10.1074/mcp.M300087-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Doherty MK, Hammond DE, Clague MJ, Gaskell SJ, Beynon RJ. J Proteome Res. 2009;8:104–112. doi: 10.1021/pr800641v. [DOI] [PubMed] [Google Scholar]
- 44.Cuerrier D, Moldoveanu T, Davies PL. J Biol Chem. 2005;280:40632–40641. doi: 10.1074/jbc.M506870200. [DOI] [PubMed] [Google Scholar]
- 45.Nixon RA, Saito KI, Grynspan F, Griffin WR, Katayama S, Honda T, Mohan PS, Shea TB, Beermann M. Ann N Y Acad Sci. 1994;747:77–91. doi: 10.1111/j.1749-6632.1994.tb44402.x. [DOI] [PubMed] [Google Scholar]
- 46.Chua BT, Guo K, Li P. J Biol Chem. 2000;275:5131–5135. doi: 10.1074/jbc.275.7.5131. [DOI] [PubMed] [Google Scholar]
- 47.Zwickl P, Voges D, Baumeister W. Philos Trans R Soc Lond B Biol Sci. 1999;354:1501–1511. doi: 10.1098/rstb.1999.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saric T, Graef CI, Goldberg AL. J Biol Chem. 2004;279:46723–46732. doi: 10.1074/jbc.M406537200. [DOI] [PubMed] [Google Scholar]
- 50.Berezniuk I, Sironi J, Callaway MB, Castro LM, Hirata IY, Ferro ES, Fricker LD. Faseb J. 2010 doi: 10.1096/fj.09-147942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lev A, Takeda K, Zanker D, Maynard JC, Dimberu P, Waffarn E, Gibbs J, Netzer N, Princiotta MF, Neckers L, Picard D, Nicchitta CV, Chen W, Reiter Y, Bennink JR, Yewdell JW. Immunity. 2008;28:787–798. doi: 10.1016/j.immuni.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stensballe A, Jensen ON. Proteomics. 2001;1:955–966. doi: 10.1002/1615-9861(200108)1:8<955::AID-PROT955>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 53.Traweek ST, Liu J, Battifora H. Am J Pathol. 1993;142:1111–1118. [PMC free article] [PubMed] [Google Scholar]
- 54.Chan R, Rossitto PV, Edwards BF, Cardiff RD. Cancer Res. 1986;46:6353–6359. [PubMed] [Google Scholar]
- 55.Churchill EN, Qvit N, Mochly-Rosen D. Trends Endocrinol Metab. 2009;20:25–33. doi: 10.1016/j.tem.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rubinstein M, Niv MY. Biopolymers. 2009;91:505–513. doi: 10.1002/bip.21164. [DOI] [PubMed] [Google Scholar]
- 57.Arkin MR, Whitty A. Curr Opin Chem Biol. 2009;13:284–290. doi: 10.1016/j.cbpa.2009.05.125. [DOI] [PubMed] [Google Scholar]
- 58.Potter DA, Tirnauer JS, Janssen R, Croall DE, Hughes CN, Fiacco KA, Mier JW, Maki M, Herman IM. J Cell Biol. 1998;141:647–662. doi: 10.1083/jcb.141.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.