Abstract
Melanization is an innate immune response in arthropods that encapsulates and kills invading pathogens. One of its rate-limiting steps is the activation of prophenoloxidase (PPO), which is controlled by an extracellular proteinase cascade and serpin inhibitors. The molecular composition of this system is largely unknown in mosquitoes with the exception of serpin-2 (SRPN2), which was previously identified as a key negative regulator of melanization. Using reverse genetic and biochemical techniques, we identified the Anopheles gambiae clip-serine proteinase CLIPB9 as a PPO-activating proteinase, which is inhibited by SRPN2. Double knockdown of SRPN2 and CLIPB9 reversed the pleiotrophic phenotype induced by SRPN2 silencing. This study identifies the first inhibitory serpin-serine proteinase pair in mosquitoes and defines a regulatory unit of melanization. Additionally, the interaction of CLIPB9 and SRPN2 affects the life span of adult female mosquitoes and therefore constitutes a well-defined potential molecular target for novel late-life acting insecticides.
Keywords: Anopheles gambiae, Innate immunity, Serpin, Serine proteinase, Malaria
Introduction
Vector-borne diseases, especially malaria, continue to be a major public health threat worldwide. In the absence of an effective vaccine, malaria control continues to mainly rely on vector control as a prevention strategy and, to a lesser extent, on drug treatment of the infected human population. Besides political and social challenges, the two currently used forms of vector control, insecticide-treated nets and indoor-residual spraying, face the technical challenge of insecticide resistance [1]. Metabolic and/or target-site resistance against all four classes of insecticides have been reported in mosquitoes. Besides resistance management that include rotations or mosaics of insecticides and improved insecticide resistance surveillance systems [2, 3], the use of novel insecticides specifically targeting old mosquitoes has recently been proposed [4]. In an ideal setting, so-called late-life acting (LLA) insecticides would kill infective females late in their reproductive cycle, thus providing minimized selective pressure on insecticide resistance. Existing biopesticides such as entomopathogenic fungi [5] show potential as LLA insecticides; however, employment of large-scale screens to identify new targets is hampered by lengthy bioassays to determine late-life killing. A potential known physiological target is the serine proteinase inhibitor, serpin (SRPN)2, from the African malaria mosquito, Anopheles gambiae. SRPN2 inhibits melanization, and its depletion from adult hemolymph shortens the life span of adult female mosquitoes significantly [6]. Our working hypothesis is that an insecticide that either depletes or inhibits SRPN2 would kill mosquitoes late in life prior to parasite transmission, minimizing the selection pressure for resistance.
Melanization is a prominent innate immune response in arthropods that encapsulates and kills invading microorganisms or pathogens. Biochemical studies in large insects, such as Bombyx mori, Manduca sexta, and Tenebrio molitor, and genetic analyses in Drosophila melanogaster have led to the current model of the melanization reaction [7–10]. Soluble pattern recognition proteins initially recognize nonself-molecular patterns. This interaction activates a clip-domain serine proteinase cascade, culminating in the activation of prophenoloxidase (PPO)-activating proteinase (PAP), also known as PPO-activating enzyme. Activated PAP directly converts inactive PPO to PO, which hydroxylates monophenols to catechols and oxidizes catechols to quinones. These in turn then polymerize to eumelanin [11]. PO activation is strictly controlled, presumably because overproduction of reactive semi-quinones and other toxic byproducts such as superoxide anion and/or other reactive oxygen species could be harmful to the insect. Additionally, the melanization response uses large quantities of aromatic amino acids, which could lead to tradeoffs with other life history traits, including longevity.
An orthologous group of the serpin superfamily of proteinase inhibitors has been identified to negatively regulate the PPO activation cascade in different insect species, including mosquitoes. This group includes serpin-3 from M. sexta [12], Spn27A from D. melanogaster [13, 14], and the aforementioned SRPN2 from An. gambiae [6] and Aedes agypti [15]. Serpins usually contain ~400 amino acid residues with an exposed reactive center loop (RCL) located at 30–40 residues from the carboxyl terminus. They function as suicide-substrate inhibitors by forming SDS-stable, covalent complexes with target proteinases after the cleavage of a scissile bond (designated P1–P1′) in the RCL [16].
Based on our previous work, we hypothesized that SRPN2 regulates the final step in PO activation by directly inhibiting one or more PAPs [17]. However, the identity of this target clip-domain serine proteinase in An. gambiae or any PAP in dipteran insects is unknown. Clip-domain serine proteinases implicated in PPO activation are synthesized as zymogens and share common structural features including one or two amino-terminal clip domains and a carboxyl-terminal serine proteinase catalytic domain [7]. The An. gambiae genome encodes 31 putative functional clip-domain serine proteinases (CLIPBs) [18]. Five of these, CLIPB3, 4, 8, 14, and 17, have been identified to affect parasite and/or bead melanization using reverse genetic methods [19–21]. It is possible that these proteinases are part of the PPO activation cascade; however, their precise contribution to melanization is unknown.
The overall aim of the current study was to identify and analyze a target proteinase of SRPN2 and to evaluate whether their interaction regulates melanization in vitro and in vivo. Using phylogenetic analysis of the CLIP protein family, we identify a potential candidate proteinase, biochemically characterize it, and describe its inhibition by SRPN2. Additionally, we show that this proteinase promotes melanization in vitro and can cleave PPO, leading to activation of the enzyme. Using reverse genetic approaches, we provide strong evidence that this serpin-proteinase pair is a key regulatory unit of melanization in An. gambiae in vivo.
Materials and methods
Insect rearing
The An. gambiae G3 strain was reared at 27°C and 80% humidity using a 12:12 light:dark cycle. After hatching, larvae were fed on baker’s yeast (Active Dry Yeast, Red Star) for 48 h and subsequently on fish food (TetraMin® Tropical Flakes, Tetra) and baker’s yeast. Adult mosquitoes were provided with sugar solution (8% fructose supplemented with 2.5 mM PABA; SIGMA) ad libitum. Heparinized horse blood (PlasVac) was used as a blood meal source and provided with an artificial glass feeder system (KSU, Chemistry Department) using stretched parafilm as the membrane.
Phylogenetic analysis of clip-serine proteinases
To identify An. gambiae serine proteinases that are related to known melanization factors, we aligned the catalytic domain sequences of all annotated, putatively active clip-domain serine proteinases from the genomes of An. gambiae, Aedes aegypti, Culex quinquefasciatus, Tribolium castaneum, D. melanogaster, and M. sexta; these were automatically aligned using the online version of MUSCLE (available at http://www.ebi.ac.uk/Tools/muscle/index.html) using default parameters [22, 23]. We examined and manually adjusted the alignment in JalView 2.5 [24, 25]. Columns, where we had low confidence that all residues in the column were related by point substitution events and any columns containing gaps were removed. We used this alignment for all subsequent phylogenetic analyses (Fig. S1).
ProtTest version 1.4 [26] was used to identify the best-fitting amino acid substitution model for the multiple sequence alignment. The guide tree was estimated using BioNJ, using the “Slow” search strategy mode, for those substitution matrices that are available in both PhyML and RAxML (JTT, MtREV, MtMam, Dayhoff, WAG, RtREV, CpREV, Blosum62, VT), using either no correction for between-site rate heterogeneity or an approximation to the gamma-distribution to model between-site heterogeneity using four different discrete rate categories (these models are described as “+G”). The WAG+G model was the optimal model under all possible orderings of the result of this ProtTest analysis.
RAxML version 7.0.4 [27] with the WAG+G model was used to (1) estimate the maximum likelihood (ML) tree for the alignment; (2) build ML trees estimated from 100 non-parametrically bootstrapped alignments based on the initial alignment; and (3) to calculate the frequency with which the split in the ML tree estimated from the initial alignment was found in the 100 non-parametrically bootstrapped ML trees. The resulting phylogeny was examined using FigTree version 1.2.3 (Rambaut 2009, http://tree.bio.ed.ac.uk/software/figtree/).
Recombinant CLIP9 production
To biochemically characterize An. gambiae CLIPB9, we produced recombinant protein using the baculovirus system. The complete coding region of proCLIPB9 was amplified by PCR from cDNA of 2-day-old female G3 mosquitoes using gene-specific primers (Table S1). The forward primer included a NotI site, and the reverse primer contained six codons for histidine residues followed by a stop codon and a HindIII site. The PCR product was digested with NotI and HindIII and then inserted into the same restriction sites in the vector pFastBac1 (Invitrogen). The resulting plasmid was used as a template to produce mutant proCLIPB9 (proCLIPB9Xa) plasmid according to the instructions of QuickChange® Multi Site-Directed Mutagenesis Kit (Stratagene). In proCLIPB9Xa, the activation site IGMR121 was replaced with IEGR121 to allow the cleavage and activation by factor Xa. The mutation was confirmed by sequencing, and the plasmid was used to generate a recombinant baculovirus according to the manufacturer’s instructions (Invitrogen). To express proCLIPB9Xa, Sf9 cells (2 × 106 cells/ml) in 1 l of Sf-900 II serum-free medium (Invitrogen) were infected with the recombinant baculovirus at multiplicity of infection of 1 and were incubated at 28°C with shaking at 150 rpm. The culture was harvested at 72 h post infection, and cells were removed by centrifugation at 5,000×g for 20 min at 4°C. The cell-free medium was mixed with 20 ml of Concanavalin A (ConA) Sepharose 4B resin (Amersham Biosciences) that had been equilibrated with buffer 1 (20 mM Tris-HCl, 0.5 M NaCl, 1 mM CaCl2, 1 mM MgCl2, 1 mM MnCl2, pH 7.5). After rotating overnight at 4°C, the mixture was packed into a column (2.0-cm i.d. × 6 cm) by gravity. Following a washing step with buffer 1 until A 280 was lower than 0.002, bound proteins were eluted with buffer 2 (20 mM Tris-HCl, 0.5 mM NaCl, 0.5 mM methyl-α-d-mannopyranoside, pH 7.5). The eluted factions were pooled and dialyzed against buffer 3 (50 mM Na2HPO4, 300 mM NaCl, pH 8.0; 2 l each time for 8 h, twice). The dialyzed sample was collected and supplemented with imidazole at a final concentration of 10 mM, and then purified by Ni-NTA agarose chromatography as described previously [28]. The eluted fractions containing recombinant proCLIPB10Xa were pooled and dialyzed against buffer 4 (20 mM Tris-HCl, 100 mM NaCl, pH 8.0). The dialyzed sample was collected and stored at −80°C.
Activation of recombinant proCLIPB9Xa
To activate proCLIPB9Xa, 5 μg of purified zymogen was incubated with 2 μg of commercial bovine factor Xa (New England Biolabs) in a total reaction volume of 110 μl (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 2 mM CaCl2) at 37°C for 6 h. Cleavage of the zymogen was determined by Western blot using 1.1 μl of the reaction.
Amidase activity of recombinant CLIPB9Xa
To measure proteinase function of CLIPB9, amidase activity was measured by incubating 3.3 μl of CLIPB9Xa with 200 μl of 50 μM acetyl-Ile-Glu–Ala-Arg-p-nitroanilide (IEARpNa) in 0.1 M Tris-HCl, pH 8.0, 0.1 M NaCl, 5 mM CaCl2 and monitoring changes in absorbance at 405 nm in a microplate reader (Bio-Tek Instrument, Inc.). One unit of amidase activity was defined as ΔA 405/min = 0.001.
Recombinant SRPN2 production
A cDNA fragment encoding the full-length mature SRPN2 was amplified using the gene-specific primers listed in Table S1. The forward primer included an NcoI site, which provided the start codon, followed by six codons for histidine residues. The reverse primer contained a HindIII restriction site at the 3′ end of stop codon. The PCR product was digested with NcoI and HindIII, and then inserted into the same restriction sites in the expression vector pET-28a (Novagen). The resulting plasmid was used to transform E. coli BL21(DE3) strain. For recombinant protein expression, these bacteria were grown at 37°C in LB medium containing 50 μg/ml of kanamycin. When the culture reached A 600 = 1.0, isopropyl β-d-thiogalactoside was added to a final concentration of 0.1 mM, and recombinant protein was expressed for 11 h at 25°C and 150 rpm. The bacteria were harvested by centrifugation and resuspended in lysis buffer (50 mM sodium phosphate, 300 mM NaCl, 10 mM imidazole, pH 8.0). Cells were lysed by sonication, and a cleared lysate was obtained by centrifugation. The soluble SRPN2 in the supernatant was purified by nickel-nitrilotriacetic acid (NTA) agarose chromatography according to Jiang et al. [28]. Subsequently, the purified protein was dialyzed against 20 mM Tris-HCl and 20 mM NaCl, pH 8.3, and applied to a pre-equilibrated Q-Sepharose™ Fast Flow column (1.0-cm i.d. × 4 cm). After washing the column with 20 mM Tris-HCl, pH 8.3, until A 280 was lower than 0.002, proteins were eluted at 1.0 ml/min with a linear gradient of 0–500 mM NaCl in 20 mM Tris-HCl, pH 8.3 for 50 min. Fractions containing recombinant protein were pooled and stored at −80°C.
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analyses
Protein samples were treated with 6× SDS sample buffer containing β-mercaptoethanol at 95°C for 5 min and then separated by 10% SDS-PAGE. Gels were stained with EZ-Run Protein Gel Staining Solution (Fisher Scientific). For immunoblot analysis, proteins were transferred onto a PVDF membrane and subjected to immunoblotting using mouse anti-His (1:2,000, GenScript), rabbit anti-M. sexta PPO (1:2,000, [29]), or rabbit anti-SRPN2 (1:2,000, [6]) as primary antibodies. Goat anti-mouse and anti-rabbit IgG HRP-conjugated secondary antibodies (Promega) were used at a 1:20,000 dilution. Antibody binding was visualized by Western Lightning Chemiluminescence Reagent Plus Kit (Perkin Elmer).
Detection of CLIPB9-SRPN2 complexes by Western blot and mass spectrometry (MS)
To detect the formation of SRPN2-CLIPB9 complex in vitro, recombinant proCLIPB9Xa was activated by factor Xa as described above and mixed with purified SRPN2 at a molar ratio of 1:10. After incubation at room temperature for 10 min, the reaction mixtures were subjected to 10% SDS-PAGE and Western blot analysis, loading the following amounts: 7 ng of factor Xa, 20 ng of CLIPB9Xa, and 150 ng of SRPN2. Bands of interest were subjected to trypsin digestion and 1D NanoLC ESI MS/MS analysis [30]. To detect the formation of serpin-proteinase complex in vivo, hemolymph was collected into PBS by proboscis-clipping from 25 2-day-old female adult An. gambiae and mixed with 50 ng of recombinant CLIPB9Xa. After incubation at 37°C for 10 min, samples were subjected to SDS-PAGE and Western blot analysis.
Inhibition of CLIPB9Xa activity by SRPN2
To directly measure the inhibitory potential of SRPN2 on CLIPB9 function, we incubated 120 ng of CLIPB9Xa with recombinant SRPN2 at different molar ratios. In control reactions, the same amount of factor Xa used to activate proCLIPB10Xa was substituted for active CLIPB10Xa. After incubation at room temperature for 10 min, residual amidase activity was measured as described above. Amidase activity of CLIPB9Xa was defined as the activity of CLIPB9Xa minus the activity of factor Xa alone. The second-order association rate constant (k a) of interactions between SRPN2 and CLIPB9 was determined according to Schick et al. [31].
PPO activation assays
To measure the ability of recombinant CLIPB9 to activate PPO in vitro, 50 ng of proCLIPB9Xa or CLIPB9Xa was incubated with 1 μl of 1:10 diluted prescreened plasma from day-2 fifth instar M. sexta larvae. Alternatively, 250 ng of proCLIPB9Xa or CLIPB9Xa was incubated with 40 ng of purified PPO (kindly provided by M. Gorman, Kansas State University). Samples were subjected to SDS-PAGE and Western blot analysis using antibodies against M. sexta PPO. To determine PO activity, 500 ng of recombinant proCLIPB9Xa or CLIPB9Xa were mixed with 3 μl of plasma. Additionally, 250 ng of recombinant proCLIPB9Xa or CLIPB9Xa was mixed with 1 μg of purified M. sexta PPO. After incubation at either at room temperature for 20 min (plasma) or 37°C for 30 min (purified PPO), PO activity in the reaction mixtures was measured using dopamine as a substrate [30].
Double-stranded RNA (dsRNA) production
SRPN2, CLIPB9, and GFP dsRNA were synthesized and injected as described previously [6, 21]. Primer pairs for template amplification are listed in Table S1.
Phenotypic analyses
To determine if CLIPB9 and SRPN2 interact, in vivo reverse genetic analyses were performed. Adult female G3 mosquitoes were injected with dsRNA within 48 h of eclosion and fed on sugar solution ad libitum throughout the experiments. To assess survival curves 30 mosquitoes were injected according to Blandin et al. [32] for each experimental group, and their survival was assessed every 24 h for 14 days. Assays were performed in four independent biological repeats. Differences in survival curves were assessed by rank log test and differences in survival proportions on specific days by paired Student’s t test, independently for each data set and in the combined data set (Fig. S3). To assess differences in melanotic psuedotumor formation, abdominal wall preparations were made 12 days post-injection according to [6]. Abdomens were examined at 40× magnification with Axio Imager A1 microscope (Zeiss) and photographed with an AxioCam MRc5 camera (Zeiss). Images were analyzed for size and number of tumors in each abdomen using ImageJ software (NIH). Differences in total melanotic area were assessed by the Mann-Whitney U test and the Kolmogorov–Smirnov (KS) test. The KS method was used as a normality test, employing the Dallal and Wilkinson approximation to Lilliefors’ method to compute the P value. All statistical anlyses were performed using GraphPad Prism software (GraphPad Software, Inc.).
Results
Phylogenetic analysis of active CLIP serine proteinases
In order to identify putative candidate proteinases that could function as PAPs in An. gambiae, we initially performed a phylogenetic analysis of CLIP serine proteinases. However, while the estimated phylogeny contained some well-supported clans [33], many of the clans have low support (Fig. S2). One clan groups An. gambiae CLIPB9 and CLIPB10 with M. sexta PAP1 to the exclusion of other An. gambiae and M. sexta sequences. This clan also included Ae. aegypti IMP_2 and D. melanogaster CG3066, which, together with PAP1, are known to function in melanization [10, 15]. However this clan had only very low bootstrap support. A similar grouping, with low, but unspecified, bootstrap support, is found in Zou et al. [15]. Support values did not show any major improvements when the analysis was repeated using more alignment columns (approximately 50% more than used in the analysis described here).
Additional analyses using another ML phylogeny estimation tool, PhyML [34], and using Bayesian analysis of phylogeny as implemented by MrBayes [35] yielded similar results to the RAxML analysis.
Production and activation of recombinant proCLIPB9Xa
In order to investigate whether An. gambiae CLIPB9 has the ability to function as a PAP, we produced active proteinase in vitro. As the protein is expressed as a zymogen (proCLIPB9) and its endogenous activating enzyme is unknown, we prepared a recombinant form of proCLIPB9 (proCLIPB9Xa), mutated to permit its activation by commercially available bovine factor Xa. SDS-PAGE analysis of purified proCLIPB9Xa suggests that the recombinant protein has a mass of ~56 kDa, approximately 14 kD larger than predicted based on its sequence (Fig. S3). The increased mass is likely due to glycosylation, as the protein sequence contains three predicted N-linked glycosylation sites, and recombinant proCLIPB9Xa can be purified using Concanavalin A resin.
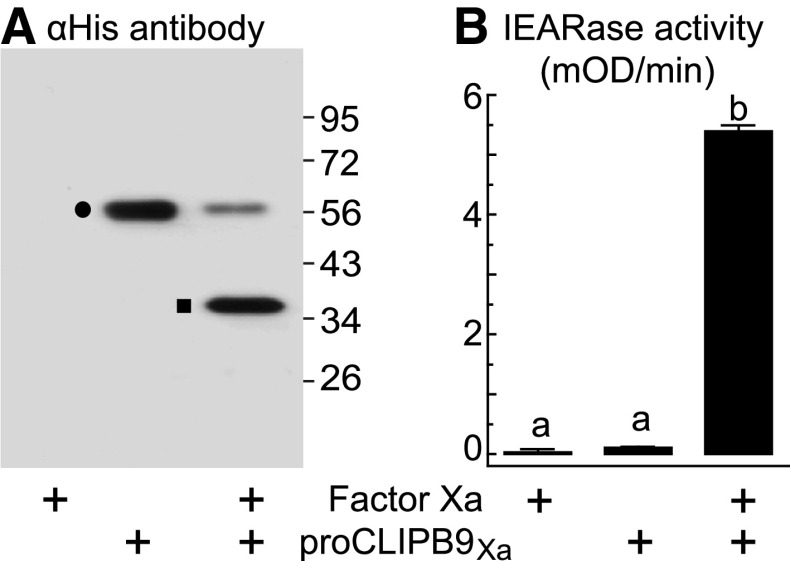
Incubation of proCLIPB9Xa with factor Xa resulted in decreased intensity of the 56-kDa zymogen band and appearance of a 35-kDa band corresponding to the catalytic domain (Fig. 1a), as expected for activation cleavage. We tested several artificial peptides as putative substrates for CLIPB9 proteinase activity (Table S2). Activation of proCLIPB9Xa by factor Xa resulted in a significant increase in activity as measured by cleavage of the artificial IEARpNA (Fig. 1b).
Fig. 1.
Activation of purified recombinant proCLIPB9Xa by factor Xa. Activation of the proteinase zymogen was analyzed by Western blot using mouse anti-His antibodies (a) and spectrophotometric assay using IEARpNA as a substrate (b). The bars represent mean ± SD (n = 3). Statistically significant differences are indicated with different letters (one-way ANOVA followed by Newman-Keuls test, P < 0.05). Circle, proCLIPB9Xa zymogen; square, catalytic domain of proCLIPB9Xa
CLIPB9 is directly inhibited by An. gambiae SRPN2
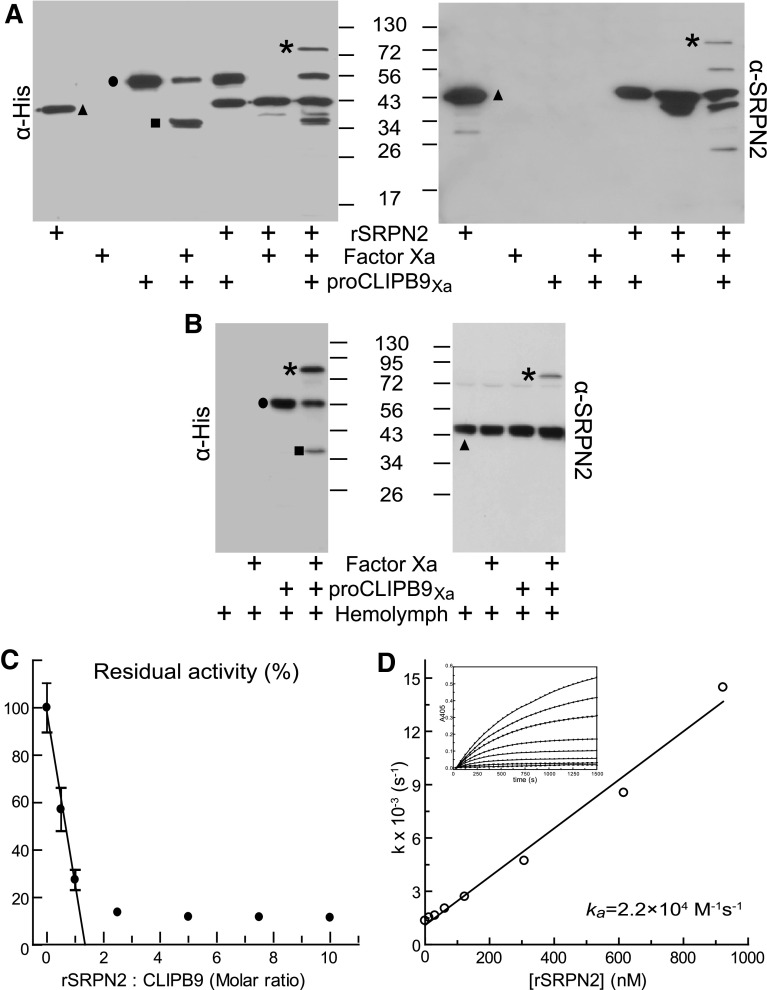
A characteristic feature of serpin inhibition is the formation of an SDS-stable complex of the serpin and its cognate proteinase [16]. In a first step to determine if CLIPB9 can be directly inhibited by SRPN2, we tested if these two proteins form such complexes in vitro. We mixed purified recombinant SRPN2 with CLIPB9Xa and detected the appearance of a high molecular weight complex by Western blot, using anti-His or anti-SRPN2 antibodies (Fig. 2a). In the absence of active CLIPB9Xa, anti-His antibodies recognized the 56-kDa CLIPB9Xa zymogen and 43-kDa recombinant SRPN2 (Fig. 2a, left panel). When SRPN2 was mixed with active CLIPB9Xa, no 34-kDa band corresponding to the CLIPB9 catalytic domain was detected. However, a new immunoreactive band at ~75 kDa (the expected mass of the SRPN2/CLIPB9Xa complex) was observed, which was also recognized by anti-SRPN2 antibodies (Fig. 2a, right panel). Analysis of tryptic peptides from the ~75-kDa band by MS/MS identified SRPN2 and CLIPB9 (Table S3), confirming that SRPN2 forms a covalent complex with active CLIPB9Xa.
Fig. 2.
An. gambiae SRPN2 binds and inhibits CLIPB9. a, b Western blot analysis of SRPN2-CLIPB9 complex. CLIPB9Xa was incubated with recombinant SRPN2 (a) or An. gambiae hemolymph (b). Western blot analysis was performed using mouse anti-His (left) or rabbit anti-SRPN2 (right) antibodies. Triangles, non-complexed SRPN2; circles, proCLIPB9Xa zymogen; squares, catalytic domain of proCLIPB9Xa; asterisks, SRPN2-CLIPB9 complex. (c) Inhibition of CLIPB9 activity by recombinant SRPN2. Purified recombinant SRPN2 was incubated with CLIPB9Xa. The residual IEARase activities of CLIPB9Xa were plotted as mean ± SD (n = 3) against the corresponding molar ratios of SRPN2 and CLIPB9Xa. (d) Kinetic analysis of the inhibition of CLIPB9 by SRPN2. CLIPB9Xa (2.8 pmol) was added to a mixture of 300 μM IEARpNa and SRPN2 at 0, 12.3, 30.8, 61.5, 123, 307.5, 615, and 922.5 nM. The progress of enzyme inactivation at each concentration of SRPN2 was followed by measuring the ΔA405 of the reaction every 49 s (inset). The rate of complex formation was calculated as described in [31]
To test if this complex can be formed in vivo, Anopheles hemolymph was mixed with recombinant CLIPB9Xa and analyzed by Western blot. Anti-His antibodies detected a higher molecular weight band of ~75 kDa in addition to the 34-kDa band corresponding to the CLIPB9 catalytic domain (Fig. 2b, left panel). This band was also recognized by anti-SRPN2 antibodies (Fig. 2b, right panel), indicating that native SRPN2 has the potential to form complexes with CLIPB9 in vivo. In the control reaction mixtures, anti-SRPN2 antibodies recognized a ~45-kDa band corresponding to naive SRPN2 and a faint ~72-kDa band, which likely represents native inhibitory complexes of SRPN2 with either CLIPB9 or other hemolymph proteinases.
To confirm that this complex formation indeed leads to the inhibition of CLIPB9, we tested SRPN2’s ability to inhibit the IEARase activity of CLIPB9Xa. CLIPB9Xa activity decreased linearly as SRPN2 concentration increased (Fig. 2c). The stoichiometry of inhibition was 1.3, indicating that under these experimental conditions SRPN2 almost exclusively acts as an inhibitor rather than a substrate of CLIPB9. The second order association rate constant (k a) for the inhibition of CLIPB9Xa by SRPN2 was 2.2 × 104 M−1s−1 (Fig. 2d), which is comparable to known inhibitory serpin-proteinase interactions [12]. SRPN2 did not inhibit factor Xa, as its amidase activity was unchanged by incubation with recombinant SRPN2 at different molar ratios (Fig. S4).
CLIPB9Xa cleaves and activates M. sexta PPO in plasma and in vitro
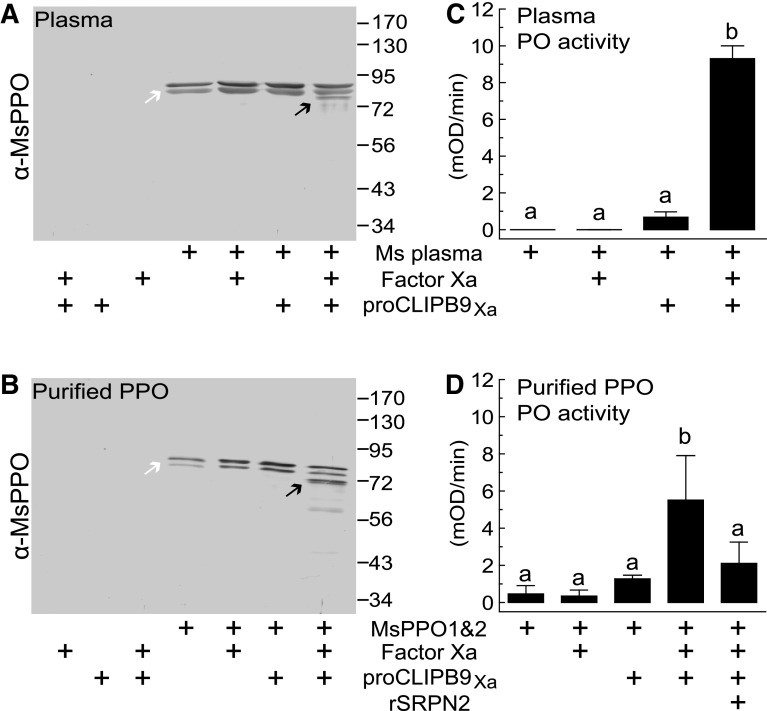
Biochemical analysis of the melanization cascade in mosquitoes is hindered by the limited amount of hemolymph that can be extracted from individual adult mosquitoes and the inability to produce functional recombinant PPO from any insect species [36]. To directly determine the putative function of CLIPB9 in PPO activation, we made use of the M. sexta model system [7]. PPO in the plasma of M. sexta is a heterodimer composed of 79-kDa PPO-p1 and 80-kDa PPO-p2 [37]. M. sexta plasma was incubated with factor Xa or proCLIPB9Xa zymogen. In subsequent Western blot analysis anti-M. sexta PO antibody identified two bands corresponding to PPO zymogen heterodimer [37], and no significant PO activity was detected (Fig. 3a, b). After M. sexta plasma was incubated with CLIPB9Xa, a 72-kDa doublet band corresponding to M. sexta PO was detected, and PO activity increased significantly (Fig. 3a, b), indicating that active CLIPB9Xa causes cleavage and activation of M. sexta PPO.
Fig. 3.
CLIPB9 cleaves and activates M. sexta PPO. CLIPB9Xa cleaves M. sexta PPO in plasma (a) and as purified protein (b), causing a significant increase in PO activity (c, d). Arrows mark the doublet bands representing PPO (white) and PO (black). Bars represent mean ± 1 SD (n = 3); statistically significantly differences are indicated by different letters (one-way ANOVA followed by Newman-Keuls test, P < 0.05)
To explore whether PPO is a substrate of CLIPB9, we incubated CLIPB9Xa with purified M. sexta PPO. As in Manduca plasma, Western blot analysis of purified PPO detected two bands with apparent molecular weight of 79 and 80 kDa. No change was observed after incubation with factor Xa or proCLIPB9Xa zymogen (Fig. 3c). However, when CLIPB9Xa was incubated with purified PPO, a new double band of ~72 kDa was observed, and PO activity of the mixture increased significantly (Fig. 3c, d). Pre-incubation of SRPN2 and CLIPB9Xa diminished the PO activity.
Taken together, these data show that CLIPB9 can act as a PAP in insect hemolymph and that this function is inhibited by SRPN2.
CLIPB9-knockdown (KD) partially reverts the SRPN2 depletion phenotype
Depletion of SRPN2 from the hemolymph of adult female An. gambiae mosquitoes causes a pleiotropic phenotype including reduced life span and spontaneous melanization as evidenced by the formation of melanotic pseudotumors ([6]; Fig. 4a). To test the extent to which CLIPB9 contributes to this phenotype in adult female mosquitoes, we performed genetic epistasis analyses of this switch regulation pathway [38]. In the absence of gene-knockout techniques in An. gambiae, double-knockdown (dKD) experiments were used measuring pseudotumor formation and life span. Adult female An. gambiae were injected with dsRNAs targeting the CLIP serine proteinase, CLIPB9, SRPN2, and both CLIPB9 and SRPN2, respectively; injection of dsGFP was used as a control. Gene KD in all cases was significant and specific as determined by quantitative RT-PCR (Fig. S5A, B). Reduction of CLIPB9 mRNA levels did not affect overall expression levels of SRPN2, and vice versa. Additionally, CLIPB9 silencing did not alter expression levels of other CLIP-serine proteinases, CLIPB3, CLIPB4, CLIPB8, CLIPB10, CLIPB14, and CLIPB17, which had previously been reported to affect melanization of malaria parasites or glass beads [10, 19–21].
Fig. 4.
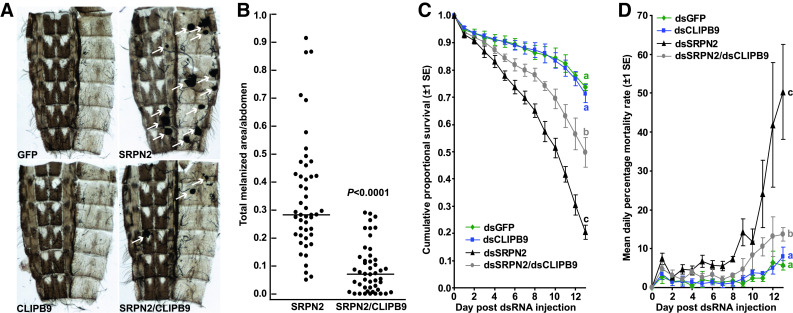
CLIPB9 knockdown partially reverts the SRPN2-depletion phenotype. a Images of abdominal wall preparations 12 days post injection; tumors are marked by white arrows. b The total melanotic area per abdomen (arbitrary unit) is significantly reduced in dsCLIPB9/dsSRPN2-treated compared to SRPN2-depleted mosquitoes (U test, P < 0.0001). No melanotic tumors were observed in dsGFP or dsCLIPB9-treated mosquitoes; thus, they were excluded from the analysis. cCLIPB9-KD alone did not affect survival, but partially rescued the mortality effect induced by SRPN2-KD. The combined data set of four independent biological repeats is shown; for individual results, see Fig. S2. Statistically significant differences between survival curves are indicated by letters (log rank test, df = 3, χ2 = 128.8, P < 0.0001; pairwise comparisons: dsGFP vs. dsCLIPB9, df = 1, χ2 = 0.351, P = 0.554; dsGFP vs. dsSRPN2, df = 1, χ2 = 99.8, P < 0.0001; dsGFP vs. dsCLIPB9/dsSRPN2, df = 1, χ2 = 19.45, P < 0.0001; dsSRPN2 vs. dsCLIPB9/dsSRPN2, df = 1, χ2 = 30.14, P < 0.0001). d Mean daily percent mortality rate from a above, calculated according to [5]
We first evaluated the effect of CLIPB9 knockdown on melanization using the appearance of melanotic tumors as a read-out (Fig. 4). Injection of dsGFP or dsCLIPB9 alone did cause melanization. In contrast, CLIPB9-KD in a SRPN2-depleted background significantly affected the SRPN2-KD melanization phenotype. Prevalence of melanotic tumors was decreased, and the average total melanotic area for each dissected mosquito abdomen was reduced four-fold (Mann-Whitney test, U = 206.5, P < 0.0001; KS test, D = 0.6672, P < 0.0001; Fig. 4a, b). Non-parametric tests were employed as data sets, and even after transformation, they deviated from normal distribution (KS test; P < 0.05).
When we compared the survival curves among the four different treatment groups, we observed significant differences (log rank test; df = 3, χ2 = 128.8, P < 0.0001; Fig. 4c, D, and S6). As previously reported [6], SRPN2 knockdown significantly shortened longevity as compared to the dsGFP-control treatment (P < 0.0001). In contrast, CLIPB9 knockdown had no effect on lifespan as compared to the control (P < 0.554). Importantly, dsCLIPB9/SRPN2 treatment substantially and significantly increased mosquito longevity as compared SRPN2-KD mosquitoes (P < 0.0001), albeit not to levels of control treatment (P < 0.0001). Likewise, the number of mosquitoes alive 14 days after dsRNA injection differed significantly among the four treatment groups (one-way ANOVA, df = 3, F = 54.7, R 2 = 0.9319, P < 0.0001, with Tukey’s multiple comparison test for pairwise analyses). While SRPN2-KD reduced the number of surviving mosquitoes by 75% (P < 0.0001, the number of CLIPB9/SRPN2-treated mosquitoes was only 30% lower than the number of control mosquitoes (P < 0.001).
Taken together, the silencing experiments strongly suggest that CLIPB9 is partially epistatic to SRPN2 and therefore either downstream of this serpin in the PPO activation cascade or required in a parallel pathway leading to activation of melanization. Intriguingly, we obtained similar results when analyzing survival instead of melanization, providing further evidence that excessive melanization can be harmful to the host.
Discussion
Melanization in An. gambiae can reduce or eliminate malaria parasite infections in allochthonous Plasmodium sp./An. gambiae combinations [6, 19, 39]. However this immune response is not elicited in autochthonous, co-adapted parasite vector combinations, due to active evasion by the parasite [40], and is rarely observed in the field [41]. Nevertheless, the melanization response is an important immune response against a variety of pathogens, and is likely to have large implications for a mosquito’s survival and reproduction. If uncontrolled, e.g., by the depletion of the inhibitor SRPN2, melanization can kill adult females late in life, and thus potentially reduce the vectorial capacity of An. gambiae. This feature makes PPO activation, which is a rate-limiting step in melanin production, a target for malaria control strategies. Specifically, an inhibitor of SRPN2, which is a key negative regulator of PPO activation, can reduce female mosquito longevity. Potentially, it would kill adult female mosquitoes after reproduction but before parasite transmission and thus act as an LLA insecticide.
In a first step to develop An. gambiae SRPN2 into an insecticide target, this study aimed to describe the inhibitory target of this serpin. To narrow down the number of putative candidate proteinases, we initially performed a detailed phylogenetic analysis of all known An. gambiae CLIPs that contain a functional catalytic triad. One clan grouped An. gambiae CLIPB9 and CLIPB10 with several proteins known to function in melanization. Additionally, CLIPB9 was previously shown to promote bead melanization [21] and therefore was a good candidate for PAP function in An. gambiae.
We produced a recombinant proCLIPB9 mutant in a baculovirus expression system and obtained active CLIPB9 in vitro. CLIPB9 indeed cleaved purified M. sexta PPO, resulting in significant levels of PO activity. Therefore, CLIPB9 can function as the terminal proteinase in the PPO activation pathway. To our knowledge, this is the first bona fide PAP identified in a dipteran species. In An. gambiae at least eight PPOs are expressed and present in the hemolymph of adult female mosquitoes [42, 43], of which seven share the same putative activation cleavage site as M. sexta PPO [18]. It is therefore possible that CLIPB9 acts as a PAP for the majority of PPOs in the An. gambiae hemolymph, which future experiments will have to validate.
The PAP function of CLIPB9 is inhibited by recombinant SRPN2 in vitro. The inhibitory complex formed at a rate comparable with those observed for other serpins acting on their physiological target proteinases [12]. The interaction of CLIPB9 and SRPN2 occurs in vivo, as reverse genetic analyses revealed that CLIPB9 is partially epistatic to SPRN2. Both aspects of the SRPN2-KD phenotype, melanotic tumor formation and decreased adult survival, are significantly reverted. However, this reversion is incomplete. A possible reason could be incomplete knockdown of CLIPB9, as its transcript levels in double-KD mosquitoes were reduced to roughly 40%, potentially allowing for residual CLIPB9 activity in these mosquitoes. An equally parsimonious explanation is that SRPN2 has other proteinase targets that contribute to its knockdown phenotype. Several lines of evidence support the second explanation: Firstly, arthropod as well as mammalian serpins commonly have several physiological proteinase targets. For example, serpin-3, the M. sexta ortholog of SRPN2, inhibits PAP-1 and PAP-3 [44], and human anti-thrombin III inhibits thrombin, factor Xa and several other plasma proteinases (reviewed in [45]). Secondly, in the An. gambiae genome, trypsin-like serine proteinases outnumber serpins by roughly 20:1. Thirdly, preliminary Western blot analysis of adult female An. gambiae hemolymph separated by 2D gel electrophoresis using anti-SRPN2 antibody highlighted several spots with higher molecular weight than the monomeric serpin. Based on our phylogenetic analysis and results in other mosquito species, putative candidates are CLIPB8 and CLIPB10. Experiments are underway to test for interaction of these proteinases with SRPN2.
The epigenetic relationship between CLIPB9 and SRPN2 also suggests that increased melanization and shortened life span have a causal relationship. Several cytotoxic byproducts, including semi-quinones and reactive oxygen intermediates (ROI) such as superoxide anion and hydroxyl radicals are produced during melanogenesis as a direct consequence of PO activity [11]. Presumably, the increase in systemic melanization caused by the SRPN2-KD leads to ROI levels that are cytotoxic and contribute to killing the mosquito. Future experiments determining hemolymph ROI concentrations will test this hypothesis.
In summary, this study identified the first regulatory unit controlling melanization in An. gambiae. It can modulate adult life expectancy, and its killing effect increases exponentially over time, killing mainly older mosquitoes. Its performance can be easily quantified using a convenient in vitro assay that is amendable to high-throughput screening of chemical libraries in order to identify small molecule inhibitors of SRPN2. Therefore, this unit potentially constitutes a well-defined target of novel LLA insecticides that aim to reduce older mosquito populations required for pathogen transmission.
Acknowledgments
We thank Dr. M. Gorman for purified M. sexta PPO and T. Graves, K. Kjos, R. Woolsey, and G. Hammon for mosquito rearing. Thanks go to Drs. J. Tomich and Y. Hiromasa at the KSU Proteomics Core Laboratory for protein digestion and mass spectrometry. Real-time PCR analyses were performed at the COBRE Core I, KSU. This work was supported by NIH grants 3P20RR017708-07S1 and P20RR017686 subawards to K.M. and GM41247 to M.K. This is contribution 10-368-J from the Kansas Agricultural Experiment Station.
Abbreviations
- dKD
Double knockdown
- IEARpNA
Acetyl-Ile-Glu-Ala-Arg-p-nitroanilide
- KD
Knockdown
- KS
Kolmogorov–Smirnov
- LLA
Late-life acting
- MS
Mass spectrometry
- PPO
Prophenoloxidase
References
- 1.Enayati A, Hemingway J. Malaria management: past, present, and future. Annu Rev Entomol. 2010;55:569–591. doi: 10.1146/annurev-ento-112408-085423. [DOI] [PubMed] [Google Scholar]
- 2.Kelly-Hope L, Ranson H, Hemingway J. Lessons from the past: managing insecticide resistance in malaria control and eradication programmes. Lancet Infect Dis. 2008;8:387–389. doi: 10.1016/S1473-3099(08)70045-8. [DOI] [PubMed] [Google Scholar]
- 3.Hemingway J, Ranson H. Insecticide resistance in insect vectors of human disease. Annu Rev Entomol. 2000;45:371–391. doi: 10.1146/annurev.ento.45.1.371. [DOI] [PubMed] [Google Scholar]
- 4.Read AF, Lynch PA, Thomas MB. How to make evolution-proof insecticides for malaria control. PLoS Biol. 2009;7:e1000058. doi: 10.1371/journal.pbio.1000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanford S, Chan BH, Jenkins N, Sim D, Turner RJ, Read AF, Thomas MB. Fungal pathogen reduces potential for malaria transmission. Science. 2005;308:1638–1641. doi: 10.1126/science.1108423. [DOI] [PubMed] [Google Scholar]
- 6.Michel K, Budd A, Pinto S, Gibson TJ, Kafatos FC. Anopheles gambiae SRPN2 facilitates midgut invasion by the malaria parasite Plasmodium berghei. EMBO Rep. 2005;6:891–897. doi: 10.1038/sj.embor.7400478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanost M, Gorman MJ (2008) In: Beckage NE (ed.) Insect immunology. Academic Press/Elsevier, San Diego, pp 69–96
- 8.Satoh D, Horii A, Ochiai M, Ashida M. Prophenoloxidase-activating enzyme of the silkworm, Bombyx mori. Purification, characterization, and cDNA cloning. J Biol Chem. 1999;274:7441–7453. doi: 10.1074/jbc.274.11.7441. [DOI] [PubMed] [Google Scholar]
- 9.Kan H, Kim CH, Kwon HM, Park JW, Roh KB, Lee H, Park BJ, Zhang R, Zhang J, Soderhall K, Ha NC, Lee BL. Molecular control of phenoloxidase-induced melanin synthesis in an insect. J Biol Chem. 2008;283:25316. doi: 10.1074/jbc.M804364200. [DOI] [PubMed] [Google Scholar]
- 10.Tang H, Kambris Z, Lemaitre B, Hashimoto C. Two proteases defining a melanization cascade in the immune system of Drosophila . J Biol Chem. 2006;281:28097. doi: 10.1074/jbc.M601642200. [DOI] [PubMed] [Google Scholar]
- 11.Nappi A, Poirie M, Carton Y. The role of melanization and cytotoxic by-products in the cellular immune responses of Drosophila against parasitic wasps. Adv Parasitol. 2009;70:99–121. doi: 10.1016/S0065-308X(09)70004-1. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Y, Wang Y, Gorman MJ, Jiang H, Kanost MR. Manduca sexta serpin-3 regulates prophenoloxidase activation in response to infection by inhibiting prophenoloxidase-activating proteinases. J Biol Chem. 2003;278:46556–46564. doi: 10.1074/jbc.M309682200. [DOI] [PubMed] [Google Scholar]
- 13.De Gregorio E, Han SJ, Lee WJ, Baek MJ, Osaki T, Kawabata S, Lee BL, Iwanaga S, Lemaitre B, Brey PT. An immune-responsive serpin regulates the melanization cascade in Drosophila . Dev Cell. 2002;3:581–592. doi: 10.1016/S1534-5807(02)00267-8. [DOI] [PubMed] [Google Scholar]
- 14.Ligoxygakis P, Pelte N, Ji C, Leclerc V, Duvic B, Belvin M, Jiang H, Hoffmann JA, Reichhart JM. A serpin mutant links Toll activation to melanization in the host defence of Drosophila . EMBO J. 2002;21:6330–6337. doi: 10.1093/emboj/cdf661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou Z, Shin SW, Alvarez KS, Kokoza V, Raikhel AS. Distinct melanization pathways in the mosquito Aedes aegypti . Immunity. 2010;32:41–53. doi: 10.1016/j.immuni.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 16.Gettins PG. Serpin structure, mechanism, and function. Chem Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 17.Michel K, Suwanchaichinda C, Morlais I, Lambrechts L, Cohuet A, Awono-Ambene PH, Simard F, Fontenille D, Kanost MR, Kafatos FC. Increased melanizing activity in Anopheles gambiae does not affect development of Plasmodium falciparum . Proc Natl Acad Sci USA. 2006;103:16858–16863. doi: 10.1073/pnas.0608033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, Hetru C, Hoa NT, Hoffmann JA, Kanzok SM, Letunic I, Levashina EA, Loukeris TG, Lycett G, Meister S, Michel K, Moita LF, Muller HM, Osta MA, Paskewitz SM, Reichhart JM, Rzhetsky A, Troxler L, Vernick KD, Vlachou D, Volz J, von Mering C, Xu J, Zheng L, Bork P, Kafatos FC. Immunity-related genes and gene families in Anopheles gambiae . Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- 19.Volz J, Muller HM, Zdanowicz A, Kafatos FC, Osta MA. A genetic module regulates the melanization response of Anopheles to Plasmodium . Cellular microbiology. 2006;8:1392. doi: 10.1111/j.1462-5822.2006.00718.x. [DOI] [PubMed] [Google Scholar]
- 20.Volz J, Osta MA, Kafatos FC, Muller HM. The roles of two clip domain serine proteases in innate immune responses of the malaria vector Anopheles gambiae . J Biol Chem. 2005;280:40161–40168. doi: 10.1074/jbc.M506191200. [DOI] [PubMed] [Google Scholar]
- 21.Paskewitz SM, Andreev O, Shi L. Gene silencing of serine proteases affects melanization of Sephadex beads in Anopheles gambiae . Insect Biochem Mol Biol. 2006;36:701–711. doi: 10.1016/j.ibmb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 25.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 27.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 28.Jiang H, Mulnix AB, Kanost MR. Expression and characterization of recombinant Manduca sexta serpin-1B and site-directed mutants that change its inhibitory selectivity. Insect Biochem Mol Biol. 1995;25:1093–1100. doi: 10.1016/0965-1748(95)00042-9. [DOI] [PubMed] [Google Scholar]
- 29.Jiang H, Wang Y, Korochkina SE, Benes H, Kanost MR. Molecular cloning of cDNAs for two pro-phenol oxidase subunits from the malaria vector, Anopheles gambiae . Insect Biochem Mol Biol. 1997;27:693–699. doi: 10.1016/S0965-1748(97)00045-3. [DOI] [PubMed] [Google Scholar]
- 30.An C, Jiang H, Kanost MR. Proteolytic activation and function of the cytokine Spatzle in the innate immune response of a lepidopteran insect, Manduca sexta . FEBS J. 2010;277:148. doi: 10.1111/j.1742-4658.2009.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schick C, Kamachi Y, Bartuski AJ, Cataltepe S, Schechter NM, Pemberton PA, Silverman GA. Squamous cell carcinoma antigen 2 is a novel serpin that inhibits the chymotrypsin-like proteinases cathepsin G and mast cell chymase. J Biol Chem. 1997;272:1849. doi: 10.1074/jbc.272.3.1849. [DOI] [PubMed] [Google Scholar]
- 32.Blandin S, Moita LF, Kocher T, Wilm M, Kafatos FC, Levashina EA. Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the Defensin gene. EMBO Rep. 2002;3:852–856. doi: 10.1093/embo-reports/kvf180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilkinson M, McInerney JO, Hirt RP, Foster PG, Embley TM. Of clades and clans: terms for phylogenetic relationships in unrooted trees. Trends Ecol Evol. 2007;22:114–115. doi: 10.1016/j.tree.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 34.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 35.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 36.Li JS, Ruyl Kim S, Christensen BM, Li J. Purification and primary structural characterization of prophenoloxidases from Aedes aegypti larvae. Insect Biochem Mol Biol. 2005;35:1269–1283. doi: 10.1016/j.ibmb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 37.Jiang H, Wang Y, Ma C, Kanost MR. Subunit composition of pro-phenol oxidase from Manduca sexta: molecular cloning of subunit ProPO-P1. Insect Biochem Mol Biol. 1997;27:835–850. doi: 10.1016/S0965-1748(97)00066-0. [DOI] [PubMed] [Google Scholar]
- 38.Huang L, Sternberg PW. In: Community TCeR (ed.) WormBook
- 39.Collins FH, Sakai RK, Vernick KD, Paskewitz S, Seeley DC, Miller LH, Collins WE, Campbell CC, Gwadz RW. Genetic selection of a Plasmodium-refractory strain of the malaria vector Anopheles gambiae . Science. 1986;234:607–610. doi: 10.1126/science.3532325. [DOI] [PubMed] [Google Scholar]
- 40.Lambrechts L, Morlais I, Awono-Ambene PH, Cohuet A, Simard F, Jacques JC, Bourgouin C, Koella JC. Effect of infection by Plasmodium falciparum on the melanization immune response of Anopheles gambiae . Am J Trop Med Hyg. 2007;76:475–480. [PubMed] [Google Scholar]
- 41.Riehle MM, Markianos K, Niare O, Xu J, Li J, Toure AM, Podiougou B, Oduol F, Diawara S, Diallo M, Coulibaly B, Ouatara A, Kruglyak L, Traore SF, Vernick KD. Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science. 2006;312:577–579. doi: 10.1126/science.1124153. [DOI] [PubMed] [Google Scholar]
- 42.Muller HM, Dimopoulos G, Blass C, Kafatos FC. A hemocyte-like cell line established from the malaria vector Anopheles gambiae expresses six prophenoloxidase genes. J Biol Chem. 1999;274:11727–11735. doi: 10.1074/jbc.274.17.11727. [DOI] [PubMed] [Google Scholar]
- 43.Pinto SB, Lombardo F, Koutsos AC, Waterhouse RM, McKay K, An C, Ramakrishnan C, Kafatos FC, Michel K. Discovery of Plasmodium modulators by genome-wide analysis of circulating hemocytes in Anopheles gambiae . Proc Natl Acad Sci USA. 2009;106:21270–21275. doi: 10.1073/pnas.0909463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang H, Wang Y, Yu XQ, Zhu Y, Kanost M. Prophenoloxidase-activating proteinase-3 (PAP-3) from Manduca sexta hemolymph: a clip-domain serine proteinase regulated by serpin-1 J and serine proteinase homologs. Insect Biochem Mol Biol. 2003;33:1049–1060. doi: 10.1016/S0965-1748(03)00123-1. [DOI] [PubMed] [Google Scholar]
- 45.Pike RN, Buckle AM, le Bonniec BF, Church FC. Control of the coagulation system by serpins. Getting by with a little help from glycosaminoglycans. FEBS J. 2005;272:4842–4851. doi: 10.1111/j.1742-4658.2005.04880.x. [DOI] [PubMed] [Google Scholar]