Abstract
Gene delivery to, and gene targeting in, stem cells would be a highly enabling technology for basic science and biomedical application. Adeno-associated viral (AAV) vectors have demonstrated the capacity for efficient delivery to numerous cells, but their application to stem cells has been limited by low transduction efficiency. Due to their considerable advantages, however, engineering AAV delivery systems to enhance gene delivery to stem cells may have an impact in stem cell biology and therapy. Therefore, using several diverse AAV capsid libraries—including randomly mutagenized, DNA shuffled, and random peptide insertion variants—we applied directed evolution to create a “designer” AAV vector with enhanced delivery efficiency for neural stem cells (NSCs). A novel AAV variant, carrying an insertion of a selected peptide sequence on the surface of the threefold spike within the heparin-binding site, emerged from this evolution. Importantly, this evolved AAV variant mediated efficient gene delivery to rat, mouse, and human NSCs, as well as efficient gene targeting within adult NSCs, and it is thus promising for applications ranging from basic stem cell biology to clinical translation.
Introduction
Stem cells—defined by the hallmark properties of self-renewal, i.e., the capacity to expand, maintain themselves in an undifferentiated state, as well as differentiation into one or more specialized lineages—have considerable biomedical potential. However, better approaches must be developed to control their behavior. Gene delivery can serve as a valuable tool for numerous applications, including modulating stem cell fates by up-/down-regulating specific genes, developing stem cell–based models of human disease, and tracking of stem cells or their progeny upon implantation in vivo.1,2
Current methods to mediate gene delivery to stem cells include plasmid- and viral vector-mediated delivery. Plasmid-based gene transfer offers typically transient gene expression and a large carrying capacity;3 however, current plasmid delivery systems for stem cells can suffer from low transfection efficiency.4 Alternatively, viral vectors based on retroviruses and lentiviruses offer high infectivity and stable gene expression and have been extensively employed in stem cell studies for genetic marking, protein or RNA overexpression, and targeted knock-down of endogenous gene expression.5,6 However, in some cases transient expression is desirable, such as for the temporary overexpression of regulatory signals to manipulate stem cells.7,8,9 Furthermore, insertional mutagenesis is a concern for potential downstream clinical application of retroviruses.10 Adenoviral vectors also have potential for efficient delivery and transient gene expression in stem cells,11 though the most commonly used adenoviral vectors carry numerous virally-encoded factors that may perturb cell function.12 An efficient vector that mediates transient gene expression in stem cells may therefore have utility in the field.
In addition to gene delivery, the capacity for gene targeting via homologous recombination at specific loci in a mammalian stem cell genome has many potential applications, including basic investigation of processes of organismal development and mechanisms of human disease, the targeted introduction of genetic material into a genome without insertional mutagenesis, and therapeutic gene correction.13 However, plasmid-mediated gene targeting is typically very inefficient. Recent approaches that use zinc finger nucleases to enhance gene targeting efficiencies are highly promising, though they require engineering a new zinc finger nuclease for each new locus one would like to manipulate.14,15
Adeno-associated virus (AAV) is a parvovirus with a 4.7 kb single-stranded DNA genome containing two genes (rep and cap), and vectors based on AAV have proven safe and effective for clinical application.16,17,18 Exposure to AAV results in persistent, latent infection in a broad range of both dividing and nondividing cells.19,20 Additionally, Russell and colleagues have demonstrated that AAV vector genomes can mediate homologous recombination with target sequences in a cellular genome at efficiencies 103–104-fold higher than plasmid constructs.21 This capability has been successfully applied to cells that AAV can effectively transduce, including fibroblasts and recently mesenchymal stem cells, which has led to the development of new animal models of human disease.22,23 Although AAV-mediated gene expression and targeting would be effective tools for stem cell studies, AAV vectors are unfortunately extremely inefficient for gene delivery numerous stem cell types.24,25 Therefore, the development of new AAV variants capable of efficient stem cell infection may have impact in stem cell biology and regenerative medicine.
Embryonic neural stem cells (NSCs) are centrally involved in the development of the nervous system,26 and adult NSCs play a fundamental role in learning and memory and may yield new therapies for neurodegenerative disorders.27 However, AAV is extremely inefficient on NSCs.25 Our recent work highlights the potential of directed evolution to create new viruses with enhanced gene delivery properties.18,28,29,30,31 In this approach, we generate large (~107) and highly diverse libraries of viral variants, and the application of an evolutionary pressure—such as the ability to infect a stem cell—selects for the “fittest” variants. Importantly, this general approach can yield successful variants in the absence of mechanistic knowledge of which step(s) in the viral entry pathway—such as cell surface binding, intracellular trafficking, and/or nuclear entry—pose(s) the most substantial barrier to infection. In this study, we have applied directed evolution to create a novel AAV vector with the capacity for high efficiency gene delivery to adult rat NSCs, and this variant was also efficient in murine and human NSCs. Furthermore, this high delivery efficiency was applied to enhance gene correction frequencies in NSCs, potentially providing a tool with potentially strong utility for both basic biology and biomedical application.
Results
AAV library generation
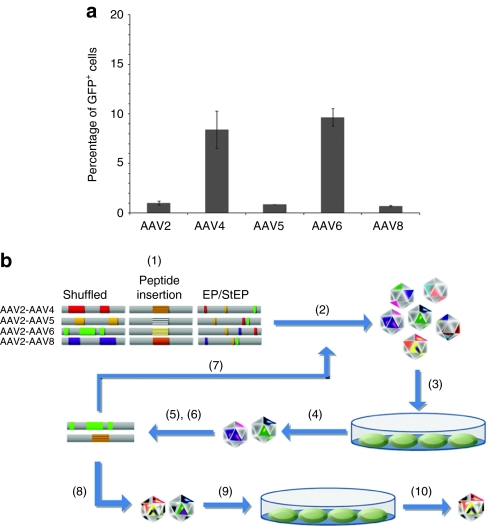
We found that several wild-type AAV serotypes (2, 4, 5, 6, and 8) exhibited low transduction efficiencies [0.7–9.7% with multiplicity of infection (MOI) 105] of NSCs (Figure 1a), as previously described.24,25 These results motivated pursuing directed evolution for the development of novel AAV variants that can more effectively infect NSCs. Three large (>106 independent clones each), diverse libraries were used: (i) random viral chimeras created via pairwise DNA shuffling of the AAV2 cap gene with that of AAV4, 5, 6, or 8, (ii) AAV2 capsid variants with a random 7 amino acid sequence inserted into a key loop region,32 and (iii) random point mutants of the cap gene of AAV2 generated via error prone PCR18 (Figure 1b). Libraries were packaged as previously described to generate a large pool of AAV mutant capsids containing the genome encoding those mutants.18 NSCs, isolated from the hippocampus of adult Fisher 344 rats,27 were infected by each AAV library, adenovirus serotype 5 was added to induce the replication and rescue of the “successful” AAV variants, the surviving AAV cap genes were recovered by PCR and cloned into the AAV genome plasmid, and the next generation library was packaged. After four such iterative selection steps, the resulting viral pool was subjected to additional mutagenesis by error prone PCR on the selected cap genes, followed by further iterative selections (Figure 1b). During the three rounds of evolution and 10 selection steps, the variant pool became progressively more infectious than wild-type AAV2 (data not shown).
Figure 1.
Rationale and strategy for engineering novel AAV variants. (a) Rat neural stem cell (NSC) infection with wild-type adeno-associated viral (AAV) 2, 4, 5, 6, and 8. Cells were infected by addition of green fluorescent protein (GFP) vectors at a multiplicity of infection (MOI) of 105. Cells were incubated with virus for 24 hours, followed by analysis of GFP expression by flow cytometry after an additional 24 hours. Even at this high MOI, the highest gene delivery efficiency was <10%. (b) Directed evolution of novel AAV vectors for enhanced NSC infection: (1) plasmid libraries were generated by DNA shuffling, peptide insertion, and error prone PCR [error prone (EP)], (2) viral libraries were packaged, (3) NSCs were infected with libraries, followed by adenovirus addition, (4) the resulting amplified AAV variants were harvested, and (5) viral genomes were isolated and cloned. The cycle of steps (2)–(5) was iterated (7) with (6) additional EP PCR after the fourth and seventh cycles. Newly evolved clones were used to produce recombinant virus (8) and analyzed for the ability to infect NSCs (9), and the final clone was selected (10).
AAV variants selected through directed evolution
DNA sequencing analysis revealed that the final pools contained seven AAV variants: two shuffled, three peptide-inserted, and two error prone clones (Figure 2, Table S1). Interestingly, the shuffled clones (r3.1, r3.10) were composed of AAV2 and AAV5 sequences, with additional point mutations located on the viral surface. Although AAV2 and 5 were less efficient than AAV4 and AAV6 (Figure 1a), the low efficiencies of AAV4 and AAV6 [ <10% green fluorescent protein positive (GFP+) NSCs at a MOI 105] may be insufficient to enable wild-type cap4 and cap6 to be represented in the selected pool. Furthermore, combinations of fragments of cap4 and cap6 with cap2 likely yield gene delivery properties that are distinct from the parental cap 4 and cap6. In addition, two peptide insertions were present in three clones (r3.3, r3.21, r3.45), which also carried several point mutations.
Figure 2.
Schematic maps of novel adeno-associated viral (AAV) variants. AAV r3.1 denotes the first AAV clone isolated after three rounds of evolution. The gray and blue coloring depicts AAV2 capsid and AAV5 capsid regions, respectively. In the peptide-inserted vectors, “LA” and “A” were inserted as linkers.
Novel AAV variant with enhanced properties for NSC infection
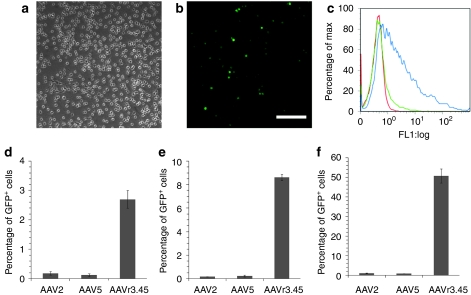
These capsid mutants were used to package recombinant vector encoding GFP driven by a cytomegalovirus (CMV) promoter, which revealed that six of the variants exhibited similar or slightly improved efficiencies compared to AAV2 or AAV5 (Supplementary Figure S1). Additional rounds of selection would likely have eliminated these variants to yield a dominant clone.30 In stark contrast, however, the variant AAV r3.45 mediated high delivery efficiencies (Figure 3a–f). At MOI of 103, 104, and 105, AAV r3.45 infected 14.4–50 fold more cells relative to AAV2 and AAV5 (Figure 3d–f). Importantly, 7 days after infection with AAV r3.45, nearly 100% of GFP+ cells maintained expression of the neural progenitor marker nestin (Figure 4a), indicating that infected cells remained in an immature state.
Figure 3.
Neural stem cell (NSC) transduction by adeno-associated virus (AAV) r3.45. (a) Phase-contrast microscopy and (b) fluorescence microscopy of cells infected by AAV r3.45 [genomic multiplicity of infection (MOI): 105] shows a high level of transduction. Bar = 100 µm. (c) Histogram of green fluorescent protein (GFP)-expressing cells: MOI 103 (red), MOI 104 (green), MOI 105 (blue). Efficiencies of recombinant AAV2, AAV5, and AAV r3.45 were quantified by flow cytometry at (d) MOI 103, (e) 104, and (f) 105.
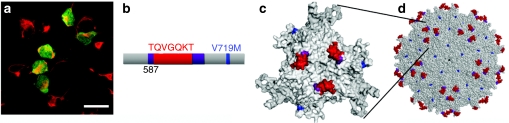
Figure 4.
Neural stem cells (NSCs) infected with adeno-associated virus (AAV) r3.45 remain undifferentiated. (a) Seven days after infection with AAV r3.45, immunostaining revealed that the majority of green fluorescent protein expressing cells were positive for nestin, a neural progenitor cell marker. Bar = 20 µm. (b) Schematic representation of the peptide sequence, point mutation, and heparin-binding residues of AAV clone r3.45. (c) Structural model of a VP3 subunit of AAV r3.45, as well as (d) the full capsid. The color scheme for panels b–d is: V719M (blue), peptide sequence at amino acid 587 (red), and heparin-binding residues (R585/R598) (purple).
AAV r3.45 carried an insertion of LATQVGQKTA (where LA and A are linker residues) at amino acid 587 with an additional V719M mutation (Figure 4b). The inserted peptide likely lies on the surface of the threefold spike within the heparin-binding site (Figure 4c,d), and the point mutation is also surface-exposed. AAV r3.3, which harbored the same peptide insertion but with three different point mutations (i.e., V118I, S537C, and G599S), exhibited considerably lower packaging titers and infectivities than AAVr3.45, indicating that slight capsid modifications can significantly impact viral properties (Figure 1 and Supplementary Figure S1). Importantly, AAV r3.45 was additionally highly infectious for both murine and human NSCs (Figure 5a–c). It was also infectious in several other nonpermissive cell types (Supplementary Figure S2), though not in human or murine embryonic stem cells (Supplementary Figure S3a,b), indicating that its benefits are somewhat cell selective. However, despite its lack of cell specificity, its enhanced gene delivery to cultured NSCs from three different species met the goals of this study and indicates that AAV r3.45 may be used in a variety of stem cell applications, such as transient expression of factors or gene targeting to generate knock-ins/-outs for basic investigation or the development of disease models.
Figure 5.
Enhanced neural stem cell tropism of AAV r3.45. Comparison of transduction efficiencies of adeno-associated virus (AAV) r3.45 versus AAV2 for (a) murine neural stem cells (NSCs), (b) human fetal NSCs, and (c) human neural progenitor cells. Cells were infected at a multiplicity of infection (MOI) of 103, 104, and 105, and green fluorescent protein (GFP) expression was analyzed via flow cytometry 48 hours after infection.
To investigate the roles of the peptide insertion and the V719M mutation for the enhancement of NSC infection, the V719M mutation was reversed via site-directed mutagenesis, and the resulting variant, which contains the peptide insertion without the V719 mutation, was packaged and employed to infect NSCs. This correction of the V719M mutation did not influence the infectivity, indicating the peptide sequence displayed on the AAV r3.45 capsid is critical to enhance gene delivery to NSCs (Supplementary Figure S4). However, although a BLAST search (http://www.ncbi.nlm.nih.gov/BLAST/) revealed that the inserted sequence bears sequence similarity to a number of intracellular enzymes (e.g., Halobacterium salinarum naphthoate synthase), it was not similar to any secreted mammalian or viral capsid proteins.
To determine whether this variant may still use heparan sulfate as its primary receptor, despite its peptide insertion within the AAV2 heparin-binding domain,33 we compared the heparin affinity of AAV r3.45 to that of wild-type AAV2. Purified AAV2 or AAV r3.45 was loaded onto a heparin column and eluted with a series of increasing NaCl concentration (150–750 mmol/l, and 1 mol/l), and a portion of each fraction was added to 293T cells to quantify infectious virus (Supplementary Figure S5). Similar to previous reports,18,29 fractions from 450 to 550 mmol/l NaCl contained the majority of wild-type AAV2. However, AAV r3.45 eluted in a sharp peak between 350 mmol/l and 450 mmol/l, indicating that heparan sulfate may still be primary receptor, but its affinity to heparan sulfate may be slightly altered.
Enhanced gene correction frequency with newly designed AAV variant
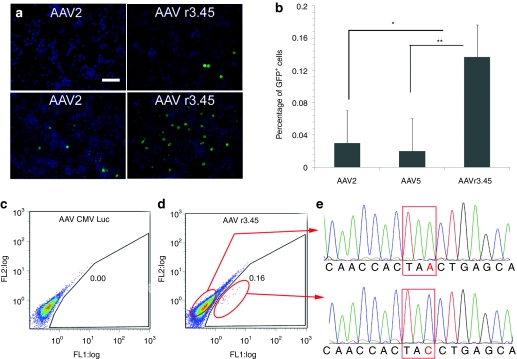
We next assessed the capacity of AAV r3.45 to mediate gene correction in NSCs. GFP cDNA harboring a stop codon (mutGFP1) was introduced into adult rat NSCs using a lentiviral vector. In parallel, a promoter-less GFP with a distinct stop codon (mutGFP2) was packaged into a recombinant AAV vector (Figure 6), and the resulting gene targeting construct carrying mutGFP2 had 586 nucleotides of homology upstream and 1,548 nucleotides downstream of the amber mutation in mutGFP1. Upon infection of NSC-mutGFP1 cells with the targeting vector, recombination with the integrated lentiviral vector corrected the amber mutation in mutGFP1, resulting in fluorescent NSCs (Figure 7a). Importantly, AAV r3.45 demonstrated a significantly improved capacity (six- to tenfold) to repair the single-base pair mutation in NSC-mutGFP1 cell line compared to AAV2 and AAV5 (Figure 7b, Table S2). Controls, including the infection of NSC-mutGFP1 cell lines by AAV CMV Luc (Figure 7c) and naive NSCs infected by the targeting vectors (data not shown), did not yield GFP expression, verifying the specificity of the process.
Figure 6.
Schematic representation of a lentiviral vector carrying UB-mutGFP1-IRES-PuroR, the adeno-associated viral (AAV)-mutGFP2 targeting vector, AAV cytomegalovirus (CMV) promoter Luc, and the corrected sequence at the target site. Genetic elements include: the ubiquitin promoter (UB), internal ribosome entry site (IRES), puromycin acetyltransferase (PuroR), mutGFP1 [a single-base substitution (C → A) 603 nucleotides (nt) from the 5′ end of green fluorescent protein (GFP)], and mutGFP2 [a single point mutation (G → T) 16 nt into GFP]. eGFP, enhanced green fluorescent protein; LTR, long terminal repeat.
Figure 7.
Adeno-associated virus (AAV) r3.45-mediated gene correction by homologous recombination with a nonfunctional green fluorescent protein (GFP) expressed in neural stem cells (NSC) and verification of gene correction. (a) Immunostaining of GFP+, gene-corrected NSC-mutGFP1 cells 72-hours postinfection with the AAV-mutGFP2 targeting vectors: GFP (green), DAPI (blue). Example images with a lower (top row) and higher (bottom row) proportion of cells with a corrected GFP are shown. Bar = 100 µm. (b) Gene targeting frequencies quantified by flow cytometry 3-days postinfection. *,**Statistical differences of P < 0.05 and P < 0.001, respectively. (c) Flow cytometry analysis of NSC-mutGFP1 cells infected by AAV cytomegalovirus (CMV) promoter Luc as a negative control. (d) Fluorescence-activated cell sorting analysis of gene targeting in NSC-mutGFP1 cells 3-days postinfection with AAV r3.45-mutGFP2, which resulted in GFP+ cells shown within the red circled regions. (e) Sequence chromatographs of “uncorrected” (top panel) and “corrected” (bottom panel) GFP DNA fragments amplified from sorted, GFP+ NSC-mutGFP1. Genomic DNA from FL1-positive population was isolated, subjected to PCR, and cloned using StrataClone PCR cloning kit (Stratagene). Nineteen of 24 clones from cells in the GFP+ population showed correction from an A to a C.
Verification of gene correction
To confirm the GFP gene correction, GFP+ cells were isolated by fluorescence-activated cell sorting 14 days after AAV infection, and cellular genomic DNA was subjected to PCR to amplify the GFP sequence specifically from the lentiviral vector. Sequence analysis of 24 cloned PCR fragments revealed 19 corrected sequences (79.2%), with the five uncorrected sequences presumably resulting from NSCs infected with multiple lentiviral vectors (Figure 7d,e). In parallel, sorted cells were expanded for an additional 26 days (total 40 days after AAV infection), and ~94.5% of cells maintained GFP and nestin expression (Figure 8).
Figure 8.
Maintenance of the undifferentiated state of gene-corrected, green fluorescent protein (GFP)-expressing neural stem cells 35-days postinfection. (Nestin: red, GFP: green). The nuclei of cells were co-stained with TOPRO3 (blue). Bar = 50 µm. (a–c: ×200, d–f: ×50, a,d: GFP, b,e: nestin/TOPRO, c,f: GFP/nestin/TOPRO3).
Discussion
AAV is a highly versatile vector with a broad-host range; however, its delivery efficiency to certain cell types, including neural or embryonic stem cells, is low.24,25 One potential reason is a lack of necessary cell surface receptors for a given serotype (e.g., heparan sulfate, integrins), resulting in low attachment affinity and cellular internalization. Alternatively, intracellular trafficking pathways can also pose barriers, as for example nuclear trafficking of AAV is a key rate-limiting step for AAV transduction of some cell types such as murine fibroblasts.34 Additionally, noncapsid related limitations including double-stranded DNA synthesis can compromise AAV-mediated gene expression.35 Our finding that a short peptide insertion into the exterior surface of AAV2 can substantially increase delivery efficiency implicates capsid-related steps such as receptor binding or intracellular trafficking as potential barriers.
The slight shift in the heparin affinity of this variant AAV r3.45 (Supplementary Figure S5) may correspond to a significant modulation of heparan sulfate binding, potentially due to the inserted peptides' conformational alteration of a key exposed loop of the heparin binding domain. However, the fact that the majority of AAV r3.45 still eluted at relatively high NaCl concentrations indicates that this variant may still utilize heparan sulfate as its primary receptor. In addition, the LATQVGQKTA peptide may thus contribute to binding to an alternate secondary receptor or to an intracellular transduction step. Future studies may elucidate this mechanism of action.
The central goal of the directed evolution in this study was to create novel vectors that can better infect NSCs; therefore, positive selection was applied for enhanced adult rat NSC transduction, and negative selection was not applied to remove tropism for other cell types. The approach successfully yielded a novel AAV variant with significantly enhanced infectivity of not only adult rat but also mouse and two human NSCs. As anticipated this enhanced infectivity was not accompanied by specificity for NSCs, as AAV r3.45 was more efficient than AAV2 for several cell types, including primary human astrocytes (Supplementary Figure S2). Accordingly, in vivo AAV r3.45 would be anticipated to transduce NSCs as well as other surrounding cell types (i.e., astrocytes, oliogodendrocytes). However, alternative selection strategies could be implemented to achieve specificity of delivery.
The development of an efficient vehicle for transient gene expression in neural stem cells has applications ranging from the overexpression of factors to induce the differentiation or “programming” of neural stem cells, as well as potentially expression of defined transcription factors to reprogram cells.36,37 Moreover, enhanced transduction and gene targeting capabilities in NSCs may facilitate stem cell biology and regenerative medicine investigations, potentially including the introduction of defined mutations for basic studies, generation of reporter cell lines, and therapeutic gene correction. By yielding a variant with high efficiency gene delivery and gene targeting in NSCs, directed evolution is further established as a powerful approach to engineer novel capabilities into AAV vectors.18,28,29,30,31 Further reverse engineering analysis of the gene delivery mechanisms of this variant may elucidate mechanisms that limit gene delivery in stem cells, as well as the other cell types efficiently infected by this variant. Finally, this general approach may in the future be applied to other cell and stem cell types, thereby extending the utility of AAV to additional biotechnology and biomedical applications.
Materials and Methods
NSC culture. Adult rat NSCs, isolated from the hippocampal region of 6-week-old female Fisher 344 rats as described,27,38 were cultured on tissue culture polystyrene dishes previously coated with poly-ornithine (10 µg/ml) (Sigma-Aldrich, St Louis, MO) and mouse laminin (5 µg/ml) (Invitrogen, Carlsbad, CA), in Dulbecco's modified Eagle's medium (DMEM)/F-12 (1:1) (Invitrogen) containing N-2 supplement (Invitrogen) and 20 ng/ml of recombinant human fibroblast growth factor 2 (Peprotech, Rocky Hill, NJ). Under these conditions, nearly 100% of cells express nestin and Sox2 (data not shown). Every 2–3 days (>60% confluency), cells were split into a fresh dish by mechanical dissociation using polished pasteur pipettes. Adult murine NSCs (a kind gift of Prof Andrew Wurmser, University of California at Berkeley) were cultured in DMEM with 20 ng/ml of recombinant fibroblast growth factor 2 (Peprotech) and 8 µg/ml of heparin. Human fetal NSCs (13 weeks of gestational age) were derived from the telencephalon (HFT13) as previously described39 and were cultured in DMEM/F-12 containing N-2 supplement, 20 ng/ml of fibroblast growth factor 2, 8 µg/ml of heparin (Sigma-Aldrich), and 10 ng/ml of leukemia-inhibitory factor (Chemicon, Temecula, CA). Human neural progenitor cells were derived from a variant of the human embryonic stem cell line BG01 (BresaGen, Athens, GA), BG01V2, using the stromal derived inducing activity involving a co-culture system of human embryonic stem cells and mouse stromal cell line PA6 (Riken BioResource Center Cell Bank, Tsukuba, Japan) (Vazin et al. manuscript in preparation) and were cultured in DMEM/F-12 supplemented with N-2, 20 ng/ml basic fibroblast growth factor, 20 ng/ml epidermal growth factor (Invitrogen), and 50 U/ml Penicillin/Streptomycin.
Plasmid library construction. Random mutagenesis libraries were generated by subjecting cap genes from AAV2 to error prone PCR as previously described.18 Peptide display libraries were generated similar to previous reports.32,40 Briefly, a unique Avr II was introduced into pSub2Cap2 between amino acid 587 and 588 by PCR mutagenesis. A random 21 base pair oligonucleotide [5′-GGAGGGCTAGCA (NNK)7 GCTAGCAAAAGCGGGGGGAGAGTGAGG-3′] was annealed to an antisense primer 5′-CCTCACTCTCCCCCCGCT-3′, followed by synthesis of the second strand. The resulting dsDNA inserts were digested with Nhe I and ligated into the unique Avr II site of pSub2Cap2. Pairwise DNA shuffling libraries of AAV2 with AAV4, AAV5, AAV6, or AAV8 (AAV2-AAV4, AAV2-AAV5, AAV2-AAV6, and AAV2-AAV8) were generated by PCR amplification of cap genes from AAV2, AAV4, AAV5, AAV6, and AAV8 and followed by insertion into pSub2. Each viral gene was then amplified via PCR, and DNA shuffling was performed as previously described.41,42 Briefly, DNase I digestion of equimolar amounts of PCR-amplified cap genes yielded fragments ranging from 50 to 500 base pair in size. Fragments were gel purified and reassembled without primers in the following conditions: 96 °C, 3 minutes; 40 cycles of 94 °C, 55 °C, and 72 °C; and 72 °C, 10 minutes. Assembled fragments were further amplified by PCR and cloned into pSub2 for replication competent AAV production.
Selection of viral libraries. The plasmid library was used to package replication competent AAV as previously described,18,43 and the resulting viral libraries were then harvested as described.44 Virus was purified using OptiPrep (60%, wt/vol iodixanol) (Sigma-Aldrich) density gradient ultracentrifugation according to the manufacturer's instructions, and genomic titers of AAV viral libraries were determined by quantitative real time PCR on a Bio-Rad iCycler (Bio-Rad, Hercules, CA) using a Taqman probe (Biosearch Technologies, Novato, CA).18
NSCs, seeded at a density of 6 × 105 per well in 6-well tissue culture plates coated with poly-ornithine/laminin one day prior to infection, were infected with either wild-type AAV2 or the AAV libraries at genomic MOIs of 10, 100, and 1,000. After a 24-hour exposure, cells were co-infected with adenovirus serotype 5 in fresh medium (i.e., DMEM/F12 + N2, fibroblast growth factor 2). Once cells exhibited cytopathic effect, after approximately two additional days of culture, they were lysed by three freeze/thaw steps, followed by the treatment with benzonase (1 unit/ml) (Sigma-Aldrich) at 37 °C for 30 minutes. Adenovirus was inactivated by incubating the viral lysates at 50 °C for 30 minutes. To quantify viral libraries rescued from the each selection, viral genomic DNA was extracted from DNase-resistant particles. At each step, the total viral genomic DNA rescued was compared for the viral libraries versus wild-type AAV2.
Evolution of viral libraries. One round of evolution is defined as cap mutagenesis followed by several selection steps, i.e., NSC infection and adenovirus rescue. After 3–4 selection steps, the rescued AAV cap genes were randomly mutated by error prone PCR as previously described.18 The resulting PCR products were digested with Hind III and Not I, and the fragments were ligated into pSub2 to create a new replication competent AAV plasmid library with at least 106 independent clones. Viral library packaging was then conducted by the calcium phosphate transient transfection method,18 and viral titration and purification were performed as with the initial library construction.
After the evolution was completed, AAV cap genes were extracted from the final pool of successful AAV variants, and genomic sequences were analyzed at the UC Berkeley DNA Sequencing Facility. The extracted viral genomes were amplified by PCR and inserted into the pXX2 Not I recombinant AAV (rAAV) packaging plasmid.22,33. Three-dimensional models of the VP3 subunit were generated using Swiss Model (http://swissmodel.expasy.org) with the coordinates of AAV2 (Protein Databank accession no. 1LP3) supplied as template, and images were rendered in Pymol (http://pymol.sourceforge.net/) and Rasmol.
Heparin column chromatography. AAV r3.45 and AAV2 binding to heparin were analyzed as previously described.18,29 Briefly, ~1011 purified genomic particles of virus were loaded onto a 1 ml HiTrap heparin column (GE Healthcare Life Sciences, Pittsburgh, PA), which was previously equilibrated with Tris buffer (50 mmol/l, pH 7.5) containing 150 mmol/l NaCl. Elutions were performed with 0.75 ml of Tris buffer containing increasing increments of 50 mmol/l NaCl up to 750 mmol/l, followed by a 1 mol/l NaCl wash. Small fractions (75 µl) of each elution were added to 2.5 × 105 293T cells cultured in 12-well dishes and employed to quantify the level of infectious virus present. Forty-eight hour postinfection, GFP expression was measured with Becton Dickinson fluorescence-activated cell sorting Caliber (Yonsei University College of Medicine Medical Research Center, Seoul, Korea).
NSC transduction. To test the infectivity of rAAV cap clones, vectors carrying cDNA encoding GFP were packaged using the transient transfection method. Briefly, an equal mass (17 µg) of three plasmids—a AAV helper plasmid (carrying a cap variant), CMV GFP vector plasmid containing ITR (pAAV CMV GFP SN), and pHelper plasmid—were transfected via calcium phosphate into AAV293 cells,18,27 and viral vectors were harvested and purified as described above. Genomic titers of each rAAV vector were determined by quantitative PCR.
One day prior to AAV infection, NSCs were plated onto 24-well dishes at a density of 30,000 cells/well. On day 1, cells were infected by the rAAV clones at genomic MOIs (103, 104, and 105) and incubated for additional 48 hours before analysis. The resulting transduction efficiencies of the individual rAAV variants on rat and murine NSCs were quantified with a Beckman–Coulter EPICS XL flow cytometer (UC Berkeley Cancer Center, Berkeley, CA) and compared to those of AAV2 and AAV5, which were chosen as controls since the two shuffled clones that emerged from the evolution carried fragments from AAV cap2 and AAV cap5 (Figure 1). Human NSC and neural progenitor cell transduction efficiencies were quantified with a Becton Dickinson fluorescence-activated cell sorting Caliber (Yonsei University College of Medicine Medical Research Center) and Beckman–Coulter FC 500 flow cytometer (Berkeley Stem Cell Center, Berkeley, CA), respectively.
Immunofluorescence staining. To analyze AAV r3.45 infection of undifferentiated NSCs, and the effects of AAV infection on NSC differentiation, immunostaining for nestin (undifferentiated NSCs), β-III-tubulin (neurons), and glial fibrillary acidic protein (astrocytes) was conducted.31 NSCs were seeded onto 8-well chamber slides and infected as indicated above. At 48 hours postinfection, cells were fixed in 4% paraformaldehyde for 15 minutes and blocked with 5% goat serum in 0.3% Triton X-100/phosphate-buffered solution for 30 minutes at room temperature. Cells were incubated overnight at 4 °C with primary antibodies. For nestin/β-III-tubulin co-staining, rabbit anti-rat nestin (1:500 dilution; Abcam) and mouse anti-β-III-tubulin (1:500 dilution; Sigma-Aldrich), and, for nestin/glial fibrillary acidic protein costaining, mouse anti-rat nestin (1:1,000 dilution; BD Biosciences) and guinea-pig anti-glial fibrillary acidic protein(1:1,000 dilution; Advanced ImmunoChemical, Long Beach, CA) were used. Subsequently, cells were rinsed thoroughly and stained with secondary fluorescent-conjugated antibodies, including goat anti-rabbit Alexa 546 (1:250 dilution; Molecular Probes, Carlsbad, CA), goat anti-mouse Cy5 (1:250 dilution; Jackson ImmunoResearch, West Grove, PA), goat anti-mouse Cy3 (1:250 dilution; Jackson ImmunoResearch), or goat anti-guinea-pig Cy5 (1:250 dilution; Jackson ImmunoResearch). Stained cells were imaged by fluorescence confocal microscopy (Leica Microsystems, Bannockburn, IL) or fluorescence microscopy (Nikon ECLIPSE TE2000-E, Nikon, Melville, NY, or Olympus IX50, Olympus America). When amplifying GFP signals, rabbit anti-GFP (1:2,000 dilution; Invitrogen, Center Valley, PA) and goat anti-rabbit Alexa 488 (1:250 dilution; Invitrogen) were used as primary and secondary antibodies, respectively.
Generation of mutant GFP NSC line. An internal ribosome entry site and puromycin resistance gene cassette was cloned into the Eco RI and Xho I sites of pFUGW (a kind gift from David Baltimore, California Institute of Technology, Pasadena, CA), which contains an enhanced GFP gene under the control of human ubiquitin-C promoter.45 To insert an amber stop codon (TAA) into the GFP sequence, the substitution of “C” to “A” was introduced 603 nucleotides from the 5′ end of GFP via QuikChange site-directed mutagenesis. The inserted point mutation was verified by DNA sequencing analysis.
A stable NSC cell population expressing the amber mutant GFP (NSC-mutGFP1) was generated by infection with lentiviral vector carrying the defective GFP. Briefly, 10 µg of pFUG carrying UB-mutGFP1-internal ribosome entry site and puromycin resistance gene cassette, 5 µg of pMDL g/p PRE, 3.5 µg of pcDNA3 IVS vesicular stomatitis virus G, and 1.5 µg of pRSV Rev, were transfected into HEK 293T cell lines (>70% confluency) by the calcium/phosphate transient transfection method.46 The lentiviral vector was harvested and concentrated by ultracentrifugation (L8-55M Ultracentrifuge; Beckman–Coulter, Brea, CA), followed by resuspension in 100 µl of phosphate-buffered solution with 20% sucrose. Infectious titer, determined as previously described,47 was 1.58 × 107 ± 4.66 × 106 IU/ml. NSCs were seeded onto 6-well tissue culture plates at a density of 3 × 105 cells/well and infected by 0, 1, 3, 5, 10, or 20 µl of the lentiviral vector. After puromycin selection,48 the well in which 70–80% of cells survived was expanded for further analysis. The integration of lentiviral genomes was verified by cellular genomic DNA extraction, PCR amplification, and sequencing analysis of the PCR products. Importantly, GFP fluorescence was not detected in the mutGFP1 NSC cells, as confirmed by both flow cytometry and immunostaining against GFP.
Gene targeting assay. A point mutation (G to T) was inserted 16 nucleotides from the 5′ end of GFP to yield a defective GFP open reading frame (mutGFP2). Subsequently, pFUG was digested with Bam HI and Xho I, and the mutGFP2-internal ribosome entry site and puromycin resistance gene cassette was subcloned into an AAV vector plasmid (i.e., pAAV CMV GFP SN), where CMV GFP was excised by Bam HI and Xho I, such that the resulting targeting vector lacked a promoter. This genetic construct was packaged into the AAV2, AAV5, and AAV r3.45 capsids, and AAV r3.45 carrying luciferase driven by CMV promoter was used as a negative control. All viral vectors were harvested and purified as described earlier.
NSCs were seeded onto 24-well tissue culture plates at a density of 50,000 cells/well 1 day prior to AAV infection. Each AAV targeting vector was added to the NSCs at a genomic MOI of 5 × 105, and the medium was replaced 24 hours later. Infected NSCs were analyzed by flow cytometry (Cytomics FC500; Beckman–Coulter) 3 days after infection, and the percentage of GFP+ cells was scored as the gene targeting frequency.
Cell sorting and sequencing. Cells continuously cultured for 14 days after AAV r3.45 infection were sorted with a Cytopeia Influx Sorter (UC Berkeley Cancer Center, Valencia, CA) to isolate FL1 (i.e., GFP) positive cells. A fraction of the sorted cells was used for DNA sequencing analysis, and the remainder was expanded for an additional 26 days, at which time 94.5% still expressed GFP (data not shown). For DNA sequencing analysis, cellular genomic DNA was extracted with the QIAamp DNA Micro Kit (Qiagen) and amplified by using 5′-GCGGCAAGAACCCAAGGTCT-3′ and 5′-CGGCTTCGGCCAGTAACGTT-3′ primers. To specifically amplify a fragment from the lentiviral vector and exclude amplification from the AAV viral genome, the forward primer was designed to amplify from the half part of UB promoter in the targeted site, which existed only in the lentiviral vectors. The resulting PCR products were cloned using the StrataClone PCR cloning kit (Stratagene, La Jolla, CA), and individual clones were sequenced to confirm the gene correction.
SUPPLEMENTARY MATERIAL Figure S1. Percentages of GFP positive neural stem cells transduced by recombinant vectors pseudotyped with capsids from AAV2, AAV5, and selected AAV variants. (a): MOI 103, (b): MOI 104, (c): MOI 105. Error bars indicate the standard deviation. Figure S2. Comparison of transduction efficiencies of selected AAV variants with wtAAV2 on a panel of cell types. (a) NIH 3T3, (b) B16F10, (c) CHO, (d) human astrocytes, and (e) HEK 293T cells. The genomic MOI used for infection was 103. Figure S3. Comparison of transduction efficiencies of AAV r3.45 versus AAV2. (a) hSF-6 human embryonic stem cells and (b) E14 mouse embryonic stem cells. The genomic MOIs for infection were 105. Figure S4. Comparison of transduction efficiencies of original AAV r3.45 (peptide insertion + V719M point mutation) versus modified AAV r3.45 (peptide insertion only, V719M correction). Figure S5. Heparin column chromatograms for wild-type AAV2 and AAV r3.45 to investigate the heparin affinity of AAV r3.45. Table S1. Genomic and infectious titers of wild-type AAV serotypes and AAV variants. Infectious titers were obtained from HEK 293T cell infection. Table S2. Genomic titers of gene targeting vectors.
Acknowledgments
This work was supported by California Institute for Regenerative Medicine Training grant number T1-00007 and grant number RT1-01021, and by National Research Foundation (NRF) grant funded by the Korea government (MEST) through the Active Polymer Center for Pattern Integration (No. R11-2007-050-00000-0). KIP was supported by Stem Cell Research Center grant and Healthcare Technology R&D Project grant funded by Korean Government. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of CIRM or any other agency of the State of California. The authors are also grateful to Prof Andrew Wurmser for kindly providing murine NSCs and Ilsun Kim for technical support on human NSC culture.
Supplementary Material
Percentages of GFP positive neural stem cells transduced by recombinant vectors pseudotyped with capsids from AAV2, AAV5, and selected AAV variants. (a): MOI 103, (b): MOI 104, (c): MOI 105. Error bars indicate the standard deviation.
Comparison of transduction efficiencies of selected AAV variants with wtAAV2 on a panel of cell types. (a) NIH 3T3, (b) B16F10, (c) CHO, (d) human astrocytes, and (e) HEK 293T cells. The genomic MOI used for infection was 103.
Comparison of transduction efficiencies of AAV r3.45 versus AAV2. (a) hSF-6 human embryonic stem cells and (b) E14 mouse embryonic stem cells. The genomic MOIs for infection were 105.
Comparison of transduction efficiencies of original AAV r3.45 (peptide insertion + V719M point mutation) versus modified AAV r3.45 (peptide insertion only, V719M correction).
Heparin column chromatograms for wild-type AAV2 and AAV r3.45 to investigate the heparin affinity of AAV r3.45.
Genomic and infectious titers of wild-type AAV serotypes and AAV variants. Infectious titers were obtained from HEK 293T cell infection.
Genomic titers of gene targeting vectors.
REFERENCES
- Hendrie PC., and, Russell DW. Gene targeting with viral vectors. Mol Ther. 2005;12:9–17. doi: 10.1016/j.ymthe.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Yates F., and, Daley GQ. Progress and prospects: gene transfer into embryonic stem cells. Gene Ther. 2006;13:1431–1439. doi: 10.1038/sj.gt.3302854. [DOI] [PubMed] [Google Scholar]
- Madeira C, Mendes RD, Ribeiro SC, Boura JS, Aires-Barros MR, da Silva CL, et al. Nonviral gene delivery to mesenchymal stem cells using cationic liposomes for gene and cell therapy. J Biomed Biotechnol. 2010;2010:735349. doi: 10.1155/2010/735349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo J., and, Tabata Y. Non-viral gene transfection technologies for genetic engineering of stem cells. Eur J Pharm Biopharm. 2008;68:90–104. doi: 10.1016/j.ejpb.2007.04.021. [DOI] [PubMed] [Google Scholar]
- Singer O, Tiscornia G, Ikawa M., and, Verma IM. Rapid generation of knockdown transgenic mice by silencing lentiviral vectors. Nat Protoc. 2006;1:286–292. doi: 10.1038/nprot.2006.44. [DOI] [PubMed] [Google Scholar]
- Marumoto T, Tashiro A, Friedmann-Morvinski D, Scadeng M, Soda Y, Gage FH, et al. Development of a novel mouse glioma model using lentiviral vectors. Nat Med. 2009;15:110–116. doi: 10.1038/nm.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Valdimarsdottir G, Larsson J, Brun A, Magnusson M, Jacobsen SE, et al. Transient disruption of autocrine TGF-beta signaling leads to enhanced survival and proliferation potential in single primitive human hemopoietic progenitor cells. J Immunol. 2002;168:755–762. doi: 10.4049/jimmunol.168.2.755. [DOI] [PubMed] [Google Scholar]
- Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW., and, Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hester ME, Song S, Miranda CJ, Eagle A, Schwartz PH., and, Kaspar BK. Two factor reprogramming of human neural stem cells into pluripotency. PLoS ONE. 2009;4:e7044. doi: 10.1371/journal.pone.0007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gropp M, Itsykson P, Singer O, Ben-Hur T, Reinhartz E, Galun E, et al. Stable genetic modification of human embryonic stem cells by lentiviral vectors. Mol Ther. 2003;7:281–287. doi: 10.1016/s1525-0016(02)00047-3. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Mitsui K, Aizawa E, Hasegawa K, Kawase E, Yamagishi T, et al. Highly efficient transient gene expression and gene targeting in primate embryonic stem cells with helper-dependent adenoviral vectors. Proc Natl Acad Sci USA. 2008;105:13781–13786. doi: 10.1073/pnas.0806976105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CE, Ehrhardt A., and, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- Conrad C, Gupta R, Mohan H, Niess H, Bruns CJ, Kopp R, et al. Genetically engineered stem cells for therapeutic gene delivery. Curr Gene Ther. 2007;7:249–260. doi: 10.2174/156652307781369119. [DOI] [PubMed] [Google Scholar]
- Lim KI, Klimczak R, Yu JH., and, Schaffer DV. Specific insertions of zinc finger domains into Gag-Pol yield engineered retroviral vectors with selective integration properties. Proc Natl Acad Sci USA. 2010;107:12475–12480. doi: 10.1073/pnas.1001402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Verma IM., and, Weitzman MD. Gene therapy: twenty-first century medicine. Annu Rev Biochem. 2005;74:711–738. doi: 10.1146/annurev.biochem.74.050304.091637. [DOI] [PubMed] [Google Scholar]
- Bennicelli J, Wright JF, Komaromy A, Jacobs JB, Hauck B, Zelenaia O, et al. Reversal of blindness in animal models of leber congenital amaurosis using optimized AAV2-mediated gene transfer. Mol Ther. 2008;16:458–465. doi: 10.1038/sj.mt.6300389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshri N, Koerber JT, Kaspar BK., and, Schaffer DV. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat Biotechnol. 2006;24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- Kaplitt MG, Leone P, Samulski RJ, Xiao X, Pfaff DW, O'Malley KL, et al. Long-term gene expression and phenotypic correction using adeno-associated virus vectors in the mammalian brain. Nat Genet. 1994;8:148–154. doi: 10.1038/ng1094-148. [DOI] [PubMed] [Google Scholar]
- Flannery JG, Zolotukhin S, Vaquero MI, LaVail MM, Muzyczka N., and, Hauswirth WW. Efficient photoreceptor-targeted gene expression in vivo by recombinant adeno-associated virus. Proc Natl Acad Sci USA. 1997;94:6916–6921. doi: 10.1073/pnas.94.13.6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DW., and, Hirata RK. Human gene targeting by viral vectors. Nat Genet. 1998;18:325–330. doi: 10.1038/ng0498-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain JR, Schwarze U, Wang PR, Hirata RK, Hankenson KD, Pace JM, et al. Gene targeting in stem cells from individuals with osteogenesis imperfecta. Science. 2004;303:1198–1201. doi: 10.1126/science.1088757. [DOI] [PubMed] [Google Scholar]
- Rogers CS, Hao Y, Rokhlina T, Samuel M, Stoltz DA, Li Y, et al. Production of CFTR-null and CFTR-DeltaF508 heterozygous pigs by adeno-associated virus-mediated gene targeting and somatic cell nuclear transfer. J Clin Invest. 2008;118:1571–1577. doi: 10.1172/JCI34773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Arica JR, Thomson AJ, Ansell R, Chiorini J, Davidson B., and, McWhir J. Infection efficiency of human and mouse embryonic stem cells using adenoviral and adeno-associated viral vectors. Cloning Stem Cells. 2003;5:51–62. doi: 10.1089/153623003321512166. [DOI] [PubMed] [Google Scholar]
- Hughes SM, Moussavi-Harami F, Sauter SL., and, Davidson BL. Viral-mediated gene transfer to mouse primary neural progenitor cells. Mol Ther. 2002;5:16–24. doi: 10.1006/mthe.2001.0512. [DOI] [PubMed] [Google Scholar]
- Temple S. The development of neural stem cells. Nature. 2001;414:112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]
- Lai K, Kaspar BK, Gage FH., and, Schaffer DV. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat Neurosci. 2003;6:21–27. doi: 10.1038/nn983. [DOI] [PubMed] [Google Scholar]
- Koerber JT, Klimczak R, Jang JH, Dalkara D, Flannery JG., and, Schaffer DV. Molecular evolution of adeno-associated virus for enhanced glial gene delivery. Mol Ther. 2009;17:2088–2095. doi: 10.1038/mt.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerber JT, Jang JH., and, Schaffer DV. DNA shuffling of adeno-associated virus yields functionally diverse viral progeny. Mol Ther. 2008;16:1703–1709. doi: 10.1038/mt.2008.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffon KJ, Koerber JT, Dickey DD, Murtha M, Keshavjee S, Kaspar BK, et al. Directed evolution of adeno-associated virus to an infectious respiratory virus. Proc Natl Acad Sci USA. 2009;106:3865–3870. doi: 10.1073/pnas.0813365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimczak RR, Koerber JT, Dalkara D, Flannery JG., and, Schaffer DV. A novel adeno-associated viral variant for efficient and selective intravitreal transduction of rat Müller cells. PLoS ONE. 2009;4:e7467. doi: 10.1371/journal.pone.0007467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller OJ, Kaul F, Weitzman MD, Pasqualini R, Arap W, Kleinschmidt JA, et al. Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors. Nat Biotechnol. 2003;21:1040–1046. doi: 10.1038/nbt856. [DOI] [PubMed] [Google Scholar]
- Opie SR, Warrington KH, Jr, Agbandje-McKenna M, Zolotukhin S., and, Muzyczka N. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J Virol. 2003;77:6995–7006. doi: 10.1128/JVI.77.12.6995-7006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J, Qing K, Kwon HJ, Mah C., and, Srivastava A. Impaired intracellular trafficking of adeno-associated virus type 2 vectors limits efficient transduction of murine fibroblasts. J Virol. 2000;74:992–996. doi: 10.1128/jvi.74.2.992-996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari FK, Samulski T, Shenk T., and, Samulski RJ. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol. 1996;70:3227–3234. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Greber B, Araúzo-Bravo MJ, Meyer J, Park KI, Zaehres H, et al. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461:649–643. doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]
- Kim JB, Sebastiano V, Wu G, Araúzo-Bravo MJ, Sasse P, Gentile L, et al. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Palmer TD, Markakis EA, Willhoite AR, Safar F., and, Gage FH. Fibroblast growth factor-2 activates a latent neurogenic program in neural stem cells from diverse regions of the adult CNS. J Neurosci. 1999;19:8487–8497. doi: 10.1523/JNEUROSCI.19-19-08487.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, Kim IS, Lee IS, Lee JP, Snyder EY., and, Park KI. Human neurospheres derived from the fetal central nervous system are regionally and temporally specified but are not committed. Exp Neurol. 2006;199:222–235. doi: 10.1016/j.expneurol.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Perabo L, Büning H, Kofler DM, Ried MU, Girod A, Wendtner CM, et al. In vitro selection of viral vectors with modified tropism: the adeno-associated virus display. Mol Ther. 2003;8:151–157. doi: 10.1016/s1525-0016(03)00123-0. [DOI] [PubMed] [Google Scholar]
- Stemmer WP. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- Zhao H., and, Arnold FH. Optimization of DNA shuffling for high fidelity recombination. Nucleic Acids Res. 1997;25:1307–1308. doi: 10.1093/nar/25.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batard P, Jordan M., and, Wurm F. Transfer of high copy number plasmid into mammalian cells by calcium phosphate transfection. Gene. 2001;270:61–68. doi: 10.1016/s0378-1119(01)00467-x. [DOI] [PubMed] [Google Scholar]
- Koerber JT, Maheshri N, Kaspar BK., and, Schaffer DV. Construction of diverse adeno-associated viral libraries for directed evolution of enhanced gene delivery vehicles. Nat Protoc. 2006;1:701–706. doi: 10.1038/nprot.2006.93. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ., and, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Yu JH., and, Schaffer DV. Selection of novel vesicular stomatitis virus glycoprotein variants from a peptide insertion library for enhanced purification of retroviral and lentiviral vectors. J Virol. 2006;80:3285–3292. doi: 10.1128/JVI.80.7.3285-3292.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard JN, Shah PS, Burnett JC., and, Schaffer DV. HIV evades RNA interference directed at TAR by an indirect compensatory mechanism. Cell Host Microbe. 2008;4:484–494. doi: 10.1016/j.chom.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J, O'Neill A., and, Schaffer DV. PI3K/Akt and CREB regulate adult neural hippocampal progenitor proliferation and differentiation. Dev Neurobiol. 2007;67:1348–1361. doi: 10.1002/dneu.20506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Percentages of GFP positive neural stem cells transduced by recombinant vectors pseudotyped with capsids from AAV2, AAV5, and selected AAV variants. (a): MOI 103, (b): MOI 104, (c): MOI 105. Error bars indicate the standard deviation.
Comparison of transduction efficiencies of selected AAV variants with wtAAV2 on a panel of cell types. (a) NIH 3T3, (b) B16F10, (c) CHO, (d) human astrocytes, and (e) HEK 293T cells. The genomic MOI used for infection was 103.
Comparison of transduction efficiencies of AAV r3.45 versus AAV2. (a) hSF-6 human embryonic stem cells and (b) E14 mouse embryonic stem cells. The genomic MOIs for infection were 105.
Comparison of transduction efficiencies of original AAV r3.45 (peptide insertion + V719M point mutation) versus modified AAV r3.45 (peptide insertion only, V719M correction).
Heparin column chromatograms for wild-type AAV2 and AAV r3.45 to investigate the heparin affinity of AAV r3.45.
Genomic and infectious titers of wild-type AAV serotypes and AAV variants. Infectious titers were obtained from HEK 293T cell infection.
Genomic titers of gene targeting vectors.