Abstract
AIM: To investigate trefoil factor (TFF) gene copy number, mRNA and protein expression as potential biomarkers in cholangiocarcinoma (CCA).
METHODS: TFF mRNA levels, gene copy number and protein expression were determined respectively by quantitative reverse transcription polymerase chain reaction (PCR), quantitative PCR and immunohistochemistry in bile duct epithelium biopsies collected from individuals with CCA, precancerous bile duct dysplasia and from disease-free controls. The functional impact of recombinant human (rh)TFF2 peptide treatment on proliferation and epidermal growth factor receptor (EGFR)/mitogen-activated protein kinase (MAPK) signaling was assessed in the CCA cell line, KMBC, by viable cell counting and immunoblotting, respectively.
RESULTS: TFF1, TFF2 and TFF3 mRNA expression was significantly increased in CCA tissue compared to disease-free controls, and was unrelated to gene copy number. TFF1 immunoreactivity was strongly increased in both dysplasia and CCA, whereas TFF2 immunoreactivity was increased only in CCA compared to disease-free controls. By contrast, TFF3 immunoreactivity was moderately decreased in dysplasia and further decreased in CCA. Kaplan-Meier analysis found no association of TFF mRNA, protein and copy number with age, gender, histological subtype, and patient survival time. Treatment of KMBC cells with rhTFF2 stimulated proliferation, triggered phosphorylation of EGFR and downstream extracellular signal related kinase (ERK), whereas co-incubation with the EGFR tyrosine kinase inhibitor, PD153035, blocked rhTFF2-dependent proliferation and EGFR/ERK responses.
CONCLUSION: TFF mRNA/protein expression is indicative of CCA tumor progression, but not predictive for histological sub-type or survival time. TFF2 is mitogenic in CCA via EGFR/MAPK activation.
Keywords: Cholangiocarcinoma, Trefoil factors, Liver fluke, Epidermal growth factor receptor, Mitogen-activated protein kinase
INTRODUCTION
Cholangiocarcinoma (CCA) is a malignant tumor arising from the biliary tract and classified according to the site of formation as either intrahepatic or extrahepatic type[1,2]. CCA accounts for 3% of all gastrointestinal cancers and is the second most common primary hepatic tumor[3]. The Southeast Asian liver fluke, Opisthorchis viverrini (O. viverrini) is a trematode parasite that infects the bile duct[4]. Previous studies have shown a strong positive correlation between CCA incidence and the prevalence of O. viverrini infection, as measured by anti-O. viverrini antibody titers in the general population residing in the northeast Thailand[5-7], where the Khon Kaen Province has shown the highest incidence of CCA in the world[8]. The truncated age-standardized incidence of CCA in Khon Kaen in age ranges older than 35 years varied between 93.8 and 317.6 per 100 000 population, depending on geographical location[5].
Several lines of evidence implicate an interaction between chemical carcinogens, especially nitrosamines, and O. viverrini infestation in the development of CCA in Thailand[9,10]. Mechanical injury to bile duct epithelial cells from feeding activity and migration of the liver fluke may also contribute to biliary damage in the human host. In addition, secretion or excretion of metabolic products from the liver fluke results in chronic irritation, hyperplasia and adenomatous changes of the bile duct epithelium[9]. Subsequent DNA damage in the biliary epithelium may drive malignant transformation[7,11].
To date, knowledge of the molecular basis of carcinogenesis and pathogenesis of CCA is limited. Previous studies in CCA patients exploring the fine mapping of chromosome region 21q22-qter showed an amplification with a frequency of more than 30% in the markers D21S1890-D21S1893, and TFF3, a member of the trefoil factor (TFF) gene family[12]. Kaplan-Meier survival curves demonstrated that patients who have amplifications at D21S1893, D21S1890 and the TFF locus had a poor prognosis, whereas patients who had deletions showed a favorable prognosis, indicating that this region may harbor candidate genes involved in the tumorigenesis and pathogenesis of CCA.
TFF genes are involved in restitution and repair of the gastric and intestinal epithelium and are rapidly upregulated in response to mucosal injury[13,14]. In particular, TFF1 has been shown to function as a gastric tumor suppressor gene[15]. However, TFF peptides are overexpressed in other solid tumors such as esophagus, breast, and pancreas and in some circumstances may function as tumor progression factors[16-18]. Prolonged inflammation caused by parasitic infection frequently occurs in liver fluke-related CCA. TFFs may exert beneficial effects during the early steps of bile duct epithelial injury and inflammation, but have undesirable effects during subsequent chronic inflammation and neoplastic progression.
The molecular basis of TFF activity remains enigmatic and a specific TFF receptor has not been identified. The epidermal growth factor receptor (EGFR) is a type-I transmembrane glycoprotein receptor with tyrosine kinase activity in its cytoplasmic domain. EGFR is activated following the binding of multiple cognate ligands and plays a significant role in initiating the signaling that directs growth, proliferation, survival, and differentiation in mammalian cells[19]. Several ligands of EGFR have been identified[20], and there is some evidence for the transactivation of EGFR by TFFs. EGFR is a key signaling pathway for TFF1- and TFF2-mediated cellular invasion in kidney and colonic cancer cells[21], suggesting the capability of TFFs to directly, or indirectly, transactivate the EGFR. While EGFR signaling has been shown to be important in CCA, there has been no report of TFF-mediated EGFR pathway activation in CCA.
With the available evidence in mind, we hypothesized that TFFs play important roles in the molecular pathogenesis of CCA, and mediate their actions, at least in part, through the transactivation of EGFR. We also hypothesized that an increase in TFF gene copy number resulting in the inappropriate overexpression of mRNA and the corresponding protein, contributes to the progression of CCA. Accordingly, we have measured TFF copy number, mRNA and protein in CCA patients, and have analyzed their association with clinicopathological parameters. In addition, TFF-mediated proliferation, EGFR transactivation and downstream signaling via the mitogen-activated protein kinase (MAPK) cascade was also investigated in a human CCA cell line.
MATERIALS AND METHODS
Patients and sample processing
Frozen liver tissues were obtained from 110 CCA patients undergoing surgical resection at Srinagarind Hospital, Faculty of Medicine, Khon Kaen University, Thailand. CCA cases were diagnosed by physicians using clinical findings, laboratory investigations and histological examinations. Clinicopathological data, such as age, gender, histological type, and tumor invasion, were obtained from medical records. The protocols were approved by the Ethical Committee of Khon Kaen University (HE480229). Informed consent was obtained from all patients who participated in the project.
Serum anti O. viverrini antibody was tested for association between liver fluke infection and CCA by enzyme-linked immunosorbent assay (ELISA) as described previously[22]. The cut-off value was 0.200 optical density for O. viverrini-positive subjects[23]. There were 20/29 (69%) O. viverrini-positive cases (Table 1), which agreed well with general screening for anti O. viverrini antibody in other sample set (68%) (unpublished data) and a previous study[5].
Table 1.
Cholangiocarcinoma samples tested for serum anti Opisthorchis viverrini antibody
| No. | Code | Anti-OV titer (OD)1 | OV positive |
| 1 | M009 | 0.804 | + |
| 2 | M012 | 0.303 | + |
| 3 | M030 | 0.820 | + |
| 4 | M043 | 0.182 | - |
| 5 | M055 | 0.185 | - |
| 6 | M068 | 0.356 | + |
| 7 | M071 | 0.257 | + |
| 8 | M097 | 0.188 | - |
| 9 | M101 | 0.235 | + |
| 10 | M111 | 0.088 | - |
| 11 | M114 | 0.285 | + |
| 12 | M117 | 0.345 | + |
| 13 | M119 | 0.168 | - |
| 14 | M132 | 0.095 | - |
| 15 | M137 | 0.330 | + |
| 16 | M139 | 0.239 | + |
| 17 | M142 | 0.344 | + |
| 18 | M148 | 0.305 | + |
| 19 | M152 | 0.108 | - |
| 20 | M155 | 0.642 | + |
| 21 | M157 | 0.482 | + |
| 22 | M208 | 0.952 | + |
| 23 | M214 | 0.188 | - |
| 24 | M229 | 0.165 | - |
| 25 | M240 | 0.669 | + |
| 26 | M265 | 0.314 | + |
| 27 | M279 | 0.502 | + |
| 28 | N004 | 0.218 | + |
| 29 | N021 | 0.232 | + |
Cut-off optical density (OD) = 0.200. OV: O. viverrini.
DNA preparation
DNA of CCA patients was prepared from frozen liver tissues using the Puregene™ DNA purification system (Gentra System, Minneapolis, MN, USA). Because establishing standard curves for analysis of TFF gene copy number by quantitative polymerase chain reaction (qPCR) required a large amount of DNA, DNA prepared from placental tissue which was collected from a patient after normal labor (postpartum) was used in this study.
Gene expression and copy number analysis
Total RNA was prepared from frozen liver tissues using RNeasy mini kits (QIAGEN, Valencia, CA, USA) according to the manufacturer’s protocols. For quantitative reverse transcription PCR (QRT-PCR), oligo-dT primed first strand cDNA was synthesized from 1 μg template RNA using Omniscript Reverse Transcriptase reagents (QIAGEN) according to the manufacturer’s protocols. RNA was also prepared from stomach and colon tissues collected from cancer patients who underwent surgical resection at Srinagarind Hospital, and was used to establish standard curves for analysis of TFF mRNA expression.
Oligonucleotide primer sequences for DNA copy number and QRT-PCR expression analysis, together with corresponding amplicon sizes are shown in Table 2. Primers sequences were designed using pDRAW32 (http://www.acaclone.com/).
Table 2.
Primer sequences of genomic and reverse transcription polymerase chain reaction, and polymerase chain reaction fragment sizes
| Primer name | Forward (5→3) | Reverse (5→3) | Product size (bp) |
| Genomic-PCR | |||
| TFF1 | CAGGGATCTGCCTGCATC | ATCGATCTCTTTTAATTTTTAGGCC | 219 |
| TFF2 | GAAGAATCTCCCGAACCAGG | GTCACACTTCAAAAACTAGAGG | 123 |
| TFF3 | CAGGCACTGTTCATCTCAGC | TATTCGTTAAGACATCAGGCTCC | 129 |
| β-actin | TCACCCACACTGTGCCCATCTACGA | CAGCGGAACCGCTCATTGCCAATGG | 375 |
| RT-PCR | |||
| TFF1 | CCCGTGAAAGACAGAATT | GATCCCTGCAGAAGTGTCT | 169 |
| TFF2 | CTCCTGGCAGCGCTCCTCGTC | GATGCCCGGGTAGCCACAGTTTCT | 223 |
| TFF3 | AACCGGGGCTGCTGCTTTG | GAGGTGCCTCAGAAGGTGC | 92 |
| RPLP0 | CTTCCCACTTGCTGAAAAG | CCAAATCCCATATCCTCGT | 168 |
PCR: Polymerase chain reaction; RT-PCR: Reverse transcription PCR; TFF: Trefoil factor.
Quantitative PCR amplification was performed on a Rotor Gene 3000 Real-time Amplification (Corbett Research, Australia) using a standard curve and SYBR Green I dye method (Amresco, Solon, OH, USA). The standard curve of each primer was generated using serial dilutions of placental DNA (for genomic TFF1, TFF2, and TFF3), stomach cDNA (for TFF1 and TFF2 mRNA), and colon cDNA (for TFF3 mRNA). The standard curve was constructed in each PCR run and the copy number or cDNA expression of genes in each sample was interpolated using these standard curves. A coefficient of variation (CV) of each sample was determined based on triplicate test. The sample with a CV higher than 15% was re-tested. The melting curves of the PCR products were performed for each reaction to demonstrate that there were no nonspecific products or primer dimers.
TFF copy number was determined from a standard curve. The relative copy number of TFF was determined in comparison to the reference gene, β-actin, which is not amplified in CCA. The normal reference range was derived as we described previously[12]. The relative copy number was interpreted as loss when the ratio was less than the mean - 2SD of the normal reference range, and as gain when the ratio was greater than mean + 2SD.
Immunohistochemistry
Tissue samples were processed for histology examination using standard procedures[24]. A 4-μm thick section was cut from each paraffin block. TFF1, TFF2 and TFF3 proteins were detected by immunohistochemical staining using rabbit polyclonal anti-TFF1 and -TFF3 antibodies (Gifts from Professor Andrew Giraud, Murdoch Children’s Research Institute, Royal Children’s Hospital, Parkville, VIC 3052, Australia) at a dilution of 1:1000, and mouse monoclonal anti-NCL-HSP (anti-TFF2) (clone GE16C, Novocastra Lab, Newcastle, UK) at a dilution of 1:200. In brief, deparaffinized sections were boiled in 0.01 mol/L citrate buffer (pH 6.0) for 3 min for antigen retrieval. Sections were then incubated with 5 mL/L hydrogen peroxide in absolute methanol for 30 min to block endogenous peroxidase activity, then incubated with 50 mL/L horse serum (Seromed Biochrom, Berlin, Germany) for 30 min and incubated overnight at 4°C with anti-TFF antibodies. The sections were incubated with the DAKO EnVisionTM+ solution (rabbit or mouse/horseradish peroxidase; DAKO Corporation, Carpinteria, CA, USA) for 30 min. Histochemical reaction for peroxidase visualization was carried out with 0.5 mL/L 3,3’-diaminobenzidine tetrahydrochloride and 1 mL/L hydrogen peroxide, and was then counterstained with hematoxylin. The amount of TFF was classified into 4 groups based on the percentage of positive staining tumor cells: score 0, for percent positive staining cells < 1%; score 1, 1% to 25%; score 2, 26% to 50%; and score 3, > 51%. Slides were examined independently by two pathologists. Results in agreement were considered to be valid. All studies were accompanied by a negative (no primary antibody) and a positive control (gastric tissues for TFF1 and TFF2, and colonic tissues for TFF3). In addition, we also observed TFF immunostaining in normal bile ducts and dysplasia of 110 CCA patients to evaluate protein expression and stepwise carcinogenesis in CCA.
Cell viability and proliferation
The KMBC cell line, derived from extrahepatic CCA, was maintained in RPMI-1640 medium supplemented with 5 mL/L fetal bovine serum (FBS); 50 IU/mL penicillin; 50 μg/mL streptomycin, at 37°C in 50 mL/L CO2/air. To determine cell viability and proliferation rates, cells were serum starved overnight, then seeded (5 × 104 cells/well) in 24-well plates containing RPMI-1640 medium supplemented with 5 mL/L FBS; 50 IU/mL penicillin; 50 μg/mL streptomycin, and incubated with recombinant human (rh)TFF2 (Gift from Professor Andrew Giraud, Murdoch Children’s Research Institute, Royal Children’s Hospital, Parkville, VIC 3052, Australia) (5, 50, 500 μg/mL) or EGF (Promega, Indianapolis, IN, USA) (5, 25, 50 ng/mL). Viable cell numbers were counted with 0.4% trypan-blue dye exclusion on a slide hemocytometer. For EGFR blockade the EGFR tyrosine kinase inhibitor, PD153035 (10 μmol/L), was added to abrogate EGFR 1 h before adding rhTFF2 or EGF. All cell viability assays were performed in triplicate for each concentration and each experiment was repeated twice (n = 6/group).
Immunoblotting
For immunoblotting, cell lysates were size fractionated by 10%-15% SDS-PAGE, transferred to nitrocellulose membranes (GE Healthcare Bio-Sciences, Piscataway, NJ, USA) and blocked in 5% non-fat milk powder/Tris-buffered saline pH 7.4, 0.1% Tween-20 as described. Membranes were incubated with primary antibodies overnight at 4°C. Detection was performed with anti-rabbit/anti-mouse IgG-HRP conjugates (DAKO Corporation, Carpinteria, CA, USA) and enhanced chemiluminescence reagents (GE Healthcare Bio-Sciences). Sources of primary antibodies: rabbit polyclonal anti-human EGFR (#2232) diluted 1:250; anti-human phospho-tyrosine 1173 EGFR (#4407) diluted 1:500; rabbit polyclonal anti-human extracellular signal-related kinase (ERK)1/2 (#9102) diluted 1:1000; anti-human phospho-threonine 202, phospho-tyrosine 204 ERK (#9101) diluted 1:1000; mouse monoclonal anti-human cyclin E (#4129) diluted 1:1000 (all Cell Signalling Technology, Danvers, MA, USA); rabbit polyclonal anti-human glyceraldehyde-3-phosphate dehydrogenase (GAPDH; #ab9485) diluted 1:3000 (Abcam, Cambridge, MA, USA).
Statistical analysis
Correlations between TFF DNA copy number and mRNA expression were analyzed using linear regression. Associations between TFF copy number, mRNA expression and protein expression, and clinicopathological parameters of CCA patients were evaluated using χ2 tests. The difference in protein expression in normal bile ducts, dysplasia and CCA was assessed by Wilcoxon’s rank sum test. Survival curves of patients (with vs without abnormal DNA copy number, normal vs high mRNA expression, and negative vs positive protein expression of TFF genes) were calculated using the Kaplan-Meier method. Differences in survival between two groups were assessed by the log-rank test. P < 0.05 was considered statistically significant.
RESULTS
Expression of TFF mRNA in CCA
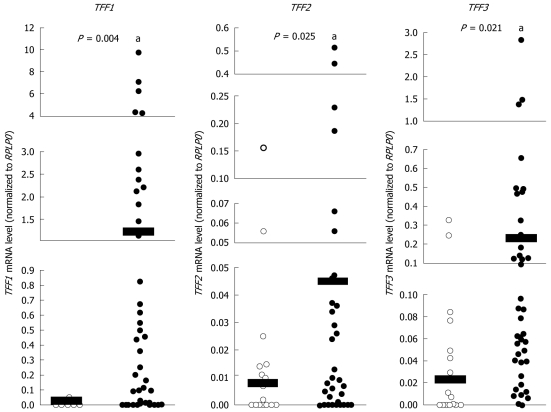
To evaluate the role of TFFs in CCA, we measured TFF mRNA levels in tumor tissues of CCA patients using quantitative reverse transcription PCR (QRT-PCR). For comparison, tissues were also collected from tumor-free margin areas of 16 individuals. TFF mRNA level was normalized to that of RPLP0 mRNA. The relative mRNA expression level of TFF1, TFF2 and TFF3 in normal samples ranged from 0.000-0.052 (mean, 0.004), 0.000-0.056 (mean, 0.008), and 0.000-0.329 (mean, 0.048), respectively. Relative mRNA expression level of TFF1, TFF2 and TFF3 in 46 tumor samples ranged from 0.000-9.773 (mean, 1.178), 0.000-0.516 (mean, 0.045), and 0.000-2.827 (mean, 0.229), respectively. The expression of TFF1, TFF2 and TFF3 mRNA in tumor tissues was significantly higher than in normal tissues (P < 0.05) (Figure 1). These results demonstrate that increased TFF gene expression correlates with tumor progression in CCA.
Figure 1.
The mRNA expression levels of Trefoil factor genes in normal and cholangiocarcinoma tissues. Quantitative reverse transcription polymerase chain reaction analysis of trefoil factor (TFF)1, TFF2 and TFF3 mRNA levels in cholangiocarcinoma tumors (n = 46) compared with normal controls (n = 16). Scatter plots show mRNA abundance for each data point normalized to the internal reference gene RPLP0. Horizontal black bars show mean mRNA levels. aP < 0.05. A statistical outlier in the TFF2 normal control data set is +3.58 standard deviations from the mean and failed a Z-test, which requires that values must fall within 3 standard deviations of the mean for statistical validity. This sample (shown in bold font) was excluded from all statistical calculations for this data set.
TFF protein expression in CCA
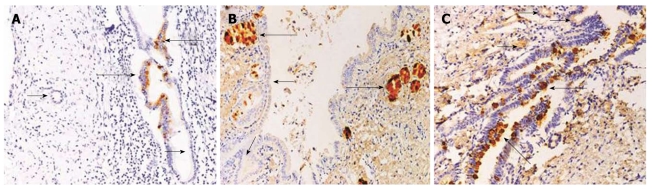
Immunohistochemistry was performed to determine TFF protein levels and cellular distribution in tissues collected from CCA patients. Examination of subcellular distribution revealed that all TFFs were present in fine cytoplasmic vesicles and at the apical/luminal surface of the epithelial cells. Faint and scattered TFF1 immunoreactivity was observed in normal bile ducts with low frequency (24.6%). However, TFF1 immunoreactivity was intense (Figure 2A) and markedly frequent in dysplasia. TFF2 immunoreactivity was present mainly in peribiliary glands and rarely expressed in normal bile ducts and dysplasia (Figure 2B). TFF3 immunoreactivity was distributed widely and expressed faintly in normal bile ducts but more intensely in dysplasia (Figure 2C) with high frequency in each case.
Figure 2.
Immunohistochemical detection of trefoil factor proteins in normal and dysplasia bile ducts (Original magnification × 400). A: Small and large normal bile ducts (short arrows) showing no trefoil factor (TFF)1 expression, whereas dysplasia showed moderately increased expression (long arrows); B: TFF2 expression was strongly positive in peribiliary glands (long arrows) but rarely present in dysplasia (short arrows); C: TFF3 was distributed widely at low level in small and large normal bile ducts (short arrows) and markedly increased in dysplasia (long arrows).
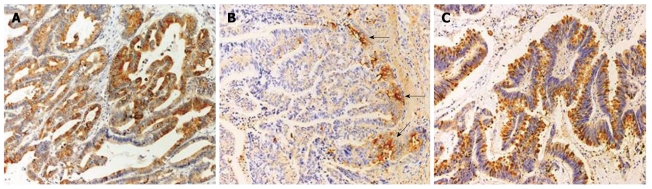
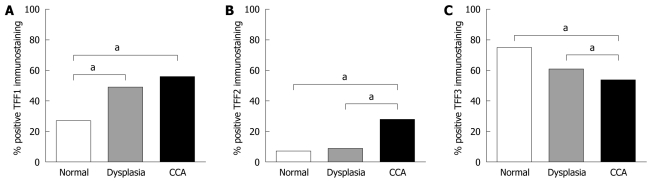
In tumor samples, TFF1 was present mostly in the cytoplasm (Figure 3A), but in three cases, TFF1 staining was also found in secreted mucus. TFF1 immunoreactivity was detected in 56/110 (50.9%). Its frequency was markedly increased compared with normal (P < 0.001) (Figure 4A). TFF2 was detected mostly in the cytoplasm with less frequency in 28/110 (25.5%). Interestingly, some positive staining was found at the invasion front (Figure 3B). TFF3 immunoreactivity was observed in 54/110 (49.1%) of patients analyzed. Two patterns of TFF3 staining were found in tumor cells: in cytoplasm as fine granules and in the apical/luminal surfaces, and as a goblet cell pattern in which TFF3 was diffusely distributed as coarse granules (Figure 3C). In addition, TFF3 immunoreactivity was highly frequent in disease-free control tissue (68.2%) but less frequent in dysplasia (55.5%) and CCA (49.1%) (Figure 4C). Although TFF2 expression in CCA was less frequent compared to those of TFF1 and TFF3, its frequency was markedly increased in comparison with those of normal bile ducts and dysplasia (P < 0.01) (Figure 4B) suggesting a role in tumor progression. Moreover, we found a significant correlation between TFF1 and TFF3 expression (P = 0.014), whereas TFF2 expression was not correlated with either TFF1 or TFF3 expression (Table 3).
Figure 3.
Immunohistochemical examinations of trefoil factor proteins in cholangiocarcinoma (Original magnification × 400). A: Trefoil factor (TFF)1 staining was markedly positive and was mostly seen in the cytoplasm; B: TFF2 staining was mostly in the cytoplasm and at the invasion front (arrows); C: TFF3 staining showing a goblet cell pattern manifested as diffusely distributed coarse granules.
Figure 4.
Frequencies of trefoil factor immunostaining in normal bile ducts, dysplasia and cholangiocarcinoma. A: Trefoil factor (TFF)1 positive immunostaining was markedly frequent in dysplasia and cholangiocarcinoma (CCA) compared to normal; B: Frequency of TFF2 positive immunostaining was significantly higher in CCA than in dysplasia and normal controls; C: TFF3 positive immunostaining was less frequent in CCA than in dysplasia and controls. aP < 0.05.
Table 3.
Associations of trefoil factor protein expression in cholangiocarcinoma
| TFF protein expression | n |
TFF2 |
TFF3 |
||||
| Negative | Positive | P-value | Negative | Positive | P-value | ||
| TFF1 | 110 | ||||||
| Negative | 54 | 42 | 12 | NS | 34 | 20 | 0.014 |
| Positive | 56 | 40 | 16 | 22 | 34 | ||
| TFF2 | 110 | ||||||
| Negative | 82 | 44 | 38 | NS | |||
| Positive | 28 | 12 | 16 | ||||
TFF: Trefoil factor.
TFF copy number in CCA
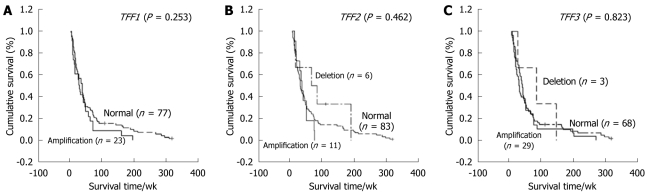
The normal range of gene copy number derived from our previous study is 0.54-1.34 under a 95% CI[12]. The percentage of CCA patients who had relative amplification at TFF1, TFF2, and TFF3 was 25%, 12%, and 30%, respectively while that of CCA patients who had relative deletion at TFF2 was 5%, and deletion of TFF3 was 4% (Table 4). Relative amplification of these genes ranged from 1.35-3.67 and deletion from 0.37-0.51. Statistical analysis showed no associations of TFF mRNA, protein and copy number with age, gender, histological type, and survival time of the patients (Figure 5).
Table 4.
Relative copy number of trefoil factor genes in cholangiocarcinoma
| No. | TFF1 | TFF2 | TFF3 |
| 1 | 2.031 | 1.691 | 2.311 |
| 2 | 1.431 | 1.371 | 1.721 |
| 3 | 1.551 | 1.371 | 2.341 |
| 4 | 2.001 | 2.191 | 2.911 |
| 5 | 1.461 | 1.731 | 1.841 |
| 6 | 1.791 | 1.781 | 3.211 |
| 7 | 1.821 | 2.741 | 3.671 |
| 8 | 1.441 | 0.96 | 1.381 |
| 9 | 1.551 | 0.98 | 1.781 |
| 10 | 1.551 | 0.99 | 1.561 |
| 11 | 1.681 | 1.30 | 2.361 |
| 12 | 1.681 | 1.10 | 1.401 |
| 13 | 1.971 | 0.85 | 2.171 |
| 14 | 1.611 | 1.33 | 2.321 |
| 15 | 1.581 | 1.00 | 1.581 |
| 16 | 1.621 | 1.22 | 2.891 |
| 17 | 1.491 | 1.17 | 1.351 |
| 18 | 1.521 | 0.97 | 2.251 |
| 19 | 1.621 | 1.21 | 1.851 |
| 20 | 1.14 | 1.10 | 1.551 |
| 21 | 1.17 | 0.98 | 1.661 |
| 22 | 0.83 | 0.92 | 1.571 |
| 23 | 1.14 | 1.03 | 1.481 |
| 24 | 0.98 | 1.14 | 1.411 |
| 25 | 1.06 | 1.15 | 1.651 |
| 26 | 1.06 | 1.33 | 2.131 |
| 27 | 1.32 | 0.91 | 1.421 |
| 28 | 1.20 | 1.01 | 1.571 |
| 29 | 1.27 | 1.631 | 1.851 |
| 30 | 1.32 | 1.891 | 1.581 |
| 31 | 1.29 | 2.681 | 2.161 |
| 32 | 1.28 | 1.381 | 1.871 |
| 33 | 0.88 | 1.481 | 1.631 |
| 34 | 1.491 | 1.421 | 1.24 |
| 35 | 1.381 | 0.71 | 0.99 |
| 36 | 1.771 | 1.03 | 1.12 |
| 37 | 1.381 | 0.92 | 1.29 |
| 38 | 1.511 | 1.17 | 0.94 |
| 39 | 1.521 | 1.02 | 1.23 |
| 40 | 1.461 | 1.12 | 1.34 |
| 41 | 1.421 | 0.97 | 1.34 |
| 42 | 1.371 | 0.73 | 0.79 |
| 43 | 0.84 | 0.472 | 0.58 |
| 44 | 0.63 | 0.372 | 0.88 |
| 45 | 0.68 | 0.532 | 0.86 |
| 46 | 0.63 | 0.512 | 0.83 |
| 47 | 0.86 | 0.382 | 0.55 |
| 48 | 0.76 | 0.422 | 0.462 |
| 49 | 0.92 | 0.68 | 0.452 |
| 50 | 0.82 | 0.85 | 0.512 |
| 51 | 1.07 | 0.79 | 0.97 |
| 52 | 1.15 | 0.59 | 1.11 |
| 53 | 0.95 | 0.84 | 1.02 |
| 54 | 0.84 | 0.79 | 0.68 |
| 55 | 0.74 | 0.77 | 1.26 |
| 56 | 0.77 | 1.03 | 0.83 |
| 57 | 1.13 | 1.31 | 1.26 |
| 58 | 1.13 | 1.03 | 1.20 |
| 59 | 1.00 | 0.88 | 0.89 |
| 60 | 1.17 | 0.70 | 1.15 |
| 61 | 0.94 | 0.91 | 1.19 |
| 62 | 0.76 | 0.72 | 0.63 |
| 63 | 1.15 | 1.04 | 1.15 |
| 64 | 1.25 | 0.95 | 0.87 |
| 65 | 1.01 | 0.77 | 0.90 |
| 66 | 1.11 | 1.27 | 1.03 |
| 67 | 0.85 | 0.62 | 0.54 |
| 68 | 1.10 | 0.96 | 1.23 |
| 69 | 1.06 | 0.92 | 0.84 |
| 70 | 1.05 | 0.88 | 0.80 |
| 71 | 1.12 | 1.09 | 1.10 |
| 72 | 1.33 | 0.87 | 1.22 |
| 73 | 0.87 | 1.01 | 0.71 |
| 74 | 0.75 | 0.88 | 0.76 |
| 75 | 1.04 | 0.74 | 1.14 |
| 76 | 1.10 | 0.67 | 0.93 |
| 77 | 1.06 | 0.98 | 1.14 |
| 78 | 1.10 | 0.97 | 1.30 |
| 79 | 1.22 | 0.87 | 1.21 |
| 80 | 1.16 | 0.95 | 1.32 |
| 81 | 1.13 | 1.07 | 1.32 |
| 82 | 1.10 | 0.88 | 1.24 |
| 83 | 0.92 | 1.05 | 1.31 |
| 84 | 1.14 | 0.97 | 1.09 |
| 85 | 1.01 | 0.98 | 1.14 |
| 86 | 1.13 | 1.07 | 1.26 |
| 87 | 0.91 | 0.59 | 1.23 |
| 88 | 0.97 | 0.69 | 1.16 |
| 89 | 0.91 | 0.78 | 0.91 |
| 90 | 1.06 | 0.71 | 1.06 |
| 91 | 1.15 | 0.69 | 0.88 |
| 92 | 0.96 | 0.58 | 0.57 |
| 93 | 1.03 | 1.00 | 1.30 |
| 94 | 1.20 | 0.93 | 1.06 |
| 95 | 1.25 | 0.76 | 0.77 |
| 96 | 1.28 | 1.21 | 1.24 |
| 97 | 1.28 | 1.15 | 0.94 |
| 98 | 1.10 | 0.82 | 0.99 |
| 99 | 1.28 | 0.73 | 0.71 |
| 100 | 0.87 | 0.59 | 0.91 |
| 101 | 1.05 | 0.78 | 1.28 |
| 102 | 1.00 | 0.70 | 0.99 |
| 103 | 0.66 | 0.61 | 0.61 |
| 104 | 0.89 | 0.74 | 0.80 |
| 105 | 0.88 | 0.57 | 0.60 |
| 106 | 0.82 | 0.56 | 0.58 |
| 107 | 1.04 | 1.05 | 1.04 |
| 108 | 0.90 | 0.98 | 0.88 |
| 109 | 0.65 | 0.65 | 0.64 |
| 110 | 1.18 | 1.21 | 1.20 |
Figures indicate relative copy number of trefoil factor (TFF) genes (TFF1, TFF2, TFF3) compared to the reference gene, β-actin.
Amplification;
Deletion. Other figures are normal.
Figure 5.
Kaplan-Meier survival curves with regard to trefoil factor copy number in cholangiocarcinoma. No association between trefoil factor (TFF) copy number and survival time (week) was found for A: TFF1; B: TFF2; C: TFF3.
TFF2 stimulates CCA cell proliferation via EGFR- mediated MAPK
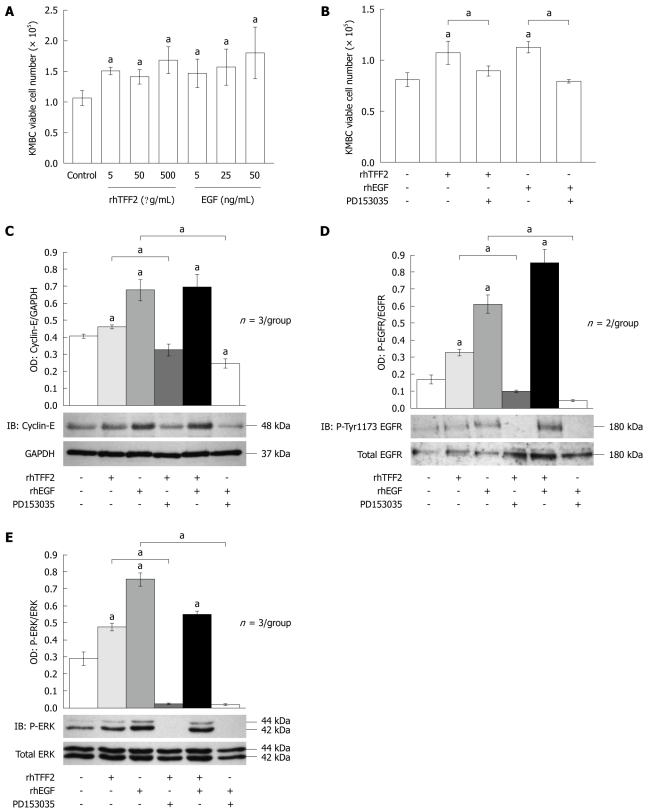
Our data showed increased TFF2 expression in CCA tumor tissue compared to normal bile ducts and dysplasia, suggesting an important role in tumor progression. To address this question in greater detail, we next utilized an in vitro culture model to manipulate TFF2 levels in CCA. The KMBC cell line is derived from extrahepatic CCA and is atypical in that it does not express significant levels of TFF2. With this in mind, we designed a series of experiments to compare the proliferation rate of KMBC cells treated with or without recombinant human (rh)TFF2 peptide. EGF is a well-known mitogen and was used to validate KMBC proliferation as a positive control. Treatment with rhTFF2 (and EGF) increased the proliferation rate of KMBC cells (Figure 6A). While signaling receptors for TFFs have not been identified, studies in other cancer cells have shown that TFF signals can be transduced by transactivation of the EGFR[21]. To determine whether rhTFF2 mediates proliferation responses via the EGFR, a specific EGFR antagonist, PD153035 (10 μmol/L), was used to abrogate EGFR activity at least 1 h before treatment with 5 μg/mL rhTFF2 or 50 ng/mL EGF. PD153035 treatment abolished both rhTFF2 and EGF dependent proliferative responses (Figure 6B). We next determined cyclin-E protein levels as a molecular endpoint of proliferation, finding that cyclin-E abundance was directly correlated with the viable cell count data (Figure 6C). Collectively, these results indicate that TFF2 stimulates CCA cell proliferation via transactivation of EGFR. To confirm this finding, we next determined the levels of EGFR activity by immunoblotting for EGFR (tyrosine-1173) phosphorylation. Consistent with expectation, rhTFF2 treatment triggered a significant increase in EGFR phosphorylation (tyrosine 1173). A similar effect was obtained by treatment with EGF. On the other hand tyrosine EGFR phosphorylation was significantly attenuated after treatment with PD153035 (Figure 6D).
Figure 6.
Trefoil factor 2 drives proliferation of KMBC cholangiocarcinoma cells via epidermal growth factor receptor tyrosine kinase and mitogen-activated protein kinase pathway activation. A: Proliferation of KMBC human cholangiocarcinoma cells determined by viable cell number counting with recombinant human trefoil factor (rhTFF)2 dose response (5, 50, 500 μg/mL) and epidermal growth factor (EGF) (5, 25, 50 ng/mL). Histograms show mean cell number per well. Error bars show standard error of the mean (SE). aP < 0.05; B: Proliferation of KMBC cells determined by viable cell number counting following treatment with combinations of
The MAP-kinase cascade is the predominant signal transduction pathway utilized by the EGFR. To confirm the identity of the intracellular mediator(s) of EGFR-dependent TFF2 activity we next investigated the response of the terminal MAP-kinases, p44/p42 or ERK 1/2 to rhTFF2 treatment in KMBC cells. Consistent with the effect on EGFR activity, exogenous rhTFF2 treatment significantly increased ERK phosphorylation compared to untreated controls (P = 0.016). A similar effect was obtained with EGF treatment, while both rhTFF2 and EGF-dependent activation of ERK was abolished by co-incubation with the EGFR antagonist, PD153035 (Figure 6E). These results suggest that increased TFF2 expression drives CCA tumor progression by increasing cell proliferation via activation of EGFR and MAPK signaling.
DISCUSSION
Previous studies have revealed differences in TFF gene expression in biliary pathologies depending upon the size of bile duct; small, medium or large[25-27]. In addition, there are variations of TFF expression observed among these studies. They showed that TFF1 and TFF3 are moderately expressed in normal bile ducts, particularly large bile ducts and markedly increased in biliary epithelial diseases[25-27], suggesting their significant roles in cytoprotection and biliary epithelial repair. Sasaki et al[28] studied TFF1 expression in normal, biliary epithelial dysplasia, and CCA associated-hepatolithiasis. It was found that TFF1 was only modestly expressed in disease-free control tissue, yet was dramatically upregulated in dysplasia and noninvasive CCA. Paradoxically, TFF1 expression was significantly decreased in invasive CCA with a positive rate of 60% due to promoter hypermethylation[28]. In this context, it seems that TFF1 expression positively regulates CCA tumor growth, but negatively regulates tumor invasion, potentially by acting as a tumor suppressor in the latter. In contrast, we found that TFF1 mRNA expression was significantly increased in tumor tissue compared to adjacent disease-free tissues. Progressive increases in TFF1 expression in dysplasia and CCA in our study strongly argues for a role in the stepwise carcinogenesis of liver fluke-associated CCA. Moreover, Vestergaard et al[29] showed that TFF1 and TFF3 promoter hypomethylation correlates with increased mRNA expression in prostate cancer compared to benign prostatic hyperplasia. It is therefore conceivable that increased TFF1 expression in our CCA cohort was due to promoter hypomethylation.
The pathological features common to liver fluke infestation and other CCA are chronic inflammation of the biliary tract, bile stasis, and increased biliary epithelial cell turnover[7]. However, the molecular mechanisms of carcinogenesis and pathogenesis of liver fluke- and non-liver fluke-associated CCA have not been well characterized and may differ according to the background of hepatobiliary lesions, and also to the stage or progression of CCA. This contradiction has been recently clarified by differential gene expression profiling of liver fluke- and non-liver fluke-associated CCA[30]. We revealed that TFF1 protein expression in liver fluke-associated CCA was 50.7% compared with 20% in non-liver fluke-associated group suggesting that different underlying etiologies may lead to different mechanism of tumorigenesis and progression.
In contrast to previous studies[25-27], we found downregulation of TFF3 during stepwise carcinogenesis in liver fluke infestation. TFF3 mRNA expression in tumors was significantly higher than in normal (P = 0.021) (Figure 1), whereas the positive rate of TFF3 expression in tumors was significantly lower than normal (P = 0.003) (Figure 4C). This suggests that TFF3 expression levels may be modified by posttranscriptional processing events. The substantially decreased TFF3 expression in CCA may reduce tumor suppressor activity, which leads to increased cell proliferation and tumor development. We found a significant association between TFF1 and TFF3 expression in our CCA cases (P = 0.014), suggesting that TFF1 and TFF3 were co-expressed in response to tumor progression and may adversely affect CCA patients. Although median survival time of CCA patients with the co-expression of TFF1 and TFF3 (29.43 wk) was shorter than that of CCA patients without co-expression (45.28 wk), no statistically significant difference was found between these two groups analyzed by Kaplan-Meier. Poulsom et al[31] reported the co-expression of TFF1 and TFF3 mRNA in normal, hyperplasia, and neoplasia human breast epithelium, implicating them in the growth and progression of mammary carcinoma.
TFF2 was rarely expressed in normal and pre-malignant bile ducts but expressed mainly in peribiliary glands and markedly increased in CCA. Srivatsa et al[26] reported that TFF1 and TFF3 are induced in biliary diseases but not TFF2. However, Sasaki et al[25] showed that TFF2 expression was correlated with the degree of small bile duct damage, not with the disease etiology. Taken together, TFF2 expression in our cases was not responsible for stepwise carcinogenesis but rather was involved in the pathogenesis of CCA.
The proliferative effects of TFF2 have been investigated in other organ systems. Most significantly TFF2-deficient mice showed reduced gastric proliferation compared to wild-type littermate controls[32]. Furthermore, it has been shown that TFF2 acting via the EGF signaling pathway promotes cell invasion and can be blocked by an EGFR inhibitor (ZD1839) in kidney and colon cell lines[21]. Recently, Dubeykovskaya et al[33] have identified ectopically expressed CXCR4 chemokine receptor as a signaling receptor for TFF2 in lymphocytic and gastric cell lines. This study also demonstrated a distinct proliferative effect of TFF2 protein in gastric cancer cells expressing CXCR4. Our study showed that rhTFF2 mediated CCA cell proliferation via the EGFR and MAPK activation, suggesting that TFF2 could act as a proliferative factor via the EGFR and intracellular MAPK pathway. An important conclusion of our study is the usefulness of these signaling molecules, acting downstream of TFF2, as potential therapeutic targets in invasive CCA.
This is the first study to directly analyze copy numbers of the three TFF genes and their relationship with mRNA and protein levels. Although a correlation among these parameters was not found, our study suggests that several mechanisms such as transcriptional and post-transcriptional processes are involved in the modulation of TFF gene expression in CCA, which plays an important role in the pathogenesis of this cancer.
Although TFF genes are involved in epithelium restitution and repair processes, the expression of TFF family members is tissue-specific. For example, TFF1 and TFF2 are expressed predominantly in the stomach, whereas TFF3 is predominantly expressed in the intestinal epithelium[34]. The differential expression of TFF genes is consistent with distinct functional roles in different cell types. However, a “switch” in TFF gene expression patterns has been observed in other cancers, as exemplified by the upregulation of TFF3 in the intestinal metaplasia lineage and its association with poor prognosis in gastric cancer[35]. Our data, showing aberrant expression and mitogenic effects of TFFs, suggests not only a mechanism for tumor progression in CCA, but also highlights the potential usefulness of TFFs as clinical biomarkers or even therapeutic targets using small molecule inhibitors.
COMMENTS
Background
Cholangiocarcinoma (CCA) in northeast Thailand is related to liver fluke infection. Previous study showed that > 30% of CCA patients have amplification of D21S1893, D21S1890 and the trefoil factor (TFF) gene family, which is associated with poor prognosis suggesting a role of TFFs in tumor progression. The authors hypothesized that an increase in TFF gene copy number resulting in the inappropriate overexpression of mRNA and corresponding protein, contributes to the progression of CCA in which TFFs mediate their actions, at least in part, through the transactivation of epidermal growth factor receptor (EGFR).
Innovations and breakthroughs
This is the first study to directly analyze copy numbers of the three TFF genes and their relationship with mRNA and protein levels. This is also the first report showing that TFF2 is mitogenic in CCA via activation of EGFR and the mitogen-activated protein kinase pathway.
Applications
Aberrant expression and mitogenic effects of TFFs suggests not only a mechanism for tumor progression in CCA, but also highlights the potential usefulness of TFFs as clinical biomarkers or even therapeutic targets using small molecule inhibitors.
Peer review
The authors clearly demonstrate that TFF2 peptide increased cell proliferation in a CCA cell line, and they first show that TFF2 activated EGFR and the ERK1/2 MAP kinase in CCA cell line, showing both the function and its mechanism of TFF2 in CCA.
Footnotes
Supported by The Thailand Research Fund through the Royal Golden Jubilee PhD program (grant PHD/0121/2547 code 5LKK/47/B1 to Kosriwong K and Limpaiboon T); Khon Kaen University Research Affairs (grant 48-03-1-01-03); and the Centre for Research and Development of Medical Diagnostic Laboratories, Faculty of Associated Medical Sciences (No. 06-01), Thailand
Peer reviewer: Masahiro Iizuka, MD, PhD, Director, Akita Health Care Center, Akita Red Cross Hospital, 3-4-23, Nakadori, Akita, 010-0001, Japan
S- Editor Sun H L- Editor Cant MR E- Editor Zheng XM
References
- 1.Khan SA, Davidson BR, Goldin R, Pereira SP, Rosenberg WM, Taylor-Robinson SD, Thillainayagam AV, Thomas HC, Thursz MR, Wasan H. Guidelines for the diagnosis and treatment of cholangiocarcinoma: consensus document. Gut. 2002;51 Suppl 6:VI1–VI9. doi: 10.1136/gut.51.suppl_6.vi1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uttaravichien T, Bhudhisawasdi V, Pairojkul C, Pugkhem A. Intrahepatic cholangiocarcinoma in Thailand. J Hepatobiliary Pancreat Surg. 1999;6:128–135. doi: 10.1007/s005340050095. [DOI] [PubMed] [Google Scholar]
- 3.Vauthey JN, Blumgart LH. Recent advances in the management of cholangiocarcinomas. Semin Liver Dis. 1994;14:109–114. doi: 10.1055/s-2007-1007302. [DOI] [PubMed] [Google Scholar]
- 4.Keiser J, Utzinger J. Food-borne trematodiases. Clin Microbiol Rev. 2009;22:466–483. doi: 10.1128/CMR.00012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sriamporn S, Pisani P, Pipitgool V, Suwanrungruang K, Kamsa-ard S, Parkin DM. Prevalence of Opisthorchis viverrini infection and incidence of cholangiocarcinoma in Khon Kaen, Northeast Thailand. Trop Med Int Health. 2004;9:588–594. doi: 10.1111/j.1365-3156.2004.01234.x. [DOI] [PubMed] [Google Scholar]
- 6.Srivatanakul P, Ohshima H, Khlat M, Parkin M, Sukarayodhin S, Brouet I, Bartsch H. Endogenous nitrosamines and liver fluke as risk factors for cholangiocarcinoma in Thailand. IARC Sci Publ. 1991:88–95. [PubMed] [Google Scholar]
- 7.Sripa B, Kaewkes S, Sithithaworn P, Mairiang E, Laha T, Smout M, Pairojkul C, Bhudhisawasdi V, Tesana S, Thinkamrop B, et al. Liver fluke induces cholangiocarcinoma. PLoS Med. 2007;4:e201. doi: 10.1371/journal.pmed.0040201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vatanasapt V, Uttaravichien T, Mairiang EO, Pairojkul C, Chartbanchachai W, Haswell-Elkins M. Cholangiocarcinoma in north-east Thailand. Lancet. 1990;335:116–117. doi: 10.1016/0140-6736(90)90591-r. [DOI] [PubMed] [Google Scholar]
- 9.Bhamarapravati N, Thammavit W, Vajrasthira S. Liver changes in hamsters infected with a liver fluke of man, Opisthorchis viverrini. Am J Trop Med Hyg. 1978;27:787–794. doi: 10.4269/ajtmh.1978.27.787. [DOI] [PubMed] [Google Scholar]
- 10.Thamavit W, Kongkanuntn R, Tiwawech D, Moore MA. Level of Opisthorchis infestation and carcinogen dose-dependence of cholangiocarcinoma induction in Syrian golden hamsters. Virchows Arch B Cell Pathol Incl Mol Pathol. 1987;54:52–58. doi: 10.1007/BF02899196. [DOI] [PubMed] [Google Scholar]
- 11.Thamavit W, Pairojkul C, Tiwawech D, Shirai T, Ito N. Strong promoting effect of Opisthorchis viverrini infection on dimethylnitrosamine-initiated hamster liver. Cancer Lett. 1994;78:121–125. doi: 10.1016/0304-3835(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 12.Muenphon K, Limpaiboon T, Jearanaikoon P, Pairojkul C, Sripa B, Bhudhisawasdi V. Amplification of chromosome 21q22.3 harboring trefoil factor family genes in liver fluke related cholangiocarcinoma is associated with poor prognosis. World J Gastroenterol. 2006;12:4143–4148. doi: 10.3748/wjg.v12.i26.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinoshita K, Taupin DR, Itoh H, Podolsky DK. Distinct pathways of cell migration and antiapoptotic response to epithelial injury: structure-function analysis of human intestinal trefoil factor. Mol Cell Biol. 2000;20:4680–4690. doi: 10.1128/mcb.20.13.4680-4690.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otto B, Wright N. Trefoil peptides. Coming up clover. Curr Biol. 1994;4:835–838. doi: 10.1016/s0960-9822(00)00186-x. [DOI] [PubMed] [Google Scholar]
- 15.Taupin D, Pedersen J, Familari M, Cook G, Yeomans N, Giraud AS. Augmented intestinal trefoil factor (TFF3) and loss of pS2 (TFF1) expression precedes metaplastic differentiation of gastric epithelium. Lab Invest. 2001;81:397–408. doi: 10.1038/labinvest.3780247. [DOI] [PubMed] [Google Scholar]
- 16.Labouvie C, Machado JC, Carneiro F, Sarbia M, Vieth M, Porschen R, Seitz G, Blin N. Differential expression of mucins and trefoil peptides in native epithelium, Barrett's metaplasia and squamous cell carcinoma of the oesophagus. J Cancer Res Clin Oncol. 1999;125:71–76. doi: 10.1007/s004320050244. [DOI] [PubMed] [Google Scholar]
- 17.May FE, Westley BR. Cloning of estrogen-regulated messenger RNA sequences from human breast cancer cells. Cancer Res. 1986;46:6034–6040. [PubMed] [Google Scholar]
- 18.Terris B, Blaveri E, Crnogorac-Jurcevic T, Jones M, Missiaglia E, Ruszniewski P, Sauvanet A, Lemoine NR. Characterization of gene expression profiles in intraductal papillary-mucinous tumors of the pancreas. Am J Pathol. 2002;160:1745–1754. doi: 10.1016/S0002-9440(10)61121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wells A. EGF receptor. Int J Biochem Cell Biol. 1999;31:637–643. doi: 10.1016/s1357-2725(99)00015-1. [DOI] [PubMed] [Google Scholar]
- 20.Harris RC, Chung E, Coffey RJ. EGF receptor ligands. Exp Cell Res. 2003;284:2–13. doi: 10.1016/s0014-4827(02)00105-2. [DOI] [PubMed] [Google Scholar]
- 21.Rodrigues S, Attoub S, Nguyen QD, Bruyneel E, Rodrigue CM, Westley BR, May FE, Thim L, Mareel M, Emami S, et al. Selective abrogation of the proinvasive activity of the trefoil peptides pS2 and spasmolytic polypeptide by disruption of the EGF receptor signaling pathways in kidney and colonic cancer cells. Oncogene. 2003;22:4488–4497. doi: 10.1038/sj.onc.1206685. [DOI] [PubMed] [Google Scholar]
- 22.Sripa B, Kaewkes S. Relationship between parasite-specific antibody responses and intensity of Opisthorchis viverrini infection in hamsters. Parasite Immunol. 2000;22:139–145. doi: 10.1046/j.1365-3024.2000.00286.x. [DOI] [PubMed] [Google Scholar]
- 23.Honjo S, Srivatanakul P, Sriplung H, Kikukawa H, Hanai S, Uchida K, Todoroki T, Jedpiyawongse A, Kittiwatanachot P, Sripa B, et al. Genetic and environmental determinants of risk for cholangiocarcinoma via Opisthorchis viverrini in a densely infested area in Nakhon Phanom, northeast Thailand. Int J Cancer. 2005;117:854–860. doi: 10.1002/ijc.21146. [DOI] [PubMed] [Google Scholar]
- 24.Cook GA, Yeomans ND, Giraud AS. Temporal expression of trefoil peptides in the TGF-alpha knockout mouse after gastric ulceration. Am J Physiol. 1997;272:G1540–G1549. doi: 10.1152/ajpgi.1997.272.6.G1540. [DOI] [PubMed] [Google Scholar]
- 25.Sasaki M, Tsuneyama K, Saito T, Kataoka H, Mollenhauer J, Poustka A, Nakanuma Y. Site-characteristic expression and induction of trefoil factor family 1, 2 and 3 and malignant brain tumor-1 in normal and diseased intrahepatic bile ducts relates to biliary pathophysiology. Liver Int. 2004;24:29–37. doi: 10.1111/j.1478-3231.2004.00883.x. [DOI] [PubMed] [Google Scholar]
- 26.Srivatsa G, Giraud AS, Ulaganathan M, Yeomans ND, Dow C, Nicoll AJ. Biliary epithelial trefoil peptide expression is increased in biliary diseases. Histopathology. 2002;40:261–268. doi: 10.1046/j.1365-2559.2002.01347.x. [DOI] [PubMed] [Google Scholar]
- 27.Kimura Y, Leung PS, Kenny TP, Van De Water J, Nishioka M, Giraud AS, Neuberger J, Benson G, Kaul R, Ansari AA, et al. Differential expression of intestinal trefoil factor in biliary epithelial cells of primary biliary cirrhosis. Hepatology. 2002;36:1227–1235. doi: 10.1053/jhep.2002.36157. [DOI] [PubMed] [Google Scholar]
- 28.Sasaki M, Tsuneyama K, Nakanuma Y. Aberrant expression of trefoil factor family 1 in biliary epithelium in hepatolithiasis and cholangiocarcinoma. Lab Invest. 2003;83:1403–1413. doi: 10.1097/01.lab.0000092230.59485.9e. [DOI] [PubMed] [Google Scholar]
- 29.Vestergaard EM, Nexø E, Tørring N, Borre M, Ørntoft TF, Sørensen KD. Promoter hypomethylation and upregulation of trefoil factors in prostate cancer. Int J Cancer. 2010;127:1857–1865. doi: 10.1002/ijc.25209. [DOI] [PubMed] [Google Scholar]
- 30.Jinawath N, Chamgramol Y, Furukawa Y, Obama K, Tsunoda T, Sripa B, Pairojkul C, Nakamura Y. Comparison of gene expression profiles between Opisthorchis viverrini and non-Opisthorchis viverrini associated human intrahepatic cholangiocarcinoma. Hepatology. 2006;44:1025–1038. doi: 10.1002/hep.21330. [DOI] [PubMed] [Google Scholar]
- 31.Poulsom R, Hanby AM, Lalani EN, Hauser F, Hoffmann W, Stamp GW. Intestinal trefoil factor (TFF 3) and pS2 (TFF 1), but not spasmolytic polypeptide (TFF 2) mRNAs are co-expressed in normal, hyperplastic, and neoplastic human breast epithelium. J Pathol. 1997;183:30–38. doi: 10.1002/(SICI)1096-9896(199709)183:1<30::AID-PATH1085>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 32.Farrell JJ, Taupin D, Koh TJ, Chen D, Zhao CM, Podolsky DK, Wang TC. TFF2/SP-deficient mice show decreased gastric proliferation, increased acid secretion, and increased susceptibility to NSAID injury. J Clin Invest. 2002;109:193–204. doi: 10.1172/JCI12529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubeykovskaya Z, Dubeykovskiy A, Solal-Cohen J, Wang TC. Secreted trefoil factor 2 activates the CXCR4 receptor in epithelial and lymphocytic cancer cell lines. J Biol Chem. 2009;284:3650–3662. doi: 10.1074/jbc.M804935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong WM, Poulsom R, Wright NA. Trefoil peptides. Gut. 1999;44:890–895. doi: 10.1136/gut.44.6.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamachika T, Werther JL, Bodian C, Babyatsky M, Tatematsu M, Yamamura Y, Chen A, Itzkowitz S. Intestinal trefoil factor: a marker of poor prognosis in gastric carcinoma. Clin Cancer Res. 2002;8:1092–1099. [PubMed] [Google Scholar]