Abstract
There is now substantial evidence that the eukaryotic nucleus consists of highly organized structures. Among such structures are transcription factories that consist of an ensemble of genes recruited by the RNA polymerase machinery. Here we suggest that antigen receptor variable regions are similarly organized. Specifically, we propose that the immunoglobulin heavy chain locus variable gene segments are anchored to the base of rosettes, wrapping around a cavity that contains the recombination machinery. We suggest that the folding of the chromatin fiber into rosettes underpins a critical mechanism by which antigen receptor diversity is generated.
Introduction
The chromatin fiber is not randomly organized but folds into elaborate patterns to allow high-density packing and long-range genomic interactions to occur with the appropriate frequencies. The chromatin fiber is organized into nucleosomes, consisting of 146 bp DNA elements that surround octamers of histones. Specifically, two copies of H2A, H2B, H3 and H4 form the core of the nucleosome. The nucleosomes themselves are organized into a 10 nm fiber, which in turn, folds into a 30 nm chromatin fiber.
Our knowledge about the folding of chromatin beyond the 30 nm fiber is still rudimentary. Distinct patterns for the folding of the chromatin fiber have been proposed. These involve helical and radial structures that permit packing at relatively high densities [1,2,3]. Studies using electron microscopy have suggested that chromosomes are organized as loops that are clustered as rosettes [4,5].
To describe chromatin topology in quantitative terms, polymer models that can be experimentally tested have been generated. Prominent among these are the Random Walk/Giant Loop (RW/GL), the Multi-Loop-Subcompartment (MLS) and Random-Loop (RL) models [6,7,8]. The RW/GL model describes the chromatin fiber as being confined to relatively large loops (2–5Mbp) [7,9]. The MLS configuration suggests that the chromatin fiber folds into bundles of loops [8]. The bundles consist of approximately ten loops and together span on average 1 Mbp of DNA. Flexible linkers of variable sizes have been proposed to separate the bundles of loops [8]. More recently, yet another design, the RL model has been proposed to underpin long-range chromatin topology [10]. The RL configuration allows both small and large loops to fold and unfold in a dynamic fashion [11]. Here we will discuss how novel computational, geometric and genome-wide approaches have provided new insights into long-range chromatin structure and propose that transcription and recombination factories have common structural features.
The structure of antigen receptor loci
Knowledge of how genetic loci are folded in 3D-space is still rudimentary. Perhaps the best-characterized structure involves the immunoglobulin heavy chain locus [12]. The Igh locus consists of distinct DNA elements encoding the variable (V), diversity (D), joining (J), and constant (C) regions. It is the largest known genetic locus. Fifteen partially dispersed VH region families, encoding for approximately 195 VH regions span approximately 3 Mbp of the murine genome. Large intergenic regions that span up to 50 kbp in size separate the individual VH regions. Located down-stream of the VH regions are 10–13 DH elements, four JH elements, and eight CH regions encoding for the various isotypes.
Using a geometric approach, named trilateration, the mean relative 3D-positions of the VH, DH, JH and CH gene segments in pre-pro-B and pro-B cells were determined [12]. In pre-pro-B cells, the DH-JH region is found within close proximity of the CH elements but away from the majority of the VH regions. The proximal and distal VH regions are separated from each other and do not seem to intermingle. In contrast, in preparation for recombination at the pro-B cell stage the proximal and distal VH regions appear to have merged and juxtaposed to the DHJH elements, providing equal opportunities for the entire VH repertoire [12].
These findings have raised the question as to whether all antigen receptor loci are organized in a similar fashion. Recent studies that involved the TCRα locus have indicated that not all antigen receptor loci are spatially organized as the Igh locus [13]. The TCRα locus encodes for approximately 100 Vα regions that span a 1.5 Mbp genomic region. The distal Vα regions are initially separated by relatively large spatial distances from the Jα gene segments but are juxtaposed to Jα gene segments during progressive rearrangements deleting proximal Vα regions [13]. Within the TCRα locus is embedded another locus encoding for antigen receptors, termed TCRδ. The TCRδ locus undergoes rearrangement in thymocyte progenitors, whereas the TCRα locus recombines in maturing thymocytes. Distal Vα regions are in a contracted state in thymocyte progenitors, but become de-contracted upon maturation. It has been proposed that the contracted conformation of the TCRα/δ locus permits efficient rearrangements of Vδ variable gene segments in early progenitors while the de-contraction in the TCRα locus initially restricts rearrangements only to the most proximal located Vα regions [13]. Thus, the more distally located Vα gene segments would only be positioned into close spatial proximity of the Jα regions upon progressive deletions of the proximal Vα elements. Overall, these studies indicate the presence of highly ordered and developmental regulated topologies that permit encounters between V, D and J regions to occur with the appropriate frequencies.
Rosettes and Anchors
As aforementioned, using a geometric approach it was recently revealed that during the transition from the pre-pro-B to the pro-B cell stage, the Igh locus undergoes large-scale conformational changes [14]. These data bring into question how the Igh fiber is folded. Comparison of experimental and simulated spatial distances as a function of genomic separation predicts that Igh locus topology is organized into clusters of loops consistent with an MLS configuration [12]. The organization of the Igh locus as clusters of loops is not unique to antigen receptor loci. Rosette-like structures have been observed in mitotic as well as interphase chromosomes, including regions such as the T helper type-2 cytokine locus as well as the bithorax complex and a genomic region involved in the development of Prader-Willi syndrome [15,16,17,18].
What are the components that promote a well-ordered yet dynamic structure of rosettes? How are the large-scale structural changes established during developmental progression? It seems likely that protein tethers mediate this function. Binding sites for putative anchors, including YY1 and CTCF, have been identified throughout the genome. YY1 deficient pro-B cells showed significant abnormalities in Igh locus rearrangement that appear to involve the distal VH regions. Further scrutiny revealed that YY1 in pro-B cells acts to promote Igh locus contraction [19]. CTCF is a multiple zinc-finger containing protein that binds to its target sites with particularly high affinity. Genome-wide studies have been illuminating in a sense that they provided a global view of CTCF occupancy and its partners named cohesins [20]. The cohesins consist of four core subunits termed Smc1, Smc3, Rad21 and Scc3. During DNA replication, the cohesins interact with sister chromatids by forming a ring-like structure, surrounding the two strands. Thus, the emerging view is that cohesins and CTCF act in concert to promote the assembly of loops. CTCF forms multimers between putative anchors and cohesins act to stabilize loop formation. Recent data have indicated that cohesins perform critical roles during transcription as well by stabilizing loop formation between enhancer and promoter elements that involve the mediator complex [21,22].
CTCF binding sites also span antigen receptor loci. Up to fifty binding sites span the entire Igh locus [23]. The CTCF binding sites span the VH region cluster but are absent within the DH - JH cluster. Other factors that have been found to affect Igh locus topology, possibly involved in tethering, are Ikaros and Pax-5. Ikaros and Pax-5 play critical roles in early B cell development. They are dispensable for the induction of DHJH and proximal VH-DHJH gene rearrangement, but are absolutely required for DNA recombination involving the distal VH regions [24,25,26,27]. Thus, it now seems settled that there are multiple molecular components that play a critical role in chromatin topology. Likely an array of additional players will be identified that perform this function. The critical challenge will be to determine how they act together to modulate chromatin structure as well as chromatin dynamics. Are the participants that appear to modulate Igh locus topology restricted to the antigen receptor loci or are they involved genome-wide? The latter possibility seems more likely. YY1 and CTCF are expressed ubiquitously and clearly perform a universal function.
Clearly much progress has been made in identifying factors that promote the folding of chromatin into rosette-like configurations. Genome-wide occupancy studies have provided a global view of the binding patterns of such factors. It should be mentioned that genome-wide binding patterns are based on populations of cells. However, it is the differences in locus topology between single cells that are particularly intriguing and perhaps more fundamental to our understanding of chromosome function than the average trajectories taken by the chromatin fiber.
Transcription factories
Nascent transcripts are not randomly localized in the nucleus. Transcripts have been detected in a limited number of nuclear structures, sensitive to inhibitors that affect transcription initiation and/or elongation. Such nuclear structures were named transcription factories because of their similarities with replication factories [28]. Transcription factories range in size between 40–100 nm in diameter. They include, based on the number of nascent RNA transcripts up to eight RNA polymerases [28,29,30,31,32]. Rather compelling data has recently been accumulated indicating that DNA is recruited to and pulled through the RNA polymerase machinery. It was shown that during transcriptional elongation, distinct genes, localized in cis and in trans, are brought into close spatial proximity, and pulled through relatively immobile RNA polymerase II complexes [33]. These remarkable studies demonstrate that transcription factories contain immobile RNA polymerase II molecules, through which transcribed genes are transported.
The number of transcription factors varies greatly, between hundreds and thousands, depending on cell context. Both intrachromosomal and interchromosomal interactomes have been observed within transcription factories [32]. Using formaldehyde-cross linking approaches it was revealed that co-regulated genes are primarily transcribed in common hubs [34]. These data raise the interesting possibility that a proportion of transcription factories become specialized because of an increase in the concentration of particular transcription factors, and suggesting that subsets of genes that are co-ordinately regulated are selectively recruited to specific nuclear structures to become transcribed. Although chromosomes are folded into distinct units, intermingling of chromosomes have been observed as well documented for nucleoli. To what extent trans-interaction between chromosomes do occur within transcriptiona factories remains to be determined. Since the number of transcription factories is limited it is to be expected that a substantial portion of transcribed loci is recruited by polymerases present in transcription factories. This might be achieved by random walk behaviour. Alternatively controlled and directed motion of chromatin fibers, through mechanisms yet to be revealed, may promote the assembly of transcription factories. Finally, now we are faced with the question as to how such factories are organized in 3D-space. 3D-FISH combined with geometric approaches as well as formaldehyde cross-linking strategies should provide further insight into the topologies of these intriguing nuclear structures [12,35].
Transcription and recombination factories
As aforementioned recent data have revealed that the immunoglobulin heavy chain locus is organized as clusters of loops, similar to those proposed by the MLS model [14,35,36]. How the loops are established and maintained remains unknown. Recently it has been proposed that the formation of clusters of loops in the Igh locus involves transcription, in a manner similar as described for transcription factories [37]. Non-coding, intergenic and antisense transcription has been observed at multiple locations throughout antigen receptor loci [38,39,40]. Such transcripts are initiated by V-region promoter and/or enhancer elements and elongate across coding, intergenic and regulatory regions. Non-coding anti-sense transcription is initiated prior to antigen receptor V(D)J gene rearrangements and is developmentally regulated. It was convincingly shown that non-coding transcription plays a critical role in antigen receptor assembly at the TCRα locus [41]. Specifically, it was demonstrated that interference with elongation of non-coding RNA transcription severely interfered with VαJα gene rearrangements [41]. This then raises the interesting possibility that RNA polymerases ‘fixed’ within transcription factories, reel in antigen receptor loci to elongate non-coding transcripts. As non-coding transcripts are pulled through the RNA polymerase complex, recombination signal sequences may become accessible to the recombination machinery [37].
Rosettes providing equal opportunities for all
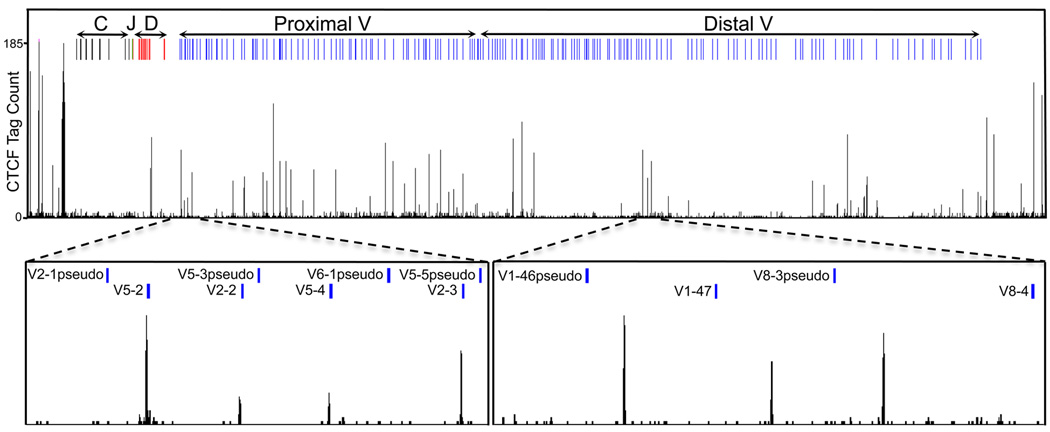
Is active non-coding transcription the only critical player in establishing clusters of loops that mediate DNA recombination? It seems unlikely. We have recently identified genome-wide CTCF occupancy in pro-B cells using ChIP-Seq [42]. As aforementioned, CTCF spans the entire Igh locus with the exception of the CH-DH region (Figure 1) [23,42]. Here we have explored further the possibility that CTCF functions to promote looping and recruit the VH regions to the recombination machinery. Specifically, we have plotted CTCF occupancy and VH region localization (Figure 1). A close relationship between CTCF occupancy and the positions of a large subset of VH regions was observed (Figure 1). Remarkably, CTCF binding sites close to the proximal VH regions immediately flank, in an asymmetric pattern, the 3’ end of recombination signal sequences (Figure 2). This pattern was not observed for pseudo VH regions. Rather they seem to be located away from the CTCF binding sites. Pseudo VH regions do not rearrange giving support for the idea that in order for proximal VH regions to recombine they need to be located within close genomic proximity to CTCF binding sites.
Figure 1.
Proximal variable regions in the immunoglobulin heavy chain locus are frequently associated with nearby CTCF occupancy. CTCF binding, determined by ChIP-sequencing is shown for Rag-1 deficient pro-B cells (42). CTCF occupancy is closely linked with the presence of proximal VH regions. Only few VH regions are highlighted but a similar pattern is observed for the entire proximal VH region cluster. Pseudo VH regions, on the other hand, are not located within close genomic proximity of the proximal VH regions.
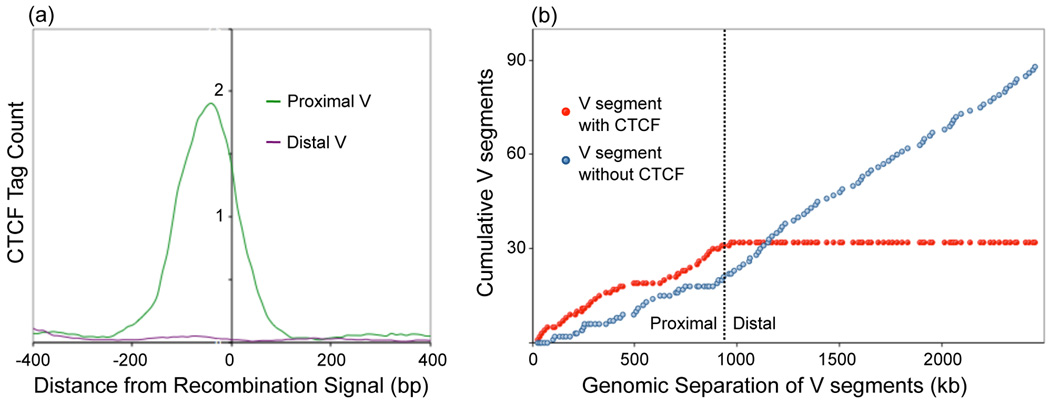
Figure 2.
Cumulative frequency distributions of VH segments located adjacent CTCF binding site. (a, b) A large fraction of proximal VH segments are located within 100 bp of CTCF binding. (b) A clear reversal of this trend is present at the border between proximal and distal VH segments suggesting that at least two separate mechanisms are involved in structuring the locus for recombination.
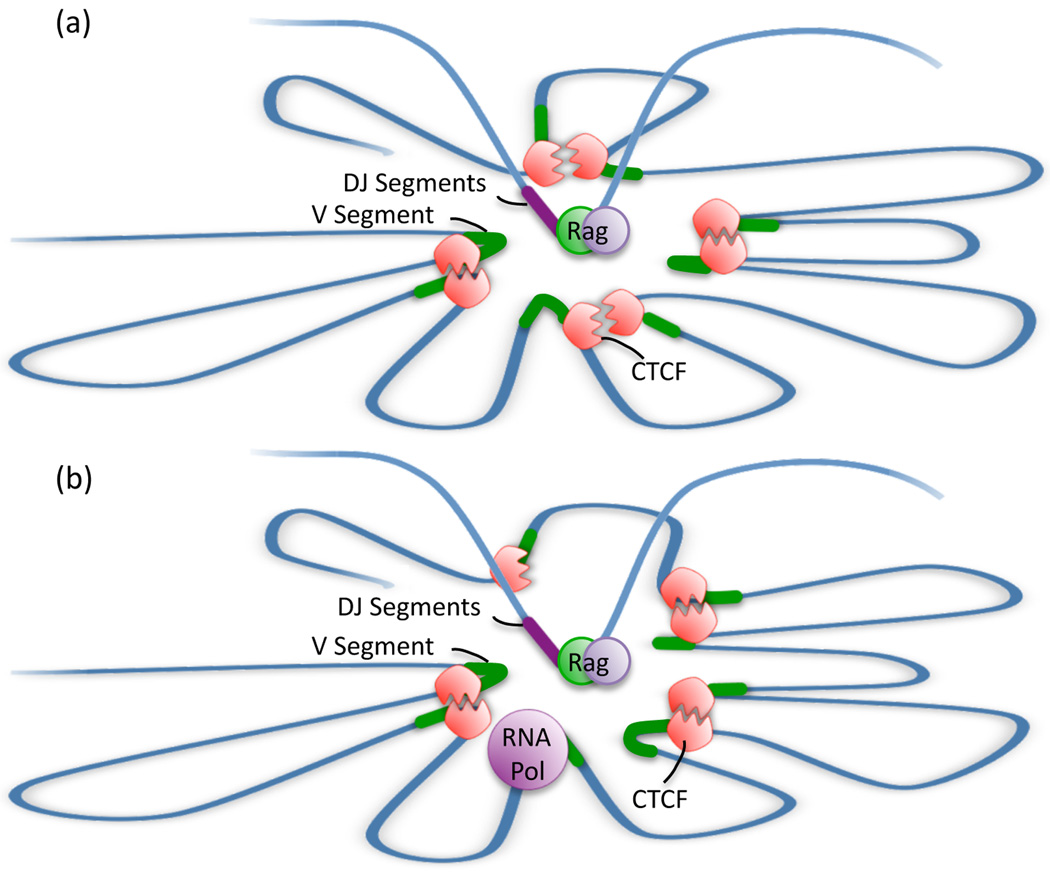
These data bring into question as to how CTCF occupancy and the presence of proximal VH regions relate to antigen receptor rearrangement. Here we would like to propose the following: As aforementioned, the spatial distances measured in the Igh locus agree well with simulated rosette-like structures [12]. Based on genome-wide occupancy studies, we suggest that the proximal VH regions are organized as rosettes by CTCF [42]. Specifically, we would like to propose that CTCF wraps the proximal VH regions around a cavity (Figure 3). The cavity itself, we suggest, contains the DH-JH elements associated with RAG1/2, previously named the recombination center (Figure 3) [43].
Figure 3.
Model describing the potential role of CTCF in Igh locus rearrangement. Spatial distances measured within the Igh locus suggest that it is folded into rosette-like structures. While it is currently unknown what proteins anchor the bases of these rosettes, the presence of CTCF adjacent to proximal VH segments makes it a key candidate. (a) In such a model, the VH segments are in close proximity, surrounding an inner cavity. At the pro-B cell stage, the DH segments bound by the recombination enzymes have the ability to move into this cavity, creating an equal probability of recombination with any of the VH segments. Incorporation of distal VH segments into these structures is likely CTCF independent. Note that in this model loop formation provides access of half of the proximal V regions to the recombinase located within the cavity whereas the other half becomes positioned within the loop and might not be accessible to the recombination machinery. (b) Transcription through the locus prior to recombination may disrupt long-range genomic interactions, allowing new interactions to form.
We note that in the proposed configuration, half of the VH regions that flank CTCF occupancy are wrapped around the cavity and thus would become accessible to the recombinase machinery. On the other hand, the other half of proximal VH regions would positioned within the loop and plausibly less likely to encounter a DHJH element (Figure 3). Then how does in such a configuration the entire repertoire of proximal VH regions encounter DHJH elements with similar frequencies? It seems likely that loop formation is dynamic, permitting loops to rapidly associate and dissociate, establishing new neighbours, permitting the entire set of proximal VH regions equal access to the recombination center.
Conclusion
While the CTCF sites flank recombination signal sequences for a substantial fraction of the Igh proximal VH regions, such a correlation was not found for the distal VH regions. As previously suggested, it may very well be that the distal VH regions are recruited to the recombination center by the RNA polymerase machinery [37]. Thus, the distal and proximal recombination signal sequences may use different mechanisms in order to be organized properly in 3D-space.
How similar are transcription and recombination factories? Do they share common features in that multiple DNA regions are recruited to a given domain containing the relevant recombination machinery? If rearrangement does occur at transcription factories, how does RNA polymerase interact with CTCF and could this allow the structure of rosettes to be constantly changing? Many questions remain. Regardless of the precise mechanism, the cardinal point of the model proposed here is that the folding of the chromatin fiber, into clusters of loops that position VH regions at the base of rosettes, permits an equal playing field for the Igh VH region repertoire.
ACKNOWLEDGEMENTS
This work was supported by funds provided by the NIH (CM). JL is supported by the Cellular and Molecular Genetics training grant. CB is supported by fellowships from the European Molecular Biology Organization and the Swiss National Science Foundation. We apologize to those not referenced because of space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Rattner JB, Lin CC. Radial loops and helical coils coexist in metaphase chromosomes. Cell. 1985;42(1):291–296. doi: 10.1016/s0092-8674(85)80124-0. [DOI] [PubMed] [Google Scholar]
- 2.Sedat J, Manuelidis A. A direct approach to the structure of eukaryotic chromosomes; Cold Spring Harbor Symp. Quant. Biol; 1978. pp. 331–350. [DOI] [PubMed] [Google Scholar]
- 3.Paulson JR. Chromosomes and Chromatin. Vol. 3. Boca Raton, Florida: CRC Press; 1988. Scaffolding and radial loops: the structural organization of metaphase chromosomes; pp. 3–36. [Google Scholar]
- 4.Paulson JR, Laemmli UK. The structure of histone depleted metaphase chromosomes. Cell. 1977;12(3):817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- 5.Okada TA, Comings DE. Higher order structure of chromosomes. Chromosoma. 1979;72(1):1–14. doi: 10.1007/BF00286426. [DOI] [PubMed] [Google Scholar]
- 6.Trask BJ, Allen S, Massa H, Fertitta A, Sachs R, van den Engh G, Wu M. Studies of metaphase and interphase chromosomes using fluorescence in situ hybridization. Cold Spring Harb Symp Quant Biol. 1993;58:767–775. doi: 10.1101/sqb.1993.058.01.084. [DOI] [PubMed] [Google Scholar]
- 7.Sachs RK, van den Engh G, Trask B, Yokota H, Hearst JE. A random-walk/giant-loop model for interphase chromosomes. Proc. Nat. Acad. Sci. USA. 1995;92(7):2710–2714. doi: 10.1073/pnas.92.7.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Münkel C, Langowski J. Chromosome structure predicted by a polymer model. Phys. Rev. E. 1998;57(5B):5888–5896. **This study provides a compelling model for long-range chromatin topology, suggesting that the eukaryotic chromatin fiber is organized into bundles of loops that are separated by flexible linkers.
- 9.Yokota H, van den Engh G, Hearst JE, Sachs RK, Trask BJ. Evidence for the organization of chromatin in megabase pair-sized loops arranged along a random walk path in the human G0/G1 interphase nucleus. J. Cell Biol. 1995;130(6):1239–1249. doi: 10.1083/jcb.130.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bohn M, Heermann DW, van Driel R. Random loop model for long polymers. Phys. Rev. 2007;76:051805-1–051805-8. doi: 10.1103/PhysRevE.76.051805. [DOI] [PubMed] [Google Scholar]
- 11. Mateos-Langerak J, Bohn M, de Leeuw W, Giromus O, Manders EM, Verschure PJ, Indemans MH, Gierman HJ, Heermann DW, van Driel R, Goetze S. Spatially confined folding of chromatin in the interphase nucleus. Proc. Natl. Acad. Sci USA. 2009;106(10):3812–3817. doi: 10.1073/pnas.0809501106. *Here a model of long-range chromatin structure is proposed in which both large and small loops interact in a dynamic fashion to modulate chromatin folding.
- 12.Jhunjhunwala S, van Zelm MC, Peak M, Cutchin S, Riblet R, van Dongen JJ, Grosveld F, Knoch TA, Murre C. The 3D-structure of the immunoglobulin heavy chain locus: Implications for long-range genomic interactions. Cell. 2008;133(2):265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shih HY, Krangel MS. Distinct contracted conformations of the TCRa/TCRd locus during Tcra and Tcrd recombination. J. Exp. Med. 2010;207(9):1835–1841. doi: 10.1084/jem.20100772. *In this study, the topology of the TCRa/d locus is revealed providing insight into the mechanism that underpins the developmental regulation of DNA recombination in two critical antigen receptor loci.
- 14.Jhujhunwala S, van Zelm MC, Peak MM, Murre C. Chromatin architecture and the generation of antigen receptor diversity. Cell. 2009;138(3):435–448. doi: 10.1016/j.cell.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 2006;38(11):1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 16.Lanzuolo C, Roure V, Dekker J, Bentignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithoroax complex. Nat. Cell Biol. 2007;9(10):1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- 17.Rauch J, Knoch TA, Solovei I, Teller K, Stein S, Buiting K, Horsthemke B, Langowski J, Cremer T, Hausmann M, Cremer C. Light optical precision measurements of the active and inactive Prader-Willi syndrome imprinted regions in human cell nuclei. Differentiation. 2008;76(1):66–82. doi: 10.1111/j.1432-0436.2007.00237.x. [DOI] [PubMed] [Google Scholar]
- 18.Fraser J, Rousseau M, Shenker S, Ferraiuolo MA, Hayashizaki Y, Blanchette M, Dostie J. Chromatin conformation signatures of cellular differentiation. Genome Biology. 2009;10(4):R37. doi: 10.1186/gb-2009-10-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu H, Schmidt-Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H, Pelanda R, Reth M, Skok J, Rajewsky K, Shi Y. Yin Yang 1 is a critical regulator of B cell development. Genes Dev. 2007;21(1):1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132(3):422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460(7253):410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467(7314):430–435. doi: 10.1038/nature09380. **In this study it is revealed that cohesins modulate gene expression by linking enhancers and core promoter elements.
- 23.Degner S, Wong TP, Jankevicius G, Feeney AJ. Developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J. Immunol. 2009;182(1):44–48. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18(4):411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roldán E, Fuxa M, Chong W, Martinez D, Novatchkova M, Busslinger M, Skok J. Locus 'decontraction' and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat. Immunol. 2005;6(1):31–41. doi: 10.1038/ni1150. *In this study evidence is presented raising the possibility that allelic exclusion is regulated by spatial proximity.
- 26.Reynaud D, Demarco IA, Reddy KL, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat. Immunol. 2008;9(8):927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sayegh C, Jhunjhunwala S, Riblet R, Murre C. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev. 2005;19(3):322–327. doi: 10.1101/gad.1254305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerases are localized within discrete transcription “factories” in human nuclei. J. Cell Sci. 1996;109(6):1427–1436. doi: 10.1242/jcs.109.6.1427. *First study that introduced transcription factories as discrete nuclear structures.
- 29.Martin S, Pombo A. Transcription factories: Quantitative studies of nanostructures in the mammalian nucleus. Chromosome Res. 2003;11(5):461–470. doi: 10.1023/a:1024926710797. [DOI] [PubMed] [Google Scholar]
- 30.Pombo A, Jackson DA, Hollinshead M, Wang Z, Roeder RG, Cook PR. Regional specialization in human nuclei: Visualization of discrete sites of transcription by RNA polymerase III. EMBO J. 1999;18(8):2241–2253. doi: 10.1093/emboj/18.8.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eskiw CH, Rapp A, Carter DR, Cook PR. RNA polymerase II activity is located on the surface of protein-rich transcription factories. J. Cell Sci. 2008;121(12):1999–2007. doi: 10.1242/jcs.027250. [DOI] [PubMed] [Google Scholar]
- 32.Chakalova L, Fraser P. Organization of transcription. Cold Spring Harbor Perspectives in Biology. 2010;2(9):a000729. doi: 10.1101/cshperspect.a000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Papantonis A, Larkin JD, Wada Y, Ohta Y, Ihara S, Kodama T, Cook PR. Active RNA polymerases: mobile or immobile molecular machines? PLoS Biol. 2010;8(7):e1000419. doi: 10.1371/journal.pbio.1000419. **This is an interesting study providing compelling evidence for relatively fixed sites for active polymerases.
- 34. Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 2010;42(1):53–61. doi: 10.1038/ng.496. *This study reveals how transcription factors act to recruit an ensemble of genes that are co-regulated to distinct transcription factories.
- 35. Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. **This is a landmark study providing insight into the fractal nature of eukaryotic chromatin folding.
- 36.Feeney A. Epigenetic regulation of V(D)J recombination. Semin Immunol. 2010;22(6):311–312. doi: 10.1016/j.smim.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corcoran AE. The epigenetic role of non-coding RNA transcription and nuclear organization in immunoglobulin repertoire generation. Semin Immunol. 2010;22(6):353–361. doi: 10.1016/j.smim.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 38.Yancopoulos G, Alt F. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985;40(2):271–281. doi: 10.1016/0092-8674(85)90141-2. [DOI] [PubMed] [Google Scholar]
- 39.Bolland DJ, Wood AL, Johnston CM, Bunting SF, Morgan C, Chakalova L, Fraser PJ, Corcoran AE. Antisense intergenic transcription in V(D)J recombination. Nat. Immunol. 2004;5(6):630–637. doi: 10.1038/ni1068. [DOI] [PubMed] [Google Scholar]
- 40.Bergman Y, Cedar H. Epigenetic control of recombination in the immune system. Semin Immunol. 2010;22(6):323–329. doi: 10.1016/j.smim.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Abarrategui I, Krangel MS. Regulation of T cell receptor-alpha gene recombination by transcription. Nat. Immunol. 2006;7(10):1109–1115. doi: 10.1038/ni1379. *Here unambiguous evidence is presented for a role of transcriptional elongation in antigen receptor assembly.
- 42.Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, Ideker T, Glass CK, Murre C. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 2010;11(7):635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141(3):419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]