Abstract
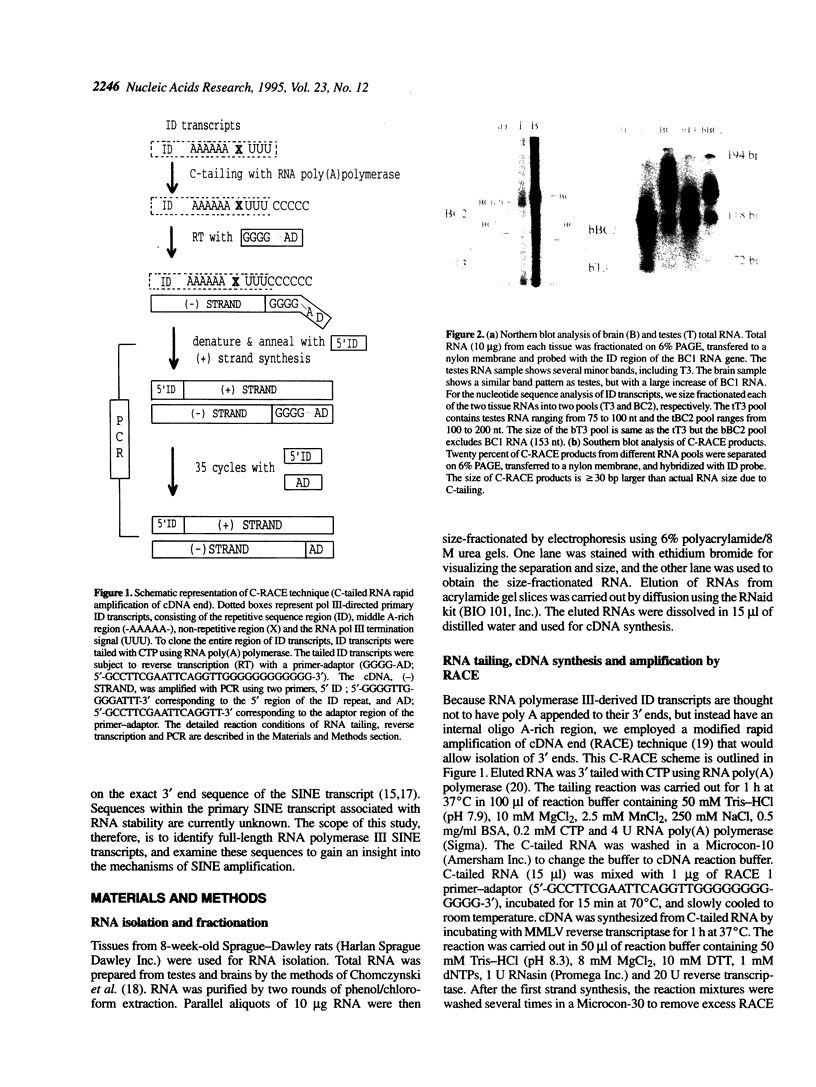
ID elements comprise a rodent SINE (short interspersed DNA repetitive element) family that has amplified by retroposition of a few master genes. In order to understand the important factors of SINE amplification, we investigated the transcription of rat ID elements. Three different size classes of ID transcripts, BC1, BC2 and T3, have been detected in various rat tissues, including brain and testes. We have analysed the nucleotide sequences of testes- and brain-derived ID transcripts isolated by size-fractionation, C-tailing and RACE. Nucleotide sequence variation of testes ID transcripts demonstrated derivation from different loci. However, the transcripts represent a preferred set of ID elements that closely match the subfamily consensus sequences. The small ID transcripts, T3, are not comprised of primary transcripts, but are instead processed polyA-transcripts generated from many different loci. These truncated transcripts would be expected to be retroposition-incompetent forms. Therefore, the amplification of ID elements is likely to be regulated at multiple steps of retroposition, which include transcription and processing. Although brain ID transcripts showed a similar pattern, with the addition of very high levels of transcription from the BC1 locus, we also found evidence that a single locus dominated the production of brain BC2 RNA species. BC1 RNA is highly stable in both germ line and brain cells, based on the low level of detection of the processing product, T3. This stability of BC1 RNA might have been a contributing factor in its role as a master gene for ID amplification.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anzai K., Kobayashi S., Suehiro Y., Goto S. Conservation of the ID sequence and its expression as small RNA in rodent brains: analysis with cDNA for mouse brain-specific small RNA. Brain Res. 1987 Apr;388(1):43–49. doi: 10.1016/0169-328x(87)90019-2. [DOI] [PubMed] [Google Scholar]
- Batzer M. A., Gudi V. A., Mena J. C., Foltz D. W., Herrera R. J., Deininger P. L. Amplification dynamics of human-specific (HS) Alu family members. Nucleic Acids Res. 1991 Jul 11;19(13):3619–3623. doi: 10.1093/nar/19.13.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Daniels G. R., Deininger P. L. Repeat sequence families derived from mammalian tRNA genes. 1985 Oct 31-Nov 6Nature. 317(6040):819–822. doi: 10.1038/317819a0. [DOI] [PubMed] [Google Scholar]
- DeChiara T. M., Brosius J. Neural BC1 RNA: cDNA clones reveal nonrepetitive sequence content. Proc Natl Acad Sci U S A. 1987 May;84(9):2624–2628. doi: 10.1073/pnas.84.9.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P. L., Batzer M. A., Hutchison C. A., 3rd, Edgell M. H. Master genes in mammalian repetitive DNA amplification. Trends Genet. 1992 Sep;8(9):307–311. doi: 10.1016/0168-9525(92)90262-3. [DOI] [PubMed] [Google Scholar]
- Devos R., van Emmelo J., Seurinck-Opsomer C., Gillis E., Fiers W. Addition by ATP: RNA adenylyltransferase from Escherichia coli of 3'-linked oligo(A) to bacteriophage Qbeta RNA and its effect on RNA replication. Biochim Biophys Acta. 1976 Oct 18;447(3):319–327. doi: 10.1016/0005-2787(76)90055-1. [DOI] [PubMed] [Google Scholar]
- Frohman M. A., Dush M. K., Martin G. R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Martignetti J. A., Shen M. R., Brosius J., Deininger P. Rodent BC1 RNA gene as a master gene for ID element amplification. Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3607–3611. doi: 10.1073/pnas.91.9.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. M., Schmid C. W. Proposed roles for DNA methylation in Alu transcriptional repression and mutational inactivation. Nucleic Acids Res. 1993 Mar 25;21(6):1351–1359. doi: 10.1093/nar/21.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia R. J., Chang D. Y., Wolffe A. P., Vorce R. L., Hsu K. The RNA polymerase III terminator used by a B1-Alu element can modulate 3' processing of the intermediate RNA product. Mol Cell Biol. 1992 Apr;12(4):1500–1506. doi: 10.1128/mcb.12.4.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia R. J., Driscoll C. T., Bilyeu T., Hsu K., Darlington G. J. Multiple dispersed loci produce small cytoplasmic Alu RNA. Mol Cell Biol. 1993 Jul;13(7):4233–4241. doi: 10.1128/mcb.13.7.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia R. J. The subset of mouse B1 (Alu-equivalent) sequences expressed as small processed cytoplasmic transcripts. Nucleic Acids Res. 1991 Oct 25;19(20):5695–5702. doi: 10.1093/nar/19.20.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martignetti J. A., Brosius J. BC200 RNA: a neural RNA polymerase III product encoded by a monomeric Alu element. Proc Natl Acad Sci U S A. 1993 Dec 15;90(24):11563–11567. doi: 10.1073/pnas.90.24.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon R. D., Danielson P., Brow M. A., Bloom F. E., Sutcliffe J. G. Expression of small cytoplasmic transcripts of the rat identifier element in vivo and in cultured cells. Mol Cell Biol. 1987 Jun;7(6):2148–2154. doi: 10.1128/mcb.7.6.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien C. A., Harley J. B. A subset of hY RNAs is associated with erythrocyte Ro ribonucleoproteins. EMBO J. 1990 Nov;9(11):3683–3689. doi: 10.1002/j.1460-2075.1990.tb07580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapienza C., St-Jacques B. 'Brain-specific' transcription and evolution of the identifier sequence. 1986 Jan 30-Feb 5Nature. 319(6052):418–420. doi: 10.1038/319418a0. [DOI] [PubMed] [Google Scholar]
- Schmid C. W. Human Alu subfamilies and their methylation revealed by blot hybridization. Nucleic Acids Res. 1991 Oct 25;19(20):5613–5617. doi: 10.1093/nar/19.20.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C., Maraia R. Transcriptional regulation and transpositional selection of active SINE sequences. Curr Opin Genet Dev. 1992 Dec;2(6):874–882. doi: 10.1016/s0959-437x(05)80110-8. [DOI] [PubMed] [Google Scholar]
- Sinnett D., Richer C., Deragon J. M., Labuda D. Alu RNA transcripts in human embryonal carcinoma cells. Model of post-transcriptional selection of master sequences. J Mol Biol. 1992 Aug 5;226(3):689–706. doi: 10.1016/0022-2836(92)90626-u. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G., Milner R. J., Gottesfeld J. M., Lerner R. A. Identifier sequences are transcribed specifically in brain. Nature. 1984 Mar 15;308(5956):237–241. doi: 10.1038/308237a0. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]