Abstract
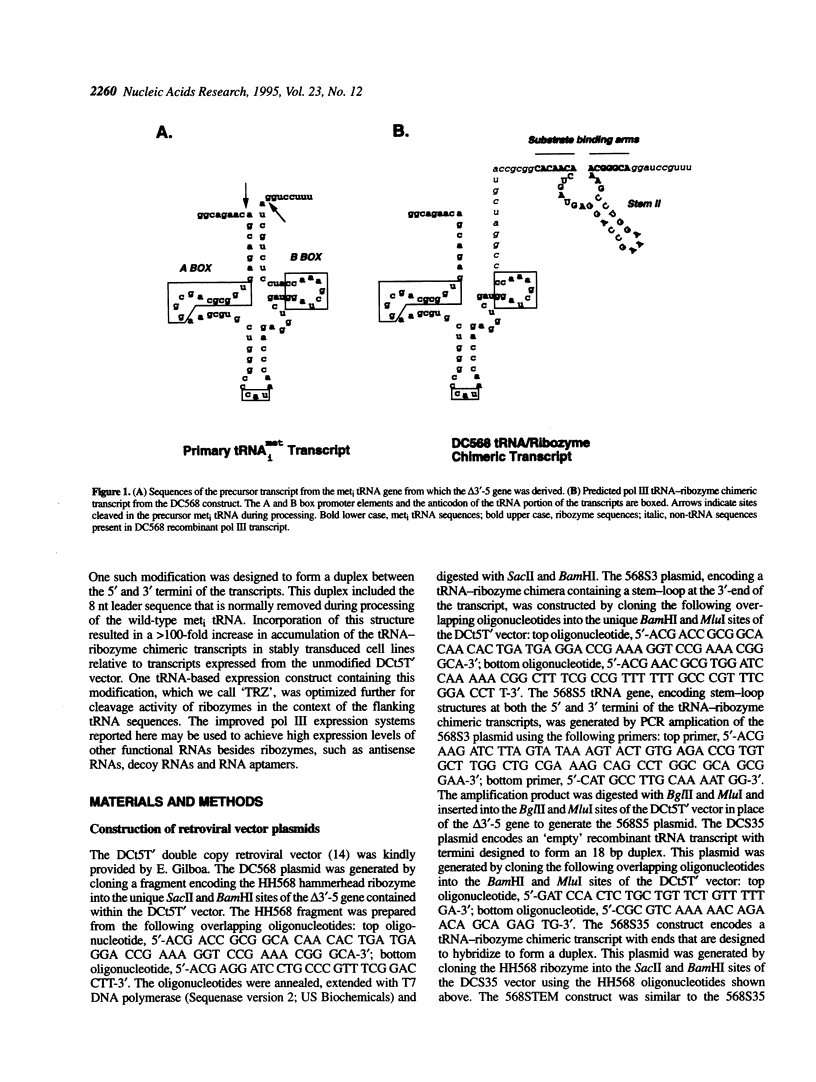
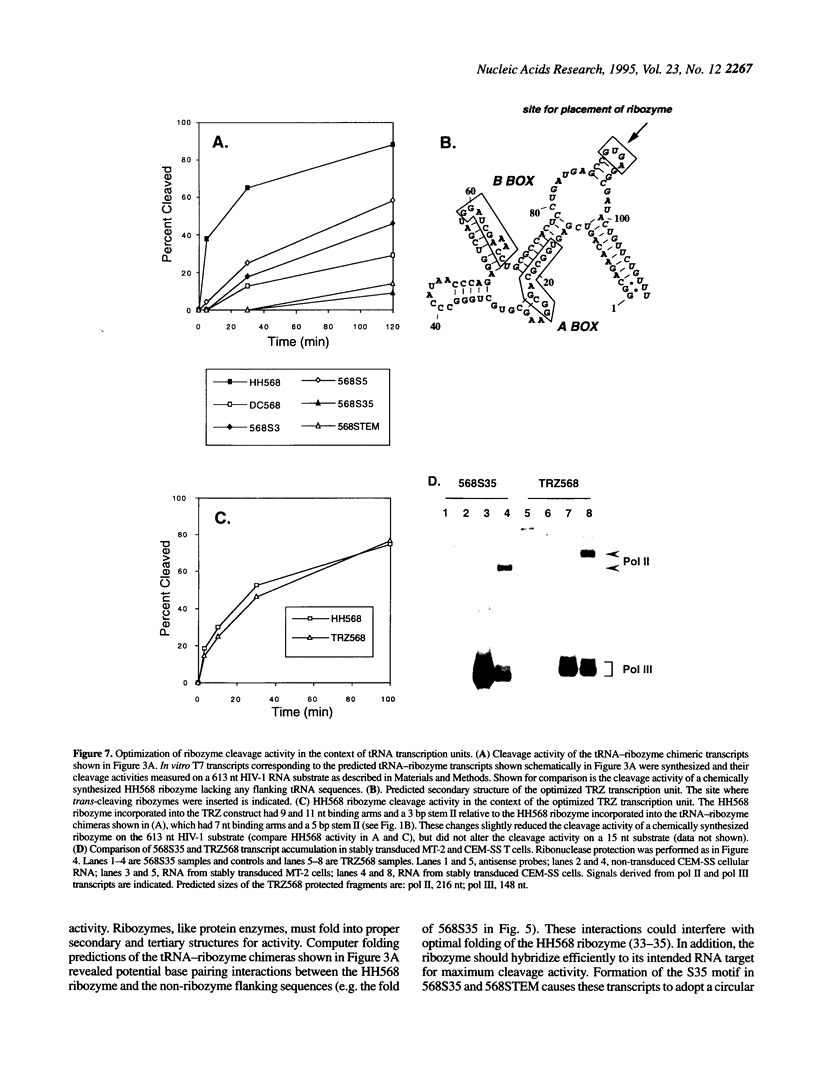
RNA polymerase III (pol III) transcripts are abundant in all cells. Therefore, pol III promoters may be ideal for expressing high levels of exogenous RNAs, such as antisense RNAs, decoy RNAs and ribozymes, in many different cell types. We have improved accumulation of recombinant RNAs expressed from a human meti tRNA-derived pol III promoter > 100-fold by modifying the 3' terminus of the transcripts to hybridize to the 5' terminus. This terminal duplex includes the 8 nt leader sequence present in the primary wild-type meti tRNA transcript that is normally removed during processing to the mature tRNA. Expression of an anti-HIV ribozyme was analyzed in cells stably transduced with retroviral vectors encoding pol III transcription units containing this modification. High accumulation of recombinant pol III ribozyme transcripts was observed in all cell lines tested. Due to the enhanced transcript accumulation, ribozyme cleavage activity was readily detectable in total RNA extracted from stably transduced human T cell lines. One pol III transcription unit, termed 'TRZ', was optimized further for ribozyme cleavage activity. The improved pol III transcription units reported here may be useful for expressing a variety of functional and therapeutic RNAs.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adeniyi-Jones S., Romeo P. H., Zasloff M. Generation of long read-through transcripts in vivo and in vitro by deletion of 3' termination and processing sequences in the human tRNAimet gene. Nucleic Acids Res. 1984 Jan 25;12(2):1101–1115. doi: 10.1093/nar/12.2.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D. Gene therapy. Intracellular immunization. Nature. 1988 Sep 29;335(6189):395–396. doi: 10.1038/335395a0. [DOI] [PubMed] [Google Scholar]
- Bertrand E., Pictet R., Grange T. Can hammerhead ribozymes be efficient tools to inactivate gene function? Nucleic Acids Res. 1994 Feb 11;22(3):293–300. doi: 10.1093/nar/22.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevec D., Volc-Platzer B., Zimmermann K., Dobrovnik M., Hauber J., Veres G., Böhnlein E. Constitutive expression of chimeric neo-Rev response element transcripts suppresses HIV-1 replication in human CD4+ T lymphocytes. Hum Gene Ther. 1994 Feb;5(2):193–201. doi: 10.1089/hum.1994.5.2-193. [DOI] [PubMed] [Google Scholar]
- Bock L. C., Griffin L. C., Latham J. A., Vermaas E. H., Toole J. J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992 Feb 6;355(6360):564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Dropulić B., Jeang K. T. Gene therapy for human immunodeficiency virus infection: genetic antiviral strategies and targets for intervention. Hum Gene Ther. 1994 Aug;5(8):927–939. doi: 10.1089/hum.1994.5.8-927. [DOI] [PubMed] [Google Scholar]
- Dropulić B., Lin N. H., Martin M. A., Jeang K. T. Functional characterization of a U5 ribozyme: intracellular suppression of human immunodeficiency virus type 1 expression. J Virol. 1992 Mar;66(3):1432–1441. doi: 10.1128/jvi.66.3.1432-1441.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of virusoid RNA is performed by the proposed 55-nucleotide active site. Cell. 1987 Jul 3;50(1):9–16. doi: 10.1016/0092-8674(87)90657-x. [DOI] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Hampel A., Tritz R., Hicks M., Cruz P. 'Hairpin' catalytic RNA model: evidence for helices and sequence requirement for substrate RNA. Nucleic Acids Res. 1990 Jan 25;18(2):299–304. doi: 10.1093/nar/18.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada S., Koyanagi Y., Yamamoto N. Infection of HTLV-III/LAV in HTLV-I-carrying cells MT-2 and MT-4 and application in a plaque assay. Science. 1985 Aug 9;229(4713):563–566. doi: 10.1126/science.2992081. [DOI] [PubMed] [Google Scholar]
- Heidenreich O., Eckstein F. Hammerhead ribozyme-mediated cleavage of the long terminal repeat RNA of human immunodeficiency virus type 1. J Biol Chem. 1992 Jan 25;267(3):1904–1909. [PubMed] [Google Scholar]
- Hélène C., Toulmé J. J. Specific regulation of gene expression by antisense, sense and antigene nucleic acids. Biochim Biophys Acta. 1990 Jun 21;1049(2):99–125. doi: 10.1016/0167-4781(90)90031-v. [DOI] [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1990;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- Jennings P. A., Molloy P. L. Inhibition of SV40 replicon function by engineered antisense RNA transcribed by RNA polymerase III. EMBO J. 1987 Oct;6(10):3043–3047. doi: 10.1002/j.1460-2075.1987.tb02610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. W., Gallardo H. F., Gilboa E., Smith C. Inhibition of human immunodeficiency virus type 1 in human T cells by a potent Rev response element decoy consisting of the 13-nucleotide minimal Rev-binding domain. J Virol. 1994 Dec;68(12):8254–8264. doi: 10.1128/jvi.68.12.8254-8264.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. C., Sullenger B. A., Gallardo H. F., Ungers G. E., Gilboa E. Overexpression of RRE-derived sequences inhibits HIV-1 replication in CEM cells. New Biol. 1992 Jan;4(1):66–74. [PubMed] [Google Scholar]
- Lisziewicz J., Sun D., Smythe J., Lusso P., Lori F., Louie A., Markham P., Rossi J., Reitz M., Gallo R. C. Inhibition of human immunodeficiency virus type 1 replication by regulated expression of a polymeric Tat activation response RNA decoy as a strategy for gene therapy in AIDS. Proc Natl Acad Sci U S A. 1993 Sep 1;90(17):8000–8004. doi: 10.1073/pnas.90.17.8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz D., Goff S., Bank A. Construction and use of a safe and efficient amphotropic packaging cell line. Virology. 1988 Dec;167(2):400–406. [PubMed] [Google Scholar]
- Nara P. L., Fischinger P. J. Quantitative infectivity assay for HIV-1 and-2. Nature. 1988 Mar 31;332(6163):469–470. doi: 10.1038/332469a0. [DOI] [PubMed] [Google Scholar]
- Noonberg S. B., Scott G. K., Garovoy M. R., Benz C. C., Hunt C. A. In vivo generation of highly abundant sequence-specific oligonucleotides for antisense and triplex gene regulation. Nucleic Acids Res. 1994 Jul 25;22(14):2830–2836. doi: 10.1093/nar/22.14.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojwang J. O., Hampel A., Looney D. J., Wong-Staal F., Rappaport J. Inhibition of human immunodeficiency virus type 1 expression by a hairpin ribozyme. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10802–10806. doi: 10.1073/pnas.89.22.10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarver N., Rossi J. Gene therapy: a bold direction for HIV-1 treatment. AIDS Res Hum Retroviruses. 1993 May;9(5):483–487. doi: 10.1089/aid.1993.9.483. [DOI] [PubMed] [Google Scholar]
- Sullenger B. A., Gallardo H. F., Ungers G. E., Gilboa E. Overexpression of TAR sequences renders cells resistant to human immunodeficiency virus replication. Cell. 1990 Nov 2;63(3):601–608. doi: 10.1016/0092-8674(90)90455-n. [DOI] [PubMed] [Google Scholar]
- Sullenger B. A., Lee T. C., Smith C. A., Ungers G. E., Gilboa E. Expression of chimeric tRNA-driven antisense transcripts renders NIH 3T3 cells highly resistant to Moloney murine leukemia virus replication. Mol Cell Biol. 1990 Dec;10(12):6512–6523. doi: 10.1128/mcb.10.12.6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N. R., Rossi J. J. Ribozyme-mediated cleavage of an HIV-1 gag RNA: the effects of nontargeted sequences and secondary structure on ribozyme cleavage activity. Antisense Res Dev. 1991 Summer;1(2):173–186. doi: 10.1089/ard.1991.1.173. [DOI] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990 Aug 3;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Ventura M., Wang P., Ragot T., Perricaudet M., Saragosti S. Activation of HIV-specific ribozyme activity by self-cleavage. Nucleic Acids Res. 1993 Jul 11;21(14):3249–3255. doi: 10.1093/nar/21.14.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. M. Antisense RNA and DNA. Sci Am. 1990 Jan;262(1):40–46. doi: 10.1038/scientificamerican0190-40. [DOI] [PubMed] [Google Scholar]
- Willis I. M. RNA polymerase III. Genes, factors and transcriptional specificity. Eur J Biochem. 1993 Feb 15;212(1):1–11. doi: 10.1111/j.1432-1033.1993.tb17626.x. [DOI] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]