Abstract
Adipose tissue contains a heterogeneous population of mature adipocytes, endothelial cells, immune cells, pericytes, and pre-adipocytic stromal/stem cells. To date, the majority of proteomic analyses have focused on intact adipose tissue or isolated adipose stromal/stem cells in vitro. In this study, human subcutaneous adipose tissue from multiple depots (arm and abdomen) obtained from female donors was separated into populations of stromal vascular fraction cells and mature adipocytes. Out of 960 features detected by 2-D gel electrophoresis, a total of 200 features displayed a 2-fold up- or down-regulation relative to each cell population. The protein identity of 136 features was determined. Immunoblot analyses comparing SVF relative to adipocytes confirmed that carbonic anhydrase II was up-regulated in both adipose depots while catalase was up-regulated in the arm only. Bioinformatic analyses of the dataset determined that cytoskeletal, glycogenic, glycolytic, lipid metabolic, and oxidative stress related pathways were highly represented as differentially regulated between the mature adipocytes and stromal vascular fraction cells. These findings extend previous reports in the literature with respect to the adipose tissue proteome and the consequences of adipogenesis. The proteins identified may have value as biomarkers for monitoring the physiology and pathology of cell populations within subcutaneous adipose depots.
Keywords: DIGE, Adipose tissue, Stem cells, Adipocytes, Proteomics, LC-MS
Introduction
Adipose tissue is a complex, heterogeneous structure composed of mature adipocytes, fibroblast pre-adipocytes or stromal/stem cells, pericytes, endothelial cells, macrophages, and T-lymphocytes 1–5. Classical studies by Rodbell and further refined by others subfractionated these cell types by collagenase digestion and differential centrifugation 6–13. These steps separated the tissue into floating mature adipocytes and pelleted stromal vascular fraction (SVF) cells. Historically, adipose tissue was thought to play a passive metabolic role, acting solely as an energy storage reservoir. This view changed with the discovery of leptin, as investigators began to appreciate adipose tissue’s dynamic role as a systemic endocrine organ 14, 15. The elevated presence of myeloid/macrophage and T-lymphocytes in the adipose tissue of obese and diabetic subjects supported its further role in immune and inflammatory processes 16–22. Studies have used RT-PCR analysis to examine the mRNA expression profile of intact human adipose tissue, mature adipocytes, and SVF cells 23. This work has demonstrated a relative enrichment of genes encoding lipogenic and lipolytic proteins in the mature adipocytes while the pro-inflammatory markers are more prevalent in SVF cells 23. Only a limited number of studies have examined the proteome or secretome of human adipocytes or adipose tissue 24–37. Previous studies have focused on the characterization of protein extracts from intact tissue 26–30 or from undifferentiated and adipocyte differentiated cultured pre-adipocytes, also known as adipose-derived stromal/stem cells (ASCs) 24, 25. However, only one group of investigators has compared the proteome of the mature adipocytes to the SVF cells in human adipose tissue using 2-dimensional gel electrophoresis and mass spectrometry 35. The current study has employed DIGE technology in combination with mass spectrometry to identify differentially expressed proteins in the adipocyte and SVF cell fractions derived from human subcutaneous adipose tissue.
Materials and Methods
Adipose Tissue Acquisition, Processing and Consent
All procedures and protocols were reviewed and approved by the Pennington Biomedical Research Center Institutional Review Board prior to implementation. Tissues were obtained with informed written consent from healthy, non-diabetic subjects undergoing elective liposuction surgery. . As a group (n = 7), the female donors displayed a mean (± S.D.) age of 41.3 ± 11.4 years and a mean (± S.D.) BMI (body mass index) of 27.4 ± 3.4. Samples were transported to the Pennington Biomedical Research Center and processed on the day of harvest using published methods 38. Lipoaspirates were washed in phosphate buffered saline (PBS), digested in PBS supplemented with 1% bovine serum albumin, 0.1% collagenase type II (Worthington, Lakewood, NJ), and 2 mM calcium chloride for 60 minutes with rocking at 37° C. The tissue digest was centrifuged at 300X g for 5 minutes at room temperature to separate the floating mature adipocytes from the stromal vascular fraction (SVF) cell pellet.
Adipose Stem Cell Culture and Differentiation
The human ASC were isolated, cultured and differentiated according to established methods 39, 40. Adipogenesis was induced with a combination of dexamethasone, insulin, methylisobutylxanthine, and rosiglitazone while osteogenesis was induced with a medium containing ascorbic acid, dexamethasone, and β-glycerophosphate 39, 40.
Adipocyte and Stromal Vascular Fraction Cell Processing
Proteins were extracted from SVF cell pellets and 1 mL of floating mature adipocytes from all seven adipose tissue samples by adding 2.5 mL of lysis buffer (5M Urea/ 2M Thiourea/ 2% CHAPS/ 2% SB3-10/ 0.2% Bio-Lyte, pH 3–10/ 2% n-dodecyl-b-d-maltoside/ 40mM Tris/ 5mM PMSF/ 2mM TBP/ 150U Benzonase). Samples were rocked for 20 min at room temperature followed by sonication and addition of 50 mM dithiothreitol (DTT). The resulting sample mixture was centrifuged for 30 min at 20,800X g to remove unsolubilized material. Proteins were further precipitated and resolubilized in the CyDye labeling buffer (30 mM Tris, 7M Urea, 2M Thiourea, 4% CHAPS, pH 8.5). The protein concentration was determined using Bradford Protein Assay (Bio-Rad, Hercules, CA).
Two Dimensional Difference In-Gel Electrophoresis (2D-DIGE)
Methods for protein labeling, electrophoresis, imaging and analysis have been described in detail previously 24, 25, 41, 42. Briefly, protein samples were covalently labeled at lysines with spectrally resolvable CyDye fluorophores (Cy2, Cy3 and Cy5). An internal control pooled sample composed of equal amounts of protein from each sample was labeled with Cy2 and included on every gel. Proteins extracted from SVF pellet and floating mature adipocytes were labeled with either Cy3 or Cy5 fluorophores. 50 µg of each sample labeled with three different labels was combined and used for each 2D-gel. Labeled protein samples were mixed with DeStreak rehydration buffer (GE Healthcare, Waukesha, WI) and 1% carrier ampholytes (pH 3–10; Bio-Rad) and loaded onto the IPG ready strips (24 cm, pH 3–10 NL; Bio-Rad) for active rehydration. Proteins were focused at a maximum of 10,000 V for a total of 70 kVh. IPG strips were equilibrated with reduction and alkylation buffers. Sodium dodecyl sulfate – polyacrylamide gel electrophoresis was performed in the Ettan DALT vertical units (GE Healthcare) at 1 Watt per gel for 1 hour and then 2 Watt per gel until the bromophenol blue dye front reached the bottom of the gel. All individual samples were evaluated on two separate gels.
Each gel was imaged at three excitation/emission wavelengths (470/530, 530/605 and 625/695 nm) using a Molecular Imager VersaDoc MP imaging system (Bio-Rad). Gel images were analyzed using PDQuest 8.0 (Bio-Rad) image analysis software. Spots that were at least 2 fold up- or down-regulated with 95% significance (student’s t-test) were selected for further analysis. Preparative gels containing 250 µg of pooled internal standard were run and post-stained with Sypro Ruby (Bio-Rad). Sypro Ruby images were matched to the analytical gel images, and spots of interest were selected for excision. 1.5 mm gel plugs were excised using ProteomeWorks spot cutter (Bio-Rad) automatically and deposited in a 96-well plate for mass spectrometric analysis.
Mass Spectrometry Analysis
In-gel tryptic digestion and liquid chromatography – tandem mass spectrometry (LC-MS/MS) analysis were performed as described in detail previously 24, 25, 41, 42. Briefly, the peptide fragments extracted from each digested spot were separated and analyzed using a capillary LC system coupled on-line with a nanospray quadrupole time-of-flight micro mass spectrometer (Waters Corp., Milford, MA). The MS was operated in a data-dependent acquisition mode in which a full survey MS scan was followed by three MS/MS scans using normalized collision energy. The instrument was operated in positive ion mode, with an electrospray voltage of 3.5 kV, sample cone voltage of 40 V and extraction cone voltage of 1.5 V. The peaklist (pkl) files were generated using ProteinLynx Global Server 2.2.5 (PLGS 2.2.5, Waters Corp.) with the following settings: background subtract type, adaptive; smoothing type, Savitzky-Golay; smoothing iterations, 2; smoothing window, 3 channels; deisotoping type, fast; minimum peak width, 4 channels and centroid top 80%. Tandem mass spectra were analyzed using the PLGS 2.2.5 (Waters Corp.) and MASCOT (version 2.2, Matrix Science, Cambridge, MA) software against a SwissProt database (release 57.3, 468,851 sequences) 24, 25. For both PLGS and MASCOT, the database search settings included one missed tryptic cleavage; precursor-ion mass tolerance, 200 ppm; fragment-ion mass tolerance, 0.1 Da and fixed carbamidomethylation of cysteine residues. Methionine oxidation of protein was allowed as a variable modification in the databank search query in PLGS and MASCOT, and automodification query was selected to identify peptides with further post-translational modifications in PLGS. The top ranking hits (score 8–13 for PLGS and p < 0.05 for Mascot assignment of “identity”) were scrutinized based on molecular weight, pI, and sequence coverage. Spectra were also examined carefully to verify that the spectra included a good number of consecutive “y” ions with high mass accuracy. If peptides matched to multiple members of a protein family with similar sequences, the protein identified was the one with the highest number of matched peptides and score.
Data Analysis
Analysis of biological pathways was performed using MetaCore software (GeneGo Inc., St. Joseph, MI) licensed through the Colorado State University Center for Bioinformatics. Lists of identified differentially abundant proteins were uploaded and analyzed using default settings for pathway, molecular function and Gene Ontology category enrichment with False Discovery Rate (FDR) correction. Each protein ID was mapped to its corresponding gene object in the Metacore database. Networks of these focus genes were then algorithmically generated based on their connectivity. The significance values for network and pathway analyses were calculated using right-tailed Fisher’s Exact Test. These values are calculated based on the number of proteins that participate in a given pathway relative to the total number of occurrences of these proteins in all pathway annotations stored in the Metacore database.
Gel Electrophoresis and Western Blot Analysis
Adipocytes and stromal vascular fraction (SVF) whole cell extracts from arm or abdominal adipose depots (two donors each) were assayed for the expression of selected proteins using western blot analysis. Polyclonal antibodies against catalase (goat, Cat # SC-34281), carbonic anhydrase II (rabbit, Cat # SC-25596), and lamin A/C (goat, N-18, Cat # SC-6215) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). An equal amount of protein from each sample (30 µg of total protein for carbonic anhydrase detection and 60 µg of total protein for catalase and lamin A/C detection) was separated using polyacrylamide (National Diagnostics, Atlanta, GA) gels containing sodium dodecyl sulfate (SDS) according to Laemmli 43 and transferred to nitrocellulose (BioRad) in 25 mM Tris, 192 mM glycine, and 20% methanol. Following transfer, the membrane was stained with MemCode reversible protein stain (Thermo Fisher/Pierce) to confirm even loading and photographed prior to reversal of the stain. The intensity of the bands in each lane was quantified using Un-Scan-It software version 6.1 (Silk Scientific) and reported as total pixels/lane. Following removal of the MemCode stain, the membrane was blocked in 4% milk in Tris-buffered saline and 0.1% Tween 20 (TBS-T) for 1 hour at room temperature. The membranes were incubated with anti-carbonic anhydrase II (1:2000), anti-catalase (1:100), or anti-lamin A/C (1:200) in TBS-T with 1% bovine serum albumin at room temperature as indicated for 1–2 hours. Following extensive washes, the results were visualized with horseradish peroxidase (HRP)-conjugated secondary antibodies and enhanced chemiluminescence (Pierce, Rockford, IL).
Results
Characterization of ASCs by Differentiation and Donor Phenotype
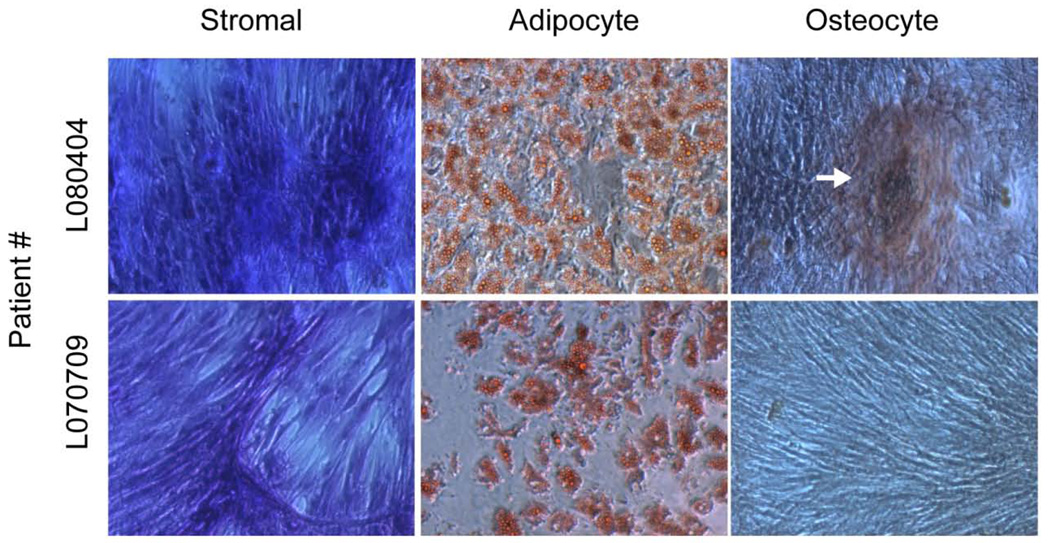
The ASCs isolated from the abdominal and arm adipose depots were capable of undergoing both adipocyte and osteoblast differentiation in vitro. A representative image of Oil Red O (adipogenic) and Alizarin Red (osteogenic) stained cultures at day 9 is displayed in Figure 1.
Figure 1.
Adipogenic and osteogenic potential of human adipose-derived stem cells. Primary cultures of ASCs derived from two donors were maintained in stromal media, adipogenic induction media or osteogenic induction media. After a period of 9–10 days, the cultures were fixed and stained with Toluidine Blue (Stromal) or for neutral lipids with Oil Red O (Adipocyte) or for mineralization of the extracellular matrix with Alizarin Red (Osteocyte). A mineralization nodule is indicated by an arrow in the osteocyte image for patient L080404. Mature adipocytes are indicated by uptake of the red stain for both patients in the adipocyte images. Cultures are displayed at 10X magnification (representative of n = 7).
Gel Analysis and Protein Identification
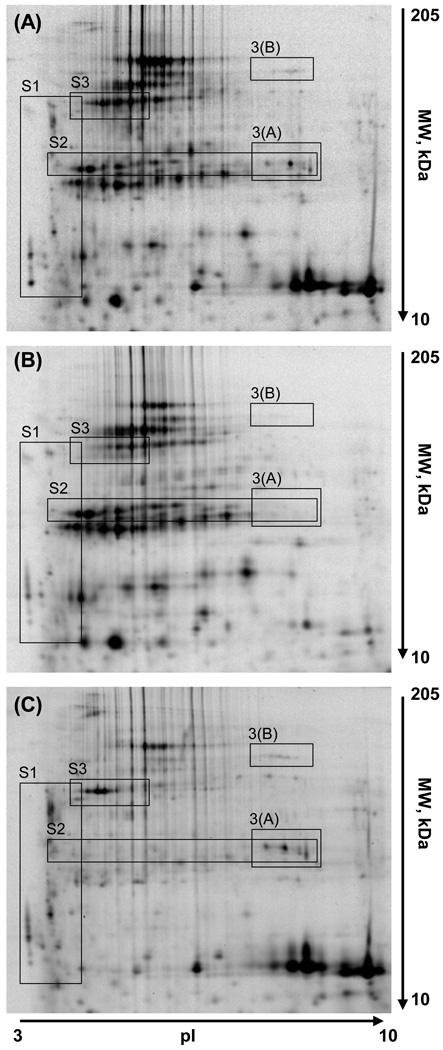
Protein extracts from mature adipocytes and stromal vascular fraction isolated from five human abdomen tissue samples and two arm samples were subjected to 2D-DIGE individually. A representative gel image from adipocytes, stromal vascular fraction and the master gel are presented in Figure 2. The adipocytes and stromal vascular fraction cells displayed 960 ± 80 features. After statistical analysis, 200 spots were identified to be at least 2-fold up- or down-regulated with 95% significance (Student’s t-test) in adipocytes vs. stromal vascular fraction cells. These spots were further matched in preparative gel images, and 147 spots were selected for excision, in-gel tryptic digestion and tandem mass spectrometric analysis. 136 spots resulted in 43 unique protein identifications. Several proteins such as carbonic anhydrase 1 and 2, catalase, vimentin, lamin A/C, fatty acid binding protein and actin, etc. were detected in multiple spots on 2D-gels (Figure 3, Supplementary Figures 1 and 2). These multiple spots represent various isoforms of these proteins. Differential effects of protein-specific isoforms are commonly observed 44, 45 and are evident in the data presented here (Figure 3, Supplementary Tables 1–3).
Figure 2.
2D-DIGE of total protein extracts from adipocytes and stromal vascular fraction cells isolated from abdomen adipose tissue. The figure presents the master gel image (A) and representative 2D-gel images from adipocytes (B) and stromal vascular fraction cell samples (C). An expanded view of the regions marked by rectangles are presented in Figures 3A, 3B and Supplementary Figures 1 – 3.
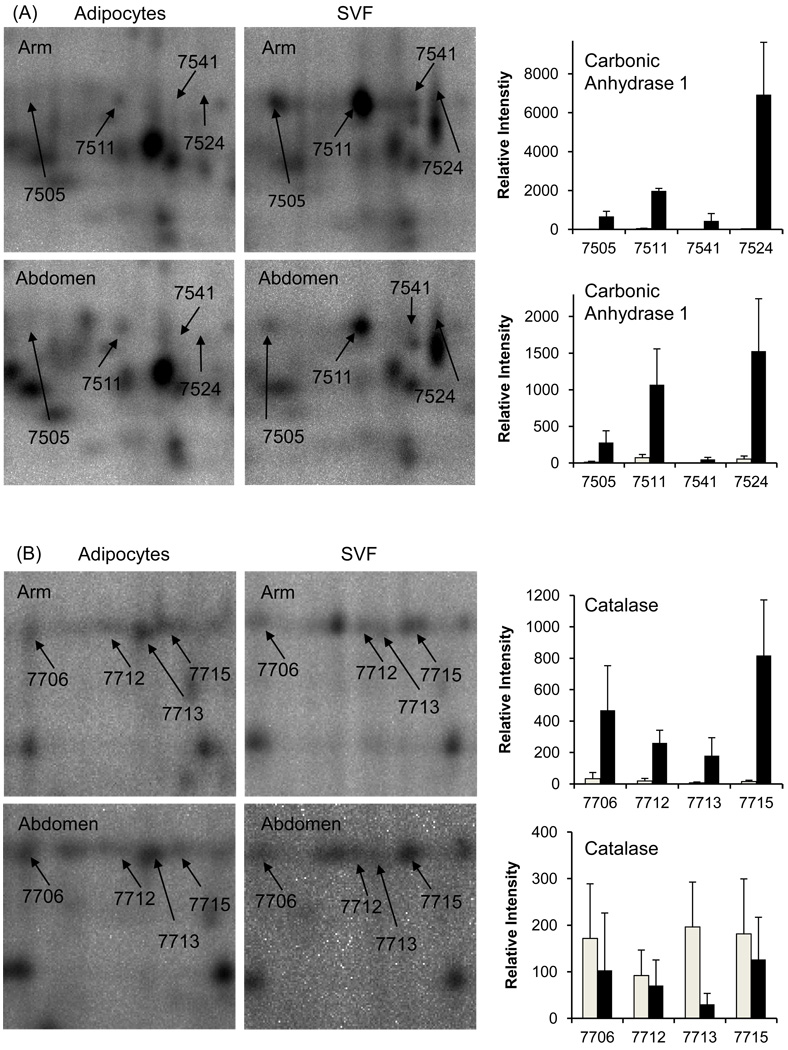
Figure 3.
Changes in relative expression levels of Carbonic Anhydrase 1 (A) and Catalase (B) in adipocytes versus stromal vascular fraction cells isolated from adipose tissue in arm and abdomen. Error bars represent standard deviation of the mean.
Representative spots identified to be differentially regulated are presented in Figure 3. The histograms in Figure 3 show relative levels of signal intensity in adipocytes and SVF cells isolated from adipose tissue. Four spots (7505, 7511, 7541 and 7524) identified as carbonic anhydrase 1 (Figure 3A) are down-regulated in mature adipocytes compared to SVF cells from both arm and abdomen adipose tissue depots. However, four spots (7706, 7712, 7713 and 7715) identified to be catalase (Figure 3B) clearly show that catalase is down-regulated in mature adipocytes compared to SVF cells from arm adipose tissue, but no statistically significant change in levels of catalase was detected in SVF and mature adipocytes isolated from abdomen. Proteins identified to be differentially regulated in SVF cells as compared to mature adipocytes isolated from both arm and adipose tissue are listed in Supplementary Tables 1–3. Supplementary Tables 1–3 also provide additional details including accession number, the number of peptides matched to each protein, their molecular weights, pI, percent coverage and score from both PLGS and MASCOT softwares. For the proteins identified based on a single peptide, the peptide sequence and tandem mass spectrum are presented in the Supplementary Table 4.
Western Blot Analysis of Selected Proteins
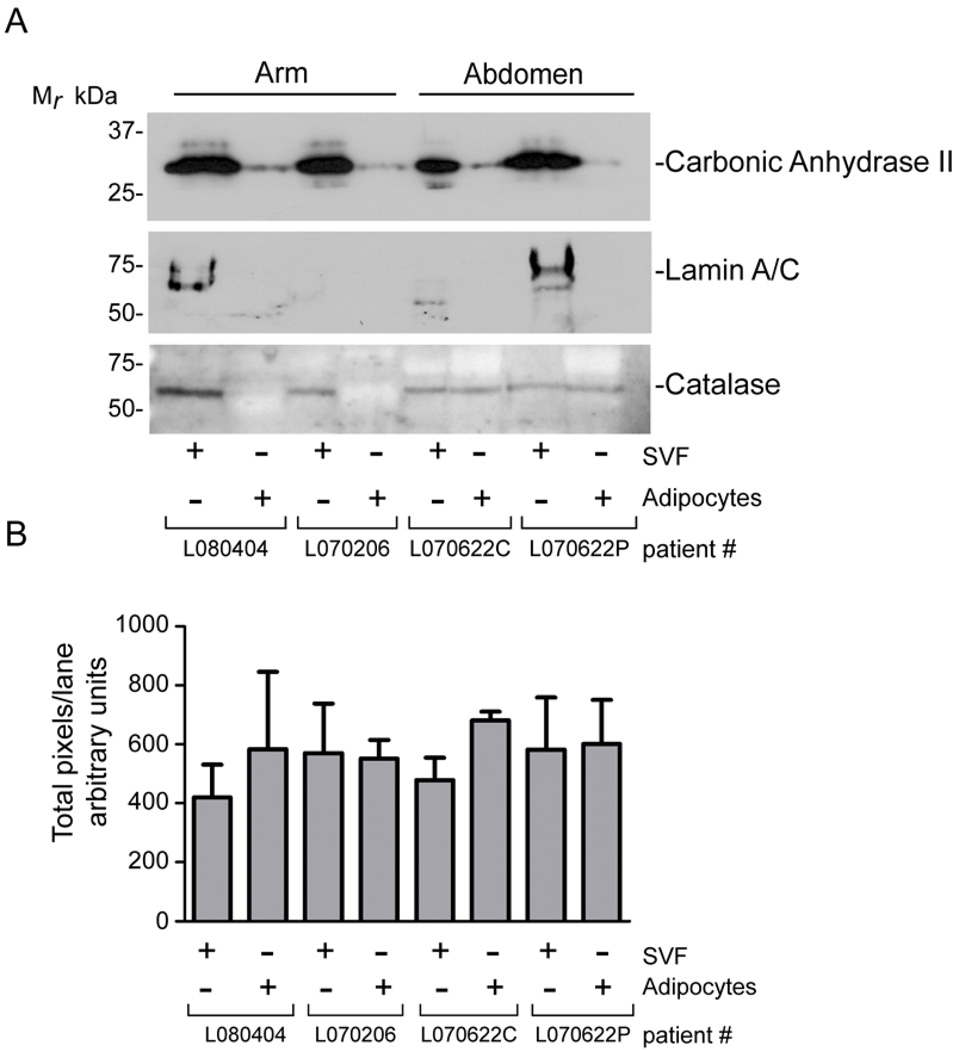
Western blot analysis of carbonic anhydrase II, catalase, and lamin A/C protein expression in mature adipocytes compared to the stromal vascular fraction is presented in Figure 4A with confirmation of even loading in each lane shown in Figure 4B. Carbonic anhydrase II is uniformly abundant in the SVF of the arm and abdominal adipose tissue depots while lamin A/C is detected in the arm or abdominal SVF in only 2 of the 4 donors. While catalase protein expression is enhanced in the SVF obtained from arm adipose tissue, there is no discernable change in the level of catalase between the SVF and the mature adipocytes obtained from abdominal adipose tissue.
Figure 4.
Western Blot Analysis of Selected Proteins. An equal amount of total protein from arm and abdominal SVF and mature adipocytes from four patients was separated by SDS-PAGE and subjected to western blot analysis. A. Carbonic Anhydrase II, Lamin A/C, and Catalase expression paralleled the results reported using 2D-DIGE. B. Protein loading in each lane was assessed using MemCode reversible staining of the nitrocellulose and quantified using Un-Scan-It software. The data are reported as the mean from two separate membranes −/+ standard deviation. An image of the stained membranes is included in the supplemental data.
Bioinformatic Pathway Analysis
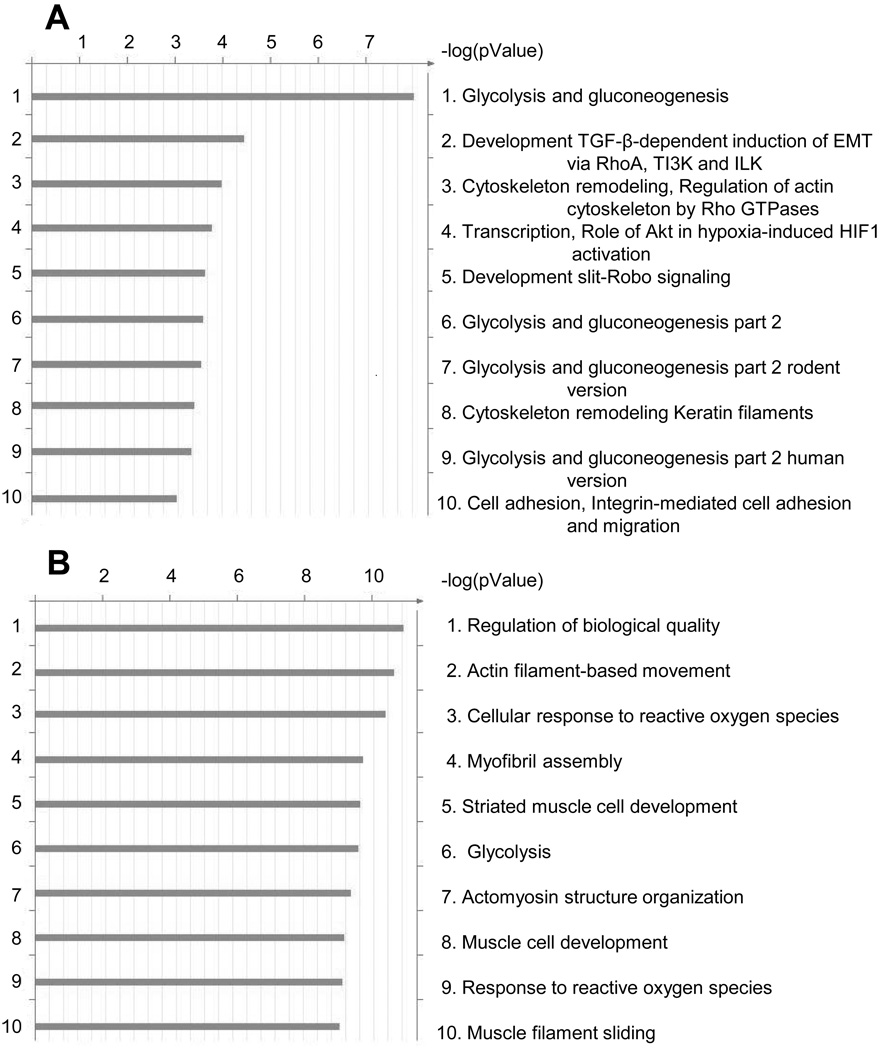
We have conducted two types of pathway analysis: the analysis of over-represented canonical pathway maps and analysis of groups (pathways) interlinked by gene interaction within the list of identified proteins. The major canonical pathways significantly over-represented in the list of identified proteins were determined using GeneGo Metacore software (Fig 5). Each analysis independently identified the glycolytic and energy catabolic pathways as over-represented protein pathways. These accounted for 60% or 80% of the top 10 pathways identified, respectively. In addition, the Metacore analysis (Figure 5A) detected a pathway involving hypoxia inducible factor 1α (HIF1α) activation and the endoplasmic reticulum stress pathway among the top 5 pathways. Previous work has determined that HIF1α activation is involved in adipogenesis 46–48 and that ER stress is associated with adipogenic induction of a CCAAT enhancer binding protein related protein, CHOP, which has also been implicated in the hypoxia response 49–53. This observation is further confirmed by analysis of connectivity among the identified proteins. The list of the top 8 interconnected groups (interaction networks) of proteins is presented in Supplementary Table 5. Gene Ontology processes (Figure 5B) associated with networks point at the same function: the list is dominated by terms “metabolic process”, “regulation of phosphate metabolic process”, “response to stress”, etc. (Supplementary Table 5).
Figure 5.
The results of the pathways analyses listing the top 10 over-represented Metacore canonic pathway maps (A) and top 10 over-represented Gene Ontology categories (B) are presented. The analysis compares all induced vs. all uninduced samples. Bars show comparative statistical significance (gauged by logarithmic p-value on abscissa) in the list of identified proteins.
Discussion
The current study demonstrated that mature (floating) adipocytes in adipose tissue shared a common proteomic profile with the SVF cells isolated from subcutaneous adipose tissue; only 200 out of a total of 960 individual spots were significantly different between adipocytes and SVF cells. The over-representation of proteins involved in glycolysis and gluconeogenesis is consistent with the increase in glycolytic intermediates and fatty acids that occurs during adipogenesis 54 and the role of adipocytes as a major source of metabolic energy 55. However, it should be noted that the over-represented proteins are also common to glyceroneogenesis, a process that occurs in adipocytes instead of gluconeogenesis. Proteins involved in acid/base balance (carbonic anhydrase), free radical oxidation (catalase, SOD), lipid binding (FABP), and intermediate filament proteins (lamin A/C, vimentin) were also present in the both mature adipocytes and SVF cells. This degree of similarity is comparable to that reported by Peinado et al. where 60% of the 800 spots identified by 2-D gel electrophoresis were shared in common between adipocytes and SVF cells isolated from subcutaneous and visceral adipose tissues using a cohort of older (mean age, 56 years), lean (mean BMI 22.8) subjects 35.
The expression level of many of these proteins was greater in the SVF cells relative to the mature adipocytes. While the overall features were similar between the two depots, the differences between the adipocytes and SVF cells were more pronounced in the arm as compared to the abdomen. Consistent with this, Schipper et al. 56 have demonstrated that ASC isolated from the arm adipose depot displayed the most robust expression of PPARγ2 and induced lipolytic activity relative to four other adipose depots. Likewise, Peinado et al. observed distinct protein expression patterns between SVF cells isolated from subcutaneous vs. visceral adipose tissues 35.
Using DIGE, the current study has identified a panel of proteins in the SVF cells and mature adipocytes that are similar to those detected in undifferentiated and adipocyte differentiated ASCs isolated from subcutaneous human adipose tissue 25. Furthermore, the relative expression profile of these proteins is consistent within the adipose tissue subfractions. Multiple proteins induced during adipogenesis of the ASCs were enriched in the adipocyte fraction of abdominal and/or arm tissue. These include carbonic anhydrase, FABP, peroxiredoxin, and SOD 25. Likewise, proteins such as annexin that were reduced during adipogenesis of the ASCs were enriched in the SVF cell fraction of the adipose depots 25.
The presence of selected proteins has been confirmed by western immunoblot analysis. This approach confirms the presence and relative subfraction expression of carbonic anhydrase, catalase, and lamin A/C. Expression of each has been evaluated previously in adipose tissue or adipogenic systems. Peinado et al. noted distinct expression profiles for annexin, carbonic anhydrase, and lamin A/C; these were among 24 individual spots differentially expressed between subcutaneous and visceral adipose tissue derived SVF cells 35, Carbonic anhydrase gene induction has been directly correlated with the adipogenic differentiation of murine 3T3-L1 pre-adipocytes 57, 58. This family of enzymes has been associated with pyruvate carboxylation in adipose metabolism 58. Adipogenesis in the 3T3-L1 model has been correlated with induced expression of catalase secondary to a PPARγ response element located within the promoter region 59, 60. Lamin A/C proteins have been found to increase with adipogenesis in primary cultures of subcutaneous human ASCs 61. Furthermore, patients with certain forms of lipodystrophy display mutations in the lamin A/C gene 62, 63. It is of interest that all three proteins based on western immunoblots are more abundant in the SVF cells relative to the mature adipocytes. This contrasts to published in vitro analyses which have compared protein extracts from pre-adipocytes in the undifferentiated and adipogenic differentiated conditions 57–59, 61. Similar observations relate to the expression of fatty acid binding protein 4 (FABP4), also known as aP2. While the FABP4 mRNA transcription is significantly induced during adipogenesis 64, a 2-dimensional gel feature associated with FABP4 was increased in the arm SVF cells relative to the adipocytes (Supplementary Table 2). This contrasts to prior proteomic studies that observed FABP4 induction with adipogenesis in primary human adipose derived stem cell cultures 25. While the current study examined tissue obtained from female subjects, the work of Delany et al. used ASC isolated from both male and female donors; this could account for the different findings 25. An alternate explanation to account for the current findings is that the heterogeneous SVF cell fraction contains pre-adipocyte cells that are already committed to the adipocyte lineage but have not yet accumulated significant lipid droplets characteristic of the mature adipocyte morphology. Additionally, it suggests that the relative level of protein expression is maximal during early events of adipogenesis and is reduced during the final stages of differentiation. This parallels the expression profile of PPARγ2 in the 3T3-L1 model, where protein levels achieve their highest level within 3 days of induction before falling off; this precedes the maximal attainment of intracellular lipid, which occurs after 6 days of induction. Further studies will be necessary to compare the SVF cell and mature adipocyte protein expression profiles relative to that of primary ASCs in culture and to compare protein expression profiles across different adipose tissue depots in a larger cohort of human subjects.
One potential confounding issue in the current data is the fact that individual proteins are represented by multiple features on the 2-dimensional gels. While one feature may be relatively enriched in the adipocyte fraction, another feature for the same protein may be relatively enriched in the SVF cell fraction. There are multiple examples of this, including collagen α3, glyceraldehydes phosphate dehydrogenase, and lamin A/C. One explanation of this observation is that adipogenic differentiation is associated with unique post-translational modifications of a particular protein. Consequently, a phosphorylated or ubiquitinated peptide may be expressed uniquely in adipocyte fraction. Alternatively, one of the many cell types in the heterogeneous SVF fraction may uniquely modify a protein expressed in common with the adipocyte, leading to the appearance of novel peptide fragment. A second potential confounding issue is the fact that multiple proteins may be associated with a single feature on the 2-dimensional gels. It is possible that the collagenase digestion used to separate the adipocytes and SVF cells could degrade proteins and increase the peptide fragment complexity. Alternatively, this may simply reflect the chemical charge of the peptides themselves. Previous proteomic analyses conducted on cultured primary human adipose-derived stem cells identified a similar pattern of features on 2-dimensional gels containing peptides derived from multiple proteins 24, 25. Finally, it should be noted that many of the proteins identified are abundant and have been reported with high frequency by others in comparative proteomic analyses 65, 66. It remains possible that cellular stresses and/or technical aspects of the 2-dimensional gel electrophoresis increase their level of detection 65, 66.
These findings have relevance in the context of a growing body of data linking the SVF cell composition to the physiology and/or pathology of adipose tissue function 16, 17, 19–22, 67, 68. The appearance of immune cells within adipose tissue as a function of obesity alters the microenvironment at the cellular level. Furthermore, the local release of inflammatory cytokines and other proteins by macrophages and T-cells may provide the mechanism accounting for the pathology of associated with obesity such as diabetes and the metabolic syndrome. The current work confirms and extends the proteomic characterization of human subcutaneous adipose tissue. Consistent with prior reports, we have identified a set of proteins relating to glucose and lipid metabolism, cytoskeletal structure, extracellular matrix, stress response, and oxidative metabolism that are differentially expressed in the mature adipocytes and stromal vascular fraction cells (Supplementary Tables 1–3). A subset of these proteins, such as catalase, have potential use in future studies on adipose depot specific differences and may serve as proteomic biomarkers for physiological stresses and pathologic conditions associated with adipose tissue. It will be necessary to use state of the art proteomic methods and evaluate multiple adipose depots obtained from substantially larger cohorts of healthy and diseased, lean and obese, young and old donors of both genders to pursue this line of research in the future.
Supplementary Material
Acknowledgements
The authors wish to thank Dr. James Wade, his office staff, and his patients for their generosity in providing adipose tissue specimens for this study. The authors would also like to thank Bio-Rad for providing the Molecular Imager VersaDoc MP System used in this work and Gary LeBlanc for his help with preparing tables and figures. This work was funded in part through the support of the Pennington Biomedical Research Foundation and the COBRE Center Grant P20-RR021945 from the National Center for Research Resources at the NIH.
Abbreviations
- BMI
body mass index
- ASC
adipose-derived stem cell
- DIGE
difference in-gel electrophoresis
Footnotes
Supporting Information Available
This information is available free of charge via the internet at http://pubs.acs.org/.
References
- 1.Mitchell JB, M K, Zvonic S, Garrett S, Floyd ZE, Kloster A, Halvorsen YD, Storms RW, Goh B, Kilroy GS, Wu X, Gimble JM. The immunophenotype of human adipose derived cells: Temporal changes in stromal- and stem cell-associated markers. Stem Cells. 2006;24:376–385. doi: 10.1634/stemcells.2005-0234. [DOI] [PubMed] [Google Scholar]
- 2.McIntosh K, Zvonic S, Garrett S, Mitchell JB, Floyd ZE, Hammill L, Kloster A, Halvorsen YD, Ting JP, Storms RW, Goh B, Kilroy G, Wu X, Gimble JM. The immunogenicity of human adipose derived cells: Temporal changes in vitro. Stem Cells. 2006;24:1245–1253. doi: 10.1634/stemcells.2005-0235. [DOI] [PubMed] [Google Scholar]
- 3.Sengenes C, Lolmede K, Zakaroff-Girard A, Busse R, Bouloumie A. Preadipocytes in the human subcutaneous adipose tissue display distinct features from the adult mesenchymal and hematopoietic stem cells. J Cell Physiol. 2005;205(1):114–122. doi: 10.1002/jcp.20381. [DOI] [PubMed] [Google Scholar]
- 4.Zannettino ACW, P S, Khor F, Itescu S, Gimble JM, Gronthos S. Multipotential Human Adipose-Derived Stromal Stem Cells Exhibit a Perivascular Phenotype In Vitro and In Vivo. Journal of Cellular Physiology. 2008;214:413–421. doi: 10.1002/jcp.21210. [DOI] [PubMed] [Google Scholar]
- 5.Traktuev DO, Merfeld-Clauss S, Li J, Kolonin M, Arap W, Pasqualini R, Johnstone BH, March KL. A population of multipotent CD34-positive adipose stromal cells share pericyte and mesenchymal surface markers, reside in a periendothelial location, and stabilize endothelial networks. Circ Res. 2008;102(1):77–85. doi: 10.1161/CIRCRESAHA.107.159475. [DOI] [PubMed] [Google Scholar]
- 6.Rodbell M. Metabolism of Isolated Fat Cells. I. Effects of Hormones on Glucose Metabolism and Lipolysis. J Biol Chem. 1964;239:375–380. [PubMed] [Google Scholar]
- 7.Rodbell M. Metabolism of isolated fat cells. II. The similar effects of phospholipase C (Clostridium perfringens alpha toxin) and of insulin on glucose and amino acid metabolism. J Biol Chem. 1966;241(1):130–139. [PubMed] [Google Scholar]
- 8.Rodbell M. The metabolism of isolated fat cells. IV. Regulation of release of protein by lipolytic hormones and insulin. J Biol Chem. 1966;241(17):3909–3917. [PubMed] [Google Scholar]
- 9.Rodbell M, Jones AB. Metabolism of isolated fat cells. 3. The similar inhibitory action of phospholipase C (Clostridium perfringens alpha toxin) and of insulin on lipolysis stimulated by lipolytic hormones and theophylline. J Biol Chem. 1966;241(1):140–142. [PubMed] [Google Scholar]
- 10.Deslex S, Negrel R, Vannier C, Etienne J, Ailhaud G. Differentiation of human adipocyte precursors in a chemically defined serum-free medium. Int J Obes. 1987;11(1):19–27. [PubMed] [Google Scholar]
- 11.Hauner H, Entenmann G, Wabitsch M, Gaillard D, Ailhaud G, Negrel R, Pfeiffer EF. Promoting effect of glucocorticoids on the differentiation of human adipocyte precursor cells cultured in a chemically defined medium. J Clin Invest. 1989;84(5):1663–1670. doi: 10.1172/JCI114345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van RL, Roncari DA. Isolation of fat cell precursors from adult rat adipose tissue. Cell Tissue Res. 1977;181(2):197–203. doi: 10.1007/BF00219980. [DOI] [PubMed] [Google Scholar]
- 13.Bjorntorp P, Karlsson M, Pertoft H, Pettersson P, Sjostrom L, Smith U. Isolation and characterization of cells from rat adipose tissue developing into adipocytes. J Lipid Res. 1978;19(3):316–324. [PubMed] [Google Scholar]
- 14.Trayhurn P. Endocrine and signalling role of adipose tissue: new perspectives on fat. Acta Physiol Scand. 2005;184(4):285–293. doi: 10.1111/j.1365-201X.2005.01468.x. [DOI] [PubMed] [Google Scholar]
- 15.Trayhurn P, Beattie JH. Physiological role of adipose tissue: white adipose tissue as an endocrine and secretory organ. Proc Nutr Soc. 2001;60(3):329–339. doi: 10.1079/pns200194. [DOI] [PubMed] [Google Scholar]
- 16.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112(12):1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, Chen H. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112(12):1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duffaut C, Galitzky J, Lafontan M, Bouloumie A. Unexpected trafficking of immune cells within the adipose tissue during the onset of obesity. Biochem Biophys Res Commun. 2009;384(4):482–485. doi: 10.1016/j.bbrc.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Duffaut C, Zakaroff-Girard A, Bourlier V, Decaunes P, Maumus M, Chiotasso P, Sengenes C, Lafontan M, Galitzky J, Bouloumie A. Interplay Between Human Adipocytes and T Lymphocytes in Obesity. CCL20 as an Adipochemokine and T Lymphocytes as Lipogenic Modulators. Arterioscler Thromb Vasc Biol. 2009;29(10):1608–1614. doi: 10.1161/ATVBAHA.109.192583. [DOI] [PubMed] [Google Scholar]
- 20.Winer S, Chan Y, Paltser G, Truong D, Tsui H, Bahrami J, Dorfman R, Wang Y, Zielenski J, Mastronardi F, Maezawa Y, Drucker DJ, Engleman E, Winer D, Dosch HM. Normalization of obesity-associated insulin resistance through immunotherapy. Nat Med. 2009;15(8):921–929. doi: 10.1038/nm.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishimura S, Manabe I, Nagasaki M, Eto K, Yamashita H, Ohsugi M, Otsu M, Hara K, Ueki K, Sugiura S, Yoshimura K, Kadowaki T, Nagai R. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat Med. 2009;15(8):914–920. doi: 10.1038/nm.1964. [DOI] [PubMed] [Google Scholar]
- 22.Feuerer M, Herrero L, Cipolletta D, Naaz A, Wong J, Nayer A, Lee J, Goldfine AB, Benoist C, Shoelson S, Mathis D. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med. 2009;15(8):930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fain JN, Buehrer B, Bahouth SW, Tichansky DS, Madan AK. Comparison of messenger RNA distribution for 60 proteins in fat cells vs the nonfat cells of human omental adipose tissue. Metabolism. 2008;57(7):1005–1015. doi: 10.1016/j.metabol.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 24.Zvonic S, Lefevre M, Kilroy G, Floyd ZE, Delany JP, Kheterpal I, Gravois A, Dow R, White A, Wu X, Gimble JM. Secretome of primary cultures of human adipose-derived stem cells (ASCs): modulation of serpins by adipogenesis. Mol Cell Proteomics. 2007;6:18–28. doi: 10.1074/mcp.M600217-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Delany J, Floyd ZE, Zvonic S, Smith A, Gravois A, Reiners E, Wu X, Kilroy G, Lefevre M, Gimble JM. Proteomic analysis of primary cultures of human adipose derived stem cells: Modulation by adipogenesis. Mol Cell Proteomics. 2005;4:731–740. doi: 10.1074/mcp.M400198-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Celis JE, Moreira JM, Cabezon T, Gromov P, Friis E, Rank F, Gromova I. Identification of extracellular and intracellular signaling components of the mammary adipose tissue and its interstitial fluid in high risk breast cancer patients: toward dissecting the molecular circuitry of epithelial-adipocyte stromal cell interactions. Mol Cell Proteomics. 2005;4(4):492–522. doi: 10.1074/mcp.M500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Corton M, Villuendas G, Botella JI, San Millan JL, Escobar-Morreale HF, Peral B. Improved resolution of the human adipose tissue proteome at alkaline and wide range pH by the addition of hydroxyethyl disulfide. Proteomics. 2004;4(2):438–441. doi: 10.1002/pmic.200300644. [DOI] [PubMed] [Google Scholar]
- 28.Roelofsen H, Dijkstra M, Weening D, de Vries MP, Hoek A, Vonk RJ. Comparison of isotope-labeled amino acid incorporation rates (CILAIR) provides a quantitative method to study tissue secretomes. Mol Cell Proteomics. 2009;8(2):316–324. doi: 10.1074/mcp.M800254-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Lavatelli F, Perlman DH, Spencer B, Prokaeva T, McComb ME, Theberge R, Connors LH, Bellotti V, Seldin DC, Merlini G, Skinner M, Costello CE. Amyloidogenic and associated proteins in systemic amyloidosis proteome of adipose tissue. Mol Cell Proteomics. 2008;7(8):1570–1583. doi: 10.1074/mcp.M700545-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmed M, Neville MJ, Edelmann MJ, Kessler BM, Karpe F. Proteomic Analysis of Human Adipose Tissue After Rosiglitazone Treatment Shows Coordinated Changes to Promote Glucose Uptake. Obesity (Silver Spring) 2009 doi: 10.1038/oby.2009.208. [DOI] [PubMed] [Google Scholar]
- 31.Kim J, Choi YS, Lim S, Yea K, Yoon JH, Jun DJ, Ha SH, Kim JW, Kim JH, Suh PG, Ryu SH, Lee TG. Comparative analysis of the secretory proteome of human adipose stromal vascular fraction cells during adipogenesis. Proteomics. 10(3):394–405. doi: 10.1002/pmic.200900218. [DOI] [PubMed] [Google Scholar]
- 32.Pérez-Pérez R, O-D F, García-Santos E, López JA, Camafeita E, Ricart W, Fernández-Real JM, Peral B. Differential proteomics of omental and subcutaneous adipose tissue reflects their unalike biochemical and metabolic properties. J Proteome Res. 2009;8(4):1682–1693. doi: 10.1021/pr800942k. [DOI] [PubMed] [Google Scholar]
- 33.Boden G, Duan X, Homko C, Molina EJ, Song W, Perez O, Cheung P, Merali S. Increase in Endoplasmic Reticulum (ER) Stress Related Proteins and Genes in Adipose Tissue of Obese, Insulin Resistant Individuals. Diabetes. 2008 doi: 10.2337/db08-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenow A, Arrey TN, Bouwman FG, Noben JP, Wabitsch M, Mariman EC, Karas M, Renes J. Identification of novel human adipocyte secreted proteins by using SGBS cells. J Proteome Res. 2010;9(10):5389–5401. doi: 10.1021/pr100621g. [DOI] [PubMed] [Google Scholar]
- 35.Peinado JR, Jimenez-Gomez Y, Pulido MR, Ortega-Bellido M, Diaz-Lopez C, Padillo FJ, Lopez-Miranda J, Vazquez-Martinez R, Malagon MM. The stromal-vascular fraction of adipose tissue contributes to major differences between subcutaneous and visceral fat depots. Proteomics. 2010;10(18):3356–3366. doi: 10.1002/pmic.201000350. [DOI] [PubMed] [Google Scholar]
- 36.Alvarez-Llamas G, Szalowska E, de Vries MP, Weening D, Landman K, Hoek A, Wolffenbuttel BH, Roelofsen H, Vonk RJ. Characterization of the human visceral adipose tissue secretome. Mol Cell Proteomics. 2007;6(4):589–600. doi: 10.1074/mcp.M600265-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Xie X, Yi Z, Bowen B, Wolf C, Flynn CR, Sinha S, Mandarino LJ, Meyer C. Characterization of the Human Adipocyte Proteome and Reproducibility of Protein Abundance by One-Dimensional Gel Electrophoresis and HPLC-ESI-MS/MS. J Proteome Res. 2010;9(9):4521–4534. doi: 10.1021/pr100268f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dubois SG, Floyd EZ, Zvonic S, Kilroy G, Wu X, Carling S, Halvorsen YD, Ravussin E, Gimble JM. Isolation of Human Adipose-derived Stem Cells from Biopsies and Liposuction Specimens. Methods Mol Biol. 2008;449:69–79. doi: 10.1007/978-1-60327-169-1_5. [DOI] [PubMed] [Google Scholar]
- 39.Yu G, W X, Dietrich MA, Polk P, Scott LK, Ptitsyn AA, Gimble JM. Yield and characterization of subcutaneous human adipose-derived stem cells by flow cytometric and adipogenic mRNA analyzes. Cytotherapy. 2010;12:538–546. doi: 10.3109/14653241003649528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bunnell BA, Estes BT, Guilak F, Gimble JM. Differentiation of adipose stem cells. Methods Mol Biol. 2008;456:155–171. doi: 10.1007/978-1-59745-245-8_12. [DOI] [PubMed] [Google Scholar]
- 41.Scherp PKG, Coleman L, Kheterpal I. Gel-based and Gel-free Proteomic Technologies for Adipose-Derived Stem Cells. Meth. Mol. Biol. 2010 doi: 10.1007/978-1-61737-960-4_13. (In-Press) [DOI] [PubMed] [Google Scholar]
- 42.Kheterpal I, Coleman L, Ku G, Wang ZQ, Ribnicky D, Cefalu WT. Regulation of insulin action by an extract of Artemisia dracunculus L. in primary human skeletal muscle culture: A proteomics approach. Phytother Res. 2010 doi: 10.1002/ptr.3093. In Press. [DOI] [PubMed] [Google Scholar]
- 43.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 44.Shah A, Singh H, Sachdev V, Lee J, Yotsukura S, Salgia R, Bharti A. Differential Serum Level of Specific Haptoglobin Isoforms in Small Cell Lung Cancer. Curr Proteomics. 7(1):49–65. doi: 10.2174/157016410790979635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stephens AN, Hannan NJ, Rainczuk A, Meehan KL, Chen J, Nicholls PK, Rombauts LJ, Stanton PG, Robertson DM, Salamonsen LA. Post-translational modifications and protein-specific isoforms in endometriosis revealed by 2D DIGE. J Proteome Res. 9(5):2438–2449. doi: 10.1021/pr901131p. [DOI] [PubMed] [Google Scholar]
- 46.Floyd ZE, Kilroy G, Wu X, Gimble JM. Effects of prolyl hydroxylase inhibitors on adipogenesis and hypoxia inducible factor 1 alpha levels under normoxic conditions. J Cell Biochem. 2007;101(6):1545–1557. doi: 10.1002/jcb.21266. [DOI] [PubMed] [Google Scholar]
- 47.Irwin R, LaPres JJ, Kinser S, McCabe LR. Prolyl-hydroxylase inhibition and HIF activation in osteoblasts promotes an adipocytic phenotype. J Cell Biochem. 2007;100(3):762–772. doi: 10.1002/jcb.21083. [DOI] [PubMed] [Google Scholar]
- 48.Yun Z, Maecker HL, Johnson RS, Giaccia AJ. Inhibition of PPAR gamma 2 gene expression by the HIF-1-regulated gene DEC1/Stra13: a mechanism for regulation of adipogenesis by hypoxia. Dev Cell. 2002;2(3):331–341. doi: 10.1016/s1534-5807(02)00131-4. [DOI] [PubMed] [Google Scholar]
- 49.Wang XZ, Lawson B, Brewer JW, Zinszner H, Sanjay A, Mi LJ, Boorstein R, Kreibich G, Hendershot LM, Ron D. Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153) Mol Cell Biol. 1996;16(8):4273–4280. doi: 10.1128/mcb.16.8.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6(3):439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 51.Sok J, Wang XZ, Batchvarova N, Kuroda M, Harding H, Ron D. CHOP-Dependent stress-inducible expression of a novel form of carbonic anhydrase VI. Mol Cell Biol. 1999;19(1):495–504. doi: 10.1128/mcb.19.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price BD, Calderwood SK. Gadd45 and Gadd153 messenger RNA levels are increased during hypoxia and after exposure of cells to agents which elevate the levels of the glucose-regulated proteins. Cancer Res. 1992;52(13):3814–3817. [PubMed] [Google Scholar]
- 53.Fornace AJ, Jr, Nebert DW, Hollander MC, Luethy JD, Papathanasiou M, Fargnoli J, Holbrook NJ. Mammalian genes coordinately regulated by growth arrest signals and DNA-damaging agents. Mol Cell Biol. 1989;9(10):4196–4203. doi: 10.1128/mcb.9.10.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roberts LD, Virtue S, Vidal-Puig A, Nicholls AW, Griffin JL. Metabolic phenotyping of a model of adipocyte differentiation. Physiol Genomics. 2009;39(2):109–119. doi: 10.1152/physiolgenomics.90365.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lafontan M. Advances in adipose tissue metabolism. Int J Obes (Lond) 2008;32 Suppl 7:S39–S51. doi: 10.1038/ijo.2008.237. [DOI] [PubMed] [Google Scholar]
- 56.Schipper BM, Marra KG, Zhang W, Donnenberg AD, Rubin JP. Regional anatomic and age effects on cell function of human adipose-derived stem cells. Ann Plast Surg. 2008;60(5):538–544. doi: 10.1097/SAP.0b013e3181723bbe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lynch CJ, Hazen SA, Horetsky RL, Carter ND, Dodgson SJ. Differentiation-dependent expression of carbonic anhydrase II and III in 3T3 adipocytes. Am J Physiol. 1993;265 (1 Pt 1):C234–C243. doi: 10.1152/ajpcell.1993.265.1.C234. [DOI] [PubMed] [Google Scholar]
- 58.Hazen SA, Waheed A, Sly WS, LaNoue KF, Lynch CJ. Differentiation-dependent expression of CA V and the role of carbonic anhydrase isozymes in pyruvate carboxylation in adipocytes. FASEB J. 1996;10(4):481–490. doi: 10.1096/fasebj.10.4.8647347. [DOI] [PubMed] [Google Scholar]
- 59.Okuno Y, Matsuda M, Kobayashi H, Morita K, Suzuki E, Fukuhara A, Komuro R, Shimabukuro M, Shimomura I. Adipose expression of catalase is regulated via a novel remote PPARgamma-responsive region. Biochem Biophys Res Commun. 2008;366(3):698–704. doi: 10.1016/j.bbrc.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 60.Hajra AK, Larkins LK, Das AK, Hemati N, Erickson RL, MacDougald OA. Induction of the peroxisomal glycerolipid-synthesizing enzymes during differentiation of 3T3-L1 adipocytes. Role in triacylglycerol synthesis. J Biol Chem. 2000;275(13):9441–9446. doi: 10.1074/jbc.275.13.9441. [DOI] [PubMed] [Google Scholar]
- 61.Lelliott CJ, Logie L, Sewter CP, Berger D, Jani P, Blows F, O'Rahilly S, Vidal-Puig A. Lamin expression in human adipose cells in relation to anatomical site and differentiation state. J Clin Endocrinol Metab. 2002;87(2):728–734. doi: 10.1210/jcem.87.2.8256. [DOI] [PubMed] [Google Scholar]
- 62.Favreau C, Dubosclard E, Ostlund C, Vigouroux C, Capeau J, Wehnert M, Higuet D, Worman HJ, Courvalin JC, Buendia B. Expression of lamin A mutated in the carboxyl-terminal tail generates an aberrant nuclear phenotype similar to that observed in cells from patients with Dunnigan-type partial lipodystrophy and Emery-Dreifuss muscular dystrophy. Exp Cell Res. 2003;282(1):14–23. doi: 10.1006/excr.2002.5669. [DOI] [PubMed] [Google Scholar]
- 63.Holt I, Clements L, Manilal S, Brown SC, Morris GE. The R482Q lamin A/C mutation that causes lipodystrophy does not prevent nuclear targeting of lamin A in adipocytes or its interaction with emerin. Eur J Hum Genet. 2001;9(3):204–208. doi: 10.1038/sj.ejhg.5200609. [DOI] [PubMed] [Google Scholar]
- 64.Graves RA, Tontonoz P, Spiegelman BM. Analysis of a tissue-specific enhancer: ARF6 regulates adipogenic gene expression. Mol Cell Biol. 1992;12(3):1202–1208. doi: 10.1128/mcb.12.3.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petrak J, Ivanek R, Toman O, Cmejla R, Cmejlova J, Vyoral D, Zivny J, Vulpe CD. Deja vu in proteomics. A hit parade of repeatedly identified differentially expressed proteins. Proteomics. 2008;8(9):1744–1749. doi: 10.1002/pmic.200700919. [DOI] [PubMed] [Google Scholar]
- 66.Wang P, Bouwman FG, Mariman EC. Generally detected proteins in comparative proteomics--a matter of cellular stress response? Proteomics. 2009;9(11):2955–2966. doi: 10.1002/pmic.200800826. [DOI] [PubMed] [Google Scholar]
- 67.Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest. 2005;115(5):1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang H, Youm YH, Vandanmagsar B, Ravussin A, Gimble JM, Greenway F, Stephens JM, Mynatt RL, Dixit VD. Obesity increases the production of proinflammatory mediators from adipose tissue T cells and compromises TCR repertoire diversity: implications for systemic inflammation and insulin resistance. J Immunol. 2010;185(3):1836–1845. doi: 10.4049/jimmunol.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.