Abstract
AIM: To investigate three isoforms of survivin in colorectal adenocarcinomas.
METHODS: We used the LightCycler Technology (Roche), along with a common forward primer and reverse primers specific for the splice variants and two common hybridization probes labeled with fluorescein and LightCycler-Red fluorophore (LC-Red 640). Real time quantitative polymerase chain reaction (PCR) was performed on cDNAs from 52 tumor specimens from colorectal cancer patients and 10 unrelated normal colorectal tissues. In the patients group, carcinoembryonic antigen (CEA) and CA19-9 tumor markers were also measured immunochemically.
RESULTS: Wild type survivin mRNA isoform was expressed in 48% of the 52 tumor samples, survivin-2b in 38% and survivin-ΔΕx3 in 29%, while no expression was found in normal tissues. The mRNA expression of wild type survivin presented a significant correlation with the expression of the ratio of survivin-2b, survivin-ΔΕx3, survivin-2b/wild type survivin and survivin-ΔΕx3/wild type survivin (P < 0.001). The mRNA expression of wild-survivin and survivin-ΔΕx3 was related with tumor size and invasion (P = 0.006 and P < 0.005, respectively). A significant difference was found between survivin-2b and morphologic cancer type. Also, the ratio of survivin-ΔEx3/wild-survivin was significantly associated with prognosis. No association was observed between the three isoforms and grade, metastasis, Dukes stage and gender. The three isoforms were not correlated with CEA and CA19-9.
CONCLUSION: Survivin isoforms may play a role in cell apoptosis and their quantification could provide information about clinical management of patients suffering from colorectal cancer.
Keywords: Survivin, mRNA isoforms, Apoptosis gene, Colorectal adenocarcinomas, Real time quantitative polymerase chain reaction, Lightcycler
INTRODUCTION
Apoptosis is a tightly controlled procedure of cellular death, which is crucial for tissue homeostasis[1]. Inhibition of apoptosis results in tumorigenesis by cell survival allowing the accumulation of genetic mutations that promote transformation of normal tissues[2]. Among key regulators of apoptosis are proteins of the bcl-2 family and the inhibitors of apoptosis (IAP) family of proteins[3-5].
Survivin was originally identified by structural homology to IAPs in human B-cell lymphomas. It is a multifunctional protein implicated in the control of cell proliferation, inhibition of apoptosis and the promotion of angiogenesis[6,7]. Survivin, as an inhibitor of apoptosis directly inhibits caspase-3 and -7 activity and regulates the cell cycle in the G2/M phase[8].
Survivin is expressed during embryonic and fetal development, is down regulated in normal adult tissues and is overexpressed in a variety of human cancers[9,10]. A strong association between expression of survivin mRNA and aggressive tumor behavior has been found in many types of cancer such as colorectal, breast, non–small lung cancer, gliomas and B-cell lymphomas[11-15].
In addition to the wild type survivin full length transcript, five other splice variants have been identified: survivin-ΔΕx3 that arises from the removal of exon 3, survivin-2b[16] that originates from the inclusion of intron 2, survivin-3b that stems from the inclusion of intron 3[17] survivin-2a made from the insertions of exon 1 and 2 at the 5’ end of intron 2[18], and the recently described survivin-2b + 32 which combines intronic sequence 2b and an insertion of 32 additional nucleotides from intron 2[19] (Table 1).
Table 1.
cDNA structure of survivin splice variants
| Transcript variants | Exons or segments |
| Wild type survivin | 1, 2, 3, 4 |
| Survivin-2b | 1, 2, 2b, 3, 4 |
| SurvivinΔΕx3 | 1, 2, 4 |
| Survivin-2a | 1, 2 |
| Survivin-3b | 1, 2, 3, 3b, 4 |
| Survivin-2b + 32 | 1, 2, 2b + 32nt, 3, 4 |
b: Sequence originated from intron.
Wild type survivin and survivin-ΔΕx3 have been shown to act as inhibitors of apoptosis while survivin-2b presents pro-apoptotic functions by dimerizing with wild type survivin and reducing the anti-apoptotic effects of wild type survivin[16,20,21]. The ratio survivin-2b/wild-survivin is higher in tumor samples compared to normal tissues. Survivin-2a is also expressed at high levels in malignant cells and attenuates the antiapoptotic effect of wild-survivin[22]. Inhibition of apoptosis by survivin predicts a poor prognosis and a shorter survival in patients suffering from carcinomas[23-28].
Till now, there has only been one other study in the literature investigating the expression of survivin’s isoforms in colorectal cancer using quantitative real time-PCR[24]. In this study, we obtained quantitative data for the distribution of the most described transcripts, which are wild type survivin, survivin-ΔΕx3 and survivin-2b in colorectal adenocarcinomas by using the accurate and sensitive real time-qPCR in the Lightcycler platform. Then, we investigated their correlation with clinicopathological characteristics of colorectal cancer, and evaluated their prognostic significance.
MATERIALS AND METHODS
Patients
Fifty-two tissues from patients with colorectal cancer and another 10 unrelated normal samples were obtained from the 4th Department of Surgery, Attikon University General Hospital. Part of the resected specimens at surgery was immediately stored in RNA Later (Ambion, USA) for at least 2 d at 4°C and then stored at -80°C until total RNA extraction. Our sample population included 21 males and 31 females [mean age: 63 years, standard deviation (SD) ± S13, range: 36-93 years]. The histological type and grade were reviewed and classified according to the World Health Organization classification criteria, and the disease stage was determined according to the Dukes staging system. The clinical characteristics of 52 patients with colorectal cancer are shown in Table 2. No patients underwent chemotherapy or radiation therapy before surgery.
Table 2.
Clinical characteristics of 52 patients with colorectal cancer n (%)
| Variables | Values |
| Gender | |
| Men | 21 (40.4) |
| Women | 31 (59.6) |
| Tumor size | |
| ≤ 5 cm | 32 (65.3) |
| > 5 cm | 17 (34.7) |
| Grade | |
| I | 3 (6.3) |
| II | 36 (75) |
| III | 7 (14.6) |
| IV | 2 (4.2) |
| Dukes stage | |
| A | 8 (16.3) |
| B | 11 (22.4) |
| C | 22 (44.9) |
| D | 8 (16.3) |
| Morphologic cancer type | |
| Polypoid (ecblastetic) | 28 (57.1) |
| Ulcerous | 21 (42.9) |
Total RNA isolation
Total RNA was extracted by the Illustra RNAspin Mini RNA isolation kit (GE Healthcare, USA) according to the manufacturer’s instructions. DNA contamination was eliminated by the use of RNAse-free DNAseI(GE Healthcare). The extracted total RNA was dissolved in free diethylpyrocarbonate (DEPC) treated water and stored at -80°C until further manipulations. RNA concentration was determined by the Quant-iT RNA Assay kit in Qubit fluorometer (Invitrogen, USA).
Reverse transcription- quantitative real-time PCR analysis
For complementary DNA (cDNA) synthesis, the Transcriptor First strand cDNA synthesis kit (Roche Applied Science, USA) was used according to the manufacturer’s instructions. In each reaction, 3 μg of quantitated total RNA was included. The cDNAs were stored at -20°C until real time quantitative PCR was performed in the LightCycler 1.5 platform (Roche Applied Science). One μL of cDNA mixture was subjected to amplification in 10 μL total volume reaction mixtures in glass capillaries. Real time PCR was performed with the FastStart DNA Master Hybprobe kit (Roche Applied Science) and cycling conditions were: initial denaturation at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 10 s, annealing for 10 s at 62°C, extension at 72°C for 10 s, respectively. Standard curves for each of the three transcripts were generated from purified and quantified amplicons in serial dilutions from total RNA extracted from MCF-7 cells, according to previous experience[29]. Primer pairs and hybridization probes for the three transcripts of survivin, survivin-2B, survivin-ΔEx3 were described previously[24] and are shown in Table 3.
Table 3.
Sequences of primers and hybridization probes
| Name | Oligonucleotide sequence, 5’-3’ | |
| Forward primer | F1 | CCACCGCATCTCTACATTCA |
| Reverse primers | WT | TATGTTCCTCTATGGGGTCG |
| Sur2B | AGTGCTGGTATTACAGGCGT | |
| SurΔΕx3 | TTTCCTTTGCATGGGGTC | |
| Hybridization probes | CAAGTCTGGCTCGTTCTCAG TGGG-FITC1 | |
| LCRed640-CAGTGGATGAAG CCAGCCTCG-Ph2 |
: 3’-end-labeled with fluorescein isothiocyanate (FITC);
: 5’-end-labeled with LC Red 640; phosphorylated on the 3’ end to avoid extension of the probe.
Real time PCR products were additionally checked for their proper size and purity by electrophoresis on 2% agarose gels containing ethidium bromide and visualized under UV transillumination. Briefly 10 μL of real-time PCR products were run along with 10 μL of a 100 bp ladder MW marker (New England Biolabs, USA).
Normalization
Real time PCR is widely used to quantify biologically relevant changes in mRNA levels but a number of problems exist and are associated with its use due to the inherent variability of RNA, variability of extraction protocols, different reverse transcription and PCR efficiencies. It is important that an accurate method of normalization is chosen to control for these errors. Unfortunately, normalization remains one of real time PCR’s most difficult problems[30]. In our protocol, we ensured that our tissues had similar size (approximately 50 mg) and performed normalization against the same amount of total RNA (3 μg), since when dealing with in vivo samples it is not possible to predict which of the housekeeping genes is stable and appropriate[31,32].
Determination of immunochemical parameters
The cancer antigens carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA 19-9) were measured using Elecsys 2010 (Roche, USA) with electro-chemiluminescent immunoassays. The reproducibility of controls 1 and 2 (Roche) were for CEA, CV1 = 3.6% and CV2 = 3.0% and for CA19-9, CV1 = 4.8% and CV2 = 3.8%. The analytical sensitivity for CEA was 0.25 ng/mL and for CA19-9 < 0.85 U/mL. The cut-off values for CEA and CA19-9 were 5.0 ng/mL and 37 U/mL, respectively.
Statistical analysis
Statistical analysis of the data was performed using SPSS® version 17 for Windows statistical software package. Initially data were assessed through simple cross-tabulations and by using χ2 test for categorical variables, t-test for normally distributed variables and Mann-Whitney U test for not normally distributed variables. Normality hypothesis was tested by Kolmogorov-Smirnov test, measures of asymmetry and Shapiro-Wilk test. In order to compare means of cases amongst different subgroups, one-way ANOVA test for normally distributed variables or Kruskal-Wallis test for not normally distributed variables were conducted. Post hoc pairwise comparisons were performed using the Bonferroni method. The Pearson or Spearman correlation coefficients (r) were used as measurements of correlation for continuous normally or not normally distributed variables respectively. In order to estimate survival functions, Kaplan-Meier curves were generated and subsequently compared using the log-rank test, the Breslow test or the Tarone-Ware test. For all tests performed, a two-sided P value of less than 0.05 was considered as significant.
RESULTS
Εxpression of mRNA survivin transcripts variants in cancer and normal tissues
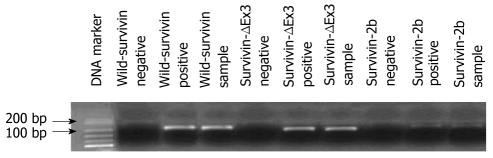
The specificity of amplification products obtained by real time qPCR was confirmed by agarose gel electrophoresis, which revealed distinct bands for all PCR products. PCR for full length survivin mRNA produced a band of 185 bp, while those of survivin-2b and survivin-ΔΕx3 were 214 and 184 bp, respectively (Figure 1). Among the 52 tumor samples, wild type survivin expression was detected in 25 samples (48%), survivin-2b expression was detected in 20 samples (38%) and survivin-ΔΕx3 was expressed in 15 samples (29%). Eighteen samples were positive for both the expression of wild type survivin and survivin-2b (34.6%), 14 samples were positive for both the expression of wild-survivin and survivin-ΔΕx3 (26.9%) and 14 samples were positive for both the expression of survivin-2b and survivin-ΔΕx3 (26.9%). All 3 survivin isoforms were found in 13 samples (25%). None of the 3 variants of survivin mRNA were expressed in the 10 unrelated normal samples, indicating a significant difference in survivin expression between cancerous and healthy tissues (P < 0.001).
Figure 1.
Electrophoresis of the survivin transcripts cDNA (wild survivin 185 bp, survivin-ΔΕx3 184 bp, and survivin-2b 214 bp).
Relationship between the levels of expressed survivin transcripts and clinicopathological parameters
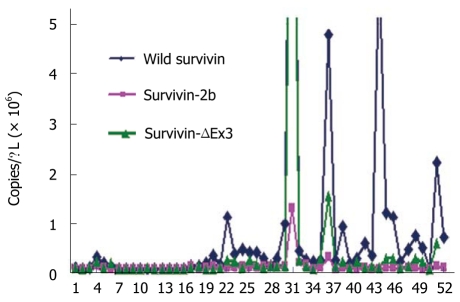
Wild type survivin was significantly more expressed than the other two splice variants in the 52 samples, as shown in Figure 2. Quantitative data from tissue mRNA and serum tumor markers from the 52 patients are shown in Table 4. The 3 survivin isoforms were not correlated with CEA and CA19-9 (Table 5). In general, no association was found between the 3 isoforms and tumor size, survival, Dukes stage, grade and gender, although wild survivin was almost present in tumors more than 5 cm in diameter (P = 0.088), and the ratio survivin-ΔΕx3/wild-survivin was almost significantly higher in tumors less than 5 cm (P = 0.097) and in advanced Dukes stage (P = 0.097) (Table 6). The mRNA expression of wild type survivin presented a significant correlation with the expression of survivin-2b and survivin-ΔΕx3 (P < 0.001) and the ratio of survivin-2b/wild-survivin was strongly related to the ratio of survivin-ΔΕx3/wild-survivin (P < 0.001). Wild type survivin and survivin-ΔΕx3 expression levels presented an association with T (P = 0.006 and P = 0.041 respectively, Table 5). Mean survivin-2b expression levels were significantly higher in ulcerous carcinomas than in polypoid (ecblastetic) ones (P = 0.030, Table 6).
Figure 2.
Expression of mRNA wild type survivin, survivin–2b and survivin ΔΕx3 isoforms in cancerous tissues (n = 52).
Table 4.
Quantitative measurements obtained from tissue mRNA and serum tumor markers from 52 patients with colorectal cancer (mean ± SD)
| Variables | Values |
| Wild-survivin (copies/μL) | 842.5 ± 2380.3 |
| Survivin-2b (copies/μL) | 58.1 ± 192.3 |
| Survivin-ΔΕx3 (copies/μL) | 393.9 ± 1988.8 |
| Survivin-2b/wild-survivin (copies/μL) | 0.12 ± 0.21 |
| Survivin-ΔΕx3/wild-survivin (copies/μL) | 0.36 ± 0.66 |
| CEA (μg/L) | 191.59 ± 934.62 |
| CA 19-9 (μg/L) | 892.48 ± 2991.78 |
CEA: Carcinoembryonic antigen; CA 19-9: Carbohydrate antigen 19-9.
Table 5.
Spearman correlation coefficients of laboratory variables in patients with colorectal cancer (n = 52)
| Variables | Sur2b | SurΔEx3 | Sur2b/wildsur | SurΔEx3/wildsur | CEA | CA19-9 | Age |
| Wildsur | 0.694b | 0.799b | 0.408b | 0.485b | 0.136 | 0.102 | 0.051 |
| Sur2b | 0.623b | 0.527b | 0.471b | 0.238 | 0.212 | 0.165 | |
| SurΔΕx3 | 0.409b | 0.651b | 0.208 | 0.063 | 0.229 | ||
| Sur2b/wildsur | 0.613b | 0.021 | 0.203 | 0.0234 | |||
| SurΔΕx3/wildsur | -0.054 | 0.084 | 0.320a | ||||
| CEA | 0.522b | 0.395a | |||||
| CA 19-9 | 0.247 |
P < 0.05,
P < 0.001. CEA: Carcinoembryonic antigen; CA 19-9: Carbohydrate antigen 19-9.
Table 6.
Association of mRNA expression survivin splice variants with clinicopathological characteristics in 52 patients suffering from colorectal cancer (mean ± SD)
| Clinical characteristics | n | Wild-sur (copies/μL) | P value wild-sur | Sur-2b (copies/μL) | P value sur-2b | Sur-ΔEx3 (copies/μL) | P value sur-ΔEx3 | Sur-2b/wildsur (copies/μL) | P value sur-2b/wildsur | Sur-ΔEx3/wildsur (copies/μL) | P value sur-ΔEx3/wildsur |
| Gender | 0.978 | 0.685 | 0.631 | 0.570 | 0.387 | ||||||
| Male | 20 | 1216.1 ± 3438.6 | 98.0 ± 299.7 | 834.8 ± 3085.6 | 0.08 ± 0.09 | 0.47 ± 0.39 | |||||
| Female | 32 | 589.5 ± 1259.1 | 31.1 ± 33.2 | 85.3 ± 117.1 | 0.17 ± 0.24 | 0.36 ± 0,41 | |||||
| Tumor size | 0.088 | 0.438 | 0.869 | 0.544 | 0.097 | ||||||
| ≤ 5 cm | 32 | 308.4 ± 491.3 | 28.6 ± 34.6 | 96.2 ± 116.1 | 0.17 ± 0.24 | 0.48 ± 0.41 | |||||
| > 5 cm | 17 | 1949.4 ± 3956.2 | 119.3 ± 330.9 | 1055.8 ± 3531.9 | 0.09 ± 0.09 | 0.23 ± 0.24 | |||||
| Dukes stage | 0.469 | 0.737 | 0.544 | 0.351 | 0.097 | ||||||
| A | 8 | 212.4 ± 403.4 | 27.7 ± 27.7 | 51.7 ± 66.1 | 0.37 ± 0.40 | 0.44 ± 0.35 | |||||
| B | 11 | 476.9 ± 743.4 | 30.8 ± 45.8 | 137.7 ± 185.7 | 0.08 ± 0.12 | 0.37 ± 0.37 | |||||
| C | 22 | 1556.8 ± 3533.1 | 95.4 ± 292.5 | 782.6 ± 3019.7 | 0.09 ± 0.10 | 0.30 ± 0.39 | |||||
| D | 8 | 226.7 ± 146.7 | 35.6 ± 32.9 | 120.7 ± 66.5 | 0.15 ± 0.15 | 0.63 ± 0.35 | |||||
| T | 0.006 | 0.125 | 0.041 | 0.103 | 0.492 | ||||||
| 0-2 | 8 | 212.4 ± 403.4 | 27.7 ± 27.7 | 51.7 ± 66.1 | 0.37 ± 0.40 | 0.44 ± 0.35 | |||||
| 3 | 29 | 340.0 ± 521.1 | 26.5 ± 35.4 | 92.9 ± 127.2 | 0.10 ± 0.13 | 0.39 ± 0.37 | |||||
| 4 | 12 | 2620.8 ± 4579.4 | 162.9 ± 389.8 | 1413.2 ± 4056.1 | 0.10 ± 0.08 | 0.40 ± 0.44 | |||||
| Histologic type | 0.584 | 0.030 | 0.731 | 0.061 | 0.892 | ||||||
| Polypoid (ecblastetic) | 28 | 520.1 ± 1006.4 | 45.7 ± 56.2 | 141.2 ± 313.7 | 0.19 ± 0.25 | 0.31 ± 0.25 | |||||
| Ulcerous | 21 | 1354.5 ± 3550.4 | 79.3 ± 298.5 | 769.5 ± 3083.3 | 0.06 ± 0.06 | 0.54 ± 0.51 |
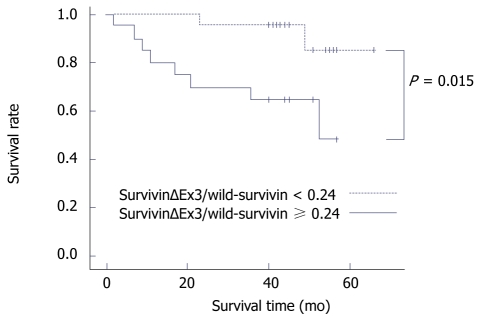
Association of survivin isoforms with prognosis
We examined the association of survivin isoforms with prognosis in 52 patients using the Kaplan-Meier survival curve. The ratio of survivin-ΔΕx3/wild-survivin presented a significant association with patient prognosis (P = 0.015, Figure 3). Specifically, when the ratio of survivinΔΕx3/wild-survivin was ≥ 0.24, patients survived a shorter time (mean survival 41 mo) compared to patients with a ratio of survivin-ΔΕx3/wild-survivin < 0.24 (mean survival 62.2 mo). No other association was found between survivin isoforms and the ratio of survivin-2b/wild-survivin with overall patient survival.
Figure 3.
Kaplan-Meier survival curve for the ratio of survivinΔΕx3/wild-survivin in 52 patients suffering colorectal adenocarcinomas.
DISCUSSION
Although survivin is cytoprotective, it is also an essential protein for cell division. In vitro survivin interacts directly with the chromosomal passenger proteins, aurora-B kinase, borealin and INCENP. These proteins are mutually dependent upon each other, and together they form a complex during mitosis, the chromosomal passenger complex, which is required for chromosome movements, proper spindle checkpoint control, and cell division[33]. The overexpression of survivin in cancer may obliterate this apoptotic checkpoint and allow aberrant progression of transformed cells through mitosis. Survivin expression has been associated with increased aggressiveness and decreased patient survival in a number of different malignancies[34]. Despite their ability to interact with wild type survivin, survivin-2b and survivin-ΔEx3 don’t act as competitors during mitosis or have an essential function[33]. They could be responsible for the fine tuning of wild type survivin’s function. Survivin-ΔEx3 exhibits pronounced antiapoptotic properties, whereas survivin-2b has largely lost its antiapoptotic activity[24].
Real-time technology has significantly extended the use and scope of Real time-PCR assays, with the potential for quantification of mRNA targets being a particular advantage[35]. Our study is the first to examine the expression of survivin and its splice variants by using quantitative Real time-PCR exclusively in colorectal adenocarcinomas. As far as the expression of the mRNA of survivin and its splice variants is concerned, previous researchers have performed mainly qualitative RT-PCR analysis or Northern blot analysis[10,36-38]. In some studies, TaqMan quantitative real Time-PCR was used with only wild type survivin mRNA determination[39-41]. Only recently, Mahotka et al[2] made the first successful quantitative analysis of survivin splice variants in renal cell carcinoma specimens. We investigated the expression of survivin’s transcript variants mRNA in patients with colorectal adenocarcinomas, while until now there have been other studies concerning colorectal cancer that used western immunoblotting, Real time PCR with SYBR Green[42,43], microarray expression analysis and immunofluorescence[44].
In our experiments, the prevalence of wild-survivin mRNA expression compared to the other splice variants is similar to the previous study that used specific hybridization probes[24] raising the question if those two isoforms might act as antagonists of wild-survivin or play a role in tumor progression. The mean copy numbers of quantitative RT-PCR amplification of wild survivin, survivin-2b and survivin-ΔΕx3 in tumor samples were respectively 842.5 ± 2380.3, 58.1 ± 192.3 and 393.9 ± 1988.8, while Suga et al[24] found 3554.1 ± 3513.7, 1233.4 ± 1280.9 and 482.0 ± 568.1. Suga et al[24] didn’t find any significant association between the expression of survivin and its splice variants with gender, location, size, age, macroscopic type, histologic type, lymphatic vessel invasion, blood vessel invasion, lymph node metastasis and serosa invasion. On the contrary, they observed a significant correlation of survivin and its splice variants with histologic disease stages, a correlation not found in our study. The differences in the results of our study with the results of Suga et al[24] might be due to race difference, genetic alterations or method performance.
The presence of transcripts of survivin isoforms has been associated with cancer. The expression of survivin in normal and cancerous samples is controversial since in previous studies data have indicated that survivin is expressed during fetal development, but not in most normal adult tissues. Using reverse transcription PCR, Mahotka et al[2] found that wild type survivin and survivin-ΔEx3 were expressed in cell lines derived from renal carcinomas but not normal renal cells. The previous isoforms are elevated in a wide variety of tumors but are not expressed to a significant extent in normal tissues[44]. Studies using sensitive quantitative Real time-PCR revealed that survivin mRNA was expressed in normal fetal tissue panels and at a very low level in the normal adult brain template. In contrast, data from a recent study have demonstrated survivin expression in normal adult tissues, such as those of skin, endometrium, endothelial cells, normal blood lymphocytes, pancreas, spleen, colon, stomach, small intestine, large intestine, lung, kidney, prostate, pancreas, heart, and thymus[45]. By using sensitive quantitative Real time-PCR analysis, low levels of total survivin mRNA were detectable in normal tissue adjacent to soft tissue sarcoma tumor cells (e.g. muscle) or in lymphocytes of blood donors. Further studies are needed in order to find out whether expression of survivin in normal tissue is due to mitotically active cells[7]. The expression of survivin mRNA by semi-quantitative Real time PCR was detected in a significantly greater proportion of colorectal carcinoma samples than in adjacent normal colorectal tissues (67.3% vs 25%, P < 0.01)[25]. With the use of tissue microarrays, survivin was detected in 147 of 230 cases of colorectal adenocarcinoma (63.9%) and no expression of survivin was observed in normal tissues[34]. Our study showed that the expression of survivin’s transcript variants mRNA was detected only in colorectal cancer tissues while it was absent in normal samples. On the contrary, Suga et al[24] found out that 60% of the normal tissues expressed survivin.
In our study, there was no significant correlation between the expression of the three isoforms and grade, Dukes stage, lymph metastasis, sex, N (TNM staging system) as well as CEA and CA19-9, two serum tumor markers not examined in any other study. Suga et al[24]. found no correlation between survivin immunoreactivity and age, sex, tumor size and site, morphologic subtype, tumor grade and clinical stage (P > 0.05). On the other hand, our study was the only one showing that the ratio of survivin-ΔEx3/wild-survivin was strongly related with age (P < 0.05), while wild-survivin and survivin-ΔEx3 were associated with T (TNM staging system) and their mRNA expression was highly elevated in patients who belonged to the group T4 (P = 0.006 and P = 0.041 respectively). Another important finding of our study is that survivin-2b was associated, for the first time in colorectal adenocarcinomas, with morphologic type, particularly overexpressed in ulcerous colorectal adenocarcinomas (P < 0.05). A differential expression of survivin-2b was also reported in gastric carcinomas with a negative correlation with disease progression. Other reports have linked the expression of survivin isoforms with poor patient prognosis[33]. In our study, we examined the association of survivin transcript variants with overall patient survival showing for the first time a significant correlation of the ratio of survivin-ΔEx3/wild-survivin with prognosis. Therefore, the ratio survivin-ΔEx3/wild-survivin might be a novel prognostic marker for colorectal carcinomas.
Despite the relative rarity of colon cancer in the Greek population and the application of expensive molecular biology methodology, we implemented a sufficiently powered study, which albeit its modest size, was adequate to generate statistically significant associations with the above variables. We also examined tissues from patients with no cancerous diseases in order to investigate the expression of survivin’s isoforms in healthy samples.
Further larger studies are required in order to examine the three other isoforms of survivin, for an overall view of survivin’s function. Moreover, other methodology could be used, such as Northern blot analysis, microarray expression analysis, nested PCR, etc to further validate the results of our study.
Recent studies showed that the phenomena of chemo-radioresistance were prevalent in survivin positive tumors and indicated that survivin may play an important role in chemoresistance of cancer cells. The survivin expression might guide chemotherapy and radiation therapy decisions. The clinical importance of survivin expression remains unclear in patients with colorectal cancer, since there are few studies on the subject. Survivin might provide important predictive and prognostic perspectives, and could offer new therapeutic alternatives for cancer[25]. Our results suggest that survivin splice variants might be related with the action of survivin in colorectal carcinogenesis but further studies are required to identify the potential role and mechanism of survivin’s isoforms in the development of colorectal cancer.
COMMENTS
Background
Survivin may provide prognostic information, because of its relation with increased tumor aggressiveness. The authors investigated three isoforms of survivin in colorectal adenocarcinomas (wild type survivin, survivin-2b and survivin-ΔΕx3).
Research frontiers
Survivin expression has been associated with increased aggressiveness and decreased patient survival in a number of different malignancies but little has been done so far in colorectal cancer. In this study, the authors used for the first time quantitative real time-polymerase chain reaction (PCR) in order to detect three isoforms of survivin in colorectal adenocarcinomas. The authors found a significant correlation of the ratio survivin-ΔEx3/wild-survivin with prognosis. Therefore, the expression level of survivin-ΔEx3/wild-survivin might be a novel prognostic marker. Another important finding was that wild-survivin and survivin-ΔEx3 were associated with TNM staging system, underscoring their association with increased tumour aggressiveness.
Innovations and breakthroughs
Till now, there is only one other study dealing with survivin’s isoforms in colorectal carcinomas using the technology of real time-qPCR. Using the same technology in colorectal adenocarcinomas the authors obtained different results.
Applications
The clinical importance of survivin expression remains unclear in patients with colorectal cancer, since there are few studies on the subject. Survivin might provide important predictive and prognostic perspectives and could offer new therapeutic alternatives for cancer. Further studies are required to identify and clarify the potential role and mechanism of survivin in the development of colorectal cancer.
Peer review
The authors have performed a nice study on the different forms of survivin and a lot of work has gone into producing the manuscript.
Footnotes
Peer reviewer: Scott Steele, MD, FACS, FASCRS, Chief, Colon and Rectal Surgery, Dept of Surgery, Madigan Army Medical Center, Fort Lewis, WA 98431, United States
S- Editor Sun H L- Editor Wang XL E- Editor Ma WH
References
- 1.Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7:979–987. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- 2.Mahotka C, Krieg T, Krieg A, Wenzel M, Suschek CV, Heydthausen M, Gabbert HE, Gerharz CD. Distinct in vivo expression patterns of survivin splice variants in renal cell carcinomas. Int J Cancer. 2002;100:30–36. doi: 10.1002/ijc.10450. [DOI] [PubMed] [Google Scholar]
- 3.Kang MH, Reynolds CP. Bcl-2 inhibitors: targeting mitochondrial apoptotic pathways in cancer therapy. Clin Cancer Res. 2009;15:1126–1132. doi: 10.1158/1078-0432.CCR-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernández-Luna JL. Apoptosis regulators as targets for cancer therapy. Clin Transl Oncol. 2007;9:555–562. doi: 10.1007/s12094-007-0103-7. [DOI] [PubMed] [Google Scholar]
- 5.Mehrotra S, Languino LR, Raskett CM, Mercurio AM, Dohi T, Altieri DC. IAP regulation of metastasis. Cancer Cell. 2010;17:53–64. doi: 10.1016/j.ccr.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambrosini G, Adida C, Sirugo G, Altieri DC. Induction of apoptosis and inhibition of cell proliferation by survivin gene targeting. J Biol Chem. 1998;273:11177–11182. doi: 10.1074/jbc.273.18.11177. [DOI] [PubMed] [Google Scholar]
- 7.Kawasaki H, Toyoda M, Shinohara H, Okuda J, Watanabe I, Yamamoto T, Tanaka K, Tenjo T, Tanigawa N. Expression of survivin correlates with apoptosis, proliferation, and angiogenesis during human colorectal tumorigenesis. Cancer. 2001;91:2026–2032. doi: 10.1002/1097-0142(20010601)91:11<2026::aid-cncr1228>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 8.Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 9.Mita AC, Mita MM, Nawrocki ST, Giles FJ. Survivin: key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res. 2008;14:5000–5005. doi: 10.1158/1078-0432.CCR-08-0746. [DOI] [PubMed] [Google Scholar]
- 10.Islam A, Kageyama H, Takada N, Kawamoto T, Takayasu H, Isogai E, Ohira M, Hashizume K, Kobayashi H, Kaneko Y, et al. High expression of Survivin, mapped to 17q25, is significantly associated with poor prognostic factors and promotes cell survival in human neuroblastoma. Oncogene. 2000;19:617–623. doi: 10.1038/sj.onc.1203358. [DOI] [PubMed] [Google Scholar]
- 11.Taubert H, Kappler M, Bache M, Bartel F, Köhler T, Lautenschläger C, Blümke K, Würl P, Schmidt H, Meye A, et al. Elevated expression of survivin-splice variants predicts a poor outcome for soft-tissue sarcomas patients. Oncogene. 2005;24:5258–5261. doi: 10.1038/sj.onc.1208702. [DOI] [PubMed] [Google Scholar]
- 12.Ryan BM, Konecny GE, Kahlert S, Wang HJ, Untch M, Meng G, Pegram MD, Podratz KC, Crown J, Slamon DJ, et al. Survivin expression in breast cancer predicts clinical outcome and is associated with HER2, VEGF, urokinase plasminogen activator and PAI-1. Ann Oncol. 2006;17:597–604. doi: 10.1093/annonc/mdj121. [DOI] [PubMed] [Google Scholar]
- 13.Xie D, Zeng YX, Wang HJ, Wen JM, Tao Y, Sham JS, Guan XY. Expression of cytoplasmic and nuclear Survivin in primary and secondary human glioblastoma. Br J Cancer. 2006;94:108–114. doi: 10.1038/sj.bjc.6602904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Virrey JJ, Guan S, Li W, Schönthal AH, Chen TC, Hofman FM. Increased survivin expression confers chemoresistance to tumor-associated endothelial cells. Am J Pathol. 2008;173:575–585. doi: 10.2353/ajpath.2008.071079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakayama K, Kamihira S. Survivin an important determinant for prognosis in adult T-cell leukemia: a novel biomarker in practical hemato-oncology. Leuk Lymphoma. 2002;43:2249–2255. doi: 10.1080/1042819021000039956. [DOI] [PubMed] [Google Scholar]
- 16.Mahotka C, Wenzel M, Springer E, Gabbert HE, Gerharz CD. Survivin-deltaEx3 and survivin-2B: two novel splice variants of the apoptosis inhibitor survivin with different antiapoptotic properties. Cancer Res. 1999;59:6097–6102. [PubMed] [Google Scholar]
- 17.Badran A, Yoshida A, Ishikawa K, Goi T, Yamaguchi A, Ueda T, Inuzuka M. Identification of a novel splice variant of the human anti-apoptopsis gene survivin. Biochem Biophys Res Commun. 2004;314:902–907. doi: 10.1016/j.bbrc.2003.12.178. [DOI] [PubMed] [Google Scholar]
- 18.Caldas H, Honsey LE, Altura RA. Survivin 2alpha: a novel Survivin splice variant expressed in human malignancies. Mol Cancer. 2005;4:11. doi: 10.1186/1476-4598-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mola G, Vela E, Fernández-Figueras MT, Isamat M, Muñoz-Mármol AM. Exonization of Alu-generated splice variants in the survivin gene of human and non-human primates. J Mol Biol. 2007;366:1055–1063. doi: 10.1016/j.jmb.2006.11.089. [DOI] [PubMed] [Google Scholar]
- 20.Caldas H, Jiang Y, Holloway MP, Fangusaro J, Mahotka C, Conway EM, Altura RA. Survivin splice variants regulate the balance between proliferation and cell death. Oncogene. 2005;24:1994–2007. doi: 10.1038/sj.onc.1208350. [DOI] [PubMed] [Google Scholar]
- 21.Ling X, Cheng Q, Black JD, Li F. Forced expression of survivin-2B abrogates mitotic cells and induces mitochondria-dependent apoptosis by blockade of tubulin polymerization and modulation of Bcl-2, Bax, and survivin. J Biol Chem. 2007;282:27204–27214. doi: 10.1074/jbc.M705161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wajapeyee N, Britto R, Ravishankar HM, Somasundaram K. Apoptosis induction by activator protein 2alpha involves transcriptional repression of Bcl-2. J Biol Chem. 2006;281:16207–16219. doi: 10.1074/jbc.M600539200. [DOI] [PubMed] [Google Scholar]
- 23.Chen WC, Liu Q, Fu JX, Kang SY. Expression of survivin and its significance in colorectal cancer. World J Gastroenterol. 2004;10:2886–2889. doi: 10.3748/wjg.v10.i19.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suga K, Yamamoto T, Yamada Y, Miyatake S, Nakagawa T, Tanigawa N. Correlation between transcriptional expression of survivin isoforms and clinicopathological findings in human colorectal carcinomas. Oncol Rep. 2005;13:891–897. [PubMed] [Google Scholar]
- 25.Tan HY, Liu J, Wu SM, Luo HS. Expression of a novel apoptosis inhibitor-survivin in colorectal carcinoma. World J Gastroenterol. 2005;11:4689–4692. doi: 10.3748/wjg.v11.i30.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng H, Lu CD, Sun YL, Dai DJ, Lee SW, Tanigawa N. Expression level of wild-type survivin in gastric cancer is an independent predictor of survival. World J Gastroenterol. 2004;10:3245–3250. doi: 10.3748/wjg.v10.i22.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 28.Ferlay J, Parkin DM, Steliarova-Foucher E. Estimates of cancer incidence and mortality in Europe in 2008. Eur J Cancer. 2010;46:765–781. doi: 10.1016/j.ejca.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 29.Kroupis C, Stathopoulou A, Zygalaki E, Ferekidou L, Talieri M, Lianidou ES. Development and applications of a real-time quantitative RT-PCR method (QRT-PCR) for BRCA1 mRNA. Clin Biochem. 2005;38:50–57. doi: 10.1016/j.clinbiochem.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 30.Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- 31.Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- 32.Tricarico C, Pinzani P, Bianchi S, Paglierani M, Distante V, Pazzagli M, Bustin SA, Orlando C. Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal Biochem. 2002;309:293–300. doi: 10.1016/s0003-2697(02)00311-1. [DOI] [PubMed] [Google Scholar]
- 33.Noton EA, Colnaghi R, Tate S, Starck C, Carvalho A, Ko Ferrigno P, Wheatley SP. Molecular analysis of survivin isoforms: evidence that alternatively spliced variants do not play a role in mitosis. J Biol Chem. 2006;281:1286–1295. doi: 10.1074/jbc.M508773200. [DOI] [PubMed] [Google Scholar]
- 34.Abd El-Hameed A. Survivin expression in colorectal adenocarcinoma using tissue microarray. J Egypt Natl Canc Inst. 2005;17:42–50. [PubMed] [Google Scholar]
- 35.Bustin SA, Benes V, Nolan T, Pfaffl MW. Quantitative real-time RT-PCR--a perspective. J Mol Endocrinol. 2005;34:597–601. doi: 10.1677/jme.1.01755. [DOI] [PubMed] [Google Scholar]
- 36.Monzó M, Rosell R, Felip E, Astudillo J, Sánchez JJ, Maestre J, Martín C, Font A, Barnadas A, Abad A. A novel anti-apoptosis gene: Re-expression of survivin messenger RNA as a prognosis marker in non-small-cell lung cancers. J Clin Oncol. 1999;17:2100–2104. doi: 10.1200/JCO.1999.17.7.2100. [DOI] [PubMed] [Google Scholar]
- 37.Sarela AI, Macadam RC, Farmery SM, Markham AF, Guillou PJ. Expression of the antiapoptosis gene, survivin, predicts death from recurrent colorectal carcinoma. Gut. 2000;46:645–650. doi: 10.1136/gut.46.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shinozawa I, Inokuchi K, Wakabayashi I, Dan K. Disturbed expression of the anti-apoptosis gene, survivin, and EPR-1 in hematological malignancies. Leuk Res. 2000;24:965–970. doi: 10.1016/s0145-2126(00)00065-5. [DOI] [PubMed] [Google Scholar]
- 39.Asanuma K, Moriai R, Yajima T, Yagihashi A, Yamada M, Kobayashi D, Watanabe N. Survivin as a radioresistance factor in pancreatic cancer. Jpn J Cancer Res. 2000;91:1204–1209. doi: 10.1111/j.1349-7006.2000.tb00906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kappler M, Köhler T, Kampf C, Diestelkötter P, Würl P, Schmitz M, Bartel F, Lautenschläger C, Rieber EP, Schmidt H, et al. Increased survivin transcript levels: an independent negative predictor of survival in soft tissue sarcoma patients. Int J Cancer. 2001;95:360–363. doi: 10.1002/1097-0215(20011120)95:6<360::aid-ijc1063>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 41.Moriai R, Asanuma K, Kobayashi D, Yajima T, Yagihashi A, Yamada M, Watanabe N. Quantitative analysis of the anti-apoptotic gene survivin expression in malignant haematopoietic cells. Anticancer Res. 2001;21:595–600. [PubMed] [Google Scholar]
- 42.Meng H, Lu C, Mabuchi H, Tanigawa N. Prognostic significance and different properties of survivin splicing variants in gastric cancer. Cancer Lett. 2004;216:147–155. doi: 10.1016/j.canlet.2003.12.020. [DOI] [PubMed] [Google Scholar]
- 43.Mandayam S, Huang R, Tarnawski AS, Chiou SK. Roles of survivin isoforms in the chemopreventive actions of NSAIDS on colon cancer cells. Apoptosis. 2007;12:1109–1116. doi: 10.1007/s10495-006-0011-2. [DOI] [PubMed] [Google Scholar]
- 44.Williams NS, Gaynor RB, Scoggin S, Verma U, Gokaslan T, Simmang C, Fleming J, Tavana D, Frenkel E, Becerra C. Identification and validation of genes involved in the pathogenesis of colorectal cancer using cDNA microarrays and RNA interference. Clin Cancer Res. 2003;9:931–946. [PubMed] [Google Scholar]
- 45.Yamada Y, Kuroiwa T, Nakagawa T, Kajimoto Y, Dohi T, Azuma H, Tsuji M, Kami K, Miyatake S. Transcriptional expression of survivin and its splice variants in brain tumors in humans. J Neurosurg. 2003;99:738–745. doi: 10.3171/jns.2003.99.4.0738. [DOI] [PubMed] [Google Scholar]