Abstract
Phytochrome B (PhyB), one of the major photosensory chromoproteins in plants, mediates a variety of light-responsive developmental processes in a photoreversible manner. To analyze the structural requirements of the chromophore for the spectral properties of PhyB, we have designed and chemically synthesized 20 analogs of the linear tetrapyrrole (bilin) chromophore and reconstituted them with PhyB apoprotein (PHYB). The A-ring acts mainly as the anchor for ligation to PHYB, because the modification of the side chains at the C2 and C3 positions did not significantly influence the formation or difference spectra of adducts. In contrast, the side chains of the B- and C-rings are crucial to position the chromophore properly in the chromophore pocket of PHYB and for photoreversible spectral changes. The side-chain structure of the D-ring is required for the photoreversible spectral change of the adducts. When methyl and ethyl groups at the C17 and C18 positions are replaced with an n-propyl, n-pentyl, or n-octyl group, respectively, the photoreversible spectral change of the adducts depends on the length of the side chains. From these studies, we conclude that each pyrrole ring of the linear tetrapyrrole chromophore plays a different role in chromophore assembly and the photochromic properties of PhyB.
Light acts not only as an energy source for life but also as an environmental signal regulating plant development and animal behavior. Organisms can sense intensity, wavelength, direction, and timing of illumination by using diverse photoreceptors. Plants contain several kinds of photoreceptors, including phytochromes (1), cryptochromes (2), phototropin, and zeaxanthin (3) to detect light signals. Phytochrome, one of the best-characterized photoreceptors in plants, was discovered as a chromoprotein controlling red/far-red reversible developmental responses (4). Phytochrome shows photoreversible absorption changes between two spectrally distinct forms—a red-light-absorbing form (Pr; λmax, 660 nm) and a far-red-light-absorbing form (Pfr; λmax, 730 nm), on sequential absorption of light. This unique photochromic property of phytochrome results from the interaction of the chromophore with apoprotein of phytochrome. The two main approaches to elucidate apoprotein–chromophore interaction are site-directed mutagenesis (5) and chromophore modification by chemical synthesis (6, 7), because the crystal structure of holoprotein has not yet been determined.
The structure of the phytochrome chromophore was investigated initially by degradation approaches. In 1969, the bile pigment produced from the Pr chromophore by oxidative degradation was first analyzed, and components derived from the B- and C-rings were identified (8). The degradation product from the A-ring was released and identified in a two-step reaction (9) after the isolation of the product from the D-ring (10). Finally, the chromophore of phytochrome was determined as phytochromobilin (PΦB), which differs from phycocyanobilin (PCB) only by substitution of the D-ring ethyl group with a vinyl group (11). In addition, the ligation site of the chromophore was investigated by proton NMR spectroscopy of phytochromopeptides isolated from sequential pepsin-thermolysin digestion of oat phytochrome in the Pr form. PΦB was shown to ligate to the A-ring via a cysteine residue located in the N-terminal half of apoprotein of phytochrome A (PHYA; ref. 11). When the chromophore of phytochrome absorbs the appropriate wavelength of light, a conformational change is induced in both the chromophore and the phytochrome, following a Z to E isomerization at the C15–C16 double bond of the chromophore (12).
Little is known about the structural requirements of the chromophore for holophytochrome functions, because of the difficult nature of chemical synthesis of the linear tetrapyrrole (bilin) chromophore. PΦB, the common chromophore of phytochrome, is very similar in structure to PCB, which is a chromophore of the light-harvesting pigment phycocyanin, although the functions of these two photoreceptors are different (13). Early studies used PCB purified from algae, which was assembled in vitro with recombinant PHYA. Li and Lagarias (6) purified PΦB, PCB, phycoerythrobilin, biliverdin, and bilirubin from algae and assembled them with recombinant oat PHYA. Gossauer and Hinze (14) reported total synthesis of the dimethyl ester derivatives of PΦB and PCB, but they were unable to assemble these analogs with phytochrome apoprotein. After the efforts of a decade, we succeeded in synthesizing PCB, PΦB, and various PCB analogs in free-acid forms by developing efficient methods for the preparation of each pyrrole ring (15–20) and a method for coupling them (18, 21–24) and palladium-catalyzed deprotection of the allyl propanoate side chains of B- and C-rings under mild conditions (15).
The development of yeast and bacterial systems for the expression of recombinant phytochrome apoprotein has allowed investigation of the biochemical and spectroscopic properties of low-abundance phytochromes and comparisons among the different phytochrome family members, phytochrome A-E in Arabidopsis (25, 26). Holophytochromes assembled with PCB showed photoreversible spectral change, although its peaks were slightly blue shifted when compared with the native phytochrome (27–29). Other strategies, including systematic N- and C-terminal truncations and site-directed mutagenesis of the apoprotein, have been used to study the structural requirements of the chromophore–apoprotein interaction in terms of photochromism (30–32).
Phytochromes have long been believed to be the photoreceptors for red/far-red reversible reactions, but it has become evident that holophytochrome A and B (PhyA and PhyB) perceive light in essentially different manners (33). PhyB photoreversibly switches responses on or off on alternate irradiation with red/far-red light, whereas PhyA triggers very low-fluence or high-irradiance responses (34). The spectral properties of PhyA have been well characterized because of the relatively high abundance of this photoreceptor in dark-grown tissue. In contrast, PhyB has not been well characterized because of its low abundance in native tissue. Recently, several groups reported in vitro assembly of PhyB and showed that it has similar spectral properties to PhyA (35–37).
In this paper, we characterized PhyB reconstituted from apoprotein of phytochrome B (PHYB) with chemically synthesized bilin analogs. The present study probed the apoprotein pocket for the chromophore and revealed that each pyrrole ring of the bilin chromophore plays different roles for photochromism of the PhyB adducts.
Materials and Methods
Chemical Syntheses of Phycocyanobilin and Its Analogs.
PCB, PΦB, and analogs were synthesized chemically as reported previously (references are summarized in Table 1). The various D-ring structures in analogs 13–20 were prepared as follows. First, the regioisomers of tosylpyrroles were prepared by Barton's method (38), followed by rearrangement reaction of the tosyl groups of 2-tosylpyrroles (19). Next, conversion of 2-tosylpyrroles to 5-tosylpyrrolinones (corresponding to the structure of the D-ring in Fig. 1) was achieved by bromination and subsequent acidic hydrolysis (24). Finally, tetrapyrrole structures using these D-rings were constructed according to the described method (15). All bilins were stored as 2-mM stock solutions in dimethyl sulfoxide under nitrogen atmosphere at −80°C.
Table 1.
Side-chain structure of chemically synthesized phycocyanobilin and analogs
| Bilins | A-ring
|
B-ring
|
C-ring
|
D-ring
|
References | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| R1* | R2 | R3 | R4 | R5 | R6 | R7 | R8 | R9 | ||
| PCB | CH3 | H | CH3 | CH3 | (CH2)2CO2H | (CH2)2CO2H | CH3 | CH3 | CH2CH3 | 15, 16, 21, 23 |
| Analog 1† | H | 20 | ||||||||
| Analog 2 | CH2CH3 | 20 | ||||||||
| Analog 3 | CH3 | 20 | ||||||||
| Analog 4 | CH2CH3 | 20 | ||||||||
| Analog 5 | (CH2)2CO2CH3 | 16 | ||||||||
| Analog 6 | (CH2)2CO2CH3 | 16 | ||||||||
| Analog 7 | (CH2)2CO2H | CH3 | 16 | |||||||
| Analog 8 | CH3 | (CH2)2CO2H | 16 | |||||||
| Analog 9 | (CH2)2CO2H | CH3 | CH3 | (CH2)2CO2H | 16 | |||||
| Analog 10 | (CH2)3CO2H | 16 | ||||||||
| Analog 11 | (CH2)3CO2H | 16 | ||||||||
| Analog 12 | (CH2)3CO2H | (CH2)3CO2H | 16 | |||||||
| Analog 13 | CH2CH2CH3 | CH3 | ‡ | |||||||
| Analog 14 | CH2(CH2)3CH3 | CH3 | ‡ | |||||||
| Analog 15 | CH2(CH2)6CH3 | CH3 | ‡ | |||||||
| PΦB | CH⩵CH2 | 22 | ||||||||
| Analog 16 | CH2CH2CH3 | ‡ | ||||||||
| Analog 17 | CH2(CH2)3CH3 | ‡ | ||||||||
| Analog 18 | CH2(CH2)6CH3 | ‡ | ||||||||
| Analog 19 | CH2CH2OCOCH3 | ‡ | ||||||||
| Analog 20 | CH2CH2SC6H4CH3-p | ‡ | ||||||||
R1–R9, side chains of PCB (see Fig. 1).
Analog structures are identical to PCB except where indicated.
Present study.
Figure 1.
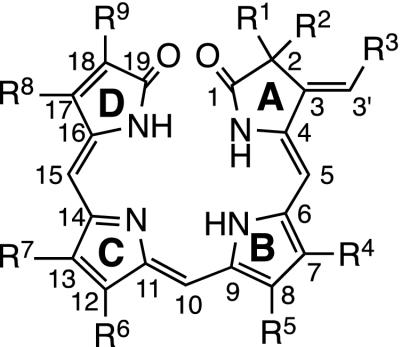
The generic chemical structure of bilin chromophores. The conventional numbering of carbon atoms on the linear tetrapyrrole backbone and lettering of the four pyrrole rings are shown on the structure. R1–R9 represent side chains of the synthesized bilins.
Plasmid Construction and Expression of PHYB in Saccharomyces cerevisiae.
For construction of the Arabidopsis PHYB expression vector, we performed PCR by using two oligonucleotide primers 5′ CGA TAT CAT GGT TTC CGG AGT CGG 3′ and 5′ ACC CGG GCT AAT ATG GCA TCA TCA 3′, and as a template, the plasmid including Arabidopsis full-length cDNA of PhyB, PHYB (generous gift from Joanne Chory). The BglII-SalI fragment of the yeast episomal plasmid pAA7 (39) was substituted by the EcoRV-SmaI fragment for the PCR product, downstream of the promoter of the GAL7 gene. Transformation and expression of PHYB was performed as reported (40).
In Vitro Assembly of PHYB with Bilins.
In vitro assembly was carried out as described (30). After disruption of the cells, the extract was clarified (12,000 × g for 30 min at 4°C). The supernatant fluid was used for reconstitution experiments. The total protein concentrations of extracts were measured by the Bio-Rad Protein Assay kit and adjusted to 6.5 mg/ml. All subsequent procedures were carried out under dim green safelight as described (41). Adduct formation was achieved by incubation of the protein extract with 5-μM bilins for 1 h on ice. Assembled fractions were used for zinc-blot analysis (Table 2) and determination of difference spectra (Figs. 2–4).
Table 2.
Chromophore-binding efficiency of adducts with synthetic bilins
| PCB Bilins | Chromophore-binding ratio* 1.00 |
|---|---|
| Analog 1 | 0.88 ± 0.08 |
| Analog 2 | 1.45 ± 0.17 |
| Analog 3 | 1.47 ± 0.14 |
| Analog 4 | 1.33 ± 0.13 |
| Analog 5 | 0.76 ± 0.11 |
| Analog 6 | 0.83 ± 0.08 |
| Analog 7 | 0.25 ± 0.08 |
| Analog 8 | 0.80 ± 0.17 |
| Analog 9 | 0.20 ± 0.09 |
| Analog 10 | 1.11 ± 0.20 |
| Analog 11 | 1.29 ± 0.26 |
| Analog 12 | 1.17 ± 0.39 |
| Analog 13 | 0.90 ± 0.16 |
| Analog 14 | 0.70 ± 0.21 |
| Analog 15 | 0.53 ± 0.15 |
| PΦB | 0.88 ± 0.11 |
| Analog 16 | 0.98 ± 0.24 |
| Analog 17 | 1.09 ± 0.37 |
| Analog 18 | 0.80 ± 0.30 |
| Analog 19 | 0.89 ± 0.25 |
| Analog 20 | 0.56 ± 0.29 |
To measure the extent of ligation efficiency to PHYB, the chromophore-binding ratio was calculated on the basis of fluorescence intensities of Zn2+ blots for adducts assembled with PCB analogs, relative to the value for PCB. Data, mean ± SE; n = 5.
Figure 2.
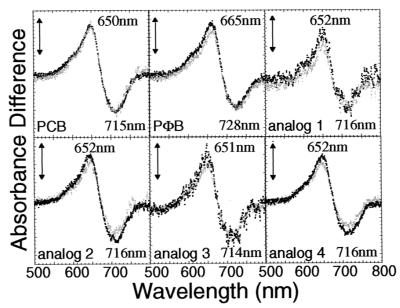
Difference absorption spectra of the PHYB adducts with PCB, PΦB, and analogs having side-chain substituents in the A-ring (analogs 1–4; see Table 1). The difference spectra were obtained by subtracting the absorption spectra determined after far-red light irradiation from that measured after red light irradiation. The sample volumes were 800 μl. Difference spectra were recorded after two consecutive red/far-red cycles. Black and gray dotted lines indicate the difference spectra obtained after the first and second cycles, respectively. The vertical bars represent 0.002 absorbance difference units.
Figure 4.
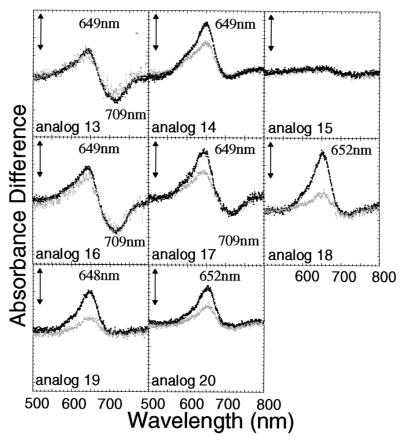
Difference absorption spectra of the PHYB adduct with PCB analogs having side chain substituents in D-ring (analogs 13–20; see Table 1). The difference spectra were obtained as described in the legend to Fig. 2. Black and gray dotted lines indicate the difference spectra obtained after the first and second cycles, respectively. The vertical bars represent 0.002 absorbance difference units.
Zinc-Blot Analysis of Adducts.
Reconstituted samples were separated electrophorically on 7.5% SDS/PAGE gels. Zn2+-induced luminescence of biliproteins was visualized (42) by using a bioimage analyzer (FMBIO-II; Hitachi Software Engineering, Kanagawa, Japan) after incubation of the gel in a buffer containing 20 mM zinc acetate and 150 mM Tris⋅HCl (pH 7.0). Ligation efficiency was determined by comparison of the band intensity to that resulting from assembly of an equivalent amount of PHYB assembled with PCB.
Measurement of Absorption and Difference Absorption Spectra of Adducts.
Absorption spectra were determined with a custom-made spectrophotometer (model Genespec V; Naka Instruments, Ibaragi, Japan) at 8°C by using quartz cuvettes with a 2-mm-wide slit and a 1-cm-long light path. The actinic beams for photoconversion were provided by light-emitting diodes (Q-beam 2200; Quantum Devices, Barneveld, WI) with peak wavelengths of 670 ± 23 nm and 742 ± 32 nm, respectively. Absorption spectra of PhyB-containing solutions were determined after actinic red light (8 mmol m−2) or far-red light (7 mmol m−2) for 10 seconds, both of which were calculated to saturate phytochrome photoconversion. Difference spectra of the PHYB adduct with analogs were obtained by subtracting the absorption spectrum recorded after red light from that recorded after far-red light.
Partial Purification of PHYB Adducts.
To measure the absorption spectra, we performed partial purification of PhyB adducts by using the IMPACT system kit (New England Biolabs). For construction of the expression vector, a PCR fragment of the full-length PHYB was generated with two primers 5′-ACA TAT GGT TTC CGG AGT CGG GGG TAG TG-3′ and 5′-AAA GCA CCC GGG CTC GAG ATA TGG CAT CAT CAG CAT- 3′, which introduced an NdeI site directly preceding the ATG codon and an SmaI site at the 3′ end. This fragment was ligated into NdeI-SmaI-digested pTYB2 (New England Biolabs). The resulting plasmid was transformed into Escherichia coli strain ER2566 (New England Biolabs), and PHYB was expressed according to the instructions of the manufacturer. The cells were broken by sonication, and then clarified cell extract was obtained by centrifugation at 12,000 × g for 30 min. For reconstitution, PCB and analogs 15 and 18 were added to extracts at a 5-μM final concentration and kept in darkness for 1 h at 4°C. The purification procedure was performed according to the manufacturer's instructions, with slight modifications. The extract was loaded onto the chitin column. Self-cleavage of the chitin-bound intein tag was achieved by keeping the tag for 16 h at 16°C with the cleavage buffer [20 mM Hepes/1 mM EDTA/50 mM DTT (pH 7.8)] after washing the column. The supernatant containing free PhyB was concentrated and applied to a DEAE-Sepharose Fast Flow (Amersham Pharmacia) column equilibrated with 50 mM Tris⋅HCl buffer (pH 7.8). The PhyB fraction was eluted by the linear gradient of NaCl (0–500 mM) and determined by zinc-blot analysis. The protein concentrations of the PhyB eluted fractions were measured by the Bio-Rad Protein Assay kit.
Results
Chemical Synthesis of Chromophore Analogs.
PCB, PΦB, and 20 analogs used in the present study are summarized in Table 1. PCB is the structural base of all analogs. Analogs 1 and 3 were designed to investigate the stereochemical requirement at the C2 position of the chromophore, because the C2 carbon is active optically in native phytochromobilin. Analogs 2 and 4 were designed to probe the spatial and chemical environment of the A-ring in the chromophore-binding pocket of PHYB. Analogs 5–12 were designed to clarify the positional and spatial requirements of the propanoic acid side chains in the B- and C-rings. Analogs 5 and 6 were regioselectively monoesterified derivatives. The positions of the methyl groups and propanoic acid side chains in the B- and C-rings were exchanged in analogs 7–9. The acid side chains were elongated further by one carbon in analogs 10–12. Modification of the D-ring is essential for analysis of photochromism, because the D-ring (position C15) is the site at which isomerization occurs during photoconversion of phytochrome. Substituents at the C17 and C18 positions of the D-ring were varied as vinyl (corresponds to PΦB), methyl, n-propyl, n-pentyl, or n-octyl groups (correspond to analogs 13–18) in a regioselective manner for structure/function analysis. In analogs 19 and 20, acetoxy and tolylthio groups were introduced at the β-position of the ethyl group at the C18 position. Chemically synthesized bilins were obtained as single [(3E), 4Z, 10Z, 15Z]-isomers with all-syn conformations confirmed by nuclear overhauser effect spectroscopy (Fig. 1; refs. 16, 23).
The difference absorption spectra of the in vitro-assembled adducts of bilins with PHYB were measured on actinic far-red and red-light irradiations. The adduct with PΦB showed a similar difference spectrum to that of the PCB adduct, except for the expected slight red shift of λΔmin and λΔmax (Fig. 2). The wavelengths of λΔmin and λΔmax were consistent with those reported (43), indicating that synthetic bilins were reconstituted with the apoprotein and were identical with the natural one extracted from algae.
Modification of Side Chains in the A-Ring.
Side chains R1, R2, and R3 of the A-ring (see Fig. 1) were substituted as shown in Table 1 (analogs 1–4). We first examined the binding efficiency (chromophore-binding ratio) of analogs 1–4 with PHYB by Zn2+ fluorescence-blot assay (Table 2). Dimethylation at C2 (analog 3) and one-carbon homologation at the C2 (analog 2) or C3 (analog 4) position of PCB increased the binding efficiency relative to PCB itself. However, demethylation at the C2 position (analog 1) of PCB did not affect the binding ratio.
The PHYB adducts assembled with analogs 1–4 showed similar difference absorption spectra and photoreversibility to those with PCB and PΦB. The results indicate that a hydrophobic environment and open space accommodate the A-ring of the tetrapyrrole at the binding site of PHYB, and that stereochemistry at the C2 position is not crucial for the ligation of the chromophore to PHYB. A slightly lower ΔΔmin/ΔΔmax ratio for the adduct containing 2-demethylated PCB (analog 1) suggests that a hydrophobic substituent such as methyl group at C2 position in PCB is responsible for stabilizing the Pfr form.
Modification of Side Chains in B- and C-Rings.
Side chains R4, R5, R6, and R7 (see Fig. 1) were varied, as shown in Table 1. Analogs 5–12 were assembled with PHYB, and the difference spectra of the resulting adducts were recorded (Fig. 3).
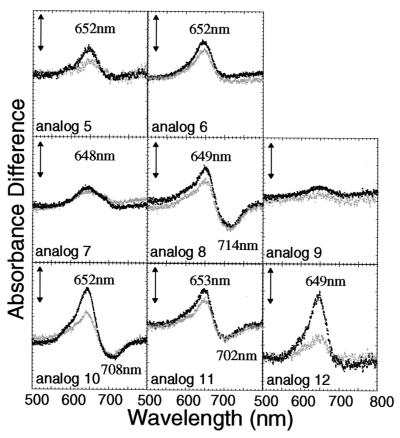
Figure 3.
Difference absorption spectra of the PHYB adducts with PCB analogs having side-chain substituents in the B- and C-rings (analogs 5–12; see Table 1). The difference spectra were obtained as described in the legend to Fig. 2. Black and gray dotted lines indicate the difference spectra obtained after the first and second cycles, respectively. The vertical bars represent 0.002 absorbance difference units.
The binding efficiency of chromophore to PHYB significantly decreased by regioselective monoesterification of the propanoic acid side chain at the C8 (analog 5) or C12 (analog 6) position of PCB (Table 2). Exchanging the methyl groups and the propanoic acid side chains of the B- and C-rings (analogs 7–9) also decreased the binding efficiency remarkably. In particular, the chromophore-binding ratios for analogs 7 and 9 were less than 30% of that of PCB. However, elongation of the acid side chains from propanoic to butanoic acid (analogs 10–12) did not affect binding efficiency (Table 2). The PHYB adducts assembled with analogs 5–9 showed relatively weak absorption bands around 650 nm corresponding to their Pr form, but virtually no absorption bands around 710 nm corresponding to the Pfr form, except for the analog 8 adduct (Fig. 3). Among analogs 10–12, PHYB adducts with analogs 10 and 11 had difference spectra somewhat similar to that of PCB, but the absorption band around 710 nm did not appear for analog 12. When PHYB adducts retained a propanoic acid as side chain at the C8 position of the B-ring, as in analogs 8 and 11, they displayed photochromism. In the case of the PHYB analog 6 adduct, reproducible photoreversibility was observed despite the disappearance of the absorption band at 710 nm.
Modification of Side Chains in the D-Ring.
Assuming that the photoreversibility of phytochrome results from the photoisomerization of the double bond at the C15 position of the chromophore, next we investigated steric requirements for the substituents R8 and R9 at the C17 and C18 positions of the D-ring. We synthesized several regioisomers (analogs 13–18) bearing longer alkyl chains at the C17 or C18 position (Table 1). The binding efficiency of analogs 13–15, substituted at the C17 position, decreased depending on the length of the alkyl chain (Table 2). On the other hand, isomers containing the corresponding substitutions at the C18 position (analogs 16–18) ligated to PHYB with an efficiency comparable to PCB and PΦB (Table 2). This result suggests that the environment of C18 within the chromophore-binding pocket of PHYB is hydrophobic and more flexible than that of C17.
The difference spectra of PHYB adducts with analogs 13–18 showed attenuated photoreversibility with increased length of the alkyl group at either the C17 or C18 position (Fig. 4). The PHYB adducts assembled with analogs 13 and 16 showed reproducible difference spectra similar to that of PCB. However, analogs 14 and 17 showed a reduction in the absorption peak at around 710 nm corresponding to the Pfr form. Adducts with analog 18, bearing the n-octyl group at C18, showed absorption peak in the red region with a maximum located around 650 nm. This peak was not observed in adducts with analog 15, which bears the same alkyl group at the C17 position. These results suggest that free space exists around C18 of the D-ring in the chromophore-binding pocket of PHYB. This suggestion was supported by the results from analogs 19 and 20, which respectively carry the 2-(acetoxy)ethyl or 2-(tolylthio)ethyl groups at the C18 position. These analogs also ligated effectively to PHYB, and the resulting adducts showed photoconversion in the red region corresponding to the Pr form (Fig. 4).
Absorption Spectra of Adducts with PCB and Analogs.
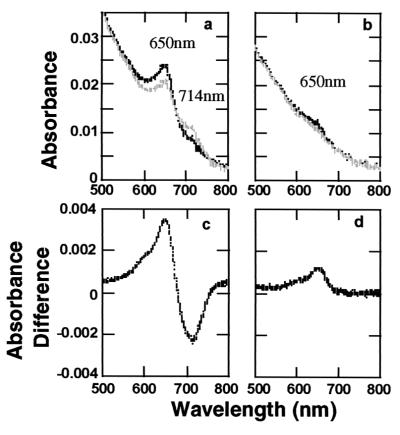
We measured the absorption spectra of adducts with PCB and analogs to elucidate the photochromic properties of the various PhyB adducts in more detail. Compared with the PCB adduct, adducts with analog 18 showed only a far-red-induced peak at around the 650 nm, and no significant red-light-induced peak was detected (Fig. 5). When the adduct with analog 18 was irradiated with sequential red and far-red light, the peak at 650 nm disappeared. Difference absorption spectra of the adduct with analog 15 showed no photoreversibility (Fig. 4), suggesting the possibility that the adducts might be locked in the Pr or Pfr form. However, no significant peak was detected in the absorption spectra or in difference absorption spectra of PhyB adduct with analog 15 (data not shown).
Figure 5.
Absorption and difference spectra of PhyB assembled with PCB and analog 18 when red and far-red light irradiated. The absorption spectra were determined after actinic red and far-red light irradiation. The difference spectra were obtained as described in the legend to Fig. 2. The sample volumes were 250 μl. Total protein concentrations were 0.82 mg/ml for the PCB adduct fraction and 0.37 mg/ml for the analog 18 adduct. Absorption spectra of adducts with PCB (a) and analog 18 (b) are shown (Upper). Black lines represent spectra measured after far-red light irradiation. Gray lines represent spectra after red light irradiation. Far-red/red difference spectra of adducts with PCB (c) and analog 18 (d) are shown (Lower).
Discussion
The capture of a photon of visible light by the phytochrome chromophore occurs within a femtosecond (44). The light-excited chromophore isomerizes to the Pfr form (45), eventually influencing the structure and surface properties of the phytochrome protein (46). Hence, characterization of the molecular interaction between the chromophore and the apoprotein is crucial for understanding the functional mechanism(s) of phytochromes. Several studies have addressed this interaction by using in vitro assembly of PHYA (5, 28, 29, 47), PHYB (35, 37, 48), PHYC and PHYE (43), or apoprotein mutants (30–32). However, in contrast to the vertebrate photoreceptor rhodopsin (49), in the phytochrome field, little has been done to examine the relationships among chromophore structure, its assembly to apoprotein, and photochromism of the holoprotein, because of the difficulty of synthesizing the chromophore and its structural analogs. This study has investigated the structural features of bilin binding to recombinant Arabidopsis PHYB and spectral properties of assembled PhyB by using systematically synthesized analogs of the natural phytochrome chromophore.
An important and as-yet-unresolved question concerns assembly of the bilin chromophore with apoprotein of phytochrome—how does the chromophore become oriented to allow ligation to the cysteine of the phytochrome apoprotein? The results of the present study give some insight into this question. First, modifications in A- and D-rings do not significantly affect chromophore binding to PHYB, suggesting that the chromophore pocket is larger and more flexible than thought previously. These results indicate that both the 3E and 3Z isomers of phytochromobilin most likely can bind to PHYB and also suggest that PHYB cannot distinguish synthetic bilins containing the 2R and 2S isomers. These hypotheses mean that it may be necessary to consider the heterogeneity in both natural and recombinant phytochrome samples when analyzing photochemical properties. Second, modification of the propanoic acid side chains of the C8 and C12 positions at the B- and C-rings has a substantial effect on chromophore binding, suggesting that free-acid elements are required for this event. A similar result has been reported already for pea PHYA (31), raising the possibility that chromophore binding is dictated by the electrostatic interaction between propanoic acid side chains and basic amino acid residues such as Arg or Lys of the phytochrome apoprotein (31), perhaps analogous to heme binding in cytochrome c (50). Our present data support this hypothesis. However, we cannot rule out a hydrogen-bonding as opposed to an ionic interaction. Experiments using analog-substituted propanoates into propanamides will make this hypothesis clear when chemical synthesis of the analogs is achieved. Third, the modification of the B- and C-ring side chains by one-carbon homologation of carboxyl groups did not affect binding efficiency suggesting that enough space may exist around the B-, C-, and D-rings to allow free movement of the chromophore in the chromophore-binding site of the apoprotein.
Two propanoate side chains not only serve as an electrostatic anchor for the chromophore in its pocket with PHYB, but also indirectly participate in photochromism. Because pea PhyA with either the R5 or the R6 methylester did not show any photoreversible spectral change (31), the chromophore–apoprotein interaction might be slightly different for PhyA and PhyB. Two propanoate side chains of all tetrapyrrole chromophores in allophycocyanin are accessible from the surface of the protein molecule (51). The two propanoic side chains of the heme in cytochromes (52), myoglobin (53), and hemoglobin (54) also are exposed from the heme pocket, but those of mitochondrial cytochrome c are buried in a crevice (50). In this regard, PhyB appears to resemble the mitochondrial cytochrome c. The phytochrome protein seems therefore to differ from other phycobiliproteins in providing a deeper, distinctly shaped chromophore pocket, which allows for photoreversibility.
The present work allows us to outline the order of events during in vitro assembly of PHYB with its chromophore. First, the ligation of chromophore to PHYB is initiated by an electrostatic interaction between the propanoic acid side chains of the B- and C-rings and a basic amino acid residue such as Arg or Lys. Second, the A-ring falls into the hydrophobic site close to the Cys of PHYB followed by nucleophilic attack of the thiol group to the exoethylene carbon at C2 to form a thioether bond. Finally, the D-ring also falls into a hydrophobic pocket where open space exists at least around the C18 position of the chromophore. Rotation around the C14/C15 single-bond axis probably occurs at this stage, forming the extended structure of the chromophore in PHYB. It is possible to replace a side chain at the C17 or C18 position of the D-ring with a bulky substituent without loss of photoconversion. These positions are suitable to be labeled with a photoreactive group for photoaffinity studies in the future.
The present collaborative study has opened an avenue of research toward understanding the nature of holophytochrome assembly and the chromophore pocket in vitro. It may be interesting to introduce the synthetic analogs described here into living plant cells to gain a better understanding of light sensing in a natural environment.
Acknowledgments
We thank Pill-Soon Song and Jim Weller for careful reading of the manuscript, Joanne Chory for providing the cDNA of Arabidopsis phytochrome B, Sadao Minagawa and Yoshio Sugiura (Naka Instruments, Ibaragi, Japan) for developing and providing a custom-built spectrophotometer, Tsuyoshi Sasaki for help with zinc-blot detection, and Chiemi Hoshina for technical assistance. This work was supported partly by a Hitachi Advanced Research Laboratory grant (B2023) and by a grant from the Program for Promotion of Basic Research Activities for Innovative Biosciences (to M.F.).
Abbreviations
- PhyA and PhyB
spectrally active holoprotein of phytochrome A and B
- PHYA and PHYB
apoprotein of phytochrome A and B
- PHYB
cDNA of phytochrome B
- Pr
phytochrome in the red light-absorbing form
- Pfr
phytochrome in the far-red light-absorbing form
- PCB
phycocyanobilin
- PΦB
phytochromobilin
References
- 1.Neff M M, Frankhauser C, Chory J. Genes Dev. 2000;14:257–271. [PubMed] [Google Scholar]
- 2.Ahmad M, Cashmore A R. Nature (London) 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- 3.Briggs R, Huala E. Annu Rev Cell Dev Biol. 1999;15:33–62. doi: 10.1146/annurev.cellbio.15.1.33. [DOI] [PubMed] [Google Scholar]
- 4.Butler W L, Norris K H, Siegelman H W, Hendricks S B. Proc Natl Acad Sci USA. 1959;45:1703–1708. doi: 10.1073/pnas.45.12.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song P S, Park M H, Furuya M. Plant Cell Environ. 1997;20:707–712. [Google Scholar]
- 6.Li L, Lagarias J C. J Biol Chem. 1992;267:19204–19210. [PubMed] [Google Scholar]
- 7.Lindner I, Knipp B, Braslavsky S, Gärtner W, Schaffner K. Angew Chem Int Ed Engl. 1998;37:1843–1846. [Google Scholar]
- 8.Rüdiger W, Correll D L. Liebigs Ann Chem. 1969;723:208–212. doi: 10.1002/jlac.19697230124. [DOI] [PubMed] [Google Scholar]
- 9.Klein G, Rüdiger W. Liebigs Ann. Chem. 1978. 267–279. [Google Scholar]
- 10.Rüdiger W, Brandlmeier T, Blos I, Gossauer A, Weller J P. Z Naturforsch C. 1980;35:763–769. [Google Scholar]
- 11.Lagarias J C, Rapoport H. J Am Chem Soc. 1980;102:4821–4828. [Google Scholar]
- 12.Rüdiger W, Thümmler F, Cmiel E, Schneider S. Proc Natl Acad Sci USA. 1983;80:6244–6248. doi: 10.1073/pnas.80.20.6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furuya M, Song P-S. In: Photomorphogenesis in Plants. Kendrick R E, Kronenberg G H M, editors. Dordrecht, The Netherlands: Kluwer; 1994. pp. 105–140. [Google Scholar]
- 14.Gossauer A, Hinze R-P. J Org Chem. 1978;43:283–285. [Google Scholar]
- 15.Kakiuchi T, Kato H, Jayasundera K P, Higashi T, Watabe K, Sawamoto D, Kinoshita H, Inomata K. Chem. Lett. 1998. , 1001–1002. [Google Scholar]
- 16.Ohta A, Sawamoto D, Jayasundera K P, Kinoshita H, Inomata K. Chem. Lett. 2000. , 492–493. [Google Scholar]
- 17.Kinoshita H, Ngwe H, Kohori K, Inomata K. Chem. Lett. 1993. , 1441–1442. [Google Scholar]
- 18.Kohori K, Hashimoto M, Kinoshita H, Inomata K. Bull Chem Soc Jpn. 1994;67:3088–3093. [Google Scholar]
- 19.Kohori K, Kinoshita H, Inomata K. Chem. Lett. 1995. , 799–800. [Google Scholar]
- 20.Sawamoto D, Nakamura H, Kinoshita H, Fujinami S, Inomata K. Chem. Lett. 2000. , 1398–1399. [Google Scholar]
- 21.Jayasundera K P, Kinoshita H, Inomata K. Chem. Lett. 1998. , 1227–1228. [Google Scholar]
- 22.Kakiuchi T, Kinoshita H, Inomata K. Synlett. 1999;S1:901–904. [Google Scholar]
- 23.Jayasundera K P, Kinoshita H, Inomata K. Bull Chem Soc Jpn. 2000;73:497–505. [Google Scholar]
- 24.Kinoshita H, Hayashi Y, Murata Y, Inomata K. Chem. Lett. 1993. , 1437–1440. [Google Scholar]
- 25.Sharrock R A, Quail P H. Genes Dev. 1989;3:1745–1757. doi: 10.1101/gad.3.11.1745. [DOI] [PubMed] [Google Scholar]
- 26.Clack T, Mathews S, Sharrock R A. Plant Mol Biol. 1994;25:413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- 27.Elich T D, Lagarias J C. J Biol Chem. 1989;264:12902–12908. [PubMed] [Google Scholar]
- 28.Wahleithner J A, Li L M, Lagarias J C. Proc Natl Acad Sci USA. 1991;88:10387–10391. doi: 10.1073/pnas.88.23.10387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deforce L, Tomizawa K, Ito N, Farrens D, Song P-S, Furuya M. Proc Natl Acad Sci USA. 1991;88:10392–10396. doi: 10.1073/pnas.88.23.10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deforce L, Furuya M, Song P-S. Biochemistry. 1993;32:14165–14172. doi: 10.1021/bi00214a014. [DOI] [PubMed] [Google Scholar]
- 31.Bhoo S H, Hirano T, Jeong H-Y, Lee J-G, Furuya M, Song P-S. J Am Chem Soc. 1997;119:11717–11718. [Google Scholar]
- 32.Remberg A, Schmidt P, Braslavsky S E, Gärtner W, Schaffner K. Eur J Biochem. 1999;266:201–208. doi: 10.1046/j.1432-1327.1999.00844.x. [DOI] [PubMed] [Google Scholar]
- 33.Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M. Proc Natl Acad Sci USA. 1996;93:8129–8133. doi: 10.1073/pnas.93.15.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shinomura T, Uchida K, Furuya M. Plant Physiol. 2000;122:147–156. doi: 10.1104/pp.122.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunkel T, Tomizawa K, Kern R, Furuya M, Chua N H, Schäfer E. Eur J Biochem. 1993;215:587–594. doi: 10.1111/j.1432-1033.1993.tb18069.x. [DOI] [PubMed] [Google Scholar]
- 36.Gärtner W, Hill C, Worm K, Braslavsky S E, Schaffner K. Eur J Biochem. 1996;236:978–983. doi: 10.1111/j.1432-1033.1996.00978.x. [DOI] [PubMed] [Google Scholar]
- 37.Ruddat A, Schmidt P, Gatz C, Braslavsky S E, Gärtner W, Schaffner K. Biochemistry. 1997;36:103–111. doi: 10.1021/bi962012w. [DOI] [PubMed] [Google Scholar]
- 38.Barton D H R, Kervagoret J, Zard S Z. Tetrahedron. 1990;21:7587–7597. [Google Scholar]
- 39.Abe A, Wada T, Handa H, Nogi Y, Fukasawa T. Agric Biol Chem. 1988;52:2035–2041. [Google Scholar]
- 40.Ito N, Tomizawa K-I, Furuya M. Plant Cell Physiol. 1991;32:981–985. [Google Scholar]
- 41.Tomizawa K-I, Sato N, Furuya M. Plant Mol Biol. 1989;12:295–299. doi: 10.1007/BF00043206. [DOI] [PubMed] [Google Scholar]
- 42.Berkelman T R, Lagarias J C. Anal Biochem. 1986;156:194–201. doi: 10.1016/0003-2697(86)90173-9. [DOI] [PubMed] [Google Scholar]
- 43.Eichenberg K, Baurle I, Paulo N, Sharrock R A, Rüdiger W, Schäfer E. FEBS Lett. 2000;470:107–112. doi: 10.1016/s0014-5793(00)01301-6. [DOI] [PubMed] [Google Scholar]
- 44.Braslavsky S E, Gärtner W, Shaffner K. Plant Cell Environ. 1997;20:700–706. [Google Scholar]
- 45.Rüdiger W, Scheer H. In: Photomorphogenesis. Shropshire W Jr, Mohr H, editors. Berlin: Springer; 1983. pp. 119–151. [Google Scholar]
- 46.Song P-S. J Biochem Mol Biol. 1999;32:215–225. [Google Scholar]
- 47.Murphy J T, Lagarias J C. Photochem Photobiol. 1997;65:750–758. doi: 10.1111/j.1751-1097.1997.tb01920.x. [DOI] [PubMed] [Google Scholar]
- 48.Elich T D, Chory J. Plant Cell. 1997;9:2271–2280. doi: 10.1105/tpc.9.12.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu S H R, Mirzadegan T. J Am Chem Soc. 1988;110:8617–8623. [Google Scholar]
- 50.Takano T, Dickerson R E. Proc Natl Acad Sci USA. 1980;77:6371–6375. doi: 10.1073/pnas.77.11.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brejc K, Ficner R, Huber R, Steinbacher S. J Mol Biol. 1995;249:424–440. doi: 10.1006/jmbi.1995.0307. [DOI] [PubMed] [Google Scholar]
- 52.Banci L, Bertini I, Rosato A. Eur J Biochem. 1997;249:270–279. doi: 10.1111/j.1432-1033.1997.t01-1-00270.x. [DOI] [PubMed] [Google Scholar]
- 53.Chu K, Vojtchovsky J, McMahon B H, Sweet R M, Berendzen J, Schlichting I. Nature (London) 2000;403:921–923. doi: 10.1038/35002641. [DOI] [PubMed] [Google Scholar]
- 54.Kolatkar P R, Ernst S R, Hackert M L, Hendrickson W A, Merritt E A, Phyzackerley R P. Acta Crystallogr B. 1992;48:191–199. doi: 10.1107/s0108768191012363. [DOI] [PubMed] [Google Scholar]