Abstract
The first step of the hydrolytic deimination of l-arginine catalyzed by arginine deiminase is examined using ab initio quantum mechanical/molecular mechanical molecular dynamics simulations. Two possible protonation states of the nucleophilic Cys406 residue were investigated and the corresponding activation free energies were obtained via umbrella sampling. Our calculations indicated a reaction free-energy barrier of 21.3 kcal/mol for the neutral cysteine, which is in reasonably good agreement with the experimental kcat of 6.3 s-1, i. e., a barrier of 16.7 kcal/mol. On the other hand, the deprotonated Cys nucleophile yields a free-energy barrier of 6.7 kcal/mol, much lower than the experimental result. The reaction free-energy barriers along with other data suggest that the Cys nucleophile is dominated by its protonated state in the Michaelis complex, and the reaction barrier corresponds largely to its deprotonation.
I. Introduction
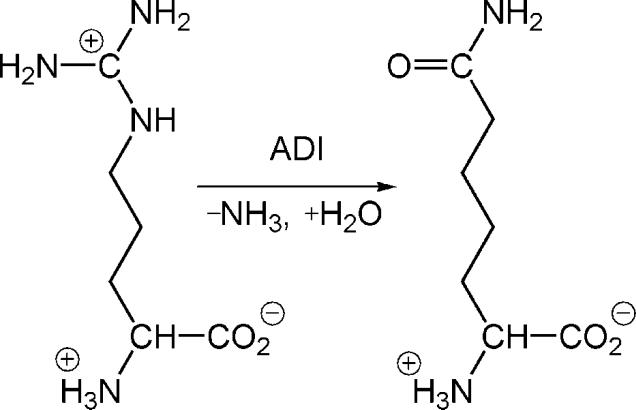
The arginine deiminase (ADI) catalyzes the hydrolytic conversion of l-arginine to l-citrulline, as shown in Scheme 1.1 It represents the initial step of the arginine dihydrolase pathway in some bacteria and parasites such as Pseudomonas aeruginosa and Giardia intestinalis.2-4 In G. intestinalis, which is the causative agent of human giardiasis, this pathway is the main mechanism for energy production,5-6 thus vitally important for the survival of the protozoan parasite. On the other hand, the ADI gene is absent in human genome, making it a potential target for anti-parasite drugs.7-9 In addition to the anti-microbial interest, ADI has also been promoted for cancer therapy, as it inhibits cancer cell growth by depleting l-arginine in circulation.10-11
Scheme 1.
The hydrolytic deimination reaction catalyzed by ADI.
There are several other enzymes that also catalyze the modification of guanidino group of l-arginine. This so-called guanidino group-modifying enzyme superfamily (GMSF),12 or more broadly pentein superfamily,13 includes dimethylarginine dimethylamino hydrolyase (DDAH) which catalyzes the hydrolysis of Nω,Nω-dimethyl-l-arginine (ADMA) to form dimethylamino and citrulline;14-15 and the peptidyl-arginine deiminase (PAD) which catalyzes the deimination of the guanidino group from carboxyl-terminal arginine residues of peptides.16 DDAH serves as a control point for regulating the concentration of ADMA and ·NO production, and its overexpression is related to an array of human diseases such as hypertension and atherogenesis.17-18 The human PADs are involved in the post-translational modification process, which plays a crucial regulatory role in development and differentiation of cells, and are also associated with the human diseases psoriasis, multiple sclerosis, and rheumatoid arthritis.19-21 A clear understanding of the catalytic mechanism of these enzymes will undoubtedly help designing novel and effective mechanism-based inhibitors.
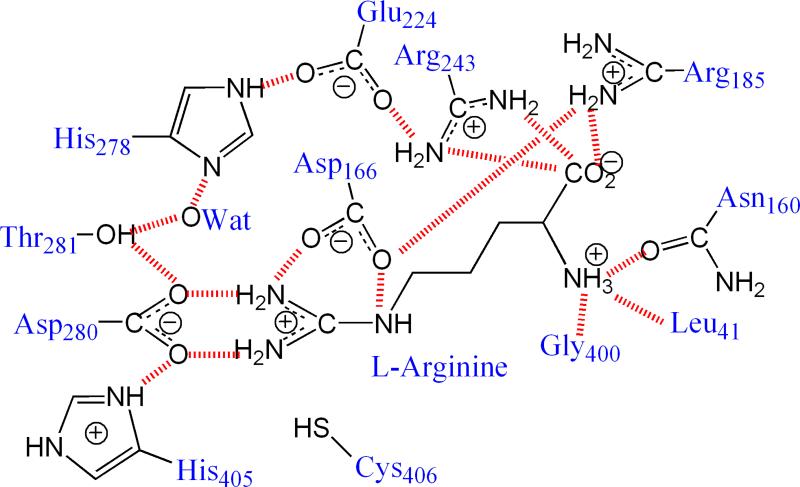
X-ray crystallographic experiments have demonstrated that the overall structure and the active-site architecture of ADI, DDAH, and PADs are quite similar,22-27 despite considerable sequence diversity.12-13 A conserved Cys-His-Glu(Asp) triad is located in the active site of GMSF enzymes, enclosed by the coil regions of five ββαβ modules. In addition, the active sites possess many charged and polar residues involved in substrate binding and catalysis. In ADI, as shown in Fig. 1, the arginine substrate is hydrogen bonded with the side chains of Asp280 and Asp166.26 The structure similarities in GMSF enzyme active sites suggest that these enzymes may utilize the same catalytic mechanism.
Fig. 1.
The hydrogen bond network in the ADI active site. The residue naming scheme is based on the ADI from P. aeruginosa.
The proposed catalytic mechanism of ADI involves two steps. In the initial deimination step, the active-site Cys attacks on the guanidinium Cζ atom, yielding a tetrahedral intermediate, which subsequently collapses to a covalent enzyme-substrate thiouronium intermediate accompanied with the release of an ammonia molecule. In the second hydrolysis step, a water molecule enters the active site to hydrolyze the Sγ-Cζ bond in the thiouronium intermediate, presumably assisted by a general base, generating the final product citrulline. For ADI, the key role of the Cys nucleophile has recently been demonstrated.28 X-ray structures and site-directed mutagenesis experiments have also identified a number of residues that play important roles in both binding and catalysis.29-31 In DDAH and PAD4, the role of the active-site Cys as a nucleophile has also been confirmed.16, 32-34
A still unresolved issue is the protonation state of the nucleophilic Cys residue. It is well established that the cysteine thiol is a poor nucleophile. In cysteine proteases,35 for example, the nucleophilic Cys is activated by a nearby His general base. Although there is indeed a His residue in the conserved ADI catalytic triad, it is however far away from the Cys nucleophile.25 In fact, X-ray structures of a mutant ADI complexed with arginine have demonstrated that these two residues are positioned in different sides of the substrate.24, 26 Several hypotheses have been proposed on the protonation state of the nucleophilic Cys residue. In the so-called substrate-mediated (SM) hypothesis,25 the thiol proton is transferred via the substrate to the His residue on the other side of the substrate. On the other hand, the substrate-assisted (SA) mechanism envisions deprotonation of the Cys residue driven by the positively charged guanidinium group brought in to the active site as the substrate binds to the enzyme.33 So far, experimental evidence is inconclusive to distinguish the two possibilities.31, 33-34
Theoretical investigations on the ADI catalysis have previously been reported by us.31, 36 The earlier classical molecular dynamics (MD) simulations of the active site of ADI confirmed the strong and exquisite hydrogen bond network between the frontline residues of the enzyme and the substrate.36 Density functional theory (DFT) calculations using a truncated active-site model suggested that deprotonation of the Cys nucleophile by the substrate occurs concomitantly with the nucleophilic addition to the substrate guanidino carbon.36 However, such a model is not expected to be quantitatively accurate as many enzyme residues were not included. A more reliable treatment of the catalysis will have to rely on a hybrid quantum mechanical/molecular mechanical (QM/MM) approach, in which the reaction region is treated with a QM Hamiltonian while the rest with a classical force field.37-43 Of particular importance for ADI is the presence of a second period element sulfur, which may not be accurately described using inexpensive semi-empirical methods such as PM3 or AM1. To avoid this problem, an ab initio quantum mechanical/molecular mechanical (QM/MM) study has been performed by us.31 Although a high-level DFT (B3LYP/6-31G**) method was used for the QM region in that work, however, only reaction paths were determined because of the high computational costs. The results suggested that the energy barrier for the deprotonated Cys nucleophilic addition is negligibly small (~2 kcal/mol), while that for the neutral Cys is quite large (~30 kcal/mol). It should be pointed out that the results obtained from reaction path calculations, while reasonable, are not considered definitive because of the neglect of protein dynamics and relaxation.44 Free-energy simulations, particularly those with an ab initio description of the QM region, are necessary to reach quantitatively reliable conclusions.41, 45 Such free-energy QM/MM studies have indeed been performed recently for the catalysis of another GMSF enzyme, PAD4.46-47 To this end, MD calculations indicated that the Michaelis complex with a neutral Cys nucleophile is more stable than that with a deprotonated Cys, and the former yielded a free energy of activation comparable to experimental data.46 In this work, we use the same ab initio QM/MM method to examine the catalysis of ADI. As demonstrated here, the free-energy barriers are quantitatively different from the previous reaction path calculations, underscoring the importance of thermal fluctuation and relaxation. In addition, both ADI and PAD4 seem to have the same reaction mechanism, involving a Michaelis complex dominated by a protonated Cys nucleophile.
II. Method
The initial structure of the enzyme-substrate complex was taken from the crystal structure of the C406A mutant of Pseudomonas aeruginosa ADI co-crystallized with l-arginine (PDB code 2A9G).26 The high resolution (2.30 Å) structure contains four asymmetric units: A, B, C, and D, but only Chain A was used in our calculations. Since residues 1 - 5, 345 – 351, and 418 were unresolved in the crystal structure, Thr6 and Asp417 were treated as N-terminal and C-terminal, while Cly344 and Arg352 are capped with methylamide and acetyl, respectively. Hydrogen atoms were added by Leap in Amber10.48
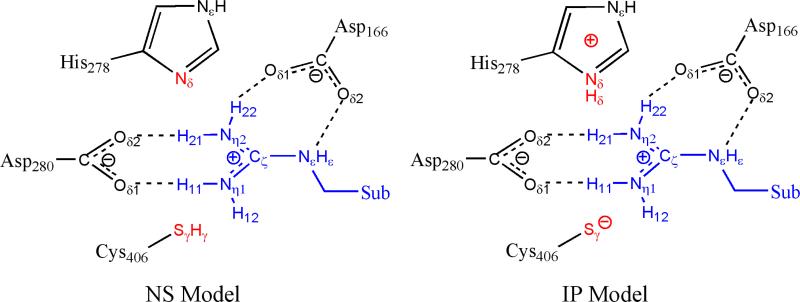
Besides His278, the other eight histidine residues in ADI were assigned as: HID11, HIE29, HIE55, HIP73, HID150, HIE199, HID218, and HIP405 based on their local hydrogen bond interactions with nearby residues. The active-site Cys406 residue was recovered by in silico mutation of Ala406. Two ionization models of ADI were prepared with the same procedure and equilibrated with the same computation protocol. The only difference between the two models is the protonation state of the active-site His and Cys residues: they exist as an imidazolium-thiolate ion-pair in the ionic pair (IP) model, while both are neutral in the neutral state (NS) model, as shown in Fig. 2. The resulting complexes were neutralized by 6 Na+ counterions and solvated in to a periodic TIP3P rectangular water box of ~77×96×81 Å3, with a buffer distance of 10 Å between each wall and the closest atom in each direction, which results in systems of approximately 50,000 atoms in total.
Fig. 2.
The two protonation models used in our calculations.
The solvated systems were first relaxed by a 50 ps NVT and a 50 ps NPT MD simulations with the heavy atoms of protein and crystal water being restrained by a force constant of 50 kcal·mol-1·Å-2. The resulting system was energy refined by reducing the constraint force constant to 20 kcal·mol-1·Å-2. Then, 200 ps production MD simulations with the periodical boundary conditions were carried out using the Amber10 molecular dynamics package.48 In the MD calculations, only the substrate and the Cys406 are constrained with a force constant of 10 kcal·mol-1·Å-2. Analysis of the subsequent 100 ps MD yielded a small (<1.5 Å) RMSD, indicating reasonably equilibrated overall structure. The AMBER99SB force field49-50 and TIP3P water model51 for water molecules were employed. The parameters and the charges for the substrate l-arginine were derived from the peptidyl arginine residue. In the MD simulations, an 8 Å cutoff was introduced for nonbonding interactions, and the particle mesh Ewald (PME) method52-53 was employed to treat long-range electrostatic interactions. The SHAKE algorithm54 was applied to constrain all covalent bonds involving hydrogen.
Using a snapshot from the classical MD simulations, the QM/MM models for both states were constructed by removing water molecules outside of a 27 Å radius sphere centered at the Cζ atom of the arginine substrate. The resulting IP and NS models had 9730 and 9742 atoms, respectively. The QM subsystem was comprised of the side chains of His278, Asp280, Cys406, and the entire substrate l-arginine molecule. The B3LYP functional with 6-31G* basis set was employed for the QM subsystem. The pseudo-bond approach was used to treat the QM/MM boundary.55-56 All the other atoms in the MM subsystem were described classically, with the ABMBER99SB force field49-50 and TIP3P model51 for water molecules. All the QM/MM calculations were performed with modified versions of QCHEM57 and TINKER programs.58 Spherical boundary conditions were applied so that only the atoms inside of 20 Å of the Cζ of l-arginine were free to move. The cutoff radii of 12 and 18 Å were employed for van der Waals interactions and electrostatic interactions among MM atoms, respectively. There was no cutoff for electrostatic interactions between QM and MM atoms.
The QM/MM complexes were first optimized with an iterative minimization procedure. This was followed by reaction coordinate driving (RCD) method59 to map out a minimum energy path. For each structure deterimined along the path, the MM subsystem was subsequently equilibrated with a 500 ps MD simulation, with the QM subsystem frozen. The resulting snapshot was used as the starting structure for the ab initio QM/MM MD calculation of the potential of mean force (PMF) using umbrella sampling,60 In particular, 18 windows were used and each was biased with a harmonic potential of 30 - 120 kcal·mol-1·Å-2 centered on the corresponding value of the reaction coordinate. For IP model, the reaction coordinate is defined as a combination of two bond lengths: ξIP = –dHδ–Nη1 – dSγ–Cζ. For the NS model, it is defined as ξNS = –dHγ–Nη1 – dSγ–Cζ – dH11–Nη1 – dH12–Nη1. In each window, a 30 ps ab initio QM/MM MD simulation was carried out, in which the forces as well as the total energy of the overall QM/MM system were calculated on-the-fly at every time step (Δt=1 fs). Newton's equations of motion were integrated with the Beeman algorithm.61 The Berendsen thermostat62 was used to maintain the system temperature at 300 K. The probability distribution along the defined reaction coordinate was determined for each window and pieced together with the weighted histogram analysis method63 (WHAM) to obtain the PMF. This approach has been successfully applied to a number of enzyme systems,64-66 including a member of the GMSF.46-47
To understand the role played by surrounding residues, we have also computed the electrostatic (ES) and van der Waals (vdW) contributions between individual residues and the QM subsystem in both reactant and transition states. The individual residue contribution was calculated as: , in which the superscripts TS and RC denote the reactant complex and transition state, respectively. Roughly, about 500 snapshots were picked up for each state to take the dynamic into account. A positive value indicates that the ith residue either destabilizes the transition state or stabilizes the reactant.
III. Results and Discussions
A. Substrate-enzyme complexes
In order to explore the protonation state of the ADI active site, we examine two (IP and NS) models. The IP model assumes His and Cys exist as an imidazolium-thiolate ion-pair, while the NS model defines both residues as neutral. Besides the classical MD simulation, the Michaelis complexes for the two models were further examined with 30 ps QM/MM MD simulations. Both complexes were stable during the MD runs, with the substrate held tightly by several active-site residues via hydrogen bonds.
In particular, the guanidino group of the substrate in the NS model forms four hydrogen bonds with Asp166 and Asp280, which is also hydrogen bonded with His405. His278 is hydrogen bonded with Glu224 and a crystal water, but it interacts weakly with the substrate being nearly parallel to the guanidino group. These hydrogen bond interactions are very similar to those in the crystal structure. The nucleophilic Cys406 is located right below the guanidino group, with the thiol proton pointing to the substrate Nη1 atom, and the average distance between Sγ and Cζ is 3.64±0.13 Å. The active-site arrangement for the NS model is quite similar to our earlier classical MD work,36 where only the NS model was examined.
In contrast to PAD4 of which the substrate was found to be unstable in the IP model in our earlier simulations,46 the ion-pair active site of ADI is stable during the 30 ps QM/MM MD simulation. This could be due to the fact that the ADI active site is shielded by a long loop, which prevents the substrate from drifting out of the active site. The hydrogen bond interactions between the substrate guanidino group and Asp166 and Asp280 are similar to that in the NS model. The deprotonated Cys thiolate is hydrogen bonded with a water molecule, Asn360, and the Sγ is placed below the substrate guanidine group with a distance of 3.34±0.33Å from the Cζ. On the other hand, the protonated His278 side chain is hydrogen bonded with Glu224 and a crystal water, and the distance between the transferring proton and the substrate Nη1 atom is 2.60±0.27 Å. There is no hydrogen bond between the protonated His278 and the substrate.
B. Reaction mechanism
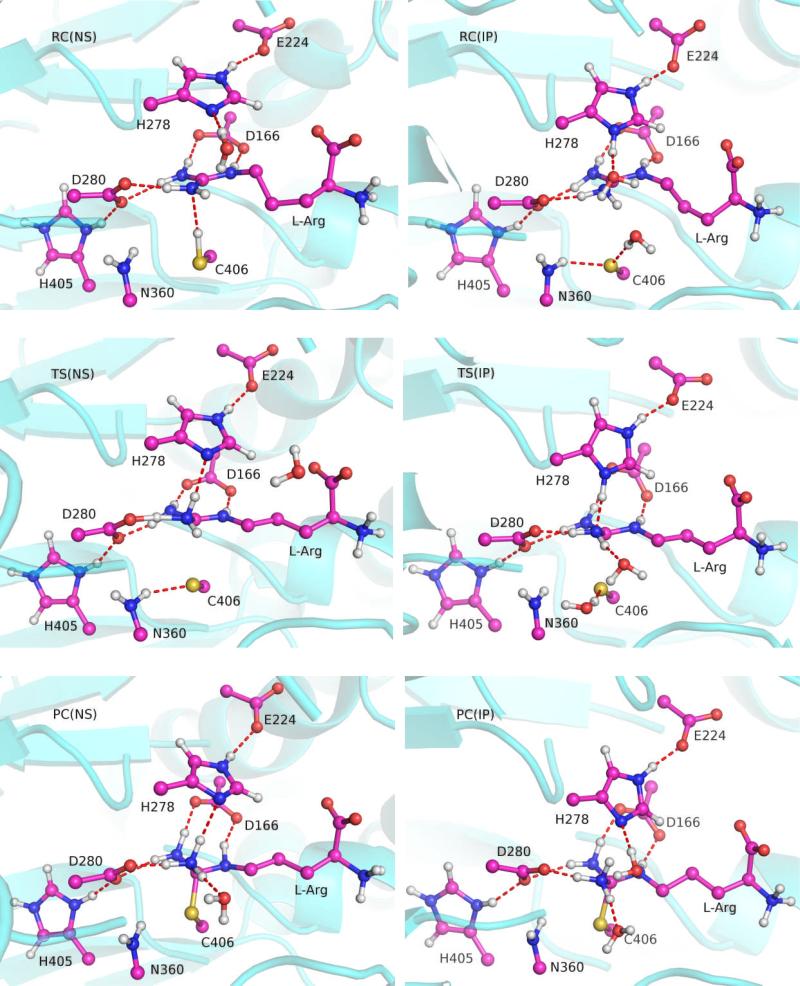
In this work, we will only consider the initial deimination step of the ADI catalysis, which is initiated by nucleophilic attack of Cys406 at the substrate guanidino carbon, leading to the formation of a tetrahedral intermediate. The elimination step, which results in the departure of an ammonia and the formation of the thiouronium intermediate, is known to have a relatively low barrier.31, 36 As a result, we will focus here only on the nucleophilic addition step. The key internuclear distances in the reactant complexes (RC), transition states (TS), and product complexes (PC) are listed in Table I, and the active-site arrangements along the reaction paths are displayed in Fig. 4. Note that PC actually represents the tetrahedral intermediate.
Table I.
Key geometric parameters (Å) of the reactant complex (RC), transition state (TS), and product complex (PC) in both the NS and IP models.
| NS Model | IP Model | |||||
|---|---|---|---|---|---|---|
| Distance (Å) | RC | TS | PC | RC | TS | PC |
| Sγ(C406)-Cζ(Sub) | 3.64±0.13 | 2.53±0.22 | 1.90±0.04 | 3.34±0.33 | 2.38±0.19 | 1.89±0.06 |
| Hγ(C406)- Sγ(C406) | 1.34±0.03 | 2.57±0.16 | 2.67±0.11 | - | - | - |
| Cγ(Sub)-Nη1(Sub) | 1.34±0.02 | 1.45±0.06 | 1.58±0.06 | 1.33±0.12 | 1.41±0.07 | 1.57±0.07 |
| Hγ(C406)- Nη1(Sub) | 2.10±0.14 | 1.02±0.05 | 1.03±0.03 | - | - | - |
| H11(Sub)-Oδ1(D280) | 1.78±0.12 | 1.07±0.11 | 1.68±0.10 | 1.74±0.20 | 1.77±0.14 | 1.60±0.11 |
| H21(Sub)- Oδ2(D280) | 1.95±0.18 | 1.99±0.19 | 1.84±0.23 | 1.93±0.24 | 1.93±0.16 | 2.01±0.14 |
| H22(Sub)- Oδ1(D166) | 1.74±0.08 | 1.73±0.10 | 1.81±0.10 | 1.69±0.16 | 1.80±0.14 | 1.81±0.12 |
| Hε(Sub)- Oδ2(D166) | 1.75±0.08 | 1.95±0.19 | 1.91±0.13 | 1.78±0.23 | 1.85±0.17 | 1.76±0.10 |
| H12(Sub)-Nδ(H278) | 3.42±0.35 | 2.06±0.17 | 1.88±0.16 | - | - | - |
| Hδ(H278)- Nη1(Sub) | - | - | - | 2.60±0.27 | 1.76±0.16 | 1.05±0.03 |
| Hδ(H278)- Nδ(H278) | - | - | - | 1.10±0.09 | 1.07±0.07 | 1.92±0.16 |
| Hε(H278)- Oε1(E224) | 1.77±0.10 | 1.75±0.20 | 1.76±0.10 | 1.69±0.16 | 1.69±0.12 | 1.72±0.09 |
Fig. 4.
Snapshots of the reactant complex (RC), transition state (TS), and product complex (PC) for the NS and IP models. Hydrogen bonds are highlighted with dashed lines.
In the neutral state, the thiol needs to be deprotonated to realize its nucleophilic potential. Indeed, the transfer of the thiol proton to Nη1 of the substrate is essential completed at the TS, as evidenced by the Hγ-Nη1 distance changing from 2.10±0.14 at RC to 1.02±0.05 Å at TS. This resulting Nη1 moiety interacts strongly with His278 and Asp280. In particular, one proton (H11) has already transferred to Asp280 at TS, as evidenced by the H11-Nη1 and H11-Oδ1(Asp280) distances of 1.59±0.19 and 1.07±0.11 Å. As shown in Table I, however, the proton transfer is temporary and it is reversed in the PC. Meanwhile, a stronger hydrogen bond is formed between H12 and Nδ of His278, evidenced by the H12-Nδ(His278) distance of 2.06±0.17 Å, which decreases substantially from 3.42±0.35 Å in the RC. In addition, total ESP charges of His278 and Asp280 change from -0.03±0.03 and -0.87±0.05 in RC to 0.04±0.04 and -0.52±0.09 in TS, indicative of effective spreading of the positive charge from the substrate to His278 and Asp280 at the TS. It is interesting to note that the guanidino group maintains its planarity at TS, suggesting a delayed nucleophilic addition. The indispensible roles played by Asp280, and by His278 to a lesser extent, are consistent with mutagenesis data that the mutants at these two sites resulted in a significant reduction in catalytic efficiency of the deimination step.28
At TS, the deprotonated sulfur atom (Sγ) is positioned right below the Cζ with a distance of 2.53±0.22 Å, poised for the nucleophilic attack. It is observed that Asn360 and a water molecule move closer to the deprotonated thiolate at TS to form hydrogen bonds with Sγ, while the transferred proton appears to have no significant interaction with the thiolate. Based on the TS structure, it can be concluded that the transition state is dominated by deprotonation of the Cys nucleophile, rather than nucleophilic addition itself. This is readily understandable as the strongly nucleophilic thiolate addition requires virtually no activation. Finally, the tetrahedral intermediate is formed in PC, which is characterized by a sp3 hybridized central Cζ.
Unlike in the neutral state, the deprotonated Cys nucleophile in the ion-pair state engages in direct attack of the substrate Cζ. At TS, the Sγ-Cζ distance is shortened to 2.38±0.19 Å, indicating a more advanced nucleophilic addition. In the mean time, the guanidino group is no longer planar. Furthermore, the Cζ-Nη1 bond is elongated from 1.33±0.12 to 1.41±0.07 Å, which presumably arises from the lost of the conjugation in the guanidine group. Accompanying with the nucleophilic attack, the hydrogen bond between His278 and the crystal water molecule broke. Then His278 formed a hydrogen bond with the substrate, as evidenced by the Hδ-Nη1 distance of 1.76±0.16 Å. That means the proton is ready to transfer from His278 to the l-arginine via the existing hydrogen bond. However, by analyzing the trajectories in the windows after the transition state, it is found that the nucleophilic attack occurs prior to the proton transfer. Thus, in contrast to the NS model, the transition state in the IP model is dominated by the nucleophilic addition, which is consistent with the significant charge transfer between atoms Sγ and Cζ, in which that the charges on the atoms Sγ and Cζ vary from -1.02±0.05 and 0.74±0.18 in RC to -0.69±0.12 and 0.31±0.24 in TS. Proton transfer starts immediately after the completion the nucleophilic attack, which finally leads to the tetrahedral intermediate. During the reaction in the IP model, the hydrogen bonds between Asp280 and the guanidino group are well kept. No temporary proton transfer to Asp280 was observed.
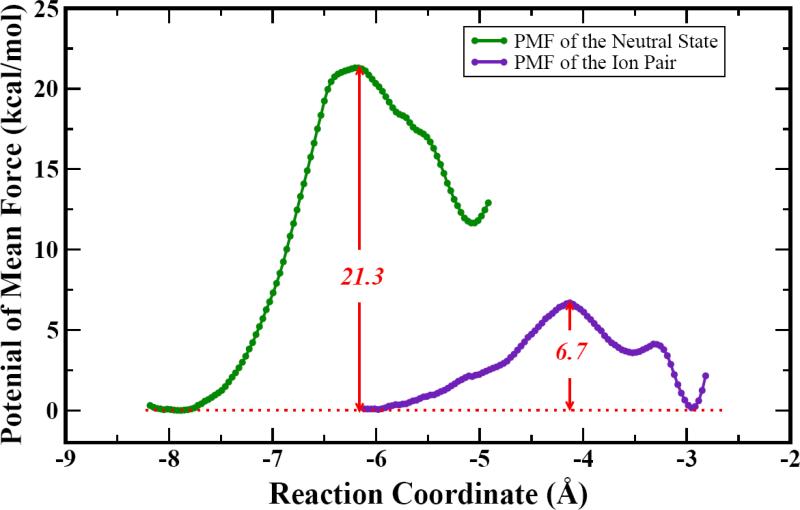
The PMFs for the two models are displayed in Fig. 4. It is readily seen that the NS model produced a free-energy barrier of 21.3 kcal/mol, while the IP model encounters a barrier of 6.7 kcal/mol. The value of the NS model is closer to the experimental result of 16.7 kcal/mol, estimated from the kcat of 6.3 s-1.28-29 The significant difference between the calculated (6.7 kcal/mol) and measured free-energy activation energies argues strongly against the involvement of the deprotonated cysteine nucleophile. A neutral Cys residue in the resting state of ADI is also consistent with our earlier PAD4 study, in which the ionized active site repels the substrate.47 Interestingly, the nucleophilic addition barriers for both free and peptidyl arginines are almost the same in our theoretical calculations (20.9 and 21.3 kcal/mol for PAD4 and ADI). The virtually identical kcat values for PAD4 (6.6 s-1) and ADI (6.3 s-1) also argue for the same catalytic mechanism.16, 29 In addition, it appears that the theory-experiment difference of ~4 kcal/mol in the free-energy barrier is systematic in both enzymes.
We note in passing that the free-energy barriers differ quantitatively from the energy barriers reported in our earlier reaction path study.31 In particular, the free-energy barrier for the NS model is about 9 kcal/mol lower than the energy barrier of 30 kcal/mol,31 underscoring the importance of protein fluctuation and rearrangement.
The mechanistic details described above provided some support for the substrate-mediated (SM) mechanism. In the NS model, we found an early transfer of the thiol proton from Cys406 to the substrate, although it is not further transferred to His278. Instead, our calculations did show a temporary proton transfer to Asp280. This is quite interesting given the lower pKa value of Asp than His. However, our simulations have shown that the hydrogen bonds between the substrate guanidino group and the carboxylate group of Asp280 are particularly strong and well aligned in RC. On the other hand, the imidazole group of His278 is only weakly interacting with the substrate. Interestingly, the proton transfer to Asp280 was initially suggested by Galkin et al.25 and also observed in our earlier DFT calculations with a truncated active-site model.36
The calculated low barrier for the IP model is consistent with our previous QM/MM study.31 Considering the high nucleophilicity of thiolate, it is not surprising that it can easily engage in nucleophilic attack with a small barrier, as found in the NS model. Typically, the thiolate group in the enzyme is stabilized by several hydrogen bonds.67 However, in ADI, only two hydrogen bonds are observed around the ionized S atom, which might not be strong enough to inactivate the thiolate nucleophile. Thus, all our calculations of the IP model for GMSF enzymes suggest that the ion-pair may not be a stable species.
Apparently, our results reported here do not preclude the existence of the ion-pair state in ADI. They merely suggest that it is unlikely that the Michaelis complex is dominated by such a species, as the corresponding reaction free-energy barrier would be inconsistent with the kinetic data. The deprotonated Cys probably exists in ADI as a minor species, whose formation might require significant energy costs. To further understand the relative stability of the two ionization states in the active site of ADI, we have attempted to determine the free-energy profile for their conversion. Unfortunately, our results (not shown) indicated that the transfer of the Cys406 proton to His278 always leads to the reaction. In other words, the conversion between the two ionization states is unavoidably coupled with the reaction path. This is understandable as the deprotonation of the Cys renders it a potent nucleophile.
C. Role of secondary residues
To understand how the enzyme environment facilitates the catalysis, we have examined the influence of secondary residues using a perturbation approach. The residues that we focused on are the ones that have been examined in a recent kinetic experiment,29 namely Glu244, Arg185, Arg243, Asn160, and Arg401. Except for Glu244, all other residues are to some extent in contact with the carboxylate and amino ends of the arginine substrate, as shown in Fig. 1. In Table II, we list the energy contributions of individual residues to catalysis, namely ΔEi for both the NS and IP models. A note of caution here, the perturbation energies should not be taken as quantitative results as the enzyme is not allowed to undergo significant conformational changes in the perturbative calculations.
Table II.
Perturbation energy contributions from five residues of ADI.
| Residue | N160 | R185 | R243 | E224 | R401 |
|---|---|---|---|---|---|
| NS Model | 2.58 | -5.33 | -0.39 | -1.76 | -7.61 |
| IP Model | -0.69 | -3.90 | -8.95 | 12.58 | 0.23 |
From the table, it is clear that all these residues except Asn160 lower the reaction barrier in the NS model, signified by their negative ΔEi values. (As shown in Fig. 1, Asn160 is primarily involved in binding of the carboxylate of the substrate and its catalytic role is probably minimal. Indeed, the Cγ-Cζ distance between Asn160 and the substrate guanidino group is about 10 Å. Thus, the small positive ΔEi for Asn160 is probably inconsequential.) The general trend in the Table is consistent with the experimental observation that mutations at these sites lead to an increase in Km and a decrease in kcat.28 It is readily understandable that the removal of binding residues such as Arg185 decreases the substrate affinity. The effect on catalysis is probably more subtle. It is conceivable that mutations will affect the precise alignment of the substrate in the active site.
The ΔEi values for the IP model are not entirely consistent with the experimental findings. The exception is Glu224, which is hydrogen bonded with His278. Our analysis indicates that this residue seems to significantly increase the IP reaction barrier, which can be understood as its hydrogen bond with the protonated His278 increases the basicity of the imidozalium. Since the proton transfer from His278 to the substrate is an important process near the IP transition state, the removal of the carboxylate side chain of Glu244 would favor the proton transfer, leading to a significantly larger kcat. However, this is in conflict with the mutagenesis data, which indicated that the Glu224 mutants results in significant reduction of kcat.28 This contradiction provided additional evidence in support of the SM mechanism.
IV. Conclusions
In this work, we examined the protonation state of the Cys nucleophile in ADI and its impact on the catalysis. Theoretical simulations offer complementary insights to experimental studies, which are often insufficient to distinguish such mechanistic issues. Our approach is more advanced than those used in earlier theoretical studies of this enzyme. First, a high level density functional theory was used in the QM/MM models, which provided a reliable description of the bond-forming and bond-breaking process in the catalysis. Second, the environmental and entropic effects have been included in the simulation by explicit calculations of the potential of mean force. The combination of the two techniques allows us to gain deeper insights into the catalysis.
Two models were studied. The neutral state (NS) model features a protonated Cys406 and unprotonated His278, while the ion-pair (IP) model envisions a deprotonated Cys406 and protonated His278. Our results indicate that the initial nucleophilic addition in the IP model has a free-energy barrier of 6.7 kcal/mol, much lower than the experimental data. On the other hand, the calculated free-energy of activation is 21.3 kcal/mol in the neutral state model, with is in reasonably good agreement with the experimental value of 16.7 kcal/mol. Thus, we conclude that the enzymatic reaction is likely to involve the protonated Cys nucleophile. This conclusion is consistent with that found in another guanidino modifying enzyme, PAD4, and with site-directed mutagenesis data on ADI.
It was found that the nucleophilic addition of the neutral Cys406 to the guanidino carbon in the substrate is preceded by the transfer of the thiol proton to a guanidino nitrogen. The Asp280 and His278 residues provide stabilization of the transition state via hydrogen bonds and transient proton transfer. In contrast, the transition state of the IP model is dominant by the nucleophilic attack, which occurs prior to the proton transfer from His278 to the substrate.
We finish with a few caveats on the results reported here. Despite the high level of theory used in our simulations, there is no guarantee that the results are error free. Our calculated free energy of activation is, for example, about 4.6 kcal/mol too high compared with experimental data. However, the comparison with the PAD4 study reported earlier46 suggests that this error is systematic for GMSF enzymes. In addition, we would like to point out that the conclusion that the ADI catalysis involves a protonated Cys nucleophile is not absolute, because the relative stability of the two ionization states in the ADI active site has not been definitively established. It is clear that more studies are needed concerning ADI catalysis.
Fig. 3.
Calculated potentials of mean force for the NS and IP models.
Acknowledgements
The work at UNM was supported by NIH (R03-AI071992). DX thanks the support of National Natural Science Foundation of China (20725312) and Chinese Ministry of Science and Technology (2007CB815201). The NYU team was supported by NIH (R01-GM079223) and NSF (CHE-CAREER-0448156). Parts of the computation were carried out at the National Center for Supercomputing Applications (NCSA). HG thanks Debra Dunaway-Mariano for many stimulating discussions.
References
- 1.Smith DW, Ganaway RL, Fahrney DE. Arginine deiminase from Mycoplasma arthritidis. J. Biol. Chem. 1978;253:6016. [PubMed] [Google Scholar]
- 2.Schimke RT, Berlin CM, Sweeney EW, Carroll WR. The generation of energy by the arginine dihydrolase pathway in Mycoplasma hominis 07. J. Biol. Chem. 1966;241:2228. [PubMed] [Google Scholar]
- 3.Schofield PJ, Edwards MR, Matthews J, Wilson JR. The pathway of arginine catabolism in Giardia intestinalis. Mol. Biochem. Parasitol. 1992;51:29–36. doi: 10.1016/0166-6851(92)90197-r. [DOI] [PubMed] [Google Scholar]
- 4.Zuniga M, Perez G, Gonzalez-Candelas F. Evolution of arginine deiminase (ADI) pathway genes. Mol. Phylogenet. Evol. 2002;25:429. doi: 10.1016/s1055-7903(02)00277-4. [DOI] [PubMed] [Google Scholar]
- 5.Knodler LA, Sekyere EO, Stewart TS, Schofield PJ, Edwards MR. Cloning and expression of a prokaryotic enzyme, arginine deiminase, from a primitive eukaryote Giardia intestinalis. J. Biol. Chem. 1998;273:4470. doi: 10.1074/jbc.273.8.4470. [DOI] [PubMed] [Google Scholar]
- 6.Palm JED, Weiland ME-L, Griffiths WJ, Ljungstrom I, Svard SG. Identification of immunoreactive proteins during acute human giardiasis. J. Infect. Dis. 2003;187:1849. doi: 10.1086/375356. [DOI] [PubMed] [Google Scholar]
- 7.Vallance P, Bush HD, Mok J, Hurtado-Guerrero R, Gill H, Rossiter S, Wilden JD, Caddick S. Inhibition of dimethylargine dimethylaminohyrolase (DDAH) and arginine deiminase (ADI) by pentafluorophenyl (PFP) sulfonates. Chem. Comm. 2005:5563–5565. doi: 10.1039/b510709a. [DOI] [PubMed] [Google Scholar]
- 8.Lu X, Li L, Feng X, Wu Y, Dunaway-Mariano D, Engen JR, Mariano PS. L-Canavanine is a time-controlled mechanism-based inhibitor of Pseudomonas aeruginosa arginine deiminase. J. Am. Chem. Soc. 2005;127:16412–16413. doi: 10.1021/ja056226p. [DOI] [PubMed] [Google Scholar]
- 9.Li L, Li Z, Chen D, Lu X, Feng X, Wright EC, Solberg NO, Dunaway-Mariano D, Mariano PS, Galkin A, Kulakova L, Herzberg O, Green-Church KB, Zhang L. Inactivation of microbial arginine deiminases by L-canavaine. J. Am. Chem. Soc. 2008;130:1918. doi: 10.1021/ja0760877. [DOI] [PubMed] [Google Scholar]
- 10.Gong H, Zolzer F, von Recklinghausen G, Havers W, Schweigerer L. Arginine deiminase inhibits proliferation of human leukemia cells more potently than asparaginase by inducing cell cycle arrest and apoptosis. Leukemia. 1999;14:826. doi: 10.1038/sj.leu.2401763. [DOI] [PubMed] [Google Scholar]
- 11.Wheatley DN. Controlling cancer by restricting arginine availability-arginine-catabolizing enzymes as anticancer agents. Anti-cancer Drugs. 2004;15:825. doi: 10.1097/00001813-200410000-00002. [DOI] [PubMed] [Google Scholar]
- 12.Shirai H, Blundell TL, Mizuguchi K. A noval superfamily of enzymes that catalyze the modification of guandino group. Trends in Biochem. Sci. 2001;26(8):465. doi: 10.1016/s0968-0004(01)01906-5. [DOI] [PubMed] [Google Scholar]
- 13.Linsky T, Fast W. Mechanistic similarity and diversity among the guanidine-modifying members of the pentein superfamily. Biochim. Biophys. Acta. 2010;1804:1943–1953. doi: 10.1016/j.bbapap.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leiper JM, Maria JS, Chubb A, MacAllister RJ, Charles IG, Whitley GSJ, Vallance P. Identification of two human dimethylarginine dimethylaminohydrolases with distinct tissue distributions and homology with microbial arginine deiminases. Biochem. J. 1999;343:209. [PMC free article] [PubMed] [Google Scholar]
- 15.Santa Maria J, Vallance P, Charles IG, Leiper JM. Identification of microbial dimethylarginine dimethylaminohydrolase enzymes. Mol. Microbiol. 1999;33:1278–9. doi: 10.1046/j.1365-2958.1999.01580.x. [DOI] [PubMed] [Google Scholar]
- 16.Kearney PL, Bhatia M, Jones NG, L. Y, Glascock MC, Catchings KL, Yamada M, Thompson PR. Kinetic characterization of protein arginine deiminase 4: A transcriptional corepressor implicated in the onset and progression of rheumetoid arthritis. Biochem. 2005;44:10570–82. doi: 10.1021/bi050292m. [DOI] [PubMed] [Google Scholar]
- 17.Vallance P, Leiper J. Blocking NO synthesis: how, where, and why? Nat. Rev. Drug Discov. 2002;1:939. doi: 10.1038/nrd960. [DOI] [PubMed] [Google Scholar]
- 18.Hong L, Fast W. Inhibition of human dimethylarginine dimethylamionhydrolase-1 by S-nitroso-L-homocysteine and hydrogen peroxide: Analysis, quantification, and implications for hyperhomocysteinemia. J. Biol. Chem. 2007;282:34684–34692. doi: 10.1074/jbc.M707231200. [DOI] [PubMed] [Google Scholar]
- 19.Vossenaar ER, Zenderman AJW, van Venrooij WJ, Pruijn GJM. PAD, a growing family of ctirullinating enzymes: genes, features, and involvment in disease. BioEssays. 2003;25:1106. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Wysocka J, Sayegh J, Lee Y-H, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, Stallcup MR, Allis CD, Coonrod SA. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 21.Thompson PR, Fast W. Histone citrullination by protein arginine deiminase: Is arginine methylation a green light or a roadblock. ACS Chem. Biol. 2006;1:433. doi: 10.1021/cb6002306. [DOI] [PubMed] [Google Scholar]
- 22.Murray-Rust J, Leiper J, McAlister M, Phelan J, Tilley S, Maria JS, Valance P, McDonald N. Structural insights into the hydrolysis of cellular nitric oxide synthase inhibitors by dimenthylaminohydrolase. Nature Struct. Biol. 2001;8:679. doi: 10.1038/90387. [DOI] [PubMed] [Google Scholar]
- 23.Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M. Structural basis for Ca2+-induced activation of human PAD4. Nat. Struc. Mole. Biol. 2004;11:777. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 24.Das K, Butler GH, Kwiatkowski V, Clark AD, Jr., Yadav P, Arnold E. Crystal structures of arginine deiminase with covalent reaction intermediates; implications for catalytic mechanism. Structure. 2004;12:657. doi: 10.1016/j.str.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 25.Galkin A, Kulakova L, Sarikaya E, Lim K, Howard A, Herzberg O. Structural insight into arginine degradation by arginine deiminase, an antibacterial and parasite drug target. J. Biol. Chem. 2004;279:14001–14008. doi: 10.1074/jbc.M313410200. [DOI] [PubMed] [Google Scholar]
- 26.Galkin A, Lu X, Dunaway-Mariano D, Herzberg O. Crystal structures representing the Michaelis complex and thiouronium reaction intermediate of Pseudomonas aeruginosa arginine deiminase. J. Biol. Chem. 2005;280:34080–34087. doi: 10.1074/jbc.M505471200. [DOI] [PubMed] [Google Scholar]
- 27.Frey D, Braun O, Briand C, Vasak M, Grutter MG. Structure of the mammalian NOS regulator dimethylarginine dimethylaminohydrolase: A basis for the design of specific inhibitors. Struct. 2006;14(5):901–911. doi: 10.1016/j.str.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 28.Lu X, Galkin A, Herzberg O, Dunaway-Mariano D. Arginine deiminase uses an active-site cysteine in nucleophilic catalysis of L-arginine hydrolysis. J. Am. Chem. Soc. 2004;126:5374. doi: 10.1021/ja049543p. [DOI] [PubMed] [Google Scholar]
- 29.Lu X, Li L, Feng X, Li Z, Yang H, Wang C, Guo H, Galkin A, Herzberg G, Mariano PS, Martin BM, Dunaway-Mariano D. Kinetic analysis of Pseudomonas aeruginosa arginine deiminase mutants and alternate substrates provides insight into structural determinants of function. Biochem. 2006;45:1162–1172. doi: 10.1021/bi051591e. [DOI] [PubMed] [Google Scholar]
- 30.Wei Y, Zhou H, Sun Y, He Y, Luo Y. Insight into the catalytic mechanism of arginine deiminase: functional studies on the crucial sites. Proteins. 2007;66:740–750. doi: 10.1002/prot.21235. [DOI] [PubMed] [Google Scholar]
- 31.Li L, Li Z, Wang C, Xu D, Mariano PS, Guo H, Dunaway-Mariano D. The electrostatic driving force for nucleophilic catalysis in L-arginine deiminase: A combined experimental and theoretical Study. Biochem. 2008;47:4721–4732. doi: 10.1021/bi7023496. [DOI] [PubMed] [Google Scholar]
- 32.Stone EM, Person MD, Costello NJ, Fast W. Characterization of a transient covalent adduct formed during dimethylarginine dimethylaminohydrolase catalysis. Biochem. 2005;44:7069–7078. doi: 10.1021/bi047407r. [DOI] [PubMed] [Google Scholar]
- 33.Stone EM, Costello AJ, Tierney DL, Fast W. Substrate-assisted cysteine deprotonation in the mechanism of dimethylargininase (DDAH) from Pseudomonas aeruginosa. Biochem. 2006;45:5618–5630. doi: 10.1021/bi052595m. [DOI] [PubMed] [Google Scholar]
- 34.Knuckley B, Bhatia M, Thompson PR. Protein arginine deiminase 4: Evidence for a reverse protonation mechanism. Biochem. 2007;46:6578. doi: 10.1021/bi700095s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brockelhust K, Watts AB, Patel M, Verma C, Thomas EW. Cysteine proteases. In: Sinnot M, editor. Comprehensive Biological Catalysis. Vol. 1. Academic Press; San Diego: 1998. [Google Scholar]
- 36.Wang C, Xu D, Zhang L, Xie D, Guo H. Molecular dynamics and density functional studies of substrate binding and catalysis of arginine deiminase. J. Phys. Chem. B. 2007;111:3267. doi: 10.1021/jp067541g. [DOI] [PubMed] [Google Scholar]
- 37.Warshel A, Levitt M. Theoretical studies of enzymatic reactions: Dielectric, electrostatic and steric stabilization of carbonium ion in the reaction of lysozyme. J. Mol. Biol. 1976;103:227–249. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- 38.Gao J. Hybrid quantum and molecular mechanical simulations: An alternative avenue to solvent effects in organic chemistry. Acc. Chem. Res. 1996;29:298–305. [Google Scholar]
- 39.Cui Q, Karplus M. Catalysis and specificity in enzymes: a study of triosephophate isomerase and comparison with methyl glyoxal synthase. Adv. Prot. Chem. 2003;66:315. doi: 10.1016/s0065-3233(03)66008-0. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y. Pseudobond ab initio QM/MM approach and its applications to enzyme reactions. Theo. Chem. Acc. 2006;116:43–50. [Google Scholar]
- 41.Hu H, Yang W. Free energies of chemical reactions in solution and in enzymes with ab initio quantum mechanics/molecular mechanics methods. Annu. Rev. Phys. Chem. 2008;59:573–601. doi: 10.1146/annurev.physchem.59.032607.093618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Senn HM, Thiel W. QM/MM methods for biomolecular systems. Angew. Chem. Int. Ed. 2009;48:1198–1229. doi: 10.1002/anie.200802019. [DOI] [PubMed] [Google Scholar]
- 43.Ranaghan KE, Mulholland AJ. Investigation of enzyme-catalyzed reactions iwth combined quantum mechanical/molecular mechanical (QM/MM) methods. Int. Rev. Phys. Chem. 2010;29:65–133. [Google Scholar]
- 44.Zhang Y, Kua J, McCammon JA. Influence of structural fluctuation on enzyme reaction barriers in combined quantum mechanical/molecular mechanical studies. J. Phys. Chem. B. 2003;107:4459–4463. [Google Scholar]
- 45.Warshel A. Computer simulations of enzyme catalysis: methods, progress, and insights. Annu. Rev. Biophys. Biomol. Struct. 2003;32:425–443. doi: 10.1146/annurev.biophys.32.110601.141807. [DOI] [PubMed] [Google Scholar]
- 46.Ke Z, Zhou Y, Hu P, Wang S, Xie D, Zhang Y. Active site cysteine is protonated in the PAD4 Michaelis complex: Evidence from Born-Oppenheimer ab initio QM/MM molecular dynamics simulations. J. Phys. Chem. B. 2009;113:12750–12758. doi: 10.1021/jp903173c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ke Z, Wang S, Xie D, Zhang Y. Born-Oppenheimer ab initio QM/MM molecular dynamics simulations of the hydrolysis reaction catalyzed by protein arginine deiminase 4. J. Phys. Chem. B. 2009;113:16705–16710. doi: 10.1021/jp9080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Case DA, Darden T, Cheatham I, E. T, Simmerling C, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz J, M. K, B. W, Hayik S, Roitberg A, Seabra G, Kolossvary I, Wong KF, Paesani F, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Bornak V, Cui G, MAthews DH, Seetin MG, Sagui C, Babin V, Kollman PA. Amber 10. University of California; San Francisco: 2008. [Google Scholar]
- 49.Cornell WD, Cieplak P, Bayly CI, Gould IR, K. M. Merz J, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of protein, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179. [Google Scholar]
- 50.Hornak V, Abel R, Okur A, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 52.Darden TA, York D, Pedersen L. Particle mesh Ewald: an NlogN method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 53.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen L. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 54.Ryckaert JP, Ciccotti G, Berendsen HJ. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 55.Zhang Y, Lee T, Yang W. A pseudobond approach to combining quantum mechanical and molecular mechanical methods. J. Chem. Phys. 1999;110:46. [Google Scholar]
- 56.Zhang Y. Improved pseudobonds for combined ab initio quantum mechanical/molecular mechanical methods. J. Chem. Phys. 2005;122:24114. doi: 10.1063/1.1834899. [DOI] [PubMed] [Google Scholar]
- 57.Shao Y, Fusti-Molnar L, Jung Y, Kussmann J, Ochsenfeld C, Brown ST, Gilbert ATB, Slipchenko LV, Levchenko SV, O'Neill DP, DiStasio RA, Jr., Lochan RC, Wang T, Beran GJO, Besley NA, Herbert JM, Lin CY, Voorhis TV, Chien SH, Sodt A, Steele RP, Rassolov VA, Maslen PE, Korambath PP, Adamson RD, Austin B, Baker J, Byrd EFC, Dachsel H, Doerksen RJ, Dreuw A, Dunietz BD, Dutoi AD, Furlani TR, Gwaltney SR, Heyden A, Hirata S, Hsu C-P, Kedziora G, Khalliulin RZ, Klunzinger P, Lee AM, Lee MS, Liang W, Lotan I, Nair N, Peters B, Proynov EI, Pieniazek PA, Rhee YM, Ritchie J, Rosta E, Sherrill CD, Simmonett AC, Subotnik JE, Woodcock HW, III., Zhang W, Bell AT, Chakraborty AK, Chipman DM, Keil FJ, Warshel A, Hehre WJ, Schaefer HF, III., Kong J, Krylov AI, Gill PMW, Head-Gordon M. QChem. 2006 doi: 10.1039/b517914a. [DOI] [PubMed] [Google Scholar]
- 58.Ponder JW. TINKER, Software Tools for Molecular Design. 2004 doi: 10.1021/acs.jctc.8b00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, Liu H, Yang W. Free energy calculation on enzyme reactions with an efficient iterative procedure to determine minimum energy paths on a combined ab initio QM/MM potential energy surface. J. Chem. Phys. 2000;112:3483–3492. [Google Scholar]
- 60.Torrie GM, Valleau JP. Non-physical sampling distributions in Monte Carlo free energy estimation: Umbrella sampling. J. Comput. Phys. 1977;23:187–199. [Google Scholar]
- 61.Beeman D. J. Comput. Phys. 1976;20:130–139. [Google Scholar]
- 62.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 63.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram analysis method for free energy calculations on biomolecules. 1 The method. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- 64.Wang S, Hu P, Zhang Y. Ab initio quantum mechanical/molecular mechanical molecular dynamics simulation of enzyme catalysis: The case of histone lysine methyltransferase SET7/9. J. Phys. Chem. B. 2007;111:3758–3764. doi: 10.1021/jp067147i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu P, Wang S, Zhang Y. How do SET-domain protein lysine methyltransferases achieve the methylation state specificity? Revisited by ab initio QM/MM molecular dynamics simulations. J. Am. Chem. Soc. 2008;130:3806–3813. doi: 10.1021/ja075896n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hu P, Wang S, Zhang Y. Highly dissociative and concerted mechanism for the nicotinamide cleavage reaction in Sir2Tm enzyme suggested by ab initio QM/MM molecular dynamics simulations. J. Am. Chem. Soc. 2008;130:16721–16728. doi: 10.1021/ja807269j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mladenovic M, Fink RF, Thiel W, Schirmeister T, Engels B. On the origin of the stabilization of the Zwitterionic resting state of cysteine proteases: A theoretical study. J. Am. Chem. Soc. 2008;130:8696–8705. doi: 10.1021/ja711043x. [DOI] [PubMed] [Google Scholar]