Abstract
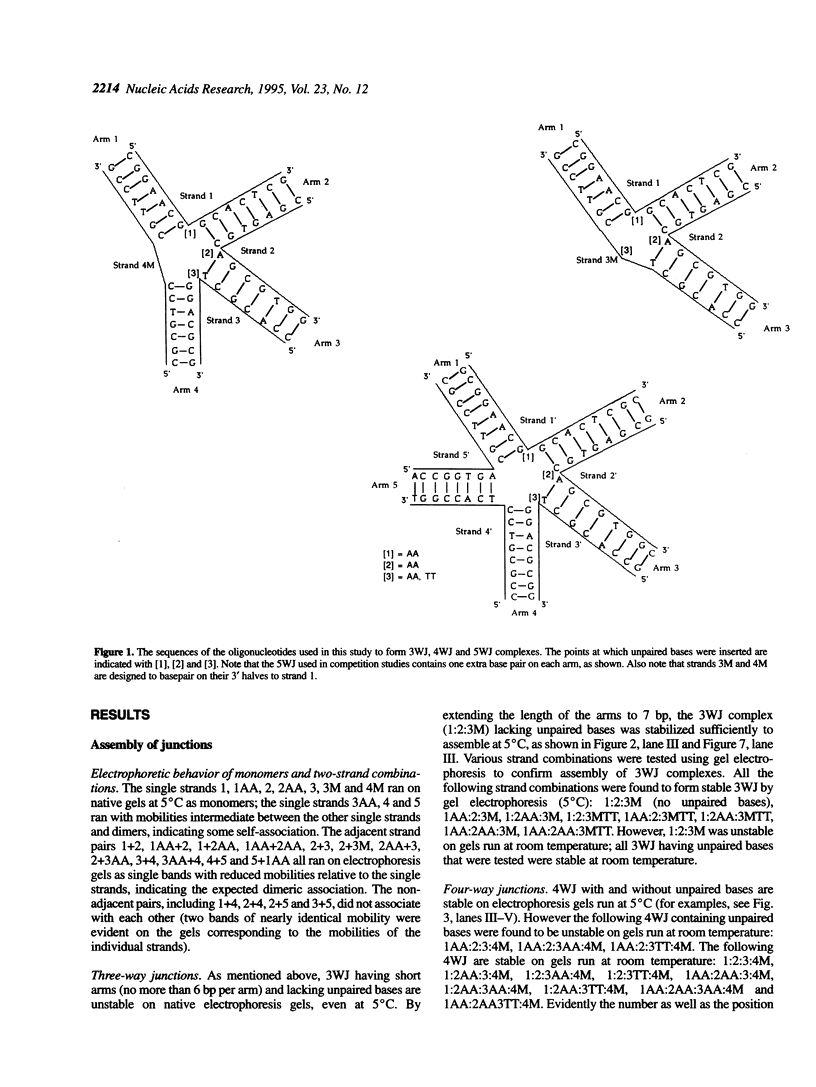
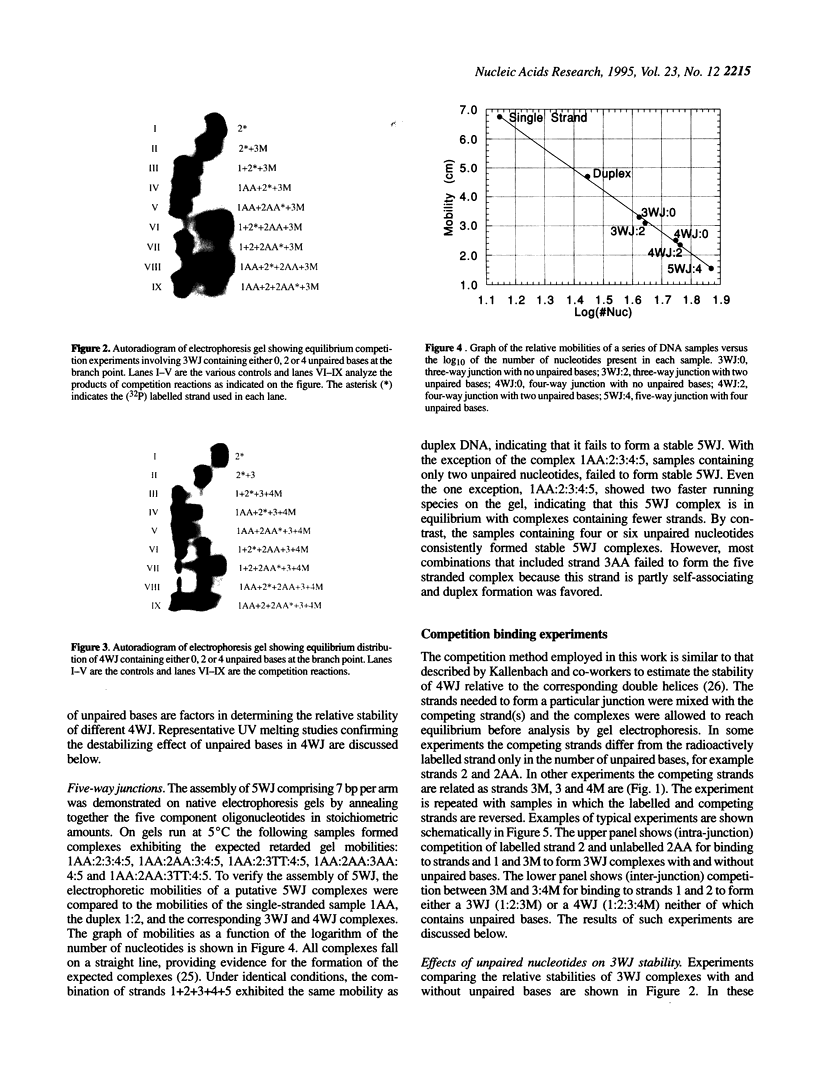
Competition binding and UV melting studies of a DNA model system consisting of three, four or five mutually complementary oligonucleotides demonstrate that unpaired bases at the branch point stabilize three- and five-way junction loops but destabilize four-way junctions. The inclusion of unpaired nucleotides permits the assembly of five-way DNA junction complexes (5WJ) having as few as seven basepairs per arm from five mutually complementary oligonucleotides. Previous work showed that 5WJ, having eight basepairs per arm but lacking unpaired bases, could not be assembled [Wang, Y.L., Mueller, J.E., Kemper, B. and Seeman, N.C. (1991) Biochemistry, 30, 5667-5674]. Competition binding experiments demonstrate that four-way junctions (4WJ) are more stable than three-way junctions (3WJ), when no unpaired bases are included at the branch point, but less stable when unpaired bases are present at the junction. 5WJ complexes are in all cases less stable than 4WJ or 3WJ complexes. UV melting curves confirm the relative stabilities of these junctions. These results provide qualitative guidelines for improving the way in which multi-helix junction loops are handled in secondary structure prediction programs, especially for single-stranded nucleic acids having primary sequences that can form alternative structures comprising different types of junctions.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chen J. H., Churchill M. E., Tullius T. D., Kallenbach N. R., Seeman N. C. Construction and analysis of monomobile DNA junctions. Biochemistry. 1988 Aug 9;27(16):6032–6038. doi: 10.1021/bi00416a031. [DOI] [PubMed] [Google Scholar]
- Chen S. M., Chazin W. J. Two-dimensional 1H NMR studies of immobile Holliday junctions: nonlabile proton assignments and identification of crossover isomers. Biochemistry. 1994 Sep 27;33(38):11453–11459. doi: 10.1021/bi00204a007. [DOI] [PubMed] [Google Scholar]
- Churchill M. E., Tullius T. D., Kallenbach N. R., Seeman N. C. A Holliday recombination intermediate is twofold symmetric. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4653–4656. doi: 10.1073/pnas.85.13.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. P., Hagerman P. J. Gel electrophoretic analysis of the geometry of a DNA four-way junction. J Mol Biol. 1987 Dec 20;198(4):711–719. doi: 10.1016/0022-2836(87)90212-9. [DOI] [PubMed] [Google Scholar]
- Cooper J. P., Hagerman P. J. Geometry of a branched DNA structure in solution. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7336–7340. doi: 10.1073/pnas.86.19.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett D. R., Murchie A. I., Diekmann S., von Kitzing E., Kemper B., Lilley D. M. The structure of the Holliday junction, and its resolution. Cell. 1988 Oct 7;55(1):79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987 Apr 24;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Helling R. B., Goodman H. M., Boyer H. W. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J Virol. 1974 Nov;14(5):1235–1244. doi: 10.1128/jvi.14.5.1235-1244.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Ladbury J. E., Sturtevant J. M., Leontis N. B. The thermodynamics of formation of a three-strand, DNA three-way junction complex. Biochemistry. 1994 Jun 7;33(22):6828–6833. doi: 10.1021/bi00188a011. [DOI] [PubMed] [Google Scholar]
- LeBlanc D. A., Morden K. M. Thermodynamic characterization of deoxyribooligonucleotide duplexes containing bulges. Biochemistry. 1991 Apr 23;30(16):4042–4047. doi: 10.1021/bi00230a031. [DOI] [PubMed] [Google Scholar]
- LeCuyer K. A., Crothers D. M. The Leptomonas collosoma spliced leader RNA can switch between two alternate structural forms. Biochemistry. 1993 May 25;32(20):5301–5311. doi: 10.1021/bi00071a004. [DOI] [PubMed] [Google Scholar]
- Leonard C. J., Berns K. I. Adeno-associated virus type 2: a latent life cycle. Prog Nucleic Acid Res Mol Biol. 1994;48:29–52. doi: 10.1016/s0079-6603(08)60852-1. [DOI] [PubMed] [Google Scholar]
- Leontis N. B., Hills M. T., Piotto M., Malhotra A., Nussbaum J., Gorenstein D. G. A model for the solution structure of a branched, three-strand DNA complex. J Biomol Struct Dyn. 1993 Oct;11(2):215–223. doi: 10.1080/07391102.1993.10508722. [DOI] [PubMed] [Google Scholar]
- Leontis N. B., Hills M. T., Piotto M., Ouporov I. V., Malhotra A., Gorenstein D. G. Helical stacking in DNA three-way junctions containing two unpaired pyrimidines: proton NMR studies. Biophys J. 1995 Jan;68(1):251–265. doi: 10.1016/S0006-3495(95)80182-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis N. B., Kwok W., Newman J. S. Stability and structure of three-way DNA junctions containing unpaired nucleotides. Nucleic Acids Res. 1991 Feb 25;19(4):759–766. doi: 10.1093/nar/19.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longfellow C. E., Kierzek R., Turner D. H. Thermodynamic and spectroscopic study of bulge loops in oligoribonucleotides. Biochemistry. 1990 Jan 9;29(1):278–285. doi: 10.1021/bi00453a038. [DOI] [PubMed] [Google Scholar]
- Lu M., Guo Q., Marky L. A., Seeman N. C., Kallenbach N. R. Thermodynamics of DNA branching. J Mol Biol. 1992 Feb 5;223(3):781–789. doi: 10.1016/0022-2836(92)90989-w. [DOI] [PubMed] [Google Scholar]
- Lu M., Guo Q., Seeman N. C., Kallenbach N. R. Parallel and antiparallel Holliday junctions differ in structure and stability. J Mol Biol. 1991 Oct 20;221(4):1419–1432. doi: 10.1016/0022-2836(91)90942-y. [DOI] [PubMed] [Google Scholar]
- Murchie A. I., Carter W. A., Portugal J., Lilley D. M. The tertiary structure of the four-way DNA junction affords protection against DNase I cleavage. Nucleic Acids Res. 1990 May 11;18(9):2599–2606. doi: 10.1093/nar/18.9.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Ouporov I. V., Leontis N. B. Refinement of the solution structure of a branched DNA three-way junction. Biophys J. 1995 Jan;68(1):266–274. doi: 10.1016/S0006-3495(95)80183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi J. D., Tinoco I., Jr Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- Rosen M. A., Patel D. J. Conformational differences between bulged pyrimidines (C-C) and purines (A-A, I-I) at the branch point of three-stranded DNA junctions. Biochemistry. 1993 Jul 6;32(26):6563–6575. doi: 10.1021/bi00077a010. [DOI] [PubMed] [Google Scholar]
- Rosen M. A., Patel D. J. Structural features of a three-stranded DNA junction containing a C-C junctional bulge. Biochemistry. 1993 Jul 6;32(26):6576–6587. doi: 10.1021/bi00077a011. [DOI] [PubMed] [Google Scholar]
- Seeman N. C., Kallenbach N. R. Design of immobile nucleic acid junctions. Biophys J. 1983 Nov;44(2):201–209. doi: 10.1016/S0006-3495(83)84292-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Freier S. M. RNA structure prediction. Annu Rev Biophys Biophys Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- Walter A. E., Turner D. H., Kim J., Lyttle M. H., Müller P., Mathews D. H., Zuker M. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc Natl Acad Sci U S A. 1994 Sep 27;91(20):9218–9222. doi: 10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. L., Mueller J. E., Kemper B., Seeman N. C. Assembly and characterization of five-arm and six-arm DNA branched junctions. Biochemistry. 1991 Jun 11;30(23):5667–5674. doi: 10.1021/bi00237a005. [DOI] [PubMed] [Google Scholar]
- Zhang S., Seeman N. C. Symmetric Holliday junction crossover isomers. J Mol Biol. 1994 May 20;238(5):658–668. doi: 10.1006/jmbi.1994.1327. [DOI] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- von Kitzing E., Lilley D. M., Diekmann S. The stereochemistry of a four-way DNA junction: a theoretical study. Nucleic Acids Res. 1990 May 11;18(9):2671–2683. doi: 10.1093/nar/18.9.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]








