Abstract
Induced pluripotent stem cells (iPSCs) can be derived from somatic cells by gene transfer of reprogramming transcription factors. Expression levels of these factors strongly influence the overall efficacy to form iPSC colonies, but additional contribution of stochastic cell-intrinsic factors has been proposed. Here, we present engineered color-coded lentiviral vectors in which codon-optimized reprogramming factors are co-expressed by a strong retroviral promoter that is rapidly silenced in iPSC, and imaged the conversion of fibroblasts to iPSC. We combined fluorescence microscopy with long-term single cell tracking, and used live-cell imaging to analyze the emergence and composition of early iPSC clusters. Applying our engineered lentiviral vectors, we demonstrate that vector silencing typically occurs prior to or simultaneously with the induction of an Oct4-EGFP pluripotency marker. Around 7 days post-transduction (pt), a subfraction of cells in clonal colonies expressed Oct4-EGFP and rapidly expanded. Cell tracking of single cell–derived iPSC colonies supported the concept that stochastic epigenetic changes are necessary for reprogramming. We also found that iPSC colonies may emerge as a genetic mosaic originating from different clusters. Improved vector design with continuous cell tracking thus creates a powerful system to explore the subtle dynamics of biological processes such as early reprogramming events.
Introduction
Differentiated somatic cells can be converted into induced pluripotent stem cells (iPSC) with properties similar to embryonic stem (ES) cells by expressing a defined set of reprogramming factors (RFs). After the first proof-of-principle was obtained and a set of four transcription factors (Oct4, Klf4, Sox2, c-Myc) was identified in the pioneering study, several reports have reproduced these findings with murine and human cells and started to investigate the underlying mechanisms.1,2,3,4,5,6,7 While the initial studies used γ-retroviral vectors for delivery of the RF, promising delivery technologies such as nonviral piggyBac transposon-based vectors, episomal vectors, tetracycline-regulated systems, stabilized mRNAs,8 and protein delivery via protein transduction domains have been developed.9,10,11,12,13 Aiming for highly efficient generation of iPSCs, polycistronic lentiviral vectors were used that co-express all RF from one construct to ensure that all RF are available in each transduced cell.14,15
Important variables guiding reprogramming factor expression in a lentiviral context are located on the transcriptional (i.e., promoter choice) and post-transcriptional level. Addressing the latter, codon-optimization of the RF (to more favored human tRNA usage) might increase their basal expression levels16,17 due to improved mRNA stability and translation. Likewise, the insertion of a woodchuck hepatitis virus post-transcriptional regulatory element (wPRE) also enhances post-transcriptional transgene expression (mRNA stability, export, and translation).18,19 However, both potentially beneficial avenues have not been systematically addressed so far.
Important components of the signalling pathways involved in reprogramming have been elucidated6,20 and small molecules21,22 or alternative cell sources23 were introduced to enhance this process or circumvent the need for some of the RF.6,20,21,24 Still, the mechanism underlying reprogramming is poorly understood and it remains enigmatic why only a minor fraction of cells expressing the RF is capable to fully convert into iPSC and where this fraction derives from. The “stochastic” model,25 in which most of the differentiated cells have the potential to become iPSC presupposing epigenetic chromatin remodelling, has only recently been substantiated by Hanna et al. using a conditional transgenic model.26 This and other groups recently investigated morphological and molecular changes associated with reprogramming,27,28 and monitored these processes every few hours or days albeit without directly marking RF expression.
In the present study, we developed fluorescence-coded lentiviral vectors that initially trigger high-level expression of the RF and are rapidly silenced in reprogramming cells. Transducing murine embryonic fibroblasts (MEFs) of an established and well-characterized Oct4-EGFP reporter mouse strain (OG2),29,30,31,32 we used continuous single-cell tracking33 with short intervals (minutes)34,35 to film the “birth” of pluripotent cells in cell clusters expressing and silencing the RF. Kinetic analyses and cell tracking provided supporting evidence for the stochastic emergence of reprogrammed cells. Our data also showed that early clonal colonies containing reprogrammed cells are frequently contaminated with cells that fail to undergo full reprogramming, and that iPSC colonies are often invaded by cells derived from surrounding clusters.
Results
Design of lentiviral vectors promoting efficient onset of reprogramming gene expression and fast epigenetic silencing in pluripotent cells
Our developed modular lentiviral vector system encodes murine or human versions of the canonical RF (Oct4, Klf4, Sox2, c-Myc). We aimed for a functional, easily interchangeable design allowing efficient coexpression of RF and a fluorescent marker (preferably dTomato or a nuclear-localized derivative for cell tracking in condensed cell clusters) on the same mRNA to monitor RF expression. We constructed either 1-factor vectors (expressing just one RF) or combinatorial (3-in-1 or 4-in-1) constructs coexpressing Oct4, Klf4, Sox2, and optionally c-Myc via different 2A-proteinase sites36 (Figure 1), that have previously been shown to mediate almost complete separation of recombinant proteins. In addition, we introduced a number of modifications to the expression cassette to improve RF and fluorescent marker expression, as conversion to pluripotency requires robust RF expression and a clear fluorescence signal is a prerequisite for imaging/cell tracking studies. Therefore, we modified mRNA processing by equipping the fully sequenced complementary DNAs (cDNAs) with a Kozak consensus element for efficient initiation of translation and added a well-characterized post-transcriptional regulatory element derived from the woodchuck hepatitis virus (wPRE).18,19 Furthermore, we tested different promoters (PGK: phosphoglycerokinase; UCOE: ubiquitous chromatin opening element; SFFV: spleen focus-forming virus U3 promoter) and introduced synthetic cDNAs for the RF in which codon usage was optimized for expression in human and murine cells.16,17 In addition, by this approach eight putative splice donor sequences were removed from the 4-in-1 vector. Codon-optimization generally resulted in enhanced expression levels of the individual murine and human RF and also increased the expression from the multicistronic 3-in-1 and 4-in-1 vectors (Figure 2a–d and Supplementary Figure S1). Most of our expression comparisons were based on dTomato fluorescence intensity measured by flow cytometry. This primarily reflects general increases in mRNA stability by codon-optimization of the RF. Immunoblot analysis of cells transduced with 1-factor and multicistronic (3-in-1 and 4-in-1) vectors showed clearly improved RF expression (Figure 2e).
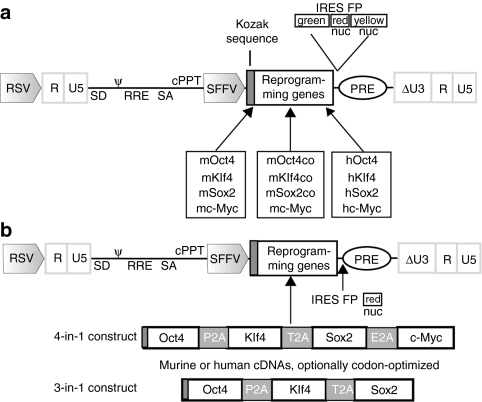
Figure 1.
Design of 1-factor and combinatorial multicistronic reprogramming vectors. (a) Modular configuration of the self-inactivating (SIN) vector backbones for expression of the murine (m) or human (h) reprogramming factor (RF) Oct4, Klf4, Sox2, and c-Myc, optionally codon-optimized (co). δ marks the SIN configuration with partially deleted U3 of the 3′ long terminal repeat. cPPT, central polypurine tract; FP, fluorescent protein (green: GFP; red: dTomato; yellow: Venus); IRES, internal ribosomal entry site; nuc, nuclear membrane-localized derivative; PRE, post-transcriptional regulatory element; RRE, rev-responsive element; RSV, Rous sarcoma virus U3 promoter; SA, splice acceptor; SD, splice donor; SFFV, spleen focus-forming virus promoter; ψ, packaging signal. (b) Multicistronic all-in-one SIN vectors expressing either 4 or 3 RF via 2A self-cleavage sites. cDNA, complementary DNA; E2A, equine rhinitis A virus 2A; P2A, porcine teschovirus 2A; T2A, thosea asigna virus 2A.
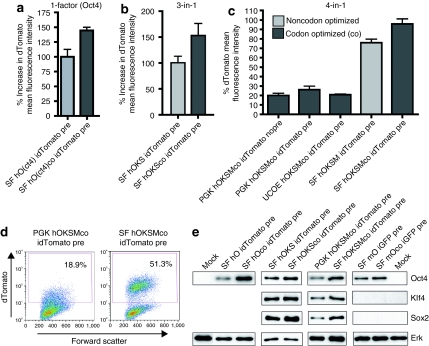
Figure 2.
Increased reprogramming factor (RF) expression by vector modifications. Effects of codon-optimization of RF on dTomato (located on the same mRNA) expression demonstrated by changes in fluorescence intensity. Changes are illustrated regarding the human (a) Oct4 1-factor vector, (b) combinatorial 3(factors)-in-1, and (c) 4(factors)-in-1 vectors. For the latter 4-in-1 reprogramming cassette also influences of different promoters (PGK, phosphoglycerokinase; UCOE, ubiquitous chromatin opening element; SF, spleen focus) and the woodchuck hepatitis virus post-transcriptional regulatory element (wPRE) are shown (PRE/wPRE). (d) Flow cytometry analysis comparing dTomato expression from codon-optimized 4-in-1 reprogramming vectors mediated by the PGK and SFFV promoter. (e) Influences of codon-optimization and promoter choice on protein levels of the RF.
We and others had previously demonstrated that addition of the wPRE element clearly increased the expression of transgenic EGFP (up to eightfold) and virus titer.19,37 As effects of the wPRE are often context dependent, we evaluated the inclusion of wPRE in the multicistronic PGK driven 4-in-1 vector. Here, increased dTomato expression (located on the same mRNA as the RFs) was visible (Figure 2c), and the titer improved more than fivefold when the wPRE was present (data not shown).
Of special importance for the present study is our choice of a retroviral promoter (SFFV), which mediates efficient expression in fibroblasts and other somatic cell types38 but is rapidly silenced in cells undergoing epigenetic remodelling,39,40,41 as indicated in differentiating ES cells (Supplementary Figure S2) as well as reprogramming cells (see below). Furthermore, SFFV mediated an at least fourfold higher transgene expression as compared to the PGK and UCOE promoter elements (see below; Figure 2c–e and Supplementary Figure S1). The SFFV promoter thus leads to an easily separable population of RF expressing cells, as required for imaging and cell tracking (Figure 2d and Supplementary Figure S1).
Taken together, the combination of transcriptional (promoter choice) and post-transcriptional modifications generated an improved vector backbone with relatively high RF expression and clearly visible fluorescent marker expression.
Evidence for efficient reprogramming by the described vector system
Using these modified vectors, we first validated our approach for iPSC generation. Supplementary Figure S3 shows the correct processing of reprogramming factor mRNAs and proteins. When transducing OG2 MEFs, we relied on EGFP expression as a sensitive indicator for the activation of a pluripotency-associated transcriptional network (see Figure 3a for overview of experimental system), as validated earlier.29,30,31,32 This reporter was also shown to correlate with SSEA-1 and Oct4 expression in iPS lines (Supplementary Figure S4a,b). Based on the detection of EGFP+ cells, reprogramming in presence of valproic acid was triggered with high efficiency (>10% Oct4-EGFP+ cells at day 11 post-transduction (pt); Figure 3b). We picked single colonies and demonstrated ES cell-like morphology and marker expression in representative iPSC colonies (Figure 3c and Supplementary Figure S4) obtained with a cocktail of RF or alternatively a “4-in-1” construct. Nanog and Oct4 promoters were widely unmethylated, as typically observed in ES cells (Supplementary Figure S5). iPSC lines were competent to form teratoma with differentiation to the three germ layers after injection into flanks of immunodeficient mice (Supplementary Figure S6). Furthermore, the developed vectors efficiently converted other cell types such as adult fibroblasts, bone-marrow–derived mesenchymal cells or blood cells into permanent iPSC lines with ES-like morphology and pluripotency marker expression (T. Cantz, E. Warlich, J. Kuehle, and A. Schambach, unpublished results).
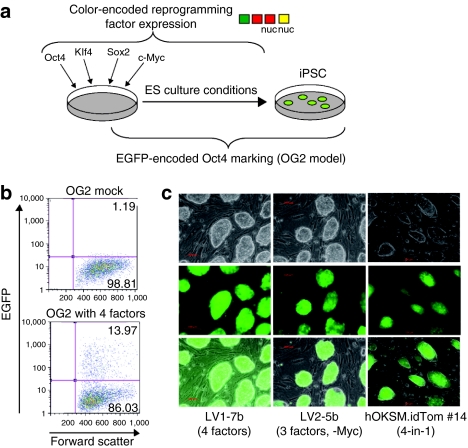
Figure 3.
Efficient reprogramming by the novel vector system. (a) Schematic illustration of reprogramming by virus-encoded transduction of Oct4-EGFP (OG2) transgenic murine embryonic fibroblasts (MEFs). (b) Representative flow cytometry analysis of Oct4-EGFP expression in OG2 MEFs 11 days post-transduction with all 4 factors (lower) and their untransduced counterparts (upper). (c) Fluorescence microscopy of selected OG2-derived induced pluripotent stem cell (iPSC) clones demonstrating embryonic stem cell morphology and Oct4-EGFP activation. Upper brightfield, middle EGFP fluorescence, lower overlay.
Kinetic analysis of early reprogramming
Next, we applied our lentiviral reprogramming vectors to monitor the kinetics of successful reprogramming. After transduction of OG2 MEFs red fluorescence indicated RF expression, while green fluorescence in combination with careful morphological analyses indicated the emergence of iPSC. EGFP half-life time had previously been determined to be in the range of 20 hours with maturation occurring within 30 minutes and our data indicate that the half-life of dTomato is likely to be in the range of hours. We co-transduced OG2 MEFs with all four single-factor reprogramming vectors (the Sox2 transgene linked to an IRES-dTomato cassette), monitoring the time window of day 4.5 to day 11.5 pt. While red fluorescent transduced cells displayed significant variance in the formation of cell clusters with a converted morphology, we identified—with some variance—the emergence of colonies containing cells with a characteristic ES-like phenotype around day 7 pt. The onset of EGFP expression occurred as little as 3 hours until >60 hours after the emergence of these colonies (Figure 4a). These putative pluripotent EGFP+ cells appeared within colonies of transduced dTomato+ cells and rapidly expanded. While tracking of individual cells expressing EGFP is difficult in the condensed structure of late colonies, the investigation of doubling times in the limited number of “earlyonset” EGFP+ cells suggested proliferation rates around 10 hours (Figure 4b) similar to the previously determined proliferation rates of ES cells.42,43 In line with this, time-lapse observation suggested the absence of frequent apoptotic events in RF transduced cells. However, an additional conversion of further cells within the colony from dTomato to EGFP expression might also contribute to the rapid expansion of EGFP+ cells. Despite this expansion the majority of iPSC-bearing colonies also contained cells that failed to be reprogrammed within the observation period and often continued to express the exogenous RF (Supplementary Videos S1a,b and S2).
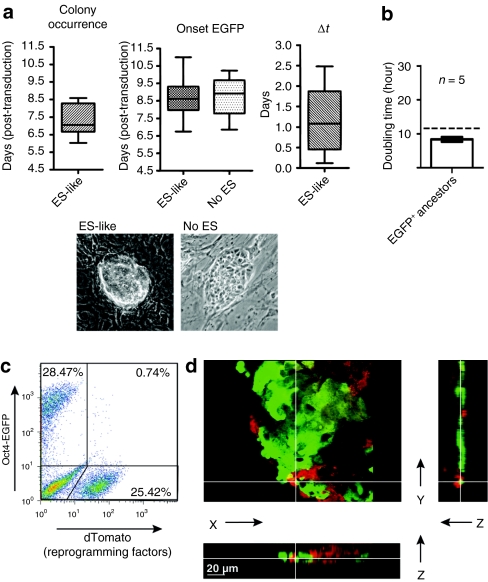
Figure 4.
Typical events in the early stages of reprogramming. (a) Colony-wise analysis of time-lapse imaging recorded over a period of 7 days (days 4.5–11.5) following co-transduction of OG2 murine embryonic fibroblasts with all four reprogramming factors. Analyzed are the emergence of embryonic stem (ES) cell-like morphology (ES-like, n = 21, left plot) and onset of Oct4-EGFP in these colonies (middle and right plot, δt = t(onset EGFP)−t(colony occurrence)). EGFP also appeared in many proliferative clusters that did not demonstrate all criteria for ES cell-like morphology (no ES, n = 33, central plot). Boxes extend from 25th to 75th percentile with line at median and whiskers spanning minimum to maximum. (b) Analysis of doubling times in early EGFP+ ancestors of reprogramming factor transduced cells by time-lapse imaging. Hatched line indicates doubling times of ES cells as previously described.42 (c) Expression levels of fluorescent markers in induced pluripotent stem cell (iPSC)-like cells as analyzed by flow cytometry 15 days post-transduction (pt) using a 4-in-1 vector linked to internal ribosomal entry site (IRES)-dTomato. A homogeneous cell population resembling the size of iPSC was gated. (d) 3D-microscopy of an iPSC-like colony (induced using a 4-in-1 vector linked to IRES-dTomato) on day 13 pt. The view from above is shown with z sections in the x and y planes (depicted on the right and below). White lines indicate where the sections have been made.
The results obtained from live-cell imaging were confirmed using a “4-in-1” vector in which we linked codon-optimized cDNAs of Oct4, Klf4, Sox2, and wild-type c-Myc, devoid of untranslated sequences, by different 2A self-cleavage sites coupled with the dTomato transgene (via an IRES site) (Figure 1b). With this vector, analysis of iPSC containing colonies by flow cytometry (Figure 4c and Supplementary Figure S7a) revealed a downregulation of SFFV-driven dTomato expression in emerging EGFP+ cells. Individual cells coexpressing EGFP and dTomato were hardly observed (<2% double positive cells) at early time points and were almost completely absent 11 days pt. Considering the necessity for dTomato degradation or dilution in proliferating cells and the requirement for EGFP accumulation, these data indicate that vector silencing typically occurred even before or at least simultaneously with the induction of the Oct4-EGFP allele.
Apoptome 3D-microscopy –based reconstruction showed that colonies were composed of both, EGFP+ potential iPSC and dTomato+ cells still expressing the RF, whereas cells coexpressing both markers were rare (Figure 4d and Supplementary Video S3). In summary, these studies underlined that we developed a system in which the onset and silencing of reprogramming factor expression can be monitored with high sensitivity, and which shows a remarkable frequency of reprogramming cassette silencing in reprogramming cells especially when using a cassette for combinatorial expression of RF.
Heterogeneity of iPSC colony composition and of induction of reprogramming
When undertaking a careful analysis of reprogramming colony composition in the first days after transduction with RF, we observed frequent heterogeneity of Oct4-EGFP expression in iPSC colonies. EGFP not only occurred with some variance after emergence of ES-like colonies, but also rather appeared at one or more distinct spots within a colony than being homogenously distributed (Supplementary Videos S1a,b, S2, S5b, and S6). Interestingly, EGFP also occurred in a number of colonies that displayed a transformed phenotype rather than matching all morphological criteria of ES cells (round, sharp border, shiny, compact structure) (Figure 4a). While early after colony formation, cells expressed at least one of the fluorescence markers, occasionally some cells within a colony neither expressed the RF nor the Oct4-EGFP reporter at later time points (Supplementary Figure S7b).
To further ease the traceability of individual cells within compacted iPS colonies, we employed nuclear Venus or dTomato (Supplementary Video S4, also see text below). Due to the high resolution of the time-lapse imaging we were able to document that many colonies did not arise from single-transduced cells, but rather incorporated neighboring transduced cells while proliferating. Also, confluence of neighboring colonies was not only often observed (Supplementary Video S1a) and most pronounced at high reprogramming efficiencies but also with remarkable frequency at relatively low plating densities. As a consequence, “colonies” were in part nonclonal, likely containing cells with different vector integration sites, and at different reprogramming levels. Both findings, colony aggregation and EGFP pattern, reflect the potentially genetic and epigenetic heterogeneity of iPSC colonies and underline the power of high-frequency imaging even for compacted and fast-dividing cells such as iPSC.
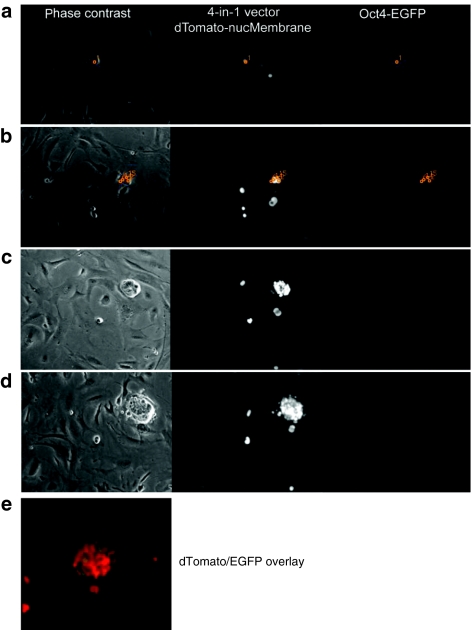
Time-lapse imaging allowed tracking of iPSC colony generation from a single cell (Figure 5, Supplementary Figure S8 and Videos S5a,b and S6) based on the software tool (TTT). Previous studies described a discrepancy between the number of genetically modified cells and successfully reprogrammed cells, suggestive of stochastic gate-keeping events. To investigate the clonal derivation of the subset of successfully reprogrammed cells in detail, we initiated continuous cell tracking reflecting RF and pluripotency marker expression under real-time conditions from very early time points (from day 4.5 until day 11.5 pt). Since a single-transduced cell gives rise to genetically identical progeny, the influence of secondary events can be investigated. In Figure 5 (sequential picture panel) and Supplementary Video S5a,b we documented such a case, in which only a fraction of the genetically identical progeny from a single-transduced (red) cell switched on Oct4-EGFP. To illustrate the clonality during iPS emergence, single colony forming cells and their progeny were tracked with TTT (Supplementary Video S5a and Figure S8). We confirmed these data in a second setting, in which we used a 4-in-1 cassette linked to the newly developed nuclear-localized dTomato to ease single cell tracking and monitored a wider time window (days 1–13 pt) (Supplementary Video S6).
Figure 5.
Exemplary sequence of induced pluripotent stem cell (iPSC)-like colony development and associated marker expression. A single fibroblast (1) (a) transduced with a 4-in-1 reprogramming vector (linked to nuclear-localized dTomato), (b) gives rise to progeny (serially numbered and tracked in orange, also see tracking tree in Supplementary Figure S8), (c) that adopts an iPSC-like morphology. (d) Consecutively, an Oct4-EGFP reporter switches on in a subset of cells. Left phase contrast; middle nuclear-localized dTomato; right Oct4-EGFP signal. (e) An overlay is shown of the dTomato-nucMembrane signal and the Oct4-EGFP of the same colony as in d.
Taken together, the combination of our vector system with high-frequency live-cell imaging allowed us to capture the conversion from RF expression to the induction of pluripotency markers, and to document the substantial heterogeneity within a clonal colony of genetically identical cells undergoing reprogramming.
Discussion
We developed and validated lentiviral vectors which express the RF from codon-optimized coexpression cassettes under control of a retroviral promoter with high activity in somatic cells but rapid silencing in pluripotent cells. Of note, we used a vector backbone with a primer binding site not sensitive to TRIM28-mediated silencing,44 because this mechanism is also potentially active in some somatic cells38 which would hamper reprogramming. Employing codon-optimized transgene sequences not only increases vector expression16,17 but also facilitates the discrimination between endogenous and exogenous RF (with identical protein sequence) based on PCR. Our system could thus, e.g., be used to determine copy numbers of integrates or to investigate the expression, silencing and reactivation of the factors even in absence of fluorescent markers.
As functional validation, we used the described vector system in the current study to visualize the early stages of reprogramming by monitoring the emergence of iPSC within cell colonies expressing and silencing the canonical RF. In MEFs with a transgenic Oct4 indicator system, we analyzed the subsequent dynamic changes by serial microscopy, including Apotome 3D reconstruction and time-lapse fluorescence microscopy, the latter enabled by an advanced live-cell imaging tool called TTT. This program has previously been validated to provide reliable kinetic analyses of cell fate dynamics over several days of observation. In these studies, TTT was successfully used for demonstrating the continuous single-cell imaging of blood generation from hemogenic endothelium34 and the instructive character of extrinsic cytokines to direct hematopoietic progenitor lineage choice.35 Thus, this was an ideal sensitive experimental system to monitor the early stages of reprogramming with high temporal resolution to confirm and expand recently published data from other groups:
(i) RF expression was efficiently downregulated simultaneously or even before the expression of endogenous Oct4 pronounced especially at late time points after transduction (>12d) and when a 4-in-1 reprogramming vector was used (Supplementary Video S3 and Figure 4c,d). (ii) Confluence of neighboring unrelated cells or colonies frequently resulted in genetic mosaics (Supplementary Video S1a), especially with increasing reprogramming efficiency. (iii) We could track the apparently random birth of iPSC-like cells in multiple locations of single cell–derived colonies expressing RF, surrounded by a majority of transduced cells that failed to express the Oct4-EGFP reprogramming marker (Supplementary Videos S5a,b and S6).
Sadelain and colleagues45 have generated human iPSCs derived from bicistronic lentiviral vectors coexpressing each single reprogramming factor with a fluorescent protein. In line with another study which suggested that the epigenetic extinction of the transgene cassette is mechanistically advantageous to acquire pluripotency,46 Sadelain et al. suggested that “pronounced (vector) silencing is a hallmark of successful reprogramming” and that it “follows the acquisition of pluripotent cell markers”.45 Due to the 20–120-fold higher temporal resolution of our approach our data suggest that vector silencing may rather be a prerequisite for the switch to the pluripotent state as prolonged ectopic RF expression may interfere with induction or maintenance of a delicately balanced pluripotency transcriptional network needed for pluripotent cell homeostasis.47
By analyzing the average output of populations of reprogrammed cells, first evidence for the epigenetic stochasticity of reprogramming was obtained.26 Here, we provide supporting evidence for stochastic reprogramming by analyzing not only anonymous populations, but also individual cells in live-cell imaging with high temporal resolution. These findings argue in support of Yamanaka's hypothesis of a stochastic model for iPSC generation,25 and our experimental system may not only be useful to address this important point but also to examine the role of small molecules and genetic determinants of the reprogramming process.
In a recent study, serial photographic imaging allowed the distinction of hiPSC from partially reprogrammed cells.28 Here, vector expression and pluripotency markers monitored every 2–3 days revealed that proviral silencing and pluripotency marker expression indicate the fully reprogrammed state. The authors also reported the conversion of partially to fully reprogrammed cells in a subset of cells within colonies. However, imaging in 2-day intervals does not rule out the possibility of a nonclonal situation, as our present investigations suggest the frequent genetic heterogeneity of iPSC colonies. Recently, Smith and colleagues used fluorescently labelled fibroblasts and analyzed their fate during the reprogramming process using live-cell imaging.27 However, the fluorescent proteins (actin fusion proteins) marked the entire cells but did not monitor the reprogramming process itself. Thus, it was still difficult to track single converting cells. In our study, the small time intervals (every 12 minutes to 2 hours) allowed the tracking of single-cell fate demonstrating that early colonies often reflect mosaics or patchworks formed on the basis of an epigenetic variability within transduced cells in conjunction with the potential confluence of neighboring transduced cells and colonies. Detailed studies aimed at the characterization of molecular events supporting the early stages of reprogramming should thus use advanced methods of cell purification to address the correct cell source, e.g., using antibody-based methods or the EOS pluripotency indicator.48
In summary, improved vector design with continuous live single-cell observation at high temporal resolution creates a powerful system to assess the subtle kinetics and morphology during biological processes such as the early stages of reprogramming. The experimental system described herein could be useful to further explore reprogramming events as well as to screen pluripotency markers, reprogramming factor candidates, roadblock inhibitors, supporting micro-environments, and other novel tools for reprogramming.
Materials and Methods
Ethics statement. All animal work has been performed strictly according to institutional guidelines and national regulations and was approved by the state office for protection of nature, environment, and consumers (LANUV) of North-Rhine Westphalia, Germany.
Vector construction. To construct a modular set of lentiviral reprogramming vectors, we used a 3rd generation lentiviral vector49 (pRRL.PPT.PGK.EGFPpre, kindly provided by L Naldini, Milano, Italy) and equipped it with NheI, AgeI and SalI sites.50 We introduced the retroviral SFFV U3 promoter amplified via PCR as NheI and AgeI fragment. The human and murine cDNAs encoding the RF Oct4, Sox2, Klf4, and c-Myc were engineered with a Kozak sequence and amplified as AgeI and SalI fragments via Pfu PCR. Furthermore, cDNAs for Oct4, Klf4, Sox2 were codon-optimized (Mr. Gene, Regensburg, Germany), thereby improving translation, mRNA half-life time, and removing cryptic splice and polyA sites.16,17 To allow tracking of reprogramming factor expression, we introduced fluorescent markers, namely EGFP, Venus (YFP derivative, here nuclear localized), and the red fluorescent protein dTomato, via an EMCV IRES (internal ribosomal entry site) sequence in the SalI site downstream of the RF. We also constructed 3-in-1 or 4-in-1 lentiviral vectors harboring Oct4, Sox2, Klf4 (3-in-1) plus optionally c-Myc (4-in-1), linked by 2A self-cleavage sites (as described in Figure 1), and equipped them with an IRES-dTomato cassette. To ease single-cell tracking, we additionally used a nuclear-localized dTomato (dTomato-nucMembrane). For promoter activity comparison we equipped the 4-in-1 constructs alternatively with the PGK and UCOE promoters. All constructs were validated by sequencing. Cloning details are available on request. See Supplementary Materials and Methods for primer details.
Cell lines, vector production, and lentiviral transduction. Murine (SC-1) and human HT1080 fibroblast lines, and the human embryonic kidney line 293T were cultured in Dulbecco's modified Eagle's medium (PAA, Pasching, Austria) supplemented with 10% fetal calf serum (PAA), 2 mmol/l -glutamine and sodium pyruvate (all PAA).
Virus production was performed as previously described.50 In short, 5 × 106 293T cells were seeded 24 hours prior to transfection in 10 cm dishes. Cells underwent transfection with 5 µg lentiviral vector, 12 µg pcDNA3.GP.4xCTE (expressing HIV-1 gag/pol), 5 µg pRSV-Rev and 1.5 µg pMD.G (encoding the vesicular stomatitis virus glycoprotein) using the calcium phosphate method in presence of 10 mmol/l HEPES and 25 µmol/l chloroquine. Supernatants were harvested at time points 24, 36, 48, and 60 hours after transfection, filtered and optionally concentrated using ultracentrifugation. Titration was performed on HT1080 and SC-1 fibroblasts 5 days after transduction as previously described.50 Fluorescence intensity was measured via flow cytometry (see below).
iPSC generation and cultivation. MEFs were prepared from pregnant (day 13.5 dpc) C3H (Charles River, Sulzfeld, Germany) and OG2 mice,29 and cultured on 0.1% gelatin-coated dishes in MEF medium (low glucose Dulbecco's modified Eagle's medium, 10% fetal calf serum, 1 mmol/l -glutamine (all from PAA), 0.1 mmol/l nonessential amino acids (Gibco, Karlsruhe, Germany), 100 µmol/l β-mercaptoethanol (Sigma, Seelze, Germany), 100 units/ml penicillin/100 µg/ml streptomycin (PAA)). To function as feeder layers, C3H MEFs were grown to confluence and inactivated with 10 µg/ml mitomycin C (Sigma). To generate iPSC, 5–7 × 104 MEFs (passage 1–3) were seeded in MEF medium in a 6-well plate 24 hours prior to transduction. MEFs were transduced with different lentiviral vectors encoding for Oct4, Klf4, Sox2, and/or c-Myc in presence of 4 µg/ml protamine sulfate for 8–16 hours. Cells were further cultured in MEF medium until day 4 pt and thereafter in ES medium (knockout Dulbecco's modified Eagle's medium (Gibco), 15% ES-tested fetal calf serum (PAA), 1 mmol/l -glutamine (PAA), 0.1 mmol/l nonessential amino acids (Gibco), 100 µmol/l β-mercaptoethanol (Sigma), 100 units/ml penicillin/100 µg/ml streptomycin (PAA) and 103 units/ml leukemia inhibitory factor (provided by the Max-Planck-Institute, Münster, Germany, and the Department of Technical Chemistry, University Hannover, Germany)). From day 2 to 9 media were supplemented with 2 mmol/l valproic acid (Sanofi-Aventis, Frankfurt, Germany) as previously described.24 Cells were either harvested via trypsinization and transferred onto mitomycin C-treated MEF feeders 4–7 days pt or reprogrammed “feeder-free” until picking of single clones (latest 15 days pt). Upon appearance of ESC-like colonies, single colonies were picked based on morphology and, in case of OG2, EGFP expression, and expanded on MEF feeder cells. ES medium was replaced every 1–3 days and iPSC was splitted every 3–4 days.
Time-lapse imaging and single-cell tracking. Long-term time-lapse imaging and single-cell tracking was done as described previously.34,35 In brief, OG2 MEFs were transduced with fluorescently labeled LV SIN (self-inactivating) vectors expressing hOct4, hSox2, hKlf4, and hc-Myc. First, we used four single-factor vectors in which hSox2 was linked to an IRES-dTomato cassette. Second, transduction was performed with a 3-in-1 construct with hOct4, hKlf4, hSox2 linked to IRES-dTomato plus a single-factor hc-Myc vector with IRES-Venus-nucMembrane cassette (nuclear-localized Venus). Third, for even better single-cell tracking, a 4-in-1 construct linked to an IRES-dTomato-nucMembrane sequence was used for reprogramming. In setup 1 and 2, transduced MEFs were transferred onto C3H feeder cells in a T12.5 flask (BD, Heidelberg, Germany) 4 days pt and further cultured in pregased medium. Live-cell imaging was performed at 37 °C from day 4.5 to day 11.5. Phase contrast pictures were acquired every 12 minutes, EGFP and dTomato fluorescence every 2.5 hours. In setup 3, transduced MEFs were maintained in situ and imaged from day 1 to day 13. Phase contrast and dTomato fluorescence pictures were taken every 12 minutes. EGFP was monitored in 2.5 hours (days 1–4) or 25-minute intervals (days 5–13).
Time-lapse microscopy was performed using a CellObserver system (Zeiss, Jena, Germany) equipped with a ×5 phase contrast objective (Zeiss), and an AxioCamHRm camera (at 1388 × 1040 pixel resolution) with Zeiss AxioVision 4.5 software. A mercury lamp (HBO103W/2) (Osram, Augsburg, Germany) was used for fluorescence illumination. EGFP and dTomato were detected using the Zeiss filter set 46HE at 1,300 ms and the Zeiss filter set 43HE at 400 ms, respectively.
Single-cell tracking was performed using a self-written computer program (TTT) on Siemens Celsius R630 workstations with 4 GB of RAM and SUSE Linux (10.1) operating systems with KDE desktop. The tracking trees served to determine doubling times (for example see Supplementary Figure S8). Videos were assembled using Quick Time Player Pro Ver7.62 (Apple, Cupertino, CA). Examples of videos are displayed in Supplementary Videos S1–S6.
Apotome fluorescence microscopy for analysis of iPS colony structure. For structural analysis OG2 MEFs were reprogrammed (using a 4-in-1 vector with IRES-dTomato cassette). Structured illumination microscopy was used to acquire optical sections of iPS-like colonies (Supplementary Video S3). For this purpose single iPS-like colonies were picked 13 days pt, transferred onto microscopical slides and embedded in Mowiol (Calbiochem, Schwalbach, Germany) for optimized optical analyses. Imaging was performed with an Axiovert 200M microscope (Zeiss) equipped with a ×40 objective (0.95 numeric aperture) and the Apotome slider (Zeiss). Imaris software (Bitplane, Zurich, Switzerland) was used for 3D visualization of raw images and colocalization analysis.
Flow cytometry. For monitoring fluorescent marker expression (Venus, EGFP, dTomato), cells were harvested, washed with phosphate-buffered saline, measured on a FACSCalibur or LSRII (Becton Dickinson, Heidelberg, Germany) using CellQuest, FACSDiva (Becton Dickinson) or FlowJo software (Tree Star, Ashland, OR). A gate was set on a homogeneous cell population, as determined by scatter characteristics, and 20,000 events were monitored. A marker gate was set to calculate the percentage and mean fluorescence intensity of positive cells.
Western blot. For protein isolation, cells were lysed in the presence of protease inhibitors (Complete Mini, Roche, Penzberg, Germany). 10–15 µg denatured proteins were separated on 12.5% sodium dodecyl sulfate–polyacrylamide gels. Proteins were transferred to a nitrocellulose membrane (Whatman, Dassel, Germany) via tank blot technique. Unspecific binding was blocked with 3% dry milk in TBS/0.05% Tween for at least 30 minutes and membranes stained with primary antibody in recommended dilution (anti-c-Myc: sc-40, anti-Oct4: sc-5279, anti-GKlf: sc-20691 and anti-Sox2: sc-17320, Santa Cruz, Heidelberg, Germany) at 4 °C overnight. After rinsing membranes were stained in appropriate peroxidase-conjugated secondary antibodies (Santa Cruz, Heidelberg, Germany) for 1 hour at room temperature, detection was performed by enhanced chemoluminescence (Thermo Scientific, Langenselbold, Germany) following manufacturer's instructions.
SUPPLEMENTARY MATERIAL Figure S1. Schematic overview of vector modifications and resulting effects. On top of the figure a schematic illustration of lentiviral vector design for 1-factor reprogramming (1factor) vectors is given. Translation initiation is enhanced by a Kozak sequence, reprogramming factors optionally codon-optimzed (co) and expression and titers improved by a wPRE element. Effects of codon-optimization of Oct4, Sox2 and Klf4 on expression of a fluorescent protein located on the same mRNA (linked via an IRES site) are shown for murine and human reprogramming factors. Single factors were combined in 3-in-1 or 4-in-1 reprogramming vectors by using various 2A sites. 4-in-1 vectors were equipped either with the PGK (phosphoglycerate kinase), UCOE (ubiquitous chromatin opening element) or SFFV (spleen focus-forming virus) promoter. Influences of the wPRE element, promoter choice and codon-optimzation measured by fluorescence intensity of co-expressed dTomato are illustrated in the bar graphs. Dot blots illustrate the different expression levels of the various promoters. Δ marks the self-inactivating (SIN) configuration with partially deleted U3 of the 3' LTR. RSV: Rous sarcoma virus U3 promoter; SD: splice donor; ψ: packaging signal; RRE: Rev response element; SA: splice acceptor; cPPT: central polypurine tract; IRES: internal ribosomal entry site; FP: fluorescent protein; PRE: woodchuck posttranscriptional regulatory element; Prom: promoter; P2A: porcine teschovirus 2A; T2A: thosea asigna virus 2A; E2A: equine rhinitis A virus 2A. Figure S2. Promoter expression characteristics in undifferentiated or differentiating ES cells. (a) Configuration of gammaretroviral (RV) and lentiviral (LV) vector plasmids harboring the PGK (phosphoglycerate kinase) or SFFV (spleen focus-forming virus) internal promoter. EGFP: enhanced green fluorescent protein; (b) Screening experiment for promoter activity. ES cells were kept under ES culture conditions or alternatively placed in embryoid body (EB) differentiation medium 4 days pt. EGFP marking was assessed by flow cytometry 2, 4 and 9 days pt, and the maximum set to 100%. Figure S3. Correct processing of reprogramming factor RNA and protein. (a) Vector RNA processing of monocistronic reprogramming constructs as shown in northern blot of HT1080 cells (total RNA). Vector RNA was detected using a radiolabeled PRE probe. Lane 1 indicates (untransduced) Mock cells, lanes 2-5 show the expected sizes for the SFFV-driven reprogramming factor RNAs. Size standards are given on the left, rehybridization was done with a probe against 18S RNA. (b) Western blot analysis of HT1080 cells transduced with reprogramming vectors and CCE ES cells serving as a positive control. In the upper part specific signals for the reprogramming factor proteins Klf4, Sox2 and Oct4 are depicted; in the lower part Erk protein is shown as loading control. Size markers are on the left. (c) Western blot analysis of murine fibroblast SC-1 cells transduced with a 4-in-1 vector (see Figure 2), expressing the reprogramming factors Oct4, Klf4, Sox2 und c-Myc. Above the specific signals of the reprogramming factors are given (label on right), below the loading control (stained for Actin). c-Myc is just weakly detectable because of the short half-life time of the protein. Figure S4. Analysis of ES cell marker expression in iPSC clones. (a) FACS analysis of SSEA-1 and Oct4-EGFP expression in representative iPS lines. The lower clone was derived from Oct4-EGFP (OG2)-MEFs, the upper from C3H-MEFs devoid of the reporter. (b) Immunofluorescence showing expression levels of Oct4 protein (red) and the Oct4-EGFP reporter (green) in an OG2-iPS line (LV1-7b) cultured under standard ES conditions. Alterations in colony morphology result from the fixation/permeablization procedure. (c) Alkaline phosphatase staining of a representative iPSC clone. Alkaline phosphatase staining was performed and was highly positive for all clones tested. (d) Real-time PCR analysis for endogenous mRNA expression of ES cell markers. iPSC were sorted based on EGFP expression and total RNA was isolated. Expression of ES cell markers was set in reference to beta-Actin (ActinB, set to 1) and displayed on a logarithmic scale. C3H MEFs served as a control. Figure S5. Endogenous Nanog and Oct4 promoters are highly unmethylated in generated iPSC clones. Analysis of the Nanog (a) and Oct4 (b) promoter methylation status based on bisulphate sequencing is shown for representative iPSC clones (LV1-7b, LV2-5b) in comparison with OG2-MEFs. Open circles are indicative for unmethylated, filled circles for methylated CpGs. Figure S6. Teratoma formation. Selected iPS cell clones LV1-7b and LV2-5b were injected subcutaneously into the flanks of NOD/SCID mice. Teratoma formation occurred between 4-8 weeks after injection. Teratomas were fixed, embedded in paraffin and stained with hematoxylin/eosin. Differentiation was shown into all 3 germ layers (endoderm/mesoderm/ectoderm) for both iPS clones (labeled on the right). Immature endodermal acinar cells (dotted circles in a) and immature endodermal epithelial structures (arrows in d) were detected demonstrating endodermal derivatives. Chondrogenic structures (dotted circles in b + e) represent mesoderm derivatives and large areas of the teratoma contained immature neuroectodermal tissue (c + f). Figure S7. Relation of Oct4-EGFP reporter and reprogramming factor expression. (a) Expression of Oct4-EGFP (green) in relation to reprogramming factor expression (dTomato signal, red) in the course of reprogramming. OG2-MEFs were reprogrammed using the 4-in-1 vector linked to dTomato, harvested at different time points after transduction and analyzed by flow cytometry. (b) Fluorescence microscopy of reprogramming colonies at early phases of reprogramming. Reprogramming of OG2-MEFs was induced using a 4-in-1 vector linked to dTomato. Reprogramming factor expression is indicated by dTomato signal, activation of the reporter by EGFP expression. Figure S8. Exemplary single-cell tracking tree of a transduced fibroblast and its progeny. Tracking of the single transduced fibroblast of Supplementary Movie S6 based on the nuclear dTomato signal (from a 4-in-1 vector) demonstrated the clonal character of the observed iPS colony. Cells are numbered according to their position in the tracking tree. The movie was started 1 day pt. Time scale after movie start is given on the left. ? indicate loss of single cell detectability due to the condensed structure of the cell cluster. ES-like morphology and the Oct4-EGFP signal (colonywise) appear roughly at the same time. Video S1a. Heterogeneity of early reprogramming. Starting 7 days after co-transduction of OG2 MEFs with vectors encoding hc-Myc, hOct4, hKlf4 and hSox2 (the latter linked to dTomato) the movies demonstrate formation of some representative iPS-like colonies (timer: d-hh:mm:ss after movie start). (a) On the left side, two transduced (red) iPS-like colonies proliferate in close proximity and in consequence merge with one another (timer: 0-20:31:03). Within this colony, EGFP (from the Oct4-EGFP reporter) appears rather inhomogeneous at one pole, reflecting a frequently observed patchwork pattern of iPS development. In contrast, another iPS-like colony emerges on the right side, that also first expresses the dTomato linked to the Sox2 factor, but switches on the Oct4-EGFP reporter rather homogeneously in the course of reprogramming. This phenomenon is observed less frequently than patchwork reprogramming. Video S1b. (b) Same experimental setup as in Supplementary Movie S1a with 3 panels: Left phase contrast, middle EGFP signal, right dTomato signal. Fibroblastic progeny start to form a colony of typical ES characteristics (round, sharp border, shiny, compact structure) around time point 2-23:26:12 after movie start. EGFP is switched on in a subset of cells (4-10:37:59) and expands in the course of the movie. Video S2. Rapid expansion of Oct4-EGFP expressing iPS-like cells. Displaying the growth behavior of an iPS-like colony the movie demonstrates that cells which continue to express the lentiviral reprogramming vectors (red) are rapidly overgrown upon emergence of reprogrammed Oct4-EGFP expressing cells (green). Video S3. Structured illumination microscopy of an induced pluripotent stem (iPS)-like colony 15 days after transduction. Apotome fluorescence microscopy provides a high precision scanning mechanism through the different layers of the iPS-like colony. Using a mathematical algorithm a 3D reconstruction of the iPS-like colony is calculated. Here we see downregulation of the red fluorescence (indicating the 4 reprogramming factors expressed from a 4-in-1 vector) in areas of green fluorescence (demonstrating the Oct4-EGFP pluripotency marker) within the iPS-like colony. The movie displays optical sections of an iPS-like colony, starting with the bottom layer of the colony, which mainly consists of reprogramming factor expressing (red) cells. After zooming in, consecutive upper layers (thickness: 2μm) are repeatedly faded in and out, demonstrating that the upper layers are largely composed of Oct4-EGFP expressing cells. Following 45° reciprocating rotations the 3-dimensional structure is illustrated, showing that most cells either express the reprogramming factors or the Oct4-EGFP pluripotency marker. Video S4. Dynamics of induced pluripotent stem cell (iPSC) generation. OG2 MEFs were transduced with the 4 reprogramming factors. Here a 3-in-1 vector expressing hOct4, hKlf4 and hSox2 linked to an IRES-dTomato cassette was used in combination with an hc-Myc vector linked to a nuclear-membrane localized Venus expression cassette. Enabled by the nuclear localization of Venus expression, colonies can be easier tracked and followed. The film depicts phase contrast (left) and EGFP/Venus fluorescence images (right). Fluorescent photos are taken along with every 12th phase contrast image. One c-Myc-IRES-Venus-nucMembrane expressing fibroblast (1) gives rise to two daughter cells (2, 3) (timer: 0-05:22:08) that segregate after division, showing mobility of transduced fibroblasts within the feeder layer. The progeny of both cells form neighboring iPS-like colonies, which fuse (2-03:02:12) upon growth. Video S5a. Stochastic character of reprogramming. (a) Oct4-EGFP murine embryonic fibroblasts (OG2 MEFs) were co-transduced with the 4 reprogramming factors (Sox2 being linked to an IRES-dTomato sequence, red) and monitored via time-lapse imaging from day 4.5-11.5 pt. Phase contrast is depicted on the left side, EGFP in the middle and dTomato signal on the right side with the movie spanning a selected period of 5.4 days. A single transduced fibroblast (1) gives rise to several daughter cells (labeled 2-15) which then compact and form a colony of typical iPS-like morphology (timer: 1-10:00:47). At time point 3-10:24:54 expression of the Oct4-EGFP reporter starts indicating activation of the transcriptional network involved in pluripotency. First, EGFP appears only in a subset of cells, arguing for a stochastic principle of reprogramming (see text for details). Note that the movie accelerates 12-fold starting at 1-15:05:29. Video S5b. (b) For a precise illustration of the kinetics of Sox2.IRES-dTomato (red) and Oct4-EGFP (green) expression, an overlay of both fluorescence markers is shown, displaying the exact same colony and time period as above. This movie is displayed in linear time course. Video S6. Stochasticity of reprogramming. In contrast to the experimental setup of Supplementary Movie S5a,b (transduced with single factor vectors) OG2 MEFs were reprogrammed using the 4-in-1 construct linked to a nuclear localized dTomato derivative, to ease the tracking of single cells in the condensed proliferative clusters emerging during reprogramming. Cells were monitored from day 1 to 13 pt with phase contrast images given on the left, Tomato signals in the middle and EGFP on the right (demonstrated in grayscale). The film displays a selected period of 4 days, demonstrating the fate of a single reprogramming factor transduced (dTomato, middle image) fibroblast (1) giving rise to its progeny (in the beginning indicated with numbers, also see Supplementary Figure S8), which first forms a cluster and then exhibits iPS-like morphology. Towards the end of the movie, Oct4-EGFP appears at multiple distinct spots within the clonal colony (EGFP, right image). Materials and Methods.
Acknowledgments
This work was supported by grants from Else Kröner-Fresenius-Stiftung, the German Academic Exchange Service (DAAD (0315187)), the German Ministry for Research and Education (PidNet, IFB-Tx (01EO0802), Carpud (016M0854)), the Deutsche Forschungsgemeinschaft (SPP1230, SPP1356, SFB566 and Cluster of Excellence REBIRTH (EXC 62/1)) and the European Union (Integrated Project PERSIST). We would like to thank Francoise Andre, Girmay Asgedom, Thomas Neumann (all Hannover Medical School) and Martina Bleidißel (MPI Münster, Germany) for technical assistance. In addition, we would like to thank Roger Tsien (San Diego, CA) for providing the Tomato cDNA, Michael Antoniou (London, UK) for the UCOE element and Shinya Yamanaka (Kyoto, Japan) for the original reprogramming factor cDNAs.
Supplementary Material
Schematic overview of vector modifications and resulting effects. On top of the figure a schematic illustration of lentiviral vector design for 1-factor reprogramming (1factor) vectors is given. Translation initiation is enhanced by a Kozak sequence, reprogramming factors optionally codon-optimzed (co) and expression and titers improved by a wPRE element. Effects of codon-optimization of Oct4, Sox2 and Klf4 on expression of a fluorescent protein located on the same mRNA (linked via an IRES site) are shown for murine and human reprogramming factors. Single factors were combined in 3-in-1 or 4-in-1 reprogramming vectors by using various 2A sites. 4-in-1 vectors were equipped either with the PGK (phosphoglycerate kinase), UCOE (ubiquitous chromatin opening element) or SFFV (spleen focus-forming virus) promoter. Influences of the wPRE element, promoter choice and codon-optimzation measured by fluorescence intensity of co-expressed dTomato are illustrated in the bar graphs. Dot blots illustrate the different expression levels of the various promoters. Δ marks the self-inactivating (SIN) configuration with partially deleted U3 of the 3' LTR. RSV: Rous sarcoma virus U3 promoter; SD: splice donor; ψ: packaging signal; RRE: Rev response element; SA: splice acceptor; cPPT: central polypurine tract; IRES: internal ribosomal entry site; FP: fluorescent protein; PRE: woodchuck posttranscriptional regulatory element; Prom: promoter; P2A: porcine teschovirus 2A; T2A: thosea asigna virus 2A; E2A: equine rhinitis A virus 2A.
Promoter expression characteristics in undifferentiated or differentiating ES cells. (a) Configuration of gammaretroviral (RV) and lentiviral (LV) vector plasmids harboring the PGK (phosphoglycerate kinase) or SFFV (spleen focus-forming virus) internal promoter. EGFP: enhanced green fluorescent protein; (b) Screening experiment for promoter activity. ES cells were kept under ES culture conditions or alternatively placed in embryoid body (EB) differentiation medium 4 days pt. EGFP marking was assessed by flow cytometry 2, 4 and 9 days pt, and the maximum set to 100%.
Correct processing of reprogramming factor RNA and protein. (a) Vector RNA processing of monocistronic reprogramming constructs as shown in northern blot of HT1080 cells (total RNA). Vector RNA was detected using a radiolabeled PRE probe. Lane 1 indicates (untransduced) Mock cells, lanes 2-5 show the expected sizes for the SFFV-driven reprogramming factor RNAs. Size standards are given on the left, rehybridization was done with a probe against 18S RNA. (b) Western blot analysis of HT1080 cells transduced with reprogramming vectors and CCE ES cells serving as a positive control. In the upper part specific signals for the reprogramming factor proteins Klf4, Sox2 and Oct4 are depicted; in the lower part Erk protein is shown as loading control. Size markers are on the left. (c) Western blot analysis of murine fibroblast SC-1 cells transduced with a 4-in-1 vector (see Figure 2), expressing the reprogramming factors Oct4, Klf4, Sox2 und c-Myc. Above the specific signals of the reprogramming factors are given (label on right), below the loading control (stained for Actin). c-Myc is just weakly detectable because of the short half-life time of the protein.
Analysis of ES cell marker expression in iPSC clones. (a) FACS analysis of SSEA-1 and Oct4-EGFP expression in representative iPS lines. The lower clone was derived from Oct4-EGFP (OG2)-MEFs, the upper from C3H-MEFs devoid of the reporter. (b) Immunofluorescence showing expression levels of Oct4 protein (red) and the Oct4-EGFP reporter (green) in an OG2-iPS line (LV1-7b) cultured under standard ES conditions. Alterations in colony morphology result from the fixation/permeablization procedure. (c) Alkaline phosphatase staining of a representative iPSC clone. Alkaline phosphatase staining was performed and was highly positive for all clones tested. (d) Real-time PCR analysis for endogenous mRNA expression of ES cell markers. iPSC were sorted based on EGFP expression and total RNA was isolated. Expression of ES cell markers was set in reference to beta-Actin (ActinB, set to 1) and displayed on a logarithmic scale. C3H MEFs served as a control.
Endogenous Nanog and Oct4 promoters are highly unmethylated in generated iPSC clones. Analysis of the Nanog (a) and Oct4 (b) promoter methylation status based on bisulphate sequencing is shown for representative iPSC clones (LV1-7b, LV2-5b) in comparison with OG2-MEFs. Open circles are indicative for unmethylated, filled circles for methylated CpGs.
Teratoma formation. Selected iPS cell clones LV1-7b and LV2-5b were injected subcutaneously into the flanks of NOD/SCID mice. Teratoma formation occurred between 4-8 weeks after injection. Teratomas were fixed, embedded in paraffin and stained with hematoxylin/eosin. Differentiation was shown into all 3 germ layers (endoderm/mesoderm/ectoderm) for both iPS clones (labeled on the right). Immature endodermal acinar cells (dotted circles in a) and immature endodermal epithelial structures (arrows in d) were detected demonstrating endodermal derivatives. Chondrogenic structures (dotted circles in b + e) represent mesoderm derivatives and large areas of the teratoma contained immature neuroectodermal tissue (c + f).
Relation of Oct4-EGFP reporter and reprogramming factor expression. (a) Expression of Oct4-EGFP (green) in relation to reprogramming factor expression (dTomato signal, red) in the course of reprogramming. OG2-MEFs were reprogrammed using the 4-in-1 vector linked to dTomato, harvested at different time points after transduction and analyzed by flow cytometry. (b) Fluorescence microscopy of reprogramming colonies at early phases of reprogramming. Reprogramming of OG2-MEFs was induced using a 4-in-1 vector linked to dTomato. Reprogramming factor expression is indicated by dTomato signal, activation of the reporter by EGFP expression.
Exemplary single-cell tracking tree of a transduced fibroblast and its progeny. Tracking of the single transduced fibroblast of Supplementary Movie S6 based on the nuclear dTomato signal (from a 4-in-1 vector) demonstrated the clonal character of the observed iPS colony. Cells are numbered according to their position in the tracking tree. The movie was started 1 day pt. Time scale after movie start is given on the left. ? indicate loss of single cell detectability due to the condensed structure of the cell cluster. ES-like morphology and the Oct4-EGFP signal (colonywise) appear roughly at the same time.
Heterogeneity of early reprogramming. Starting 7 days after co-transduction of OG2 MEFs with vectors encoding hc-Myc, hOct4, hKlf4 and hSox2 (the latter linked to dTomato) the movies demonstrate formation of some representative iPS-like colonies (timer: d-hh:mm:ss after movie start). (a) On the left side, two transduced (red) iPS-like colonies proliferate in close proximity and in consequence merge with one another (timer: 0-20:31:03). Within this colony, EGFP (from the Oct4-EGFP reporter) appears rather inhomogeneous at one pole, reflecting a frequently observed patchwork pattern of iPS development. In contrast, another iPS-like colony emerges on the right side, that also first expresses the dTomato linked to the Sox2 factor, but switches on the Oct4-EGFP reporter rather homogeneously in the course of reprogramming. This phenomenon is observed less frequently than patchwork reprogramming.
(b) Same experimental setup as in Supplementary Movie S1a with 3 panels: Left phase contrast, middle EGFP signal, right dTomato signal. Fibroblastic progeny start to form a colony of typical ES characteristics (round, sharp border, shiny, compact structure) around time point 2-23:26:12 after movie start. EGFP is switched on in a subset of cells (4-10:37:59) and expands in the course of the movie.
Rapid expansion of Oct4-EGFP expressing iPS-like cells. Displaying the growth behavior of an iPS-like colony the movie demonstrates that cells which continue to express the lentiviral reprogramming vectors (red) are rapidly overgrown upon emergence of reprogrammed Oct4-EGFP expressing cells (green).
Structured illumination microscopy of an induced pluripotent stem (iPS)-like colony 15 days after transduction. Apotome fluorescence microscopy provides a high precision scanning mechanism through the different layers of the iPS-like colony. Using a mathematical algorithm a 3D reconstruction of the iPS-like colony is calculated. Here we see downregulation of the red fluorescence (indicating the 4 reprogramming factors expressed from a 4-in-1 vector) in areas of green fluorescence (demonstrating the Oct4-EGFP pluripotency marker) within the iPS-like colony. The movie displays optical sections of an iPS-like colony, starting with the bottom layer of the colony, which mainly consists of reprogramming factor expressing (red) cells. After zooming in, consecutive upper layers (thickness: 2μm) are repeatedly faded in and out, demonstrating that the upper layers are largely composed of Oct4-EGFP expressing cells. Following 45° reciprocating rotations the 3-dimensional structure is illustrated, showing that most cells either express the reprogramming factors or the Oct4-EGFP pluripotency marker.
Dynamics of induced pluripotent stem cell (iPSC) generation. OG2 MEFs were transduced with the 4 reprogramming factors. Here a 3-in-1 vector expressing hOct4, hKlf4 and hSox2 linked to an IRES-dTomato cassette was used in combination with an hc-Myc vector linked to a nuclear-membrane localized Venus expression cassette. Enabled by the nuclear localization of Venus expression, colonies can be easier tracked and followed. The film depicts phase contrast (left) and EGFP/Venus fluorescence images (right). Fluorescent photos are taken along with every 12th phase contrast image. One c-Myc-IRES-Venus-nucMembrane expressing fibroblast (1) gives rise to two daughter cells (2, 3) (timer: 0-05:22:08) that segregate after division, showing mobility of transduced fibroblasts within the feeder layer. The progeny of both cells form neighboring iPS-like colonies, which fuse (2-03:02:12) upon growth.
Stochastic character of reprogramming. (a) Oct4-EGFP murine embryonic fibroblasts (OG2 MEFs) were co-transduced with the 4 reprogramming factors (Sox2 being linked to an IRES-dTomato sequence, red) and monitored via time-lapse imaging from day 4.5-11.5 pt. Phase contrast is depicted on the left side, EGFP in the middle and dTomato signal on the right side with the movie spanning a selected period of 5.4 days. A single transduced fibroblast (1) gives rise to several daughter cells (labeled 2-15) which then compact and form a colony of typical iPS-like morphology (timer: 1-10:00:47). At time point 3-10:24:54 expression of the Oct4-EGFP reporter starts indicating activation of the transcriptional network involved in pluripotency. First, EGFP appears only in a subset of cells, arguing for a stochastic principle of reprogramming (see text for details). Note that the movie accelerates 12-fold starting at 1-15:05:29.
(b) For a precise illustration of the kinetics of Sox2.IRES-dTomato (red) and Oct4-EGFP (green) expression, an overlay of both fluorescence markers is shown, displaying the exact same colony and time period as above. This movie is displayed in linear time course.
Stochasticity of reprogramming. In contrast to the experimental setup of Supplementary Movie S5a,b (transduced with single factor vectors) OG2 MEFs were reprogrammed using the 4-in-1 construct linked to a nuclear localized dTomato derivative, to ease the tracking of single cells in the condensed proliferative clusters emerging during reprogramming. Cells were monitored from day 1 to 13 pt with phase contrast images given on the left, Tomato signals in the middle and EGFP on the right (demonstrated in grayscale). The film displays a selected period of 4 days, demonstrating the fate of a single reprogramming factor transduced (dTomato, middle image) fibroblast (1) giving rise to its progeny (in the beginning indicated with numbers, also see Supplementary Figure S8), which first forms a cluster and then exhibits iPS-like morphology. Towards the end of the movie, Oct4-EGFP appears at multiple distinct spots within the clonal colony (EGFP, right image).
REFERENCES
- Meissner A, Wernig M., and, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K.et al. (2007Induction of pluripotent stem cells from adult human fibroblasts by defined factors Cell 131861–872. [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A.et al. (2008Disease-specific induced pluripotent stem cells Cell 134877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S.et al. (2007Induced pluripotent stem cell lines derived from human somatic cells Science 3181917–1920. [DOI] [PubMed] [Google Scholar]
- Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C., and, Hochedlinger K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M.et al. (2009Suppression of induced pluripotent stem cell generation by the p53-p21 pathway Nature 4601132–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya A, Rodríguez-Pizà I, Guenechea G, Vassena R, Navarro S, Barrero MJ.et al. (2009Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells Nature 46053–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F.et al. (2010Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA Cell Stem Cell 7618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Nakagawa M, Hyenjong H, Ichisaka T., and, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II.et al. (2009Human induced pluripotent stem cells free of vector and transgene sequences Science 324797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T.et al. (2009Generation of induced pluripotent stem cells using recombinant proteins Cell Stem Cell 4381–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schambach A, Cantz T, Baum C., and, Cathomen T. Generation and genetic modification of induced pluripotent stem cells. Expert Opin Biol Ther. 2010;10:1089–1103. doi: 10.1517/14712598.2010.496775. [DOI] [PubMed] [Google Scholar]
- Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hämäläinen R.et al. (2009piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells Nature 458766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer CA, Sommer AG, Longmire TA, Christodoulou C, Thomas DD, Gostissa M.et al. (2010Excision of reprogramming transgenes improves the differentiation potential of iPS cells generated with a single excisable vector Stem Cells 2864–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Hanna J, Saha K, Gao Q, Mitalipova M.et al. (2009Reprogramming of murine and human somatic cells using a single polycistronic vector Proc Natl Acad Sci USA 106157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng YY, Baert MR, Pike-Overzet K, Rodijk M, Brugman MH, Schambach A.et al. (2010Correction of B-cell development in Btk-deficient mice using lentiviral vectors with codon-optimized human BTK Leukemia 241617–1630. [DOI] [PubMed] [Google Scholar]
- Moreno-Carranza B, Gentsch M, Stein S, Schambach A, Santilli G, Rudolf E.et al. (2009Transgene optimization significantly improves SIN vector titers, gp91phox expression and reconstitution of superoxide production in X-CGD cells Gene Ther 16111–118. [DOI] [PubMed] [Google Scholar]
- Hope T. Improving the post-transcriptional aspects of lentiviral vectors. Curr Top Microbiol Immunol. 2002;261:179–189. doi: 10.1007/978-3-642-56114-6_9. [DOI] [PubMed] [Google Scholar]
- Schambach A, Bohne J, Baum C, Hermann FG, Egerer L, von Laer D.et al. (2006Woodchuck hepatitis virus post-transcriptional regulatory element deleted from X protein and promoter sequences enhances retroviral vector titer and expression Gene Ther 13641–645. [DOI] [PubMed] [Google Scholar]
- Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D.et al. (2009A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog Cell Stem Cell 5491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Ng JH, Heng JC., and, Ng HH. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell. 2009;4:301–312. doi: 10.1016/j.stem.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J.et al. (2010Vitamin C enhances the generation of mouse and human induced pluripotent stem cells Cell Stem Cell 671–79. [DOI] [PubMed] [Google Scholar]
- Kim JB, Sebastiano V, Wu G, Araúzo-Bravo MJ, Sasse P, Gentile L.et al. (2009Oct4-induced pluripotency in adult neural stem cells Cell 136411–419. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE.et al. (2008Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds Nat Biotechnol 26795–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460:49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP.et al. (2009Direct cell reprogramming is a stochastic process amenable to acceleration Nature 462595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Nachman I, Regev A., and, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol. 2010;28:521–526. doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, Huo H.et al. (2009Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells Nat Biotechnol 271033–1037. [DOI] [PubMed] [Google Scholar]
- Szabó PE, Hübner K, Schöler H., and, Mann JR. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech Dev. 2002;115:157–160. doi: 10.1016/s0925-4773(02)00087-4. [DOI] [PubMed] [Google Scholar]
- Boiani M, Gentile L, Gambles VV, Cavaleri F, Redi CA., and, Schöler HR. Variable reprogramming of the pluripotent stem cell marker Oct4 in mouse clones: distinct developmental potentials in different culture environments. Stem Cells. 2005;23:1089–1104. doi: 10.1634/stemcells.2004-0352. [DOI] [PubMed] [Google Scholar]
- Han DW, Do JT, Gentile L, Stehling M, Lee HT., and, Schöler HR. Pluripotential reprogramming of the somatic genome in hybrid cells occurs with the first cell cycle. Stem Cells. 2008;2644:5–454. doi: 10.1634/stemcells.2007-0553. [DOI] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V.et al. (2008Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors Nature 454646–650. [DOI] [PubMed] [Google Scholar]
- Schroeder T. Imaging stem-cell-driven regeneration in mammals. Nature. 2008;453:345–351. doi: 10.1038/nature07043. [DOI] [PubMed] [Google Scholar]
- Eilken HM, Nishikawa S., and, Schroeder T. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature. 2009;457:896–900. doi: 10.1038/nature07760. [DOI] [PubMed] [Google Scholar]
- Rieger MA, Hoppe PS, Smejkal BM, Eitelhuber AC., and, Schroeder T. Hematopoietic cytokines can instruct lineage choice. Science. 2009;325:217–218. doi: 10.1126/science.1171461. [DOI] [PubMed] [Google Scholar]
- Szymczak AL., and, Vignali DA. Development of 2A peptide-based strategies in the design of multicistronic vectors. Expert Opin Biol Ther. 2005;5:627–638. doi: 10.1517/14712598.5.5.627. [DOI] [PubMed] [Google Scholar]
- Schambach A, Wodrich H, Hildinger M, Bohne J, Kräusslich HG., and, Baum C. Context dependence of different modules for posttranscriptional enhancement of gene expression from retroviral vectors. Mol Ther. 2000;2:435–445. doi: 10.1006/mthe.2000.0191. [DOI] [PubMed] [Google Scholar]
- Baum C, Hegewisch-Becker S, Eckert HG, Stocking C., and, Ostertag W. Novel retroviral vectors for efficient expression of the multidrugresistance (mdr-1) gene in early hematopoietic cells. J Virol. 1995;69:7541-7547. doi: 10.1128/jvi.69.12.7541-7547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentilin L, Qin G, Tafuro S, Dinauer MC, Baum C., and, Giacca M. Variegation of retroviral vector gene expression in myeloid cells. Gene Ther. 2000;7:153–166. doi: 10.1038/sj.gt.3301057. [DOI] [PubMed] [Google Scholar]
- Zhang F, Thornhill SI, Howe SJ, Ulaganathan M, Schambach A, Sinclair J.et al. (2007Lentiviral vectors containing an enhancer-less ubiquitously acting chromatin opening element (UCOE) provide highly reproducible and stable transgene expression in hematopoietic cells Blood 1101448–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A.et al. (2010Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease Nat Med 16198–204. [DOI] [PubMed] [Google Scholar]
- Ogawa K, Saito A, Matsui H, Suzuki H, Ohtsuka S, Shimosato D.et al. (2007Activin-Nodal signaling is involved in propagation of mouse embryonic stem cells J Cell Sci 120Pt 155–65. [DOI] [PubMed] [Google Scholar]
- Lee J, Go Y, Kang I, Han YM., and, Kim J. Oct-4 controls cell-cycle progression of embryonic stem cells. Biochem J. 2010;426:171–181. doi: 10.1042/BJ20091439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D., and, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- Papapetrou EP, Tomishima MJ, Chambers SM, Mica Y, Reed E, Menon J.et al. (2009Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation Proc Natl Acad Sci USA 10612759–12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta A., and, Ellis J. Retroviral vector silencing during iPS cell induction: an epigenetic beacon that signals distinct pluripotent states. J Cell Biochem. 2008;105:940–948. doi: 10.1002/jcb.21912. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J., and, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Hotta A, Cheung AY, Farra N, Vijayaragavan K, Séguin CA, Draper JS.et al. (2009Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency Nat Methods 6370–376. [DOI] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D.et al. (1998A third-generation lentivirus vector with a conditional packaging system J Virol 728463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schambach A, Bohne J, Chandra S, Will E, Margison GP, Williams DA.et al. (2006Equal potency of gammaretroviral and lentiviral SIN vectors for expression of O6-methylguanine-DNA methyltransferase in hematopoietic cells Mol Ther 13391–400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic overview of vector modifications and resulting effects. On top of the figure a schematic illustration of lentiviral vector design for 1-factor reprogramming (1factor) vectors is given. Translation initiation is enhanced by a Kozak sequence, reprogramming factors optionally codon-optimzed (co) and expression and titers improved by a wPRE element. Effects of codon-optimization of Oct4, Sox2 and Klf4 on expression of a fluorescent protein located on the same mRNA (linked via an IRES site) are shown for murine and human reprogramming factors. Single factors were combined in 3-in-1 or 4-in-1 reprogramming vectors by using various 2A sites. 4-in-1 vectors were equipped either with the PGK (phosphoglycerate kinase), UCOE (ubiquitous chromatin opening element) or SFFV (spleen focus-forming virus) promoter. Influences of the wPRE element, promoter choice and codon-optimzation measured by fluorescence intensity of co-expressed dTomato are illustrated in the bar graphs. Dot blots illustrate the different expression levels of the various promoters. Δ marks the self-inactivating (SIN) configuration with partially deleted U3 of the 3' LTR. RSV: Rous sarcoma virus U3 promoter; SD: splice donor; ψ: packaging signal; RRE: Rev response element; SA: splice acceptor; cPPT: central polypurine tract; IRES: internal ribosomal entry site; FP: fluorescent protein; PRE: woodchuck posttranscriptional regulatory element; Prom: promoter; P2A: porcine teschovirus 2A; T2A: thosea asigna virus 2A; E2A: equine rhinitis A virus 2A.
Promoter expression characteristics in undifferentiated or differentiating ES cells. (a) Configuration of gammaretroviral (RV) and lentiviral (LV) vector plasmids harboring the PGK (phosphoglycerate kinase) or SFFV (spleen focus-forming virus) internal promoter. EGFP: enhanced green fluorescent protein; (b) Screening experiment for promoter activity. ES cells were kept under ES culture conditions or alternatively placed in embryoid body (EB) differentiation medium 4 days pt. EGFP marking was assessed by flow cytometry 2, 4 and 9 days pt, and the maximum set to 100%.
Correct processing of reprogramming factor RNA and protein. (a) Vector RNA processing of monocistronic reprogramming constructs as shown in northern blot of HT1080 cells (total RNA). Vector RNA was detected using a radiolabeled PRE probe. Lane 1 indicates (untransduced) Mock cells, lanes 2-5 show the expected sizes for the SFFV-driven reprogramming factor RNAs. Size standards are given on the left, rehybridization was done with a probe against 18S RNA. (b) Western blot analysis of HT1080 cells transduced with reprogramming vectors and CCE ES cells serving as a positive control. In the upper part specific signals for the reprogramming factor proteins Klf4, Sox2 and Oct4 are depicted; in the lower part Erk protein is shown as loading control. Size markers are on the left. (c) Western blot analysis of murine fibroblast SC-1 cells transduced with a 4-in-1 vector (see Figure 2), expressing the reprogramming factors Oct4, Klf4, Sox2 und c-Myc. Above the specific signals of the reprogramming factors are given (label on right), below the loading control (stained for Actin). c-Myc is just weakly detectable because of the short half-life time of the protein.
Analysis of ES cell marker expression in iPSC clones. (a) FACS analysis of SSEA-1 and Oct4-EGFP expression in representative iPS lines. The lower clone was derived from Oct4-EGFP (OG2)-MEFs, the upper from C3H-MEFs devoid of the reporter. (b) Immunofluorescence showing expression levels of Oct4 protein (red) and the Oct4-EGFP reporter (green) in an OG2-iPS line (LV1-7b) cultured under standard ES conditions. Alterations in colony morphology result from the fixation/permeablization procedure. (c) Alkaline phosphatase staining of a representative iPSC clone. Alkaline phosphatase staining was performed and was highly positive for all clones tested. (d) Real-time PCR analysis for endogenous mRNA expression of ES cell markers. iPSC were sorted based on EGFP expression and total RNA was isolated. Expression of ES cell markers was set in reference to beta-Actin (ActinB, set to 1) and displayed on a logarithmic scale. C3H MEFs served as a control.
Endogenous Nanog and Oct4 promoters are highly unmethylated in generated iPSC clones. Analysis of the Nanog (a) and Oct4 (b) promoter methylation status based on bisulphate sequencing is shown for representative iPSC clones (LV1-7b, LV2-5b) in comparison with OG2-MEFs. Open circles are indicative for unmethylated, filled circles for methylated CpGs.
Teratoma formation. Selected iPS cell clones LV1-7b and LV2-5b were injected subcutaneously into the flanks of NOD/SCID mice. Teratoma formation occurred between 4-8 weeks after injection. Teratomas were fixed, embedded in paraffin and stained with hematoxylin/eosin. Differentiation was shown into all 3 germ layers (endoderm/mesoderm/ectoderm) for both iPS clones (labeled on the right). Immature endodermal acinar cells (dotted circles in a) and immature endodermal epithelial structures (arrows in d) were detected demonstrating endodermal derivatives. Chondrogenic structures (dotted circles in b + e) represent mesoderm derivatives and large areas of the teratoma contained immature neuroectodermal tissue (c + f).
Relation of Oct4-EGFP reporter and reprogramming factor expression. (a) Expression of Oct4-EGFP (green) in relation to reprogramming factor expression (dTomato signal, red) in the course of reprogramming. OG2-MEFs were reprogrammed using the 4-in-1 vector linked to dTomato, harvested at different time points after transduction and analyzed by flow cytometry. (b) Fluorescence microscopy of reprogramming colonies at early phases of reprogramming. Reprogramming of OG2-MEFs was induced using a 4-in-1 vector linked to dTomato. Reprogramming factor expression is indicated by dTomato signal, activation of the reporter by EGFP expression.
Exemplary single-cell tracking tree of a transduced fibroblast and its progeny. Tracking of the single transduced fibroblast of Supplementary Movie S6 based on the nuclear dTomato signal (from a 4-in-1 vector) demonstrated the clonal character of the observed iPS colony. Cells are numbered according to their position in the tracking tree. The movie was started 1 day pt. Time scale after movie start is given on the left. ? indicate loss of single cell detectability due to the condensed structure of the cell cluster. ES-like morphology and the Oct4-EGFP signal (colonywise) appear roughly at the same time.
Heterogeneity of early reprogramming. Starting 7 days after co-transduction of OG2 MEFs with vectors encoding hc-Myc, hOct4, hKlf4 and hSox2 (the latter linked to dTomato) the movies demonstrate formation of some representative iPS-like colonies (timer: d-hh:mm:ss after movie start). (a) On the left side, two transduced (red) iPS-like colonies proliferate in close proximity and in consequence merge with one another (timer: 0-20:31:03). Within this colony, EGFP (from the Oct4-EGFP reporter) appears rather inhomogeneous at one pole, reflecting a frequently observed patchwork pattern of iPS development. In contrast, another iPS-like colony emerges on the right side, that also first expresses the dTomato linked to the Sox2 factor, but switches on the Oct4-EGFP reporter rather homogeneously in the course of reprogramming. This phenomenon is observed less frequently than patchwork reprogramming.
(b) Same experimental setup as in Supplementary Movie S1a with 3 panels: Left phase contrast, middle EGFP signal, right dTomato signal. Fibroblastic progeny start to form a colony of typical ES characteristics (round, sharp border, shiny, compact structure) around time point 2-23:26:12 after movie start. EGFP is switched on in a subset of cells (4-10:37:59) and expands in the course of the movie.
Rapid expansion of Oct4-EGFP expressing iPS-like cells. Displaying the growth behavior of an iPS-like colony the movie demonstrates that cells which continue to express the lentiviral reprogramming vectors (red) are rapidly overgrown upon emergence of reprogrammed Oct4-EGFP expressing cells (green).
Structured illumination microscopy of an induced pluripotent stem (iPS)-like colony 15 days after transduction. Apotome fluorescence microscopy provides a high precision scanning mechanism through the different layers of the iPS-like colony. Using a mathematical algorithm a 3D reconstruction of the iPS-like colony is calculated. Here we see downregulation of the red fluorescence (indicating the 4 reprogramming factors expressed from a 4-in-1 vector) in areas of green fluorescence (demonstrating the Oct4-EGFP pluripotency marker) within the iPS-like colony. The movie displays optical sections of an iPS-like colony, starting with the bottom layer of the colony, which mainly consists of reprogramming factor expressing (red) cells. After zooming in, consecutive upper layers (thickness: 2μm) are repeatedly faded in and out, demonstrating that the upper layers are largely composed of Oct4-EGFP expressing cells. Following 45° reciprocating rotations the 3-dimensional structure is illustrated, showing that most cells either express the reprogramming factors or the Oct4-EGFP pluripotency marker.
Dynamics of induced pluripotent stem cell (iPSC) generation. OG2 MEFs were transduced with the 4 reprogramming factors. Here a 3-in-1 vector expressing hOct4, hKlf4 and hSox2 linked to an IRES-dTomato cassette was used in combination with an hc-Myc vector linked to a nuclear-membrane localized Venus expression cassette. Enabled by the nuclear localization of Venus expression, colonies can be easier tracked and followed. The film depicts phase contrast (left) and EGFP/Venus fluorescence images (right). Fluorescent photos are taken along with every 12th phase contrast image. One c-Myc-IRES-Venus-nucMembrane expressing fibroblast (1) gives rise to two daughter cells (2, 3) (timer: 0-05:22:08) that segregate after division, showing mobility of transduced fibroblasts within the feeder layer. The progeny of both cells form neighboring iPS-like colonies, which fuse (2-03:02:12) upon growth.
Stochastic character of reprogramming. (a) Oct4-EGFP murine embryonic fibroblasts (OG2 MEFs) were co-transduced with the 4 reprogramming factors (Sox2 being linked to an IRES-dTomato sequence, red) and monitored via time-lapse imaging from day 4.5-11.5 pt. Phase contrast is depicted on the left side, EGFP in the middle and dTomato signal on the right side with the movie spanning a selected period of 5.4 days. A single transduced fibroblast (1) gives rise to several daughter cells (labeled 2-15) which then compact and form a colony of typical iPS-like morphology (timer: 1-10:00:47). At time point 3-10:24:54 expression of the Oct4-EGFP reporter starts indicating activation of the transcriptional network involved in pluripotency. First, EGFP appears only in a subset of cells, arguing for a stochastic principle of reprogramming (see text for details). Note that the movie accelerates 12-fold starting at 1-15:05:29.
(b) For a precise illustration of the kinetics of Sox2.IRES-dTomato (red) and Oct4-EGFP (green) expression, an overlay of both fluorescence markers is shown, displaying the exact same colony and time period as above. This movie is displayed in linear time course.
Stochasticity of reprogramming. In contrast to the experimental setup of Supplementary Movie S5a,b (transduced with single factor vectors) OG2 MEFs were reprogrammed using the 4-in-1 construct linked to a nuclear localized dTomato derivative, to ease the tracking of single cells in the condensed proliferative clusters emerging during reprogramming. Cells were monitored from day 1 to 13 pt with phase contrast images given on the left, Tomato signals in the middle and EGFP on the right (demonstrated in grayscale). The film displays a selected period of 4 days, demonstrating the fate of a single reprogramming factor transduced (dTomato, middle image) fibroblast (1) giving rise to its progeny (in the beginning indicated with numbers, also see Supplementary Figure S8), which first forms a cluster and then exhibits iPS-like morphology. Towards the end of the movie, Oct4-EGFP appears at multiple distinct spots within the clonal colony (EGFP, right image).