Abstract
A newly identified bacterial strain that can grow in the presence of arsenate, and possibly in the absence of phosphate, has raised much interest, but also fueled an active debate. Can arsenate substitute for phosphate in some, or possibly in most, of the absolutely essential phosphate-based biomolecules, including DNA? If so, then the possibility of alternative, arsenic-based life forms must be considered. The physicochemical similarity of these two oxyanions speaks in favor of this idea. However, arsenate-esters, and arsenate-diesters in particular, are extremely unstable in aqueous media. Here we explore the potential of arsenate to be used as substrate by phosphate-utilizing enzymes. We review the existing literature on arsenate enzymology, that intriguingly, dates back to the 1930s, and Otto Warburg. We address the issue of how and to what degree proteins can distinguish between arsenate and phosphate, and what is known in general about oxyanion specificity. And, we also discuss how phosphate-arsenate promiscuity may affect evolutionary transitions between phosphate and arsenate based biochemistry. Finally, we highlight potential applications of arsenate as a structural and mechanistic probe of enzymes whose catalyzed reactions involve the making or breaking of phosphoester bonds.
A recently published article in Science describes a newly discovered bacterium isolated from the unique environment of California's Mono Lake. The authors provide data that support the presence of arsenate in nucleic acids and proteins. They thereby argue that arsenate can be a viable substitute for phosphate in the DNA of the Halomonadaceae (a family halophilic protobacteria) GFAJ-1 strain (1). Following the four most abundant elements - carbon, hydrogen, nitrogen and oxygen, phosphorus comprises one of the major elements of life. Foremost, phosphorus is a key element in the metabolic currencies of all known life forms. In addition, phosphate provides the connecting bridge between the nucleobases in RNA and DNA. That arsenic can potentially take over any of the roles of phosphorus is therefore a fundamental discovery with far-fetched implications. This discovery raises the possibility that the onset of life on Earth, and of other possible life forms outside this planet, can be based on an entirely different chemistry (2). Indeed, Wolfe-Simon et al. have raised the intriguing possibility that arsenate-base life forms may have existed at the early stages of evolution of life on this and potentially on other planets (3).
Intriguing yet controversial findings
Why is arsenate an attractive option? Phosphate is not an uncommon compound in the earth's crust, but it is relatively scarce in oceans (where the first life forms may have emerged). And, unlike the major elements (C, N, O and H), there exists no common gas form of phosphorus that could have made it available to those niches where life emerged. Even for present life forms, phosphorous is a rate-limiting factor for growth. For example, some soil bacteria have evolved to exploit phosphate-containing pesticides (organophosphates), presumably to supplement shortages in inorganic phosphate (4, 5). Arsenic and phosphorous share key chemical properties, including oxidative states. In their most common 5+ (V) oxides arsenate (HAsO4−2) exhibits very similar pKa values to phosphate (HPO4−2), and forms analogous esters. Arsenic rich environments are available on Earth, and these locations may have been particularly relevant when life originated (3). Why then did Nature choose phosphorus? The reasons for this preference are quite clear (6): The reduction of As(V) to As(III) is much easier than for phosphorous, allowing phosphorus to be more stable in the preferred higher oxidation state. In particular, as further discussed below, arsenate esters are notoriously unstable in water, and rapid hydrolysis of the corresponding high-energy arsenate esters leads to wasteful “futile cycles” (7). This higher reactivity with water would also create serious problems for the long-term stability of the repository of genetic information for life forms containing arsenate-linked nucleic acids.
Whilst Wolfe-Simon et al. raise an intriguing hypothesis about possible alternative life chemistries, their supporting data is rather slim. Several scientists have already criticized the published data (8, 9), and our reaction upon reading the paper has been similar. The isolated strain grows faster on arsenate on its own than on phosphate on its own (Fig. 1A in Ref. (1)). However, growth rate under the condition which might be optimal for this strain, namely, high concentration of arsenate plus low phosphate concentration, has not been reported. DNA preparations from the newly isolated strain have been shown to contain arsenate, but these preparations also contain phosphate. Arsenate may be associated with the DNA, but no direct evidence has been provided to support the hypothesis that arsenate substitutes for phosphate in the DNA's base-linking diester bonds. In addition, whereas total nucleic acid preparation (DNA plus RNA samples analyzed for As and P content) from the cells grown on phosphate appear normal by gel-electrophoresis, the arsenate grown bacteria yielded only one band of discrete size (possibly chromosomal DNA) and no low molecular weight smear that usually corresponds to RNA (Fig. 2A in Ref. (1)). It could therefore be that growth under arsenate induces drastic change in the nucleic acid content and/or properties, as suggested by the authors. However, these nucleic acid samples extracted with standard methods, and examined in aqueous solution, are not necessarily relevant.
The hydrolytic instability of arsenate esters
Compared to the extensive literature on the hydrolysis of phosphate esters, data on the hydrolysis of arsenate esters is quite limited. It is known, however, that arsenate esters, including tri- and di-, and mono-esters, and pyro-arsenate, hydrolyze many orders of magnitude faster than the equivalent phosphate esters. The mechanisms of hydrolysis also differ, with arsenate esters hydrolyzing via mechanisms with a more associative character (Ref. (10) and references therein). High reactivity and the use of a more associative mechanism, result in a fundamental difference in the hydrolysis kinetics between phosphate and arsenate esters. Phospho-triesters are hydrolyzed ~105-fold faster than the corresponding diesters. The latter comprise the connecting bonds in DNA, and their remarkable stability is critical for genome integrity (6). Arsenate-triesters are highly labile, the half-life of trimethyl-arsenate in water, at neutral pH and ambient temperature, is in the range of 0.02 sec. However, unlike the analogous phosphate esters, the resulting dimethyl ester of arsenate hydrolyzes even more rapidly (extrapolating from the data of Ref. (10)(10)). The 5'and 3'-ribosyl groups of DNA bases exhibit about a 10-fold lower rate of hydrolysis than methyl groups (diethyl-arsenate hydrolyzes ~5-fold slower than dimethyl-arsenate) (10), but beyond that, no fundamental differences in reactivity between the dialkyl-arsenates used as a model and the diester bonds of DNA are expected. The corresponding arsenate esters of small metabolites such as NTPs and dNTPs (nucleotide, and deoxy-nucleotide triphosphates) are expected to be even more labile and will not sustain in aqueous environments.
So how would an arsenate-based DNA maintain its vital genetic information along generations? Wolfe-Simon et al. suggest that DNA, and perhaps other key molecules might be stored in vacuole-like regions, and that these non-aqueous environments may prevent, or at least slow down hydrolysis (1). However, arsenate esters are labile even when water is present at low concentrations: Baer et al. report for trimethyl-arsenate half-lives of <0.1 sec at pH ~9 with 0.25M water in organic solvent, i.e., <0.5% water) (10). Another possibility is that: “Arsenic-based life simply has a higher turnover for molecular disintegration and assembly than does conventional life” as stated by one of the authors (Paul Davies, Arizona State University) (8). This is not unprecedented - the extremophilic bacterium Deinococcus radiodurans that can resist extreme levels of ionizing radiation possesses a highly robust DNA repair system capable of reassembling its constantly fragmented genome. The reassembly of the fragmented DNA in this organism is based on diploidy and the annealing and elongation of overlapping fragments (11). Still, repairing frequent strand breaks, and dealing with the rapid hydrolysis of nearly every base-connecting bond, are two quite different scenarios.
Other complications for the incorporation of arsenate-based compounds in living organisms arise from the low reduction potential of arsenate (6). Free thiols can readily reduce arsenate to give As(III) tri-thiolates and oxidized di-thiols (12). Although the arsenite-thiolates hydrolyze at pH >7, this chemistry implies a highly oxidizing environment where reduced cysteines, and other free thiols, would be hard to maintain.
The arsenate-phosphate analogy
Whether or not Halomonadaceae GFAJ-1 uses arsenate instead of phosphate in its macromolecules (RNA, DNA, proteins), and/or small molecules, remains controversial. But this work does provide preliminary indications for such a possibility, and will thus inspire many to examine various aspects of arsenate biochemistry. Skepticism is an essential trait of every good scientist, and making scientists doubtful, or even angry, is a proven inspirational tool for many previous paradigm shifts (13).
From our own point of view, beyond the above-discussed issues, this work opens several interesting questions with respect to the oxyanion promiscuity of enzymes, and specifically, regarding the potential of arsenate to be used as substrate by phosphate utilizing enzymes. How readily can this replacement occur? What are the possible consequences of such replacement? We are obviously not the first to be interested in this problem. Some of the 20th Century's most notable enzymologists, Otto Warburg, Frank Weistheimer and Henry Dixon, have each addressed this issue (Ref. (6, 7), and references therein). However, these first raised interests in the enzymology of arsenate, and other arsenic derivatives, have not been sustained, and at present, the enzymology of arsenic derivates is rarely explored.
Here, we review the sparse existing literature on arsenate enzymology, and discuss the relevance of these studies to the hypothesis of arsenate-based life forms. We address the issue of to what degree, and how, can proteins distinguish between arsenate and phosphate, and what is known in general about oxyanion specificity. And, we also discuss how the lack of ion specificity (i.e., ion promiscuity) may affect evolutionary transitions between phosphate and arsenate based biochemistry. Finally, we highlight potential applications of arsenate as a structural and mechanistic probe of enzymes whose catalyzed reactions involve the making or breaking of phosphoester bonds.
Enzymes promiscuity with arsenate
Arsenate and phosphate share many physicochemical properties. However, their atomic radii and electron shells differ, hence the below discussed different reactivities of their esters. The bond lengths and relative charges on the arsenic and oxygens of arsenate also vary from the corresponding phosphate: P-O bond lengths are 1.52–1.54Å versus 1.68–1.71 for As-O (7, 14). The partial negative charge on oxygen atoms is −0.952 for phosphate vs. −0.892 for arsenate (14). This difference, although relatively small, also affects the nucleophilicity of these ions. The overall sizes of these ions are, however, quite similar: the thermochemical radius for phosphate is 2.38 Å, and of arsenate is 2.48 Å (15).
Could enzymes that utilize phosphate, or phospho-esters, bind the equivalent arsenates, and if so would they utilize them as substrates? The main hurdle for exploring this question is stability. Arsenate (HAsO4−2) can be used as a substitute for inorganic phosphate, but because most phosphate liberating processes are effectively irreversible (e.g. ATP→ADP + Pi), the number of enzymes that use inorganic phosphate as substrate is limited (for example, only about a dozen such enzymes could be identified in the E. coli genome using EcoCyC). Nevertheless, several key phosphate utilizing enzymes have been explored with arsenate (7, 14, 16, 17), and two detailed examples are discussed below.
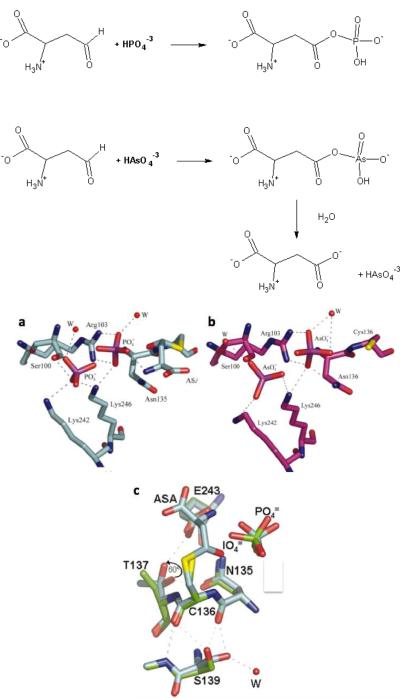
The protein data bank (PDB) contains two enzyme structures with a bound arsenate that replaces phosphate. One structure is L-aspartate-β-semialdehyde dehydrogenase (H. influenzae ASADH), an enzyme that reversibly catalyzes the reductive dephosphorylation of β-aspartyl phosphate to give L-aspartate- β-semialdehyde. The position of the bound arsenate, and those of its protein ligands, are identical for both phosphate and arsenate. Critical hydrogen-bonds with catalytic residues, and with a water molecule ligand, are also maintained (18) (Figure 1). In accordance, arsenate comprises a surprisingly good substrate for this enzyme, both in terms of KM (1.6 mM, versus 2.9 mM for phosphate), and kcat (510 and 710 min−1) (14). The primary difference between the phosphate vs. the arsenate reaction is the product: whereas phosphate yields β-aspartyl-phosphate, a somewhat unstable acyl phosphate that can nevertheless be easily isolated, the corresponding arsenate product rapidly hydrolyses to yield aspartic acid and free arsenate. In fact, ASADH was examined with a variety of potential oxyanions. Several ions were found to be effective inhibitors of this enzyme, including tungstate and iodate. The latter has been shown to bind in ASADH's active-site in an altered manner that explains the inhibitory effect (Figure 1). Of those oxyanions examined only vanadate and arsenate comprised alternative substrates, with arsenate being as good a substrate as phosphate, while vanadate showed a 2-fold lower kcat, but a 20-fold lower KM, and hence ~10-fold higher kcat/KM than phosphate (14).
Figure 1.
The reaction catalyzed by L-aspartate-β-semialdehyde dehydrogenase with phosphate and arsenate (top panel), and structures of the enzyme complexes with different oxyanions (bottom panel; PDB codes 1NX6, 1TA4, and 1TB4; adopted from Ref. 18). a. The phosphate-bound structure with the substrate, aspartate-β-semialdehyde (ASA), as a covalent adduct with active-site Cys136. b. The arsenate-bound structure. The positions of the bound arsenate ions, and the interacting protein ligands, are identical for phosphate and arsenate. c. An ovelay of the periodate complex (in green) and phsophate complex (in blue). The two oxyanions are bound in a very similar mode. However, the side-chains of Thr137 is ~60° rotated in the preiodate structure, resulting in the alteration of a hydrogen bond with Glu243 (a residue involved in substrate binding and catalysis). The side-chain rotamer of Ser139, which is hydrogen-bonded to the nucleophilic Cys136, is also roated in the periodate structure. These two relatively subtle changes mediate the shift from a substrate (phosphate, or arsenate) to inhibitor (periodate).
Arsenate and vanadate, as well as molybdate, comprise alternative substrates for another well-characterized dehydrogenase, glyceraldehyde-3-phosphate dehydrogenase (GAPDH). This enzyme had been explored with arsenate by Otto Warburg, as early as 1939. As is the case with ASADH, the acyl-arsenate product rapidly decomposes (estimated half-life is <2.5 sec), resulting in a `futile cycle' and the irreversibility of the reaction (Ref. (7) and references therein).
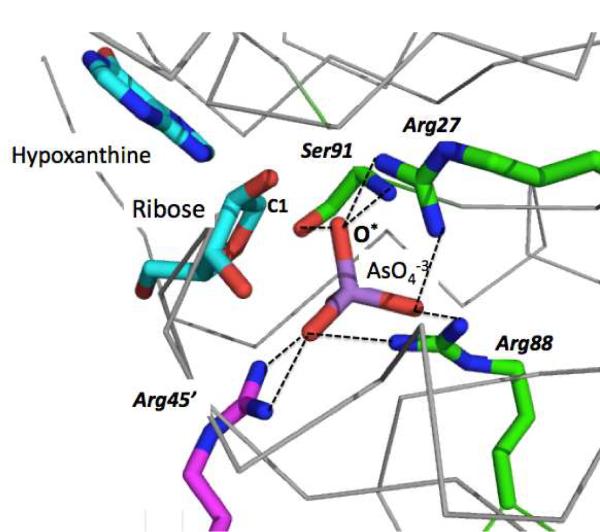
Another case of an arsenate-bound structure is purine nucleoside phosphorylase (PNP). This enzyme catalyzes the reversible phosphorolysis of various nucleosides including inosine and guanosine, to yield the free bases and D-ribose-1-phosphate. PNP also catalyzes the arsenolysis of inosine and guanosine to yield the corresponding arsenic-containing product. However, in oppose to D-ribose-1-phosphate, ribose 1-arsenate, is unstable and rapidly hydrolyzes to ribose and arsenate, thus rendering the reaction irreversible (17). As in other cases of arsenate-ester products, the hydrolysis of ribose 1-arsenate is not catalyzed by the enzyme, but occurs spontaneously (17). The KM values for PNP (human erythorcyte) are similar for arsenate and phosphate (1.8 mM, and 0.8 mM, respectively) (19). Whilst we could not identify a reference for the kcat values, the microscopic parameters measured with arsenate as a substrate indicate that the rate-limiting step is dissociation of the enzyme-free-base complex, suggesting that the rate of chemistry with arsenate is not significantly slower than that with phosphate. The structure of Plasmodium falciparum PNP was solved with inosine and arsenate, and was found to contain a mixture of products and reactants – hypoxanthine, ribose and arsenate (20). The arsenate is ligated in a prototypic phosphate binding site, through bidentate interactions to 2–3 arginyl residues along with additional side chain and backbone donor atoms, and is seen to be in an attacking position with respect to C1 of the ribose (Figure 2).
Figure 2.
Plasmodium falciparum PNP's active site with bound hypoxanthine (the free base, in cyan), ribose (cyan), and arsenate (purple) (PDP code: 3ENZ). The arsenate position corresponds to the nucleophilic attack by one of the oxygens (marked as O*) on the CI of ribose (O*-C1 distance is 2.3 Å). The arsenate ion is bound via hydrogen bonds (dashed lines) to three arginine side chains (Arg 88, Arg 27, and Arg 45' of the neighboring subunit). Other hydrogen donors to the bound arsenate include the hydroxyl group of Ser 91, and its backbone amide nitrogen.
Enzyme promiscuity with arsenate esters
In contrast to arsenate, the repertoire of arsenate analogues of phosphate esters that can be explored is limited because arsenate esters are unstable in aqueous solution. Nonetheless, several enzymes that act on phospho-monoesters have been examined via the in situ enzymatic synthesis of the arsenate-monoester, and found to readily accept the corresponding R-O-AsO3−2 esters (e.g. glucose-6-arsenate; see Ref. (7) and references therein). In addition, alkylarsonic acids (R-CH2-AsO3−2) are stable, and can be used, similarly to phosphonates (R-CH2-PO3−2), as non-hydrolizable analogs of phospho-monoesters (Ref. (7) and references therein). However, the replacement of oxygen by methylene generally renders these analogs inactive, since enzyme pockets appear to be highly specific towards oxygen atoms at the reaction's center. Thus, for example, the arsonomethyl analogue of ADP is a very poor substrate for enzymes for which ADP reacts via the β-phosphate group (7).
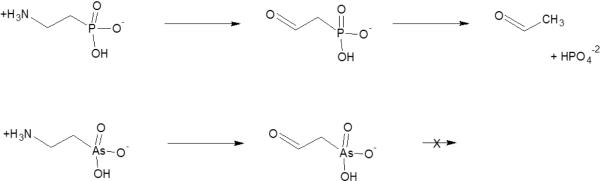
The above said, an interesting precedent for accepting an R-CH2-AsO3−2 instead of R-O-PO3−2 relates directly to the issue of whether nucleic acids may include arsenate. Dixon et al. examined the reverse reaction of E. coli RNA polymerase - i.e., the reaction of RNA and pyrophosphate to give dNTPs. Pyrophosphate was replaced with arsonomethyl-phosphonic acid (−2O3P-CH2-AsO3−2) and the reaction did occur, indicating that the polymerase accepted the CH2-AsO3−2 replacement of phosphate. However, because the corresponding arsenic analog of dNTPs rapidly hydrolyses, the isolated products were dNMPs (deoxynucleotide mono phosphates) (21). The polymerase therefore acted as a nuclease with these alternative substrates (7). As discussed above, such `futile cycles' underline all enzyme reactions with arsenate esters as the substrate instead of phosphate esters.
Promiscuity towards other arsenic-based substrates
Replacements of -O-P- with CH2-As- are not strictly isosteric, and are rarely of potential metabolic relevance. Nevertheless, the RNA polymerase and other cases, provide clear indications that enzymes tend to mistake arsenate for phosphate, not just in binding, but in some cases also for catalysis. The relatively few cases where C-P- bonds are relevant do provide interesting insights with respect to ion selectivity. In cases where C-PO3−2 (a phosphonate group) comprises part of the substrate, but the C-P bond is not transformed, the analogous arsonate substrate (C-AsO3−2) seems to be well accepted. For example, a transaminase, that convert 2-aminoethyl-phosphonate to the 2-oxoethyl-phosphonate while releasing ammonia, enables certain bacteria to utilize the former as a sole nitrogen source. The enzyme readily accepts 2-aminoethyl-arsonate as substrate, thus enabling growth on this compound as nitrogen source (Figure 3). However, the subsequent reaction that involves breakage of the C-P bond (converting 2-oxoethylphosphonate into acetaldehyde and phosphate by phosphonoacetaldehyde hydrolase) does not occur in the case of the arsonate analogue (22).
Figure 3.
The reactions catalyzed by P. araginusa transaminase and phosphonoacetaldehyde hydrolase on 2-aminoethyl-phosphate, and on its arsenic analogue 2-aminoethyl-phosphonate.
This lack of distinction between arsenate and phosphate is reflected not only in the promiscuity of this enzyme, but also in the growth of bacteria on 2-aminoethylarsonate instead of 2-aminoethylphosphate (22). The latter indicates the promiscuity of transporters and of regulatory proteins (transcription factors, etc'). Indeed, it is generally assumed that arsenate penetrates cells via phosphate transporters that exhibit similar affinities to both ions, although certain mammalian transporters have been shown to exhibit 10–40-fold lower affinities to arsenate than for phosphate (23).
Another enzyme that transforms a C-P bond - phosphoenolpyruvate mutase, has been examined with the analogous C-As substrate. The KM value was similar with this alternative substrate, suggesting that recognition was not adversely affected. However, kcat for the synthesis of the 3-arsonopyruvate analogue was reduced by 6000-fold (24).
The above examples indicate that in contrast to the oxygens of arsenate and phosphate, which are poorly distinguished when carbon-phosphorus bonds are directly involved in the enzymatic reaction, enzymes can readily distinguish between phosphorus and arsenic.
Structural aspects of arsenate-phosphate promiscuity
What about the structural protein features that underline arsenate-phosphate binding, and the above discussed lack of discrimination? There are very few examples where arsenate has been specifically added to form enzyme complexes for structural studies. However, cacodylate (dimethylarsenic acid), a common buffer that has been incorporated into many crystallization screening kits, is seen in numerous crystal structures within specific binding sites. Examining the binding interactions seen with cacodylate may provide some insights into the nature of arsenate and other oxyanion recognition. Due to its two C-As bonds, cacodylate is not a precise mimic of natural phosphate containing compounds, yet the fact that it can selectively bind to enzyme binding sites further supports the hypothesis that proteins hardly discriminate between the oxides of these closely related elements.
Our analysis of nearly 90 structures in the PDB in which the nature of the interactions to cacodylate have been described, indicates that almost half are monodentate interactions, either to a wide range of amino acid side chains (with histidine, arginine and carboxylate groups being most prominent), or to a bound metal ion (mostly Zn2+). The next most prominent examples are bidentate interactions with a metal ion and an amino acid side chain, followed by bidentate interactions to different amino acid side chain or backbone amide groups. It is likely that the high concentrations of cacodylate used (typically 100 mM) leads to non-specific binding to sites with only one or two well defined protein ligands. However, there are several examples of potentially more selective oxyanion binding sites in which cacodylate is interacting through its oxygen atoms to three or more amino acid side chains or backbone amide groups.
An even more common oxyanion used in protein crystallization is sulfate, that is present in high concentrations as part of the use of ammonium sulfate for ionic precipitation of proteins. Over 8000 structures in the PDB list sulfate as a bound ligand, clearly showing the high likelihood of non-specific binding of oxyanions to proteins sites that do not normally bind oxyanions, or to sites that normally bind other oxyanions such as phosphate.
Arsenate - a highly useful mechanistic probe
In addition to the discussion of alternative life chemistries and oxyanion promiscuity, we also wish to highlight the utility of arsenate as a structural and mechanistic probe for phosphate-utilizing enzymes. The irreversibility of the arsenate reaction catalyzed by PNP (purine nucleoside phosphorylase) was used to help decipher the catalytic mechanism and the microscopic rates for various steps of the enzymatic reaction (17). In an earlier example of the application of this approach, Fitting and Doudoroff used arsenate to distinguish between the mechanism of action of sucrose phosphorylase (which acts via a glucosyl-enzyme intermediate) and maltose phosphorylase that acts via direct attack of phosphate on maltose (16). Similar mechanistic approaches are still being used with glycosyltransferases that use phospho-sugars as substrates (25, 26). Arsenate-based structures can provide a glimpse of the nature of reaction intermediates, as demonstrated with PNP (20) (Figure 3), and can also shed some light on the ion binding mode in phosphate-utilizing enzymes such as ASADH (18) (Figure 2).
Also of interest as potential mechanistic probes are alkylaronates, not only as non-hydrolizable analogues of phospho-esters, but also as substrate analogues for enzymes acting on phosphonate substrates. The latter are becoming increasingly interesting, as phosphonates have been shown to comprise a key component of the dissolved marine organic phosphorus pool (27). For example, phosphonates are a major component of total cellular phosphorus in the marine cyanobacterium Trichodesmium erythraeum (28). The enzymology of marine phosphonates is still substantially unexplored, but alkylaronates are likely to be useful as inhibitors in the isolation and characterization of the relevant enzymes involved in phosphonate metabolism.
Ion selectivity and promiscuity
Judging by the above examples, and others that have not been discussed here, it appears that enzymes, and others proteins, show very low phosphate-arsenate binding selectivity. As is the case with other protein traits, this lack of discrimination may can be due to two unrelated reasons: (i) Physicochemical constrains may limit the ability of proteins to differentiate between these two highly similar ions with significant (orders-of-magnitude) differences in affinity; (ii) The absence of evolutionary pressure to prevent arsenate competition on phosphate sites results in little discrimination when arsenate is introduced.
How physicochemically feasible is a strict distinction between arsenate and phosphate? And, what, in general, dictates the specificity of binding of small ions? Proteins can easily distinguish between small charged atoms such as H+, Na+ and K+, or F−, Cl− and Br−, primarily by virtue of considerable differences in size, charge density, and hydration energy. For example, ion channels show high specificity for the closely related Na+ and K+. However, these ions differ considerably in their radii (>20%), and dramatically in their “hardness”. The latter is reflected in 106-fold differences of the solubility of Na+ and K+ in certain hydrophobic organic molecules (15). Relatively high selectivity is also observed with larger metal ions, for example, with metallo-enzymes that may accept a range of transition metals, yet with affinities that differ by orders of magnitude, and different catalytic activities (for examples, Refs. (29, 30)). Here, ligand donor atom type, and coordination geometry preferences, offer additional selectivity criteria.
For larger oxyanions such as arsenate and phosphate size differences are relatively small (the thermochemical radius for phosphate is 2.38 Å, arsenate 2.48 Å, and sulfate 2.30 Å) (15). Binding selectivity is therefore unlikely to be based on size differences, and differences in geometry and charge distribution are more likely to be important (14). For example, at neutral pH, carbonate, which is abundant in living media, has the same net charge as phosphate (−2), and essentially the same partial charge on the charged oxygens (−0.964 versus −0.952 for phosphate). However, is size is much smaller (1.85 Å), and in particular, its geometry is planar as opposed to tetrahedral for phosphate. A study of the oxyanion interactions with ASADH indicates that ions that are tetrahedral and exhibit high negative charge densities on their oxygens are either good substrates, or effective inhibitors, for the enzyme. For example, sulfate is neither a substrate nor an inhibitor - while it is tetrahedral, the partial negative charge on its oxygens (− 0.861) is significantly lower than that of phosphate (−0.952). The only exception observed for ASADH is periodate, which is tetrahedral, but exhibits very low negative charge (−0.719). Interestingly, periodate binds is the most tightly bound ion, but it disrupts an important hydrogen-bonding with Glu243, thus rendering the enzyme inactive (Figure 2) (14, 18). It appears therefore, that the relatively small differences in size between phosphate and arsenate (~4%), and in the partial negative charges of the oxygens (−0.952 for phosphate, −0.892 for arsenate), suggest that a strict distinction between arsenate and phosphate might be rather challenging.
Can very high oxyanion binding selectivity be achieved, and, if so, then which structural features may mediate it? The recently described ultra-high resolution (0.88–0.98 Å) structure of a phosphate binding protein from Pseudomonas fluorescens affords an interesting insight by virtue of the electronic density that is observed for certain hydrogen atoms. This structure suggests that oxyanion discrimination may involve some unique interactions, such as low-barrier hydrogen bonds (LBHBs) to selected donor atoms. The bound HPO4−2 ion in this protein seems to accept as many as 11 hydrogen atoms in binding interactions. But it only donates a single hydrogen atom, to an aspartate side chain, whilst forming a LBHB to this functional group. This LBHB is likely to mediate the dibasic phosphate binding specificity, allowing the protein to discriminate phosphate from sulfate by a factor of 105 (31). Thus, beyond the type and geometry of the donor atoms in an oxyanion binding site, the specific polarity of the hydrogen-bonding pattern, and the incorporation of special binding interactions, may also provide a possible basis for ion discrimination.
As for evolutionary pressures, arsenic and arsenate are toxic for most organisms. The toxicity of arsenic(III) oxide, the favorite murder weapon of Italian Renaissance aristocrats and murder mystery novels, relates to the As(III) state. But other toxic effects may stem from its oxidation to arsenate, with the latter occupying essential phosphate sites. This may result in inhibiting essential protein functions, or yielding enzymatic products that are rapidly hydrolyzed. Arsenate itself is also toxic to most organisms. By virtue of its similarity to phosphate, it uncouples organismal metabolism by impairing ATP synthesis (32). Since mechanisms for arsenic and arsenate detoxification have been identified in various organisms, evolutionary pressures to minimize arsenate binding to phosphate sites may exist. However, the relatively few enzymes that have been analyzed with arsenate come from a very narrow range of organisms, and these enzymes are not likely to have been under selection for high phosphate-arsenate selectivity. It appears, therefore, that the lack of discrimination in these enzymes stems from both factors - namely from the physicochemical similarity of these ions and from the absence of evolutionary selection pressure.
The evolution of arsenate based biochemistry
Promiscuity generally provides the starting points for the evolution of new protein functions (33). Arsenate-phosphate cross-reactivities may therefore serve as the basis for evolutionary transitions from a phosphate-based organism into an arsenate-based one, or vice-versa. On the other hand, the extreme sensitivity of arsenate esters to hydrolysis suggests that many metabolites and macromolecules must remain phosphate based. Thus, in an organism that does utilize arsenate, enzymes which need to work with phosphate-based substrates and products must evolve to incorporate high phosphate selectivity. Thus, a bacterium that dwells in environments containing high concentrations of arsenate and low concentrations of phosphate faces a paramount challenge, even if arsenate does not get incorporated into its metabolites and/or macromolecules. Selective transport to maximize phosphate uptake and minimize entry of arsenate, and/or active transport to export arsenate, and storage in specialized compartments, including insoluble precipitates, are essential for an organism that dwells in an arsenic rich environment to protect itself from toxic effects. But these mechanisms alone cannot eliminate arsenate altogether. Numerous proteins, including transporters and enzymes, must evolve high selectivity towards phosphate. The DNA repair system in such organisms might also be unique in having to deal with strand breakages that may occur due to the accidental (or non-accidental) incorporation of arsenate instead of phosphate.
Alternatively, we need not exclude the possibility that arsenic-based life forms do not involve a simple replacement of phsophate-esters by arsenate-esters. Alternative chemistries, such as stable C-As bonds (see the methylene-arsonates discussed above), could replace O-P bonds, or even a completely different chemistry that has no precedents in known life forms. Needless to say, however, these chemistries present their own difficulties (how, for example, can “DNA” polymerization and editing readily occur when C-As bonds must be made and broken).
Concluding remarks
Whatever the ultimate case may be regarding the role of arsenic, the in-depth study of the Halomonadaceae strain described by Wolfe-Simon et al. should yield intriguing insights, as well as the motivation to study other bacterial strains that resist extremely high arsenate levels (e.g. see Ref (34)). The notion of life without phosphate might be proven wrong, but life with arsenate presents extreme challenges, as well as intriguing research opportunities. Specifically, as discussed here, the distinction between arsenate and phosphate ions represents a challenging problem in molecular recognition. The study of the structural, functional, and evolutionary aspects of arsenate-phosphate discrimination, and of the mechanisms for the discrimination of similar ions, may therefore yield key insights into the possibilities of alternative life chemistries.
Acknowledgments
D.S.T. is thankful to Mikael Elias for inspirational discussions, and to the Israel Science Foundation, and the Nella and Leon Benoziyo Professorship for financial support. R.E.V. is supported by a grant from the National Institutes of Health (AI077720).
References
- 1.Wolfe-Simon F, Blum JS, Kulp TR, Gordon GW, Hoeft SE, Pett-Ridge J, Stolz JF, Webb SM, Weber PK, Davies PC, Anbar AD, Oremland RS. A Bacterium That Can Grow by Using Arsenic Instead of Phosphorus. Science. 2010 doi: 10.1126/science.1197258. [DOI] [PubMed] [Google Scholar]
- 2.Woolfson A. Life without genes: The histroy and future of genomes. HarperCollins; 2000. [Google Scholar]
- 3.Wolfe-Simon F, Davies PC, Anbar AD. Did nature also use arsenic? International Journal of Astrobiology. 2009;8:69–74. [Google Scholar]
- 4.Afriat L, Roodveldt C, Manco G, Tawfik DS. The latent promiscuity of newly identified microbial lactonases is linked to a recently diverged phosphotriesterase. Biochemistry. 2006;45:13677–13686. doi: 10.1021/bi061268r. [DOI] [PubMed] [Google Scholar]
- 5.Yang K, Metcalf WW. A new activity for an old enzyme: Escherichia coli bacterial alkaline phosphatase is a phosphite-dependent hydrogenase. Proc Natl Acad Sci U S A. 2004;101:7919–7924. doi: 10.1073/pnas.0400664101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Westheimer FH. Why nature chose phosphates. Science. 1987;235:1173–1178. doi: 10.1126/science.2434996. [DOI] [PubMed] [Google Scholar]
- 7.Dixon HBF. The biochemical action of arsonic acids especially as phosphate analogues. Advances in Inorganic Chemistry. 1997;44:191–227. [Google Scholar]
- 8.Katsnelson A. Microbe gets toxic response. Nature. 2010;468:741. doi: 10.1038/468741a. [DOI] [PubMed] [Google Scholar]
- 9.Drahl H. Arsenic bacteria breed backlash. Chemical & Engineerin News. 2010 Dec. 13:7. [Google Scholar]
- 10.Baer CD, Edwards JO, Rieger PH. Kinetics of the hydrolysis of arsenate(V) triesters. Inorganic Chemistry. 1981;20:905–907. [Google Scholar]
- 11.Slade D, Lindner AB, Paul G, Radman M. Recombination and replication in DNA repair of heavily irradiated Deinococcus radiodurans. Cell. 2009;136:1044–1055. doi: 10.1016/j.cell.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 12.Delnomdedieu M, Basti MM, Otvos JD, Thomas DJ. Reduction and binding of arsenate and dimethylarsinate by glutathione: a magnetic resonance study. Chem Biol Interact. 1994;90:139–155. doi: 10.1016/0009-2797(94)90099-x. [DOI] [PubMed] [Google Scholar]
- 13.Perutz MF. I Wish I'd Made You Angry Earlier: Essays on Science, Scientists, and Humanity. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; new York: 2003. [Google Scholar]
- 14.Kish MM, Viola RE. Oxyanion Specificity of Aspartate-beta-semialdehyde Dehydrogenase. Inorg Chem. 1999;38:818–820. doi: 10.1021/ic981082j. [DOI] [PubMed] [Google Scholar]
- 15.Fraústo da Silva JJR, Williams RJP. The Biological Chemistry of the Elements: The Inorganic Chemistry of Life. 1st ed. Oxford University Press; Oxford: 1997. [Google Scholar]
- 16.Fitting C, Doudoroff M. Phosphorolysis of maltose by enzyme preparations from Neisseria meningitidis. J Biol Chem. 1952;199:153–163. [PubMed] [Google Scholar]
- 17.Kline PC, Schramm VL. Purine nucleoside phosphorylase. Catalytic mechanism and transition-state analysis of the arsenolysis reaction. Biochemistry. 1993;32:13212–13219. doi: 10.1021/bi00211a033. [DOI] [PubMed] [Google Scholar]
- 18.Faehnle CR, Blanco J, Viola RE. Structural basis for discrimination between oxyanion substrates or inhibitors in aspartate-beta-semialdehyde dehydrogenase. Acta Crystallogr D Biol Crystallogr. 2004;60:2320–2324. doi: 10.1107/S0907444904026411. [DOI] [PubMed] [Google Scholar]
- 19.Park RE, Jr., Agrawal RP. Purine nucleoside phosphorylase. In: Boyer PD, editor. The Enzymes. Academic Press; New York: 1972. pp. 483–514. [Google Scholar]
- 20.Chaikuad A, Brady RL. Conservation of structure and activity in Plasmodium purine nucleoside phosphorylases. BMC Struct Biol. 2009;9:42. doi: 10.1186/1472-6807-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rozovskaya TA, Rechinsky VO, Bibilashvili RS, Karpeisky M, Tarusova NB, Khomutov RM, Dixon HB. The mechanism of pyrophosphorolysis of RNA by RNA polymerase. Endowment of RNA polymerase with artificial exonuclease activity. Biochem J. 1984;224:645–650. doi: 10.1042/bj2240645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lacoste AM, Dumora C, Ali BR, Neuzil E, Dixon HB. Utilization of 2-aminoethylarsonic acid in Pseudomonas aeruginosa. J Gen Microbiol. 1992;138:1283–1287. doi: 10.1099/00221287-138-6-1283. [DOI] [PubMed] [Google Scholar]
- 23.Villa-Bellosta R, Sorribas V. Arsenate transport by sodium/phosphate cotransporter type IIb. Toxicol Appl Pharmacol. 2010;247:36–40. doi: 10.1016/j.taap.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 24.Chawla S, Mutenda EK, Dixon HB, Freeman S, Smith AW. Synthesis of 3-arsonopyruvate and its interaction with phosphoenolpyruvate mutase. Biochem J. 1995;308(Pt 3):931–935. doi: 10.1042/bj3080931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eis C, Nidetzky B. Characterization of trehalose phosphorylase from Schizophyllum commune. Biochem J. 1999;341(Pt 2):385–393. [PMC free article] [PubMed] [Google Scholar]
- 26.Elbein AD, Pastuszak I, Tackett AJ, Wilson T, Pan YT. Last step in the conversion of trehalose to glycogen: a mycobacterial enzyme that transfers maltose from maltose 1-phosphate to glycogen. J Biol Chem. 2010;285:9803–9812. doi: 10.1074/jbc.M109.033944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White AE. New insights into bacterial acquisition of phosphorus in the surface ocean. Proc Natl Acad Sci U S A. 2009;106:21013–21014. doi: 10.1073/pnas.0912475107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dyhrman ST, Chappell PD, Haley ST, Moffett JW, Orchard ED, Waterbury JB, Webb EA. Phosphonate utilization by the globally important marine diazotroph Trichodesmium. Nature. 2006;439:68–71. doi: 10.1038/nature04203. [DOI] [PubMed] [Google Scholar]
- 29.Cox JD, Hunt JA, Compher KM, Fierke CA, Christianson DW. Structural influence of hydrophobic core residues on metal binding and specificity in carbonic anhydrase II. Biochemistry. 2000;39:13687–13694. doi: 10.1021/bi001649j. [DOI] [PubMed] [Google Scholar]
- 30.Shim H, Raushel FM. Self-assembly of the binuclear metal center of phosphotriesterase. Biochemistry. 2000;39:7357–7364. doi: 10.1021/bi000291o. [DOI] [PubMed] [Google Scholar]
- 31.Liebschner D, Elias M, Moniot S, Fournier B, Scott K, Jelsch C, Guillot B, Lecomte C, Chabriere E. Elucidation of the phosphate binding mode of DING proteins revealed by subangstrom X-ray crystallography. J Am Chem Soc. 2009;131:7879–7886. doi: 10.1021/ja901900y. [DOI] [PubMed] [Google Scholar]
- 32.Ventura-Lima J, Bogo MR, Monserrat JM. Arsenic toxicity in mammals and aquatic animals: A comparative biochemical approach. Ecotoxicol Environ Saf. 2010 doi: 10.1016/j.ecoenv.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 33.Khersonsky O, Tawfik DS. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu Rev Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 34.Huang A, Teplitski M, Rathinasabapathi B, Ma L. Characterization of arsenic-resistant bacteria from the rhizosphere of arsenic hyperaccumulator Pteris vittata. Can J Microbiol. 2010;56:236–246. doi: 10.1139/w10-005. [DOI] [PubMed] [Google Scholar]