Abstract
In-depth knowledge of bodily fluid phosphoproteomes, such as whole saliva, is limited. To better understand the whole saliva phosphoproteome, we generated a large-scale catalog of phosphorylated proteins. To circumvent the wide dynamic range of phosphoprotein abundance in whole saliva, we combined dynamic range compression using hexapeptide beads, strong cation exchange HPLC peptide fractionation, and immobilized metal affinity chromatography prior to mass spectrometry. In total, 217 unique phosphopeptides sites were identified representing 85 distinct phosphoproteins at 2.3% global FDR. From these peptides, 129 distinct phosphorylation sites were identified of which 57 were previously known, but only 11 of which had been previously identified in whole saliva. Cellular localization analysis revealed salivary phosphoproteins had a distribution similar to all known salivary proteins, but with less relative representation in “extracellular” and “plasma membrane” categories compared to salivary glycoproteins. Sequence alignment showed that phosphorylation occurred at acidic-directed kinase, proline-directed, and basophilic motifs. This differs from plasma phosphoproteins, which predominantly occur at Golgi casein kinase recognized sequences. Collectively, these results suggest diverse functions for salivary phosphoproteins and multiple kinases involved in their processing and secretion. In all, this study should lay groundwork for future elucidation of the functions of salivary protein phosphorylation.
Keywords: Phosphoproteomics, Whole Saliva, Dynamic Range Compression, Mass Spectrometry
Introduction
Phosphorylation is a common post-translational modification vital to the function and regulation of proteins. The human genome alone contains several hundred kinases and phosphatases with diverse functions and specificities 1. Reversible phosphorylation is a common mechanism controlling several cellular processes including growth, proliferation, apoptosis, senescence, and metabolism among others 2.
Although cellular phosphorylation has been extensively studied, much less is known about phosphoproteomes from secreted bio-fluids including whole saliva. A logical approach to initiate investigation in this new field is to identify phosphorylated proteins along with their specific phosphosites (p-sites). This can then lead to functional analyses of specific sites and can also lead to determination of which types of kinases and phosphatases may act on these sites and potentially regulate their function. The specific phosphorylation state of a polypeptide may also facilitate a better understanding of disease mechanisms or the discovery of biomarkers. Whole saliva is particularly attractive for biomarker studies due to its diagnostic potential and because of the ease and non-invasive nature of sample collection 3–5.
Recent studies of bio-fluids have reported phosphorylated peptides and proteins in plasma, serum, cerebral spinal fluid (CSF), urine, and saliva 6–10. Newly available sensitive, fast-scanning mass spectrometers coupled with phosphopeptide enrichment using immobilized metal ion affinity chromatography (IMAC) have facilitated these studies and have led to high confidence identification of 127 p-sites from 70 phosphoproteins in plasma 7, and 56 p-sites from 38 phosphoproteins in CSF 6. Only a handful of salivary polypeptides had been had been previously found to be phsophorylated (salivary acidic proline-rich phosphoprotein 1/2, basic salivary proline-rich protein 2, statherin, histatin-1, and cystatins S and SA-III) 9, 11, 12. However, recently Salih et al. undertook a mass spectrometry-based phosphoproteomics study in whole saliva, identifying 61 unique phosphopeptides corresponding to 28 phosphoproteins at 2% false discovery rate 13. Although a good start for characterizing the whole saliva phosphoproteome, this study employed an older generation mass spectrometer lacking the mass accuracy and mass resolution of current instruments, which have the improved ability to confidently identify more phosphopeptides from complex mixtures 14. Additionally, employing emerging methods for processing, fractionating, and enriching phosphopeptides could extend the current knowledge of the whole saliva phosphoproteome.
In this study, we sought to expand the catalog of phosphoproteins in whole saliva. We employed advanced methods including dynamic range compression (DRC) using Proteominer hexapeptide library beads and IMAC for phosphopeptide enrichment along with high mass accuracy tandem mass spectrometry using an LTQ-Orbitrap mass spectrometer to generate a high confidence (2.3% FDR) catalog of salivary phosphoproteins and their p-sites. A total of 217 unique phosphopeptides were found pertaining to 85 distinct proteins. Manual spectral analysis determined 126 distinct and confident p-site localizations. Analysis of sequence alignment of determined p-sites showed that salivary phosphoproteins had unique kinase recognition motifs suggesting a complex protein processing and secretory pathway for this bio-fluid.
Material and Methods
Collection and Preparation of Saliva
Unstimulated whole saliva was collected from healthy volunteers between 9:00 and 11:00 AM who had refrained from eating, drinking, using chewing gum, etc. for at least 2hr prior to collection. Samples were obtained by requesting subjects to swallow first, tilt their head forward, and expectorate all saliva into a cold 50 mL centrifuge tubes for 10-15 min without swallowing. About 10 mL of total saliva from each subject was collected. After collection, all samples were immediately placed on ice in the presence of both 1x PhosSTOP phosphatase inhibitor (Roche) and 1x (final concentration) Complete EDTA-free protease inhibitor (Roche). Saliva was separately centrifuged at 12,000 g at 4°C for 5 min twice. Clarified supernatants were pooled to a final volume of 55 mL. Total protein was quantified using the BCA assay (Thermo Pierce) and was 1.03 mg/mL.
Dynamic Range Compression Using Hexapeptide Beads
Proteominer beads (Bio-Rad) from 1 column (100 μL) were washed 3 times and resuspended with 1 mL of deionized water. The prepared beads were added to 50 mL (51.5 mg) of clarified saliva and were incubated overnight at 4°C with rotation. The next day beads were centrifuged at 2,000 × g for 4 min, the supernatant was removed, and the pelleted beads were transferred to a 1.5 mL microcentrifuge tube. Beads were washed three times with 1 mL of standard PBS and then once with 1 mL of deionized water. Each wash step consisted of 5 min of end to end rotation followed by centrifugation at 2000 × g for 2 min. Polypeptides bound to the Proteominer beads were eluted with addition of 125 μL 1x SDS-PAGE sample buffer (100mM Tris-HCl, 4% SDS, 20% glycerol, 200mM dithiothreitol, pH6.8) for 10 min at 70°C with regular vortexing. Eluted proteins were collected as the supernatant after centrifugation at 1,000x g for 2 min. The elution procedure was repeated once more at 95°C and the supernatants were pooled. Recovered proteins were precipitated with addition of 4x volume of ice-cold acetone and incubated overnight at −20°C. Precipitates were reconstituted in 50 mM Tris pH 8.0, 5 mM EDTA, and 0.5% SDS and protein concentration was determined by BCA assay.
SDS-PAGE
Protein samples were divided into aliquots of 20 μg and were treated with 1 unit of alkaline phosphatase (AP) (Sigma) for 1 hr at 37°C. The AP was either native or heat inactivated with prior boiling for 10 min. After addition of SDS-PAGE sample buffer containing bromphenol blue, protein samples were boiled. Proteins were resolved using 12% precast gels (Bio-Rad) and were stained first with Pro-Q diamond (Invitrogen) followed by Sypro Ruby (Invitrogen) in accordance to manufacturer’s instructions. Gels were visualized using a Typhoon 8610 fluorescent scanner using appropriate excitation and emission wavelengths.
In solution digestion, strong cation exchange chromatography, and immobilized metal ion affinity chromatography
Protein samples were reduced with addition of DTT (10 mM) for 60 min at 56°C. Samples were then diluted 10-fold in 50 mM Tris pH 8.0, 5 mM EDTA to reduce the SDS concentration to 0.05%. Proteomics grade trypsin (Promega) was added at an enzyme:substrate ratio of 1:50 and digestion proceeded overnight at 37°C. Digested peptides were purified via MCX columns (Waters) and samples were dried with vacuum centrifugation. Samples were dissolved in 250 μL of 10 mM KH2PO4 pH 2.7 with phosphoric acid containing 20% ACN and subjected to strong cation exchange (SCX) chromatography which was performed as described 15. After SCX, selected fractions were desalted using SepPak cartridges (Waters) and dried with vacuum centrifugation. Phosphopeptides from each fraction were enriched using immobilized metal ion affinity chromatography with Fe3+ beads (PHOS-Select affinity gel, Sigma) essentially as described 16. Eluted peptides were desalted using “Stage” tips 17 and were dried with vacuum centrifugation.
Mass spectrometry, database searching, and protein identification
Column loading, capillary reversed-phase HPLC, and mass spectrometry all were performed essentially as described 15. Mass analysis of Proteominer treated saliva samples were performed in two technical replicates using CID. Mass analysis of non-Proteominer treated samples was performed in two replicates where one analysis was performed with CID and the other with ETD. AGC settings for ETD reagent ions were 30 ms and 2 × 105 ions, the activation time was set to 100 ms, and supplemental activation was disabled. Data dependent settings excluded +1 charges for CID fragmentation and both +1 and +2 charges for ETD fragmentation. Mass spectral data were acquired and saved as .raw files using Xcalibur software v. 2.0.7 on an LTQ-orbitrap XL (ThermoScientific) mass spectrometer. Data were searched with SEQUEST V27.0 software using a database consisting of the forward and reversed NCBI human database V200806 and common contaminants totaling 70711 entries. Search parameters included variable modification of Met oxidation and Ser and Thr phosphorylation, semi-tryptic enzyme specificity, and parent and fragment ion mass tolerances of 1 and 0.8 amu, respectively. Identified peptide probabilities 18 and protein probabilities 19 were calculated within Scaffold software (www.proteomesoftware.com). Identification outputs were filtered using Scaffold 3 viewer software (www.proteomesoftware.com) in two ways to contain either strictly fully tryptic peptides or to include semi-tryptic peptides. The list of fully tryptic peptides was filtered with settings of 5% peptide probability, 7 ppm parent mass tolerance, and 80% protein probability. The list of semi-tryptic peptides was filtered with settings of 5% peptide probability, 10 ppm parent mass tolerance, 20% protein probability, and at least 2 unique peptides per identification. For CID data, only +2, +3, and +4 parent ion charge states were considered. For ETD data, +3 charge states and up were considered. Protein false discovery rates were determined by dividing the number of reversed database identifications by total number of identifications. All identified phosphopeptides were verified by manual inspection. For CID data, all identified phosphopeptides had to include a prominent neutral loss peak corresponding to -98 amu. If non-existent, the identification was discarded.
Synthetic peptides
The peptide MISADpSHEKR from histatin 1 and MIGADpSpSEEKFLR from statherin were synthesized in crude using the FasTrackTM service from ThermoScientific. Approximately 1 pmol of each was subjected to reversed-phase HPLC, mass spectrometry using CID, and database searching essentially as described above.
Determination of cellular localization
Associated GI numbers were obtained from total identified proteins from whole saliva from 15, phosphoproteins from whole saliva (this study), and N-linked glycoproteins of whole saliva from 20. These lists of GI numbers were imported into Ingenuity Pathway Analysis (Ingenuity Systems, Inc., Redwood City, CA) as dataset files. The location descriptions of each identified protein were grouped and counted. Percentages for each location description were calculated after dividing by total number of proteins within each list.
Phosphosite determination
Localization of p-sites for peptides containing multiple potential sites was performed by manual inspection. P-site determinations were based on the detection of flanking b or y-ions. In cases where site distinguishing b or y-ions were at noise level, then at least 2 flanking ions were required for definitive localization. Sites meeting these criteria for localization were scored as “1” (true) and those that did not meet these criteria were scored as “0” (unverified).
Results and Discussion
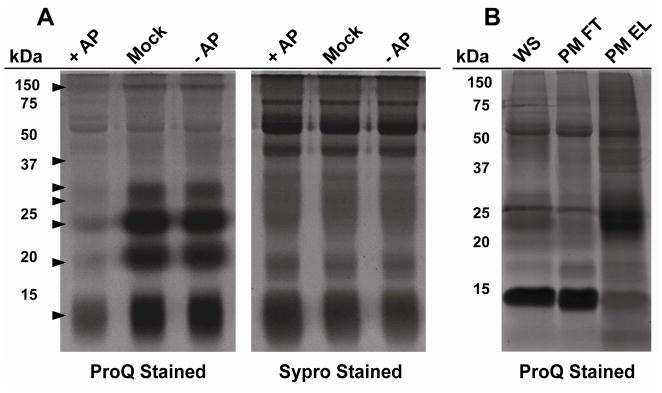
To obtain an initial visual profile of whole saliva protein phosphorylation, salivary proteins were characterized by SDS-PAGE analysis and phosphoprotein-specific staining. Saliva was collected and either treated with or without alkaline phosphates (AP) treatment. Afterwards, aliquots were separated by SDS-PAGE and gels were stained either with ProQ Diamond (phosphoprotein-specific) or Sypro Ruby (total protein-specific) fluorescent dyes. As shown in Figure 1A, significantly different staining patterns were observed when comparing the total salivary proteins to the ProQ Diamond treated lanes. Importantly, the intensity of the ProQ Diamond stained bands were generally diminished after AP treatment compared to mock or no AP treatment as marked by the arrows in the left panel of Figure 1A, which supported the specificity of the stain for detecting numerous phosphoproteins in saliva. Notably, highly intense phosphoprotein bands were observed in the molecular weight region around 25 kDa and below. Phosphoproteins were also detected at higher molecular weight regions, although the staining intensity of these bands was generally faint, suggesting that these proteins were present at relatively low abundance. The vast difference in intensity between bands stained with ProQ Diamond in Figure 1A suggested a large dynamic range of abundance for salivary phosphoproteins.
Figure 1.
SDS-PAGE of phosphorylated proteins whole saliva. (A) Whole saliva (20 μg) was pre-treated with 1 unit of alkaline phosphatase (+AP, lane 1), heat-inactivated alkaline phosphatase (mock, lane 2), or without alkaline phosphatase (-AP, lane 3). The samples were separated by SDS-PAGE and stained with either the phospho-specific ProQ diamond dye (Left Panel) or for total protein with Sypro Ruby dye (Right Panel). Arrows indicate bands sensitive to alkaline phosphatase treatment. (B) Whole saliva was treated with Proteominer (PM) beads and incubated overnight with shaking. The unbound fraction was collected and protein bound to the beads was washed and eluted. Whole saliva (WS) starting material, the unbound fraction (Proteominer, FT), and the eluted fraction (Proteominer, EL) were separated by SDS-PAGE and stained with ProQ Diamond dye.
To circumvent the large dynamic range of salivary phosphoproteins, we treated whole saliva proteins with hexapeptide libraries immobilized on beads (Proteominer, Bio-Rad). Their ability to attenuate levels of high-abundance proteins in various biological samples, including saliva, has been previously described ,15, 21, 22. Figure 1B shows the results of DRC of whole saliva proteins on the detection of phosphoproteins. The eluate from the hexapeptide beads was subjected to SDS-PAGE and stained for phosphorylation or total protein with or without prior addition of AP. DRC using the hexapeptide beads resulted in the detection of numerous phosphoproteins that were either not detected or detected very faintly in the non-DRC treated sample.
Based on the encouraging results from SDS-PAGE, we chose to use DRC to maximize the identification of phosphoproteins by mass spectrometry. However, although the phosphoprotein detection results after DRC treatment looked promising, we had concerns that the hexapapetide beads may not effectively bind all phosphoproteins leading to some phosphoproteins being missed. Therefore we undertook two analyses, one with DRC treatment and one without. Proteins from each of these collections were digested with trypsin, fractionated by SCX HPLC, enriched for phosphorylated peptides with IMAC, and were subjected to nanoflow reversed-phase HPLC in-line with the mass spectrometer. To maximize phosphoprotein detection, fractions were analyzed using CID or ETD on the LTQ-Orbitrap mass spectrometer.
To obtain the most accurate results, the identification data was obtained with specific parameters. Only Ser and Thr were considered for phosphorylation. Initial searches including Tyr yielded inconclusive results for Tyr phosphorylation. This was consistent with studies in other biofluids as little evidence of Tyr phosphorylation has been reported 6, 7, 13 even after affinity enrichment with pTyr specific antibodies 10. Identification data were filtered separately in two ways: with semi-tryptic specificity and with full tryptic specificity. Semi-tryptic specificity was necessary since several salivary-derived polypeptides are known to be post-translationally processed resulting in non-tryptic ends. However, introduction of multiple possible variable modifications with semi-trypsin specificity greatly increases the global false discovery rates (i.e., potential for decoy database matches). As a result, protein identifications in this scenario were required to include at least 2 unique peptides for high confidence. The peptides considered included both non-phosphorylated peptides that were still retained with IMAC as well as phosphorylated peptides. On the other hand, limiting identifications to have 2 tryptic termini is in itself a strong filter and identifications from this dataset were allowed to include single peptide hits. In both cases, global protein FDR was <2.3%. All mass spectral data can be viewed through Tranche (https://proteomecommons.org) using the following URL: tQ/jD7djpa1NPhDkeDo5o3LttmlZV442+uI0fne4bp5YxaOjpAaVsDVoU7wurvv5C59UV8HLdBshyUxwXlae15JyoRQAAAAAAAAC4w==
In sum, 217 unique phosphopeptides were identified corresponding to 85 phosphoproteins. The identification report can be found in Supplemental Table 1. Representative annotated spectra of each distinct phosphopeptitde can be found in Supplemental Figure 1. Use of Proteominer for DRC resulted in identification of 36 unique phosphopeptides that otherwise would not have been identified. This pool of unique phosphopeptides derived from a total of 30 proteins, 26 of which would not have been identified without incorporation of DRC in this dataset. Additionally, the repeated analysis of the IMAC enriched SCX fractions from the same whole saliva sample using ETD allowed for identification of 22 unique phosphopeptides not previously identified using CID, and corroborated identification of 30 phosphopeptides also identified with CID. Therefore, the use of CID and ETD on tryptic digests complemented each other, expanding the total number of salivary proteins identified.
We next aimed to examine individual phosphorylation sites within the dataset. The spectra from all identified phosphopeptides in Supplemental Table 1 were manually analyzed for p-site determination. Localization confidence was based on the presence of b and y ions flanking the proposed site of modification as described in Materials and Methods. From this investigation, 126 unique p-sites were determined with high confidence corresponding to 81 proteins. Of these, only 11 were previously identified from saliva, even when compared to the most recent phosphoproteomics study in saliva 13. However, 51 p-sites derived from 42 different proteins had been previously identified from analysis of other biological sources according to the UniProt protein knowledge database, which added validity to their identities. The 11 previously determined sites from saliva were from histatin 1, statherin, basic proline rich proteins 2 and 3, and acidic proline rich protein. The detection of specific phosphopeptides has been shown to depend on the specific sample preparation and enrichment methods used 23, which can preclude identification of even highly abundant phosphopeptides identified in previous studies using alternative methods. As a consequence, not all previously determined sites from the abundant salivary peptides were seen. However, several new sites were found. These included sites identified in acidic proline rich protein, basic proline rich protein 2, basic proline rich protein 4 cystatin C, histatin 1, and histatin 3. A list of these newly identified p-sites can be found in Table 1.
Table 1.
Newly identified phosphorylation sites from abundant salivary peptides
| Protein name | Accession No. | P-Site | Flanking Sequence | Number of unique peptides containing p-site |
|---|---|---|---|---|
| cystatin C precursor | gi|4503107|ref|NP_000090.1| | Ser-43 | GGPMDAsVEEEGV | 3 |
| histatin 1 | gi|4504529|ref|NP_002150.1| | Ser-39 | FHEKHHsHREFPF | 2 |
| histatin 3 | gi|4557653|ref|NP_000191.1| | Ser-45 | SHRGYRsNYLYDN | 2 |
| proline-rich protein BstNI subfamily 2 | gi|117168265|ref|NP_006239.2| | Ser-391 | PQGGRPsRPPQ- | 1 |
| proline-rich protein BstNI subfamily 4 precursor | gi|37537692|ref|NP_002714.2| | Ser-19 | LSSAESsSEDVSQ | 4 |
| proline-rich protein BstNI subfamily 4 precursor | gi|37537692|ref|NP_002714.2| | Ser-20 | SSAESSsEDVSQE | 1 |
| proline-rich protein BstNI subfamily 4 precursor | gi|37537692|ref|NP_002714.2| | Ser-24 | SSSEDVsQEESLF | 13 |
| proline-rich protein BstNI subfamily 4 precursor | gi|37537692|ref|NP_002714.2| | Ser-28 | DVSQEEsLFLISG | 2 |
| proline-rich protein HaeIII subfamily 1/2 | gi|158966674|ref|NP_001103683.1| | Ser-164 | GPPQGQsPQ- | 1 |
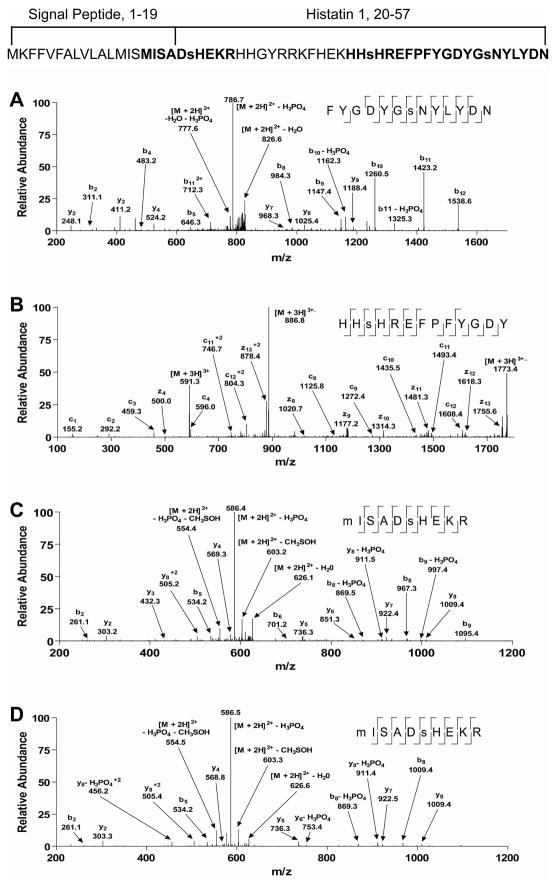
Interestingly, all 3 Ser residues in the mature histatin 1 polypeptide were shown to be phosphorylated. It had been previously shown with mass spectrometric analysis that Ser-51 is phosphorylated 13, which was confirmed by the CID spectrum shown in Figure 2A. In addition, we found that the previously unknown site, Ser-39, is phosphorylated as revealed by the ETD spectrum in Figure 2B. This site lies in a region that may also be tyrosine sulfated 24. However, the modification here appeared to be phosphorylation and not tyrosine sulfation based on the c and z ion pattern. Furthermore, the triply charged parent ion mass determined from the orbital trap was 591.2399. This mass is very close to that expected for phosphorylation (2 ppm mass error) complared to the expected mass for sulfation (7 ppm mass error). Also, a CID spectrum identifying the same modification site displayed a neutral loss peak pertaining to -98 amu, further supporting phosphorlylation (data not shown). Finally, Ser-21 was also shown to be phosphorylated (Figure 2C). This site has been known to be commonly modified 25, 26. Of note, the phosphopeptide identified by the spectrum in Figure 2C contained amino acid residues within the signal sequence preceding the mature form of histatin 1 suggesting the presence of previously unknown mature forms of this gene product in whole saliva. This is the first report of identification of this site by mass spectrometry following trypsin digestion likely because digestion of the common mature form would result in a peptide too small for detection by mass spectrometry 13. The CID spectrum of a synthetic version of this peptide (Figure 2D) had a very similar fragmentation pattern to that of the whole saliva derived peptide. Database searching of this spectrum against the entire human protein database also identified the predicted histatin sequence as the best match. Several of the same fragment ions were matched as shown in the respective fragmentation table in Supplemental Figure 1. Also, the relative intensities of the fragment ions were quite similar, providing conclusive confirmation of this unique identification.
Figure 2.
Representative tandem mass spectra of histatin 1 phosphopeptides. (A) CID spectrum identified as FYGDYGsNYLYDN, where Ser-51 is phosphorylated. (B) ETD spectrum identified as HHsHREFPFYGDY, where Ser-39 is phosphorylated. (C) CID spectrum identified as MISADsHEKR, where Ser-21 is phosphorylated. (D) CID spectrum of a synthesized form of MISADsHEKR for comparison, where Ser-21 is phosphorylated. Also displayed is the full amino acid sequence of the histatin 1 gene product with signal peptide and mature sequences marked. Residues in bold were identified by the spectra from the figure and residues in lower case are identified phosphorylation sites. Also shown at the top of the figure is the complete primary amino acid sequence of histatin 1 including with the signal peptide and mature protein sequence.
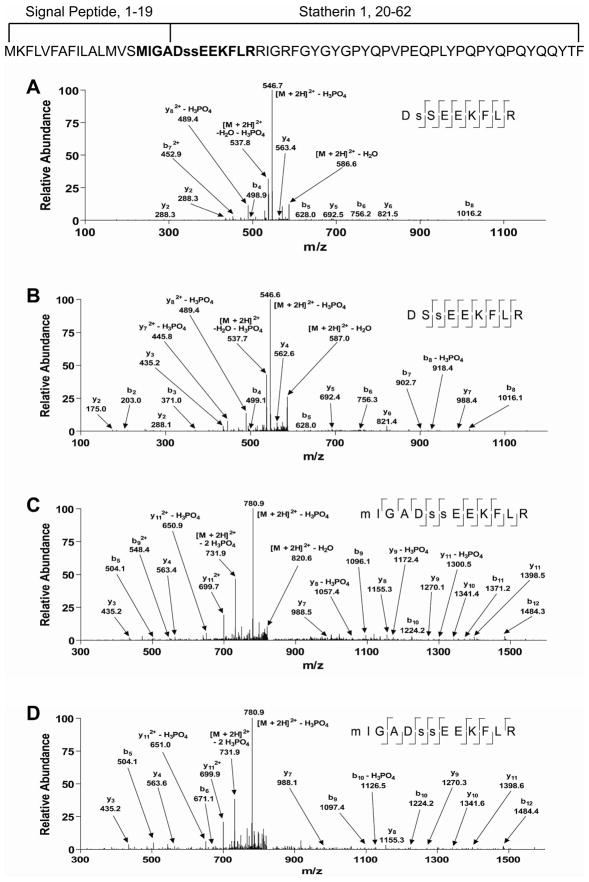
Similar to histatin 1, we also identified a unique phosphopeptide of statherin containing a sequence falling outside of the previously known mature form of this salivary polypeptide. Figure 3 shows spectra of statherin peptides identified in its monophosphorylated states (Figures 3A and B), and diphosphorylated state (Figure 3C). Phosphorylation of statherin at both of these Ser residues had previously been identified 27. However, in this study the diphosphorylated peptide identified from Figure 3C was also shown to include amino acid residues within the signal peptide region. Both Met oxidized and unoxidized peptides were identified for this sequence (data not shown). A CID spectrum from a synthesized version of the peptide identified from Figure 3C was quite similar in terms matched fragment ions and relative intensities of matched fragment ions (Figure 4 and Supplemental Figure 1). Finally, the predicted sequence was identified as the best match after database searching, which provided strong confirmation of its identification and thus expression in whole saliva.
Figure 3.
Representative tandem mass spectra generated by CID of phosphophoryaltion states of statherin peptides. (A) Spectrum identified as DsSEEKFLR, where Ser-21 is phosphorylated. (B) Spectrum identified as DSsEEKFLR, where Ser-22 is phosphorylated. (C) Spectrum identified as MIGADssEEKFLR, where Ser-21 and Ser-22 are phosphorylated. (D) Spectrum of a synthesized form of MIGADssEEKFLR for comparison, where Ser-21 and Ser-22 are phosphorylated. Also displayed is the full amino acid sequence of the statherin gene product with signal peptide and mature sequences marked. Residues in bold were identified by the spectra from the figure and residues in lower case are identified phosphorylation sites. The complete primary amino acid sequence of statherin including that of the signal peptide and mature protein is displayed at the top.
Figure 4.
Cellular localization comparison of whole saliva proteins. Total identified proteins (black bars), phosphoproteins (open bars), and N-linked glycoproteins (gray bars) from whole saliva were organized into cellular compartments using Ingenuity Pathways AnalysisTM. The number of proteins and percentage of total are listed next to each grouping. The total salivary proteins are from 15. The salivary glycoproteins are from 20.
The presence of histatin 1 and statherin peptides containing residues not previously known to exist in their mature forms may be due to alternative translational initiation or signal sequence cleavage. Further studies are necessary to understand this mechanism. It is interesting to note that the first 24 residues of histatin 1 and statherin are 75% identical. By analogy, the initial 24 residues of histatin 3 are 79% identical to histatin 1 suggesting that histatin 3 may have similar differential expression, however no peptide of histatin 3 was found in this study to support this possibility.
We next sought to characterize the cellular distribution of the identified phosphoproteins. For this, the salivary phosphoproteins were grouped into ‘cellular components’ (i.e. cellular localization) using Ingenuity Pathway AnalysisTM. We also included in this analysis a previously generated list of total salivary proteins 15, and a catalog of salivary glycoproteins generated in a separate study 20. As shown in Figure 4, the total proteins and phosphoproteins in saliva had a similar distribution of cellular component categories. Conversely, salivary glycoproteins had relatively higher representations in the “extracellular” (45%) and “plasma membrane” (22%) categories than both the total salivary proteins (extracellular 15%, plasma membrane 11%) and salivary phosphoproteins (extracellular 16%, plasma membrane 13%). Additionally, salivary glycoproteins had lower relative representations in the “cytoplasm” (25%) and “nucleus” (2%) categories. In contrast, 48% of total and 43% of phosphorylated proteins were grouped in the “cytoplasm” category, and 13% of total and 15% of phosphorylated proteins were grouped in the “nucleus” category. These results suggested that unlike glycoproteins, which are mostly secreted and/or localized to the plasma membrane, salivary phosphoproteins have a wider distribution of cellular localizations, and potentially a wider distribution of functions.
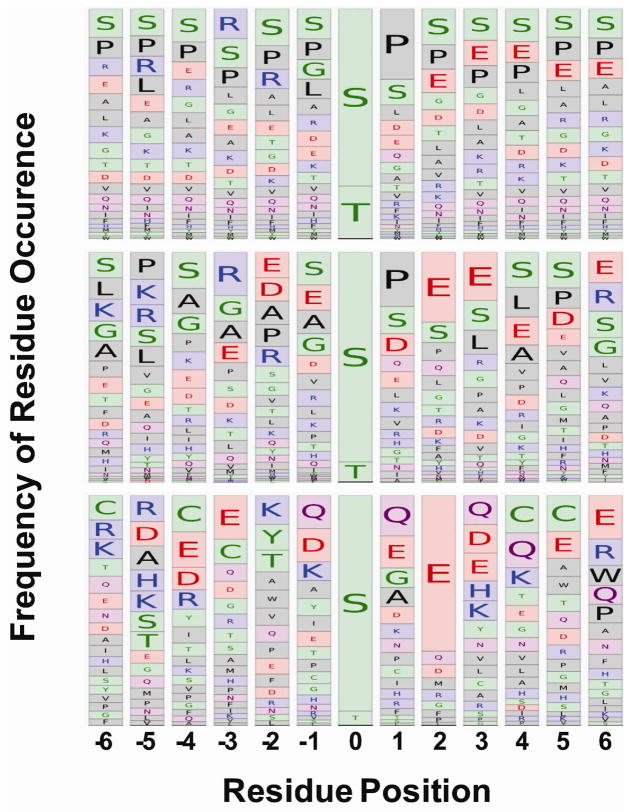
As a final analysis, we aligned sequences preceding (N-terminal side) and following (C-terminal side) of identified p-sites to look for potentially conserved consensus sequence patterns within salivary phosphoproteins. Only manually verified sequences of identified p-sites in this study were considered for alignment. This was compared to p-sites determined from phosphoproteomics analysis of plasma (n = 47) 7 and p-sites of all known human protein p-sites (n ~ 38,000) from www.phosphosite.org 28. For whole saliva (Figure 5, middle panel), there was a high level of acidic directed sites where Glu was frequently found either 2 or 3 positions downstream of the p-site. There was also a high level of proline directed sites shown by a high occurrence of Pro immediately following the p-site. Finally, there was also a high occurrence of basic directed sites shown by the high frequency of Arg at the third residue upstream of the modification site. Residue frequencies from whole saliva sequence alignments appeared to be diverse and resembled that of the entire known phosphoproteome (Figure 5, upper panel). Conversely, plasma (Figure 5, lower panel) was dominated by the high occurrence of a Glu residue 2 positions upstream of the p-site, which is characteristic of a Golgi casein kinase (GCK) recognition motif 29, 30. In whole saliva, the prevalence of downstream acidic residues at 2 and 3 positions from the p-site were consistent with both a (GCK) and a broader casein kinase II-like recognition motif. The wide array of residue frequencies near the p-site in salivary proteins suggested the possibility of a large number of potential kinase recognition motifs. Whole saliva likely contains proteins that are shed from epithelial cells lining the oral cavity as well as from salivary gland secretions. As such, the kinase network governing the phoshorylation of proteins found in whole saliva may be quite complex.
Figure 5.
Sequence alignment comparison salivary protein phosphorylation sites. The sequence windows from 6 residues upstream and downstream surrounding the phosphorylation sites were gathered for the entire human phosphoprotein from www.phosphosite.org, (upper panel), the salivary phosphoproteome where the validity of localization was determined manually (middle panel), and the plasma phosphoproteome from 7 (lower panel). Sequences were aligned and amino acid occurrence frequencies are represented by the size of the box containing the respective 1 letter code.
Conclusions
Overall, our catalog contains numerous salivary proteins previously not known to be phosphorylated, providing a significantly expanded view into the nature of protein phosphorylation in whole saliva. This dataset expands upon and compliments another recently published dataset of phosphoproteins in whole saliva13. We have increased our knowledge on phosphorylation in several well-studied salivary polypeptides (histatin 1, histatin 3, statherin, basic proline rich protein 2, and acidic proline rich protein). The relatively even distribution of cellular localization of the salivary phosphoproteins (Figure 4) suggested a broad array of functions. The diversity of sequence motifs in whole saliva suggested a complex pattern of kinase activity towards proteins found in saliva (Figure 5). Our catalog should serve as a guide for future investigations in functional consequences of salivary protein phosphorylation, shedding new light on the role of protein phosphorylation in the biochemistry of the oral environment and its potential ties to oral disease.
Supplementary Material
Acknowledgments
This research was funded in part by NIH grant 1R01DE017734 and X.C. was funded in part by International Program of Project 985, Sun Yat-Sen University, PR China. We thank Mark Nelson, John Chilton, Dr. Pratik Japtap at the Minnesota Supercomputing Institute for computational support. We thank Todd Markowski for SCX fractionation and the Center for Mass Spectrometry and Proteomics at the University of Minnesota for instrumental access and maintenance.
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T. Signaling--2000 and beyond. Cell. 2000;100(1):113–27. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 3.Nagler RM. Saliva as a tool for oral cancer diagnosis and prognosis. Oral Oncol. 2009;45(12):1006–10. doi: 10.1016/j.oraloncology.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Hu S, Arellano M, Boontheung P, Wang J, Zhou H, Jiang J, Elashoff D, Wei R, Loo JA, Wong DT. Salivary proteomics for oral cancer biomarker discovery. Clin Cancer Res. 2008;14(19):6246–52. doi: 10.1158/1078-0432.CCR-07-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Jong EP, Xie H, Onsongo G, Stone MD, Chen XB, Kooren JA, Refsland EW, Griffin RJ, Ondrey FG, Wu B, Le CT, Rhodus NL, Carlis JV, Griffin TJ. Quantitative proteomics reveals myosin and actin as promising saliva biomarkers for distinguishing pre-malignant and malignant oral lesions. PLoS One. 2010;5(6):e11148. doi: 10.1371/journal.pone.0011148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahl JM, Jensen SS, Larsen MR, Heegaard NH. Characterization of the human cerebrospinal fluid phosphoproteome by titanium dioxide affinity chromatography and mass spectrometry. Anal Chem. 2008;80(16):6308–16. doi: 10.1021/ac800835y. [DOI] [PubMed] [Google Scholar]
- 7.Carrascal M, Gay M, Ovelleiro D, Casas V, Gelpi E, Abian J. Characterization of the human plasma phosphoproteome using linear ion trap mass spectrometry and multiple search engines. J Proteome Res. 2010;9(2):876–84. doi: 10.1021/pr900780s. [DOI] [PubMed] [Google Scholar]
- 8.Hu L, Zhou H, Li Y, Sun S, Guo L, Ye M, Tian X, Gu J, Yang S, Zou H. Profiling of endogenous serum phosphorylated peptides by titanium (IV) immobilized mesoporous silica particles enrichment and MALDI-TOFMS detection. Anal Chem. 2009;81(1):94–104. doi: 10.1021/ac801974f. [DOI] [PubMed] [Google Scholar]
- 9.Cirulli C, Chiappetta G, Marino G, Mauri P, Amoresano A. Identification of free phosphopeptides in different biological fluids by a mass spectrometry approach. Anal Bioanal Chem. 2008;392(1–2):147–59. doi: 10.1007/s00216-008-2266-7. [DOI] [PubMed] [Google Scholar]
- 10.Zhou W, Ross MM, Tessitore A, Ornstein D, Vanmeter A, Liotta LA, Petricoin EF., 3rd An initial characterization of the serum phosphoproteome. J Proteome Res. 2009;8(12):5523–31. doi: 10.1021/pr900603n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helmerhorst EJ, Oppenheim FG. Saliva: a dynamic proteome. J Dent Res. 2007;86(8):680–93. doi: 10.1177/154405910708600802. [DOI] [PubMed] [Google Scholar]
- 12.Messana I, Inzitari R, Fanali C, Cabras T, Castagnola M. Facts and artifacts in proteomics of body fluids. What proteomics of saliva is telling us? J Sep Sci. 2008;31(11):1948–63. doi: 10.1002/jssc.200800100. [DOI] [PubMed] [Google Scholar]
- 13.Salih E, Siqueira WL, Helmerhorst EJ, Oppenheim FG. Large-scale phosphoproteome of human whole saliva using disulfide-thiol interchange covalent chromatography and mass spectrometry. Anal Biochem. 2010;407(1):19–33. doi: 10.1016/j.ab.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mann M, Kelleher NL. Precision proteomics: the case for high resolution and high mass accuracy. Proc Natl Acad Sci U S A. 2008;105(47):18132–8. doi: 10.1073/pnas.0800788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bandhakavi S, Stone MD, Onsongo G, Van Riper SK, Griffin TJ. A dynamic range compression and three-dimensional peptide fractionation analysis platform expands proteome coverage and the diagnostic potential of whole saliva. J Proteome Res. 2009;8(12):5590–600. doi: 10.1021/pr900675w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villen J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc. 2008;3(10):1630–8. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75(3):663–70. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 18.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 19.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 20.Bandhakavi S, Van Riper SK, Tawfik P, Stone MD, Haddad T, Rhodus N, Carlis JV, Griffin TJ. Hexapeptide libraries for enhanced protein PTM identification and relative abundance profiling in whole human saliva. J Proteome Res. doi: 10.1021/pr100857t. ePub 2010 Dec 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mouton-Barbosa E, Roux-Dalvai F, Bouyssie D, Berger F, Schmidt E, Righetti PG, Guerrier L, Boschetti E, Burlet-Schiltz O, Monsarrat B, de Peredo AG. In-depth exploration of cerebrospinal fluid by combining peptide ligand library treatment and label-free protein quantification. Mol Cell Proteomics. 2010;9(5):1006–21. doi: 10.1074/mcp.M900513-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Righetti PG, Boschetti E, Zanella A, Fasoli E, Citterio A. Plucking, pillaging and plundering proteomes with combinatorial peptide ligand libraries. J Chromatogr A. 2010;1217(6):893–900. doi: 10.1016/j.chroma.2009.08.070. [DOI] [PubMed] [Google Scholar]
- 23.Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R. Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat Methods. 2007;4(3):231–7. doi: 10.1038/nmeth1005. [DOI] [PubMed] [Google Scholar]
- 24.Cabras T, Fanali C, Monteiro JA, Amado F, Inzitari R, Desiderio C, Scarano E, Giardina B, Castagnola M, Messana I. Tyrosine polysulfation of human salivary histatin 1. A post-translational modification specific of the submandibular gland. J Proteome Res. 2007;6(7):2472–80. doi: 10.1021/pr0700706. [DOI] [PubMed] [Google Scholar]
- 25.Oppenheim FG, Yang YC, Diamond RD, Hyslop D, Offner GD, Troxler RF. The primary structure and functional characterization of the neutral histidine-rich polypeptide from human parotid secretion. J Biol Chem. 1986;261(3):1177–82. [PubMed] [Google Scholar]
- 26.Oppenheim FG, Xu T, McMillian FM, Levitz SM, Diamond RD, Offner GD, Troxler RF. Histatins, a novel family of histidine-rich proteins in human parotid secretion. Isolation, characterization, primary structure, and fungistatic effects on Candida albicans. J Biol Chem. 1988;263(16):7472–7. [PubMed] [Google Scholar]
- 27.Schlesinger DH, Hay DI. Complete covalent structure of statherin, a tyrosine-rich acidic peptide which inhibits calcium phosphate precipitation from human parotid saliva. J Biol Chem. 1977;252(5):1689–95. [PubMed] [Google Scholar]
- 28.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4(6):1551–61. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 29.Meggio F, Boulton AP, Marchiori F, Borin G, Lennon DP, Calderan A, Pinna LA. Substrate-specificity determinants for a membrane-bound casein kinase of lactating mammary gland. A study with synthetic peptides. Eur J Biochem. 1988;177(2):281–4. doi: 10.1111/j.1432-1033.1988.tb14374.x. [DOI] [PubMed] [Google Scholar]
- 30.Salvi M, Cesaro L, Tibaldi E, Pinna LA. Motif analysis of phosphosites discloses a potential prominent role of the Golgi casein kinase (GCK) in the generation of human plasma phospho-proteome. J Proteome Res. 2010;9(6):3335–8. doi: 10.1021/pr100058r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.