Abstract
Tourette Syndrome (TS) is diagnosed based upon clinical criteria including motor and vocal tics. We hypothesized that differences in exon expression and splicing might be useful for pathophysiology and diagnosis. To demonstrate exon expression and alternatively spliced gene differences in blood of individuals with TS compared to healthy controls (HC), RNA was isolated from the blood of 26 un-medicated TS subjects and 23 healthy controls (HC). Each sample was run on Affymetrix Human Exon 1.0 ST (HuExon) arrays and on 3’ biased U133 Plus 2.0 (HuU133) arrays. To investigate the differentially expressed exons and transcripts, analyses of covariance (ANCOVA) were performed, controlling for age, gender and batch. Differential alternative splicing patterns between TS and HC were identified using analyses of variance (ANOVA) models in Partek. 376 exon probe sets were differentially expressed between TS and HC (raw p<0.005, fold change >|1.2|) that separated TS and HC subjects using hierarchical clustering and Principal Components Analysis. The probe sets predicted TS compared to HC with a >90% sensitivity and specificity using a 10-fold cross validation. 90 genes (transcripts) had differential expression of one exon (raw p<0.005) and were predicted to be alternatively spliced (raw p<0.05) in TS compared to HC. These preliminary findings might provide insight into the pathophysiology of TS and potentially provide prognostic and diagnostic biomarkers. However, the findings are tempered by the small sample size and multiple comparisons and require confirmation using PCR or deep RNA sequencing and a much larger patient population.
Keywords: Tourette Syndrome, gene expression profiling, exon expression profiles, alternative splicing, blood biomarker
Tourette Syndrome (TS) is a complex neurodevelopmental disorder characterized by multiple motor and one or more vocal tics that begin before age 18, often peak during adolescence, and typically decrease in adulthood (APA, 2000; Leckman et al., 2006; Muller, 2007; Singer, 2005; Swain et al., 2007). The diagnostic criteria for TS are defined by the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, Text Revision (DSM-IV-TR) which considers the history and pattern of the clinical symptoms (APA, 2000; Leckman, 2002; Stillman A et al., 2009a). However, the subjective nature of the diagnostic criteria may lead to a misdiagnosis or delay accurate diagnosis. Tics may be missed because they wax and wane. Additionally, TS may be misdiagnosed as another movement disorder (Mejia and Jankovic, 2005; Wand et al., 1992). Finally, a diagnosis of TS may mask the accurate diagnosis and treatment of a number of other disorders associated with tics (Mejia and Jankovic, 2005).
Gene expression changes in peripheral blood can reflect brain injury, genetic diseases and psychiatric disorders (Rollins et al., ; Sharp et al., 2006a; Sharp et al., 2006b; Tang et al., 2005). Distinct disease-specific gene expression fingerprints have been demonstrated in several neurological and psychiatric diseases including stroke (Moore et al., 2005; Tang et al., 2006; Xu et al., 2008), Down Syndrome, Tuberous Sclerosis, Neurofibromatosis (Tang et al., 2004a; Tang et al., 2004b) and Huntington’s Disease (Borovecki et al., 2005). Our previous study showed a subgroup of TS patients over-expressed natural killer cell genes in blood (Lit et al., 2007; Tang et al., 2005).
Alternative splicing occurs during the transcription of a gene (a primary gene transcript or pre-mRNA) when individual exons are reconnected in multiple ways during RNA splicing. The resulting different mRNAs are translated into different protein isoforms (Black, 2003; Cuperlovic-Culf et al., 2006; Matlin et al., 2005), which have distinct properties and different functions. Moreover, differences in exon splicing potentially provide qualitative splicing-specific diagnostic and prognostic biomarkers.
The 3’ biased U133 Affymetrix microarrays employed in previous TS studies (Hong et al., 2004; Lit et al., 2007; Lit et al., 2009; Tang et al., 2005) did not assess expression of the entire transcript, and did not assess pre-mRNA splicing. Affymetrix Human Exon 1.0 ST (HuExon) Arrays, which cover the entire length of the transcripts, allow the investigator to study expression at the exon level and to predict whether there is likely to be alternative splicing of a given gene. In the current study we used the 3’ biased U133 Plus 2.0 arrays (HuU133) to assess 3’ biased mRNA expression. In addition, using the same samples, we employed HuExon arrays to assess expression of individual exons, expression of full length transcripts (mRNAs) as well as to assess predicted alternative splicing in TS compared to HC. Only un-medicated TS patients were examined in the current study and compared to healthy controls (HC), since our previous studies showed that medications markedly affect gene expression (Liao, et al., submitted). Due to the small sample size and the lack of PCR verification, this represents a discovery type study.
Materials and Methods
Subjects
Un-medicated TS subjects were recruited via the Tourette Syndrome Association, clinical referrals, local advertisements, physician referrals, and through the University of California at Davis. TS subjects were recruited as part of a functional magnetic resonance imaging study of tic severity and cognitive control conducted by Dr. S Bunge and colleagues (Baym et al., 2008). TS diagnosis was based on DSM-IV-TR criteria (APA, 2000). Tic severity was assessed based on direct child and parent interview using the Yale Global Tic Severity Scale (YGTSS) (Leckman et al., 1989). HC children were recruited through area schools, fliers and recreational centers. Following initial contact, potential participants were screened via parent interview for the absence of neurodevelopmental disorders. Individuals with a history of serious physical illness (e.g. endocrine, cardiovascular or neurological disorders) were excluded from enrollment in the study. Protocols were approved by the institutional review boards at the University of California at Davis. Informed consent was obtained from the parent or legal guardian of each participant.
Sample collection and RNA isolation
Blood sample collection and RNA isolation were performed as described previously (Tian et al., 2009). Whole blood (15ml) was collected from each subject via antecubital fossa venipuncture into six PAXgene Vacutainer tubes (Qiagen, Valencia, CA, USA). These tubes contain a solution that immediately lyses all of the cells in whole blood and stabilizes the RNA without measurable degradation. The RNA represents genes expressed in all white blood cells and immature red blood cells and immature platelets. Blood samples were stored frozen at −70°C until processed.
Total RNA was isolated using the PAXgene Blood RNA Kit (Qiagen) according to the manufacturer’s protocol. RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Foster City, CA, USA) and quantified using fiberoptic spectrophotometry using the Nanodrop ND-1000 (Nanodrop Inc., Wilmington, DE, USA). RNA yielding both an A260/A280 absorbance ratio greater than 2.0 and a 28s/18s rRNA ratio equal to or exceeding 1.8 was utilized.
HuU133 microarray processing
HuU133 microarray processing was performed according to the manufacturer’s protocol. The Ovation RNA Amplification System V2 kit and the Ovation® WB Reagent (NuGEN, San Carlos,CA) were used for an optimized whole blood amplification starting with 50 ng total RNA. The amplified cDNA was fragmented and labeled using NuGEN’s FL-Ovation™ cDNA Biotin Module V2 (NuGEN, San Carlos,CA). Hybridization, washing and scanning were performed according to the Affymetrix Human U133 plus2 protocol (Affymetrix, Santa Clara, CA).
HuExon microarray processing
20 ng RNA samples were amplified using the WT-Ovation™ Pico RNA Amplification System (NuGEN, San Carlos, CA) with the exon Module. These were then fragmented and labeled using the FL-Ovation™ cDNA Biotin Module V2 (NuGEN, San Carlos, CA). Hybridization, washing and scanning were performed according to the Affymetrix Human Exon 1.0 ST protocol (Affymetrix, Santa Clara, CA).
To minimize batch effects, TS and HC samples were randomized in the order that they were processed. In addition, similar numbers of TS and HC samples were included in each of the batches for both the HuU133 arrays and for the HuExon arrays.
Data analysis
Raw data in the form of Affymetrix CEL files was imported into Partek Genomics Suite 6.4 (Partek Inc., St. Louis, MO). Probe summarization and probe set normalization were performed using Robust Multi-Chip Average (RMA), which included background correction, Quantile Normalization, log2-transformation and Median Polish probe set summarization. For exon array analysis, only the core probe sets were considered (about 228,000 probe sets), and transcript-level expression analysis was assessed by averaging all the core probe sets for that gene. For HuU133 analysis, one array from a healthy control was found to be an outlier, as its average intensity was significantly different from other arrays, and was excluded.
To demonstrate that exons and transcripts were differentially expressed between TS and HC, an analysis of covariance (ANCOVA) was performed, controlling for the effects of age, gender and batch. To predict alternative splicing in TS compared to HC samples, the interaction of diagnosis and exon was assessed using the Partek alternative splicing ANOVA, controlling for the effects of diagnosis, exon, patient, and the interaction of diagnosis and patient. An uncorrected p<0.05 was considered significant for alternative splicing events, since it is unclear how to appropriately correct for the multiple comparisons for alternative splicing (Partek, Manual). However, to decrease the false positives in the alternative splicing analysis, several approaches were used: 1) Probe sets with a log2 value < 3.0 in all samples were excluded except for those cases where there was a significant difference in expression of a single exon between the groups (p<0.05)(Partek Manual) ; 2) genes whose transcript expression was significantly different between TS and HC were filtered out, since an ideal splicing event assumes equal gene expression between samples, and most exons belong to genes that are constitutively expressed while a single exon is alternatively spliced (Affymetrix, white paper); and 3) genes without significant (p<0.05, fold change > |1.2|) differences in expression of single exons were excluded.
Ingenuity Pathways Analysis (IPA 8.0, Ingenuity® Systems) was used to identify statistically significant functional categories in the data set using the Fisher’s exact test. A 10-fold leave one out cross-validation model in Prediction Analysis of Microarrays (PAM) was used to predict TS compared to HC. This method of cross-validation generates a model on 90% of the samples to predict the remaining 10% of samples. The procedure is repeated 10 times to compute the overall error in the model.
Results
Subject demographics and medications
Subject demographics of the 26 TS and 23 HC subjects are summarized in Table 1. There was no significant difference for age (p=0.39) and race (p=0.07) between the two groups. Gender was significantly different between the groups (p=0.04).
Table 1.
Demographics of Tourette Syndrome (TS) and Healthy Control (HC) subjects.
| Description | TS | HC | |
|---|---|---|---|
| Number | Subjects | 26 | 23 |
| Age | Mean±SD (Years) | 10.2 ± 2.2 | 9.7 ± 2.1 |
| Range (Years) | 7–15 | 7–13 | |
| Gender | Male | 20 (77%) | 11 (48%) |
| Female | 6 (23%) | 12 (52%) | |
| White | 18 | 21 | |
| Race | Hispanic | 2 | 0 |
| Black | 0 | 1 | |
| Others | 6 | 1 | |
| YGTSS | Mean ± SD | 20.9 ± 10.9 | - |
| Range | 0–41 | - |
YGTSS=Yale Global Tic Severity Scale
3’ Transcript Expression using the HuU133 array
Using the data from the U133 array, the expression of 90 genes (represented by 126 probe sets) was considered significantly different between TS and HC (p<0.01, fold change > |1.2|; Supplementary Table 1). Using the Benjamini and Hochberg False Discovery Rate (FDR) correction for multiple comparisons, the 90 genes had FDR corrected p values between 0.22 and 0.73. The 5 probe sets with FDR corrected p<0.5 are listed in Supplementary Table 1. The unsupervised hierarchical cluster analysis and Principal Components Analysis (PCA) for these 90 genes did not separate these TS subjects from the HC subjects (not shown).
Full-length Transcript Expression in HuExon
Only 17 genes (represented by 19 probe sets) were differentially expressed between TS compared to HC (p<0.01, fold change > |1.2|; Supplementary Table 2) using the Affymetrix Exon arrays. Again, using the FDR correction for multiple comparisons, none of the genes had a corrected p<0.9. The unsupervised hierarchical cluster analysis and PCA for these 17 genes did not separate the TS subjects from the HC subjects (not shown). Two genes (killer cell lectin-like receptor subfamily D, member 1 [KLRD1] and cytochrome P450, family 27, subfamily A, polypeptide 1 [CYP27A1]) over lapped between the 17 HuExon gene list and the 90 HuU133 gene list.
Exon-Level Expression Differences in TS compared to HC
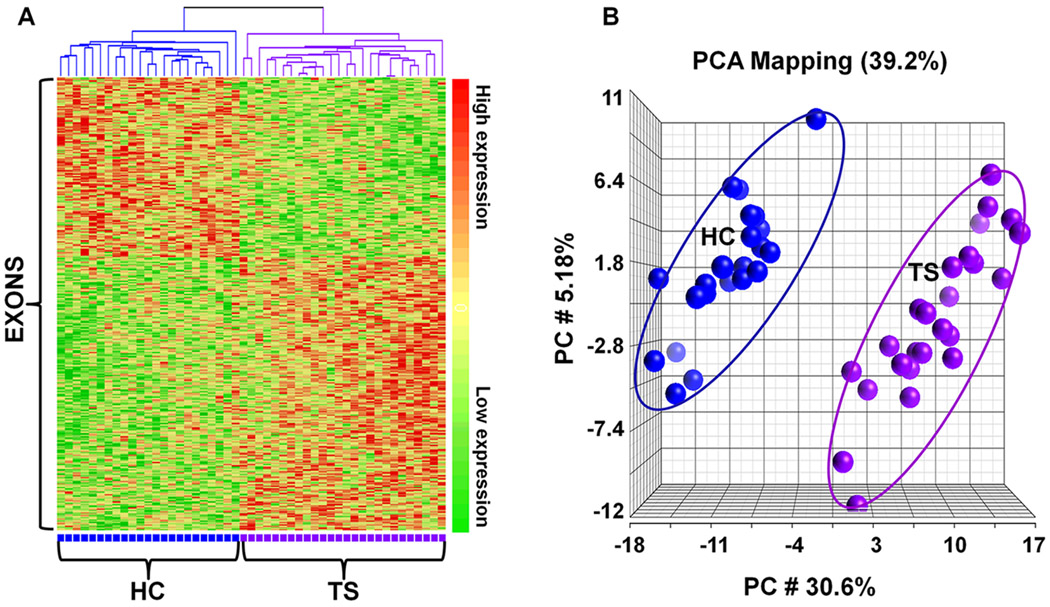
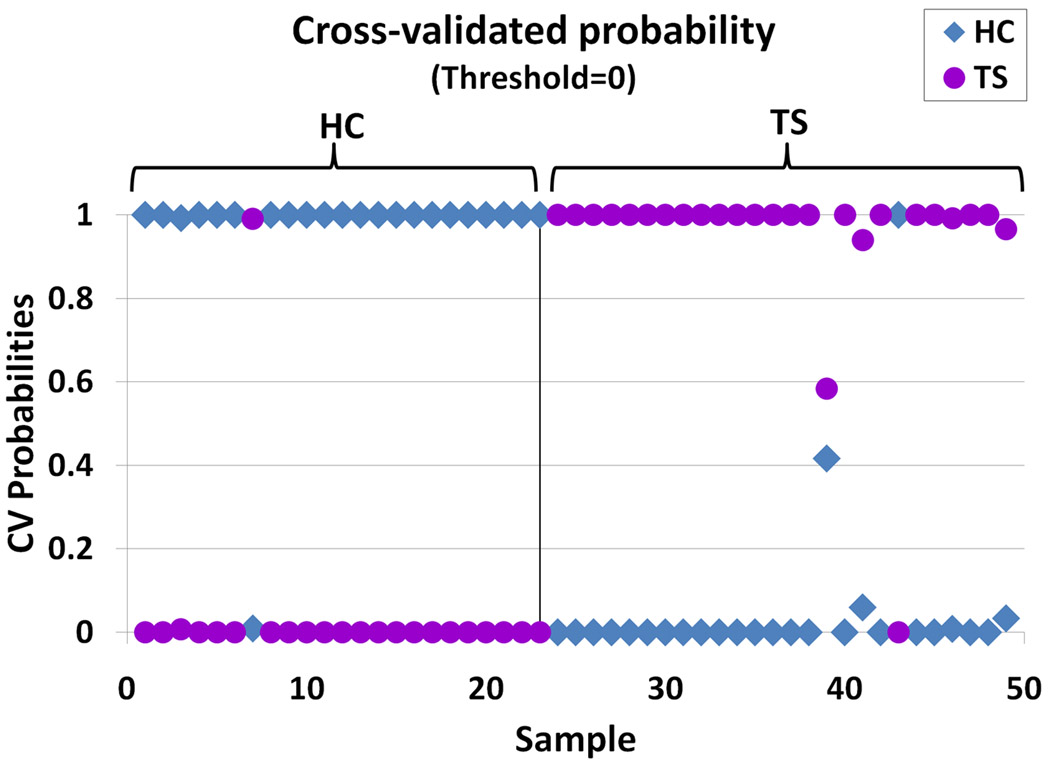
There were 6700 exon probe sets whose expression was significantly different between TS and HC (p<0.05, fold change > |1.2|). A total of 1403 exon probesets were significantly different at p <0.01 (fold change > |1.2|; Supplementary Table 3). There were 376 exons whose expression was significantly different with a p<0.005 and fold change > |1.2|, and without any significant differences for the other factors (page>0.05, pgender>0.05, pbatch>0.05; Supplementary Table 4). These results help support the primary data for differences between TS and HC as they were not affected by significant technical batch, gender or age effects. However, using the FDR correction for multiple comparisons, only two exons had a corrected p<0.9 for either the 1403 or 376 exon list. Even so, the unsupervised hierarchical cluster analysis (Figure 1A) and PCA (Figure 1B) for the 376 exons (p<0.005) showed TS patients completely separated from HC. The three principal components accounted for 39.2% of the total variance, and the first principal component (30.6% of variance) alone separated the TS from HC (Figure 1B). The ability of the 376 exons to predict TS from HC was evaluated using PAM. Ten-fold leave-one-out cross validation showed that these 376 exon probe sets were able to correctly classify 24 out of the 26 TS samples (92.3%), and 22 out of the 23 HC samples (95.7%) (Figure 2). Using the less stringent 1403 exon list (p<0.05, fold change > |1.2|), a similar separation of TS and HC samples was obtained following a cluster analysis (Supplementary Figure 1) and PCA (Supplementary Figure 2).
Figure 1.
A total of 376 exons were differentially expressed in blood (ANOVA, p<0.05, fold change > |1.2|) of Tourette Syndrome (TS) patients compared to healthy controls (HC) using Affymetrix Human Exon arrays. (a) An unsupervised hierarchical clustering analysis of these 376 probe sets (Y axis) showed complete separation of TS and HC patients (X axis). High gene expression is shown in bright red, low gene expression in bright green, and no difference in yellow. (b) The Principal Components Analysis (PCA) also showed TS patients (purple on the right) separated from the HC patients (blue on the left) based on 376 exon probe sets. The three principal components accounted for 39.2% of the total variance.
Figure 2.
Prediction Analysis of Microarrays (PAM) results of the 376 exon probe sets identified using the human Affymetrix Exon arrays (p<0.05, fold change > |1.2|). The X axis shows individual samples from TS and HC subjects. The Y axis shows the predicted probability for each sample to be either HC (on the left) or TS (on the right) subjects.
Differential Alternative splicing in TS compared to HC
A maximum of 706 genes were predicted (p<0.05) to have alternative splicing in TS compared to HC subjects. Using a Benjamini and Hochberg FDR of <0.05 (5% false positives) for multiple comparison correction, three genes had a corrected p<0.05. These included ubiquitously transcribed tetratricopeptide repeat gene, Y-linked (UTY), myosin XVIIIB (MYO18B) and potassium voltage-gated channel, subfamily H (eag-related), member 4 (KCNH4). There were 55 genes that were significantly different when a FDR corrected p<0.5 was used (Supplementary Table 5). Notably, of the 706 genes predicted to be alternatively spliced, and of the genes associated with the 6700 exons (p<0.05, fold change > |1.2|) found to be differentially expressed in TS compared to HC, a total of 545 genes were common to both lists (Supplementary Table 6). Of the 706 genes predicted to be alternatively spliced (p<0.05), and of the genes associated with the 1403 exons (p<0.01, fold change > |1.2|) found to be differentially expressed in TS compared to HC (Supplementary Table 3), a total of 235 genes were common to both lists (Supplementary Table 7). Note that only two out of three genes with a FDR corrected p<0.05, MYO18B and KCNH4, were in both the 545 and 235 alternatively spliced gene lists. Similarly, for the more stringent list of 376 exons (p<0.005) that were differentially expressed in TS compared to HC and of the 706 genes predicted to be alternatively spliced, 90 genes were common to both lists (Supplementary Table 8). These 90 gene, 235 gene and 545 gene lists which were predicted to be alternatively spliced, and had differentially expressed exons in TS compared to HC, probably represent the more reliable alternatively spliced genes at least based upon statistics alone (see discussion).
The biological functional analysis of the 706 transcripts predicted to be alternatively spliced showed that the communication between innate and adaptive immune cells (p= 3.6 × 10−2) and acute phase response signaling (p= 3.8 × 10−2) were among the top canonical pathways. The top molecular and cellular functions for these genes included cellular development (p value range 2.3 × 10−5 to 3.5 × 10−2), cell morphology (p value range 8.0 × 10−4 to 3.5 × 10−2), molecular transport (p value range 8.5 × 10−4 to 3.5 × 10−2), amino acid metabolism (p value range 1.2 × 10−3 to 3.5 × 10−2), and cell cycle (p value range 1.4 × 10−3 to 3.5 × 10−2)(Supplementary Table 9). The functional analysis results for the most stringent 90 alternatively spliced genes are shown in Supplementary Table 10. The communication between innate and adaptive immune cell pathways identified in the 706 alternatively spliced gene list was not in this more stringent functional analysis.
Discussion
This is the first study to show that there are large numbers of single exons differentially expressed between TS and HC. These differentially expressed exons may be useful for developing a diagnostic test for TS. In contrast to the large numbers of differentially regulated exons, there are few changes in gene-level expression in TS compared to HC subjects, which was supported by both 3’ and exon expression data. However, in spite of the few differences of gene expression, there appear to be hundreds of genes predicted to be differentially spliced in TS compared to HC which may help in understanding the causes and pathogenesis of TS.
Few Changes in Gene-level Expression in TS compared to HC
Gene-level analyses using either the 3’ (HuU133) or exon (HuExon) arrays confirmed that there were few differences of gene expression in TS compared to HC subjects. This result is comparable to our previous medicated TS versus HC study which identified only 56 genes differentially expressed between medicated TS and HC. These genes could not differentiate medicated TS from HC (Lit et al., 2009). Thus, there are relatively few differences in gene expression in TS compared to HC independent of whether the TS subjects were treated or not. Alternatively, this lack of difference could be explained by low power to detect such changes since the sample sizes were very small. It is notable that the KLRD1- natural killer cell gene was down-regulated in TS compared to HC using the 3’ and exon microarrays, and that this same gene was up-regulated in two different TS cohorts in our previous studies of medicated TS subjects (Lit et al., 2007). Since KLRD1 is down regulated in un-medicated TS subjects compared to controls in this study, and was up-regulated following treatment of TS subjects compared to controls in our previous studies, we assume the drug increases expression of KLRD1 from below control levels to above control levels and that this correlates with improvement of TS symptoms with the drug treatment. Thus, since the medication induced expression of the KLRD1 natural killer gene likely correlates with improved TS symptoms, KLRD1 could be involved in the pathogenesis of TS.
Exon biomarkers differentiated TS and HC
Large numbers of single exons were differentially expressed between TS and HC. Given that gene expression did not differ in many genes for TS compared to HC, it is not immediately apparent why there might be so many differences in expression of exons. We interpret this finding to mean that exon skipping, that is splicing a single exon out of a transcript, does not significantly alter expression of the spliced transcript compared to the un-spliced transcript. This would account for many differences in expression of single exons between TS and HC, with very few differences of “gene expression” (full length transcripts) between TS and HC. Thus, exon expression potentially could be used as a biomarker for TS, though it is important to emphasize that the current data must be interpreted with caution since no exon probe set had a multiple comparison corrected p<0.05 in TS compared to HC. We consider the genes that were predicted to be alternatively spliced (see below), AND in which there were significant differences of expression of exons within those genes to be more reliable. The large number of exons that had significant differences in expression between TS and HC but were not associated with genes that were predicted to be alternatively spliced may represent mostly false positives related to the multiple comparisons performed using the exon microarrays.
Alternative Splicing of genes in TS compared to HC
Alternative splicing is the major source of protein diversity for higher eukaryotic organisms, given that the numbers of genes are very similar for all mammals. Current estimates indicate over 50 percent of human genes exhibit alternative splicing (Black, 2003; Cuperlovic-Culf et al., 2006; Matlin et al., 2005). The most common splicing mode in mammalian pre-mRNAs is cassette exon (exon skipping), which is defined as an exon present in one mRNA transcript but absent in another isoform of the transcript. As mentioned above, we predict that differences in exon skipping (of a single exon) between TS and HC samples does not ordinarily produce any detectable “change of gene expression” in two transcripts from the same gene. However, as hypothesized above, exon skipping could result in significant differences of expression of single exons as detected in the present study. Given that there are large numbers of genes predicted to be alternatively spliced in the TS compared to HC subjects, the explanation for this differential splicing in TS compared to HC is unclear. This could relate in part to the genetics of TS – gene polymorphisms resulting in changes of splicing, to environmental factors that might affect alternative splicing (Black, 2003; Faustino and Cooper, 2003), and other genetic and epigenetic factors that modulate splicing.
One of the top canonical pathways represented by the 706 alternatively spliced transcripts in TS compared to HC subjects was the communication between innate and adaptive immune cell pathways. This is consistent with the many studies that have suggested abnormalities in the immune response in TS patients (Martino et al., 2005; Stillman A et al., 2009b; Swain et al., 2007; Walkup et al., 1996). Increased plasma levels of specific pro-inflammatory cytokines including Interleukin-12 (IL-12) and tumor necrosis factor alpha (TNF-α) were observed in TS patients and correlated with disease severity (Leckman et al., 2006). Moreover, the numbers of specific lymphocyte subtypes appear to be altered in some TS subjects (Kawikova et al., 2007; Moller et al., 2008) and our previous studies found that a subgroup of TS patients over-expressed natural killer cell genes (Lit et al., 2007). It is interesting that some neurotransmitter receptors which associated with TS were in the alternative splicing gene list, including Gamma-aminobutyric acid (GABA) receptors (GABRA4, GABRG1), acetylcholine receptor (CHRNA4), adrenergic receptor (ADRA1B), serotonin receptor (HTR3B), and the dopamine DRD4 receptor (Supplementary Tables 5). Although alternative splicing of these neurotransmitter receptors could mirror the changes of those in the brain (Achiron and Gurevich, 2006; Glatt et al., 2005; Sharp et al., 2006a; Sharp et al., 2006b; Sullivan et al., 2006), these neurotransmitter genes expressed in the blood cells are involved in the regulation of immune response (Kawashima and Fujii, 2003; Kohm and Sanders, 2001; Martino et al., 2009; Rane et al., 2005; Tian et al., 2004; Yang et al., 2006). The different isoforms of these genes could result in dys-regulated immune function in TS, and susceptibility to specific infections such as Group A β-streptococcus, viral infections, and others that might stimulate autoimmune reactions to basal ganglia. Alternatively, the abnormal immune dysfunction could result in immune-mediated basal ganglia dysfunction via cross-talk between the blood-brain-barrier and neuronal transmission in the absence of infection (Martino et al., 2009).
Limitations of this study
The largest limitation of the study is that, in spite of the many genes predicted to be alternatively spliced in TS compared to HC, these were not confirmed using an independent method. The sensitivity and specificity of the Affymetrix Exon arrays for detecting exon splicing across the whole genome is not known, at least based upon current published data. Thus, confirmation of the exon splicing will require PCR that uses primers to flank the exons of interest or RNA deep sequencing to demonstrate consistent differential splicing between TS and HC subjects based upon blood samples.
In addition, there are limitations related to the study itself. The analyses did not consider the influences of attention deficit/hyperactivity disorder, obsessive-compulsive disorder (OCD), and other co-morbidities that also affect gene expression because this would have decreased the sample sizes and decreased power even more (in preparation). Secondly, there is a major problem in the analyses related to the multiple comparisons. When a False Discovery Rate (FDR) of 5% was used (allows 5% false positives), only three alternatively spliced genes passed this FDR. However, the FDR correction for multiple comparisons assumes all genes (exons) are independently expressed, which is not the case (Zhang, 2006) and it is unclear how to correct for the multiple comparisons for alternative splicing (Partek manual).
Finally, this is a preliminary study using very small numbers of subjects (26 TS and 23HC) which limited power. The data must be validated in an independent follow-up study with much larger sample sizes, which is the only true way to confirm the results of expression microarray studies or whole genome studies of any kind.
Supplementary Material
Supplementary Figure 1. The unsupervised hierarchical clustering of the 1403 probe sets that were differentially expressed between Tourette Syndrome (TS) and Healthy Control (HC) subjects identified using the human Affymetrix Exon arrays (p<0.01, fold change > |1.2|). The Y axis shows genes regulated in the blood and the X axis shows the individual TS and HC subjects. High gene expression is shown in bright red, low gene expression in bright green, and no difference in yellow.
Supplementary Figure 2. The Principal Components Analysis (PCA) completely separated Tourette Syndrome (TS) patients (purple on the right) from the Healthy Control (HC) (blue on the left) using the 1403 differentially expressed exon probesets identified by human Affymetrix Exon array (P<0.01, fold change > |1.2|) The three principal components accounted for 37.3% of the total variance.
Acknowledgements
This study was supported by a gift from Ron and Darin Mittelstaedt (FRS); mentored clinical research award NIMH K08 MH072958 (BC); previous support from the Tourette Syndrome Association (TSA) (FRS); and the MIND Institute. We thank Silvia A. Bunge, Carol L. Baym, Samantha B. Wright, and Debra Galik for subject recruitment and data collection. We also thank Ryan R. Davis and Jeffrey P. Gregg for processing the microarrays.
References
- Achiron A, Gurevich M. Peripheral blood gene expression signature mirrors central nervous system disease: the model of multiple sclerosis. Autoimmun Rev. 2006;5:517–522. doi: 10.1016/j.autrev.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Affymetrix, white paper. identifying and validating alternative splicing variants. Vol. ed.^eds., http://www.affymetrix.com/products_services/arrays/specific/exon.affx. [Google Scholar]
- APA. Diagnostic and Statistical Manual of Mental Disorders. 4th edn. Washington, DC: American Psychiatric Association; 2000. text revision. ed.^eds. [Google Scholar]
- Baym CL, Corbett BA, Wright SB, Bunge SA. Neural correlates of tic severity and cognitive control in children with Tourette syndrome. Brain. 2008;131:165–179. doi: 10.1093/brain/awm278. [DOI] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Borovecki F, Lovrecic L, Zhou J, Jeong H, Then F, Rosas HD, Hersch SM, Hogarth P, Bouzou B, Jensen RV, Krainc D. Genome-wide expression profiling of human blood reveals biomarkers for Huntington's disease. Proc Natl Acad Sci U S A. 2005;102:11023–11028. doi: 10.1073/pnas.0504921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperlovic-Culf M, Belacel N, Culf AS, Ouellette RJ. Microarray analysis of alternative splicing. OMICS. 2006;10:344–357. doi: 10.1089/omi.2006.10.344. [DOI] [PubMed] [Google Scholar]
- Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- Glatt SJ, Everall IP, Kremen WS, Corbeil J, Sasik R, Khanlou N, Han M, Liew CC, Tsuang MT. Comparative gene expression analysis of blood and brain provides concurrent validation of SELENBP1 up-regulation in schizophrenia. Proc Natl Acad Sci U S A. 2005;102:15533–15538. doi: 10.1073/pnas.0507666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JJ, Loiselle CR, Yoon DY, Lee O, Becker KG, Singer HS. Microarray analysis in Tourette syndrome postmortem putamen. J Neurol Sci. 2004;225:57–64. doi: 10.1016/j.jns.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Kawashima K, Fujii T. The lymphocytic cholinergic system and its contribution to the regulation of immune activity. Life Sci. 2003;74:675–696. doi: 10.1016/j.lfs.2003.09.037. [DOI] [PubMed] [Google Scholar]
- Kawikova I, Leckman JF, Kronig H, Katsovich L, Bessen DE, Ghebremichael M, Bothwell AL. Decreased numbers of regulatory T cells suggest impaired immune tolerance in children with tourette syndrome: a preliminary study. Biol Psychiatry. 2007;61:273–278. doi: 10.1016/j.biopsych.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Kohm AP, Sanders VM. Norepinephrine and beta 2-adrenergic receptor stimulation regulate CD4+ T and B lymphocyte function in vitro and in vivo. Pharmacol Rev. 2001;53:487–525. [PubMed] [Google Scholar]
- Leckman JF, Riddle MA, Hardin MT, Ort SI, Swartz KL, Stevenson J, Cohen DJ. The Yale Global Tic Severity Scale: initial testing of a clinician-rated scale of tic severity. J Am Acad Child Adolesc Psychiatry. 1989;28:566–573. doi: 10.1097/00004583-198907000-00015. [DOI] [PubMed] [Google Scholar]
- Leckman JF. Tourette's syndrome. Lancet. 2002;360:1577–1586. doi: 10.1016/S0140-6736(02)11526-1. [DOI] [PubMed] [Google Scholar]
- Leckman JF, Bloch MH, Scahill L, King RA. Tourette syndrome: the self under siege. J Child Neurol. 2006;21:642–649. doi: 10.1177/08830738060210081001. [DOI] [PubMed] [Google Scholar]
- Lit L, Gilbert DL, Walker W, Sharp FR. A subgroup of Tourette's patients overexpress specific natural killer cell genes in blood: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:958–963. doi: 10.1002/ajmg.b.30550. [DOI] [PubMed] [Google Scholar]
- Lit L, Enstrom A, Sharp FR, Gilbert DL. Age-related gene expression in Tourette syndrome. J Psychiatr Res. 2009;43:319–330. doi: 10.1016/j.jpsychires.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino D, Church AJ, Defazio G, Dale RC, Quinn NP, Robertson MM, Livrea P, Orth M, Giovannoni G. Soluble adhesion molecules in Gilles de la Tourette's syndrome. J Neurol Sci. 2005;234:79–85. doi: 10.1016/j.jns.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Martino D, Dale RC, Gilbert DL, Giovannoni G, Leckman JF. Immunopathogenic mechanisms in tourette syndrome: A critical review. Mov Disord. 2009;24:1267–1279. doi: 10.1002/mds.22504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Mejia NI, Jankovic J. Secondary tics and tourettism. Rev Bras Psiquiatr. 2005;27:11–17. doi: 10.1590/s1516-44462005000100006. [DOI] [PubMed] [Google Scholar]
- Moller JC, Tackenberg B, Heinzel-Gutenbrunner M, Burmester R, Oertel WH, Bandmann O, Muller-Vahl KR. Immunophenotyping in Tourette syndrome--a pilot study. Eur J Neurol. 2008;15:749–753. doi: 10.1111/j.1468-1331.2008.02159.x. [DOI] [PubMed] [Google Scholar]
- Moore DF, Li H, Jeffries N, Wright V, Cooper RA, Jr, Elkahloun A, Gelderman MP, Zudaire E, Blevins G, Yu H, Goldin E, Baird AE. Using peripheral blood mononuclear cells to determine a gene expression profile of acute ischemic stroke: a pilot investigation. Circulation. 2005;111:212–221. doi: 10.1161/01.CIR.0000152105.79665.C6. [DOI] [PubMed] [Google Scholar]
- Muller N. Tourette's syndrome: clinical features, pathophysiology, and therapeutic approaches. Dialogues Clin Neurosci. 2007;9:161–171. doi: 10.31887/DCNS.2007.9.2/nmueller. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partek, Manual. Alternative splice analysis of exon data in Partek Genomics Suite [Google Scholar]
- Rane MJ, Gozal D, Butt W, Gozal E, Pierce WM, Jr, Guo SZ, Wu R, Goldbart AD, Thongboonkerd V, McLeish KR, Klein JB. Gamma-amino butyric acid type B receptors stimulate neutrophil chemotaxis during ischemia-reperfusion. J Immunol. 2005;174:7242–7249. doi: 10.4049/jimmunol.174.11.7242. [DOI] [PubMed] [Google Scholar]
- Rollins B, Martin MV, Morgan L, Vawter MP. Analysis of whole genome biomarker expression in blood and brain. Am J Med Genet B Neuropsychiatr Genet. doi: 10.1002/ajmg.b.31062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp FR, Lit L, Xu H, Apperson M, Walker W, Wong B, Gilbert DL, Hershey A, Glauser TA. Genomics of brain and blood: progress and pitfalls. Epilepsia. 2006a;47:1603–1607. doi: 10.1111/j.1528-1167.2006.00809.x. [DOI] [PubMed] [Google Scholar]
- Sharp FR, Xu H, Lit L, Walker W, Apperson M, Gilbert DL, Glauser TA, Wong B, Hershey A, Liu DZ, Pinter J, Zhan X, Liu X, Ran R. The future of genomic profiling of neurological diseases using blood. Arch Neurol. 2006b;63:1529–1536. doi: 10.1001/archneur.63.11.1529. [DOI] [PubMed] [Google Scholar]
- Singer HS. Tourette's syndrome: from behaviour to biology. Lancet Neurol. 2005;4:149–159. doi: 10.1016/S1474-4422(05)01012-4. [DOI] [PubMed] [Google Scholar]
- Stillman A, Ercan-Sencicek AG, MW S. Tourette Disorder Overview. In: Pagon RA, Bird TC, Dolan CR, Stephens K, editors. GeneReviews [Internet] Vol. Seattle (WA): University of Washington, Seattle; 2009a. ed.^eds. [Google Scholar]
- Stillman A, Ercan-Sencicek AG, MW S. Developmentally regulated and evolutionarily conserved expression of SLITRK1 in brain circuits implicated in Tourette syndrome. J Comp Neurol. 2009b;Vol. 513:21–37. doi: 10.1002/cne.21919. ed.^eds. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Fan C, Perou CM. Evaluating the comparability of gene expression in blood and brain. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:261–268. doi: 10.1002/ajmg.b.30272. [DOI] [PubMed] [Google Scholar]
- Swain JE, Scahill L, Lombroso PJ, King RA, Leckman JF. Tourette syndrome and tic disorders: a decade of progress. J Am Acad Child Adolesc Psychiatry. 2007;46:947–968. doi: 10.1097/chi.0b013e318068fbcc. [DOI] [PubMed] [Google Scholar]
- Tang Y, Lu A, Ran R, Aronow BJ, Schorry EK, Hopkin RJ, Gilbert DL, Glauser TA, Hershey AD, Richtand NW, Privitera M, Dalvi A, Sahay A, Szaflarski JP, Ficker DM, Ratner N, Sharp FR. Human blood genomics: distinct profiles for gender, age and neurofibromatosis type 1. Brain Res Mol Brain Res. 2004a;132:155–167. doi: 10.1016/j.molbrainres.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Tang Y, Schapiro MB, Franz DN, Patterson BJ, Hickey FJ, Schorry EK, Hopkin RJ, Wylie M, Narayan T, Glauser TA, Gilbert DL, Hershey AD, Sharp FR. Blood expression profiles for tuberous sclerosis complex 2, neurofibromatosis type 1, and Down's syndrome. Ann Neurol. 2004b;56:808–814. doi: 10.1002/ana.20291. [DOI] [PubMed] [Google Scholar]
- Tang Y, Gilbert DL, Glauser TA, Hershey AD, Sharp FR. Blood gene expression profiling of neurologic diseases: a pilot microarray study. Arch Neurol. 2005;62:210–215. doi: 10.1001/archneur.62.2.210. [DOI] [PubMed] [Google Scholar]
- Tang Y, Xu H, Du X, Lit L, Walker W, Lu A, Ran R, Gregg JP, Reilly M, Pancioli A, Khoury JC, Sauerbeck LR, Carrozzella JA, Spilker J, Clark J, Wagner KR, Jauch EC, Chang DJ, Verro P, Broderick JP, Sharp FR. Gene expression in blood changes rapidly in neutrophils and monocytes after ischemic stroke in humans: a microarray study. J Cereb Blood Flow Metab. 2006;26:1089–1102. doi: 10.1038/sj.jcbfm.9600264. [DOI] [PubMed] [Google Scholar]
- Tian J, Lu Y, Zhang H, Chau CH, Dang HN, Kaufman DL. Gamma-aminobutyric acid inhibits T cell autoimmunity and the development of inflammatory responses in a mouse type 1 diabetes model. J Immunol. 2004;173:5298–5304. doi: 10.4049/jimmunol.173.8.5298. [DOI] [PubMed] [Google Scholar]
- Tian Y, Green PG, Stamova B, Hertz-Picciotto I, Pessah IN, Hansen R, Yang X, Gregg JP, Ashwood P, Jickling G, Van de Water J, Sharp FR. Correlations of Gene Expression with Blood Lead Levels in Children with Autism Compared to Typically Developing Controls. Neurotox Res. 2009 doi: 10.1007/s12640-009-9126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkup JT, LaBuda MC, Singer HS, Brown J, Riddle MA, Hurko O. Family study and segregation analysis of Tourette syndrome: evidence for a mixed model of inheritance. Am J Hum Genet. 1996;59:684–693. [PMC free article] [PubMed] [Google Scholar]
- Wand R, Shady G, Broder R, Furer P, Staley D. Tourette syndrome: issues in diagnosis. Neurosci Biobehav Rev. 1992;16:449–451. doi: 10.1016/s0149-7634(05)80186-1. [DOI] [PubMed] [Google Scholar]
- Xu H, Tang Y, Liu DZ, Ran R, Ander BP, Apperson M, Liu XS, Khoury JC, Gregg JP, Pancioli A, Jauch EC, Wagner KR, Verro P, Broderick JP, Sharp FR. Gene expression in peripheral blood differs after cardioembolic compared with large-vessel atherosclerotic stroke: biomarkers for the etiology of ischemic stroke. J Cereb Blood Flow Metab. 2008;28:1320–1328. doi: 10.1038/jcbfm.2008.22. [DOI] [PubMed] [Google Scholar]
- Yang GB, Qiu CL, Zhao H, Liu Q, Shao Y. Expression of mRNA for multiple serotonin (5-HT) receptor types/subtypes by the peripheral blood mononuclear cells of rhesus macaques. J Neuroimmunol. 2006;178:24–29. doi: 10.1016/j.jneuroim.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Zhang A. Advanced analysis of gene expression microarray data. Vol. Singapore; Hackensack, N.J.: World Scientific Press; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. The unsupervised hierarchical clustering of the 1403 probe sets that were differentially expressed between Tourette Syndrome (TS) and Healthy Control (HC) subjects identified using the human Affymetrix Exon arrays (p<0.01, fold change > |1.2|). The Y axis shows genes regulated in the blood and the X axis shows the individual TS and HC subjects. High gene expression is shown in bright red, low gene expression in bright green, and no difference in yellow.
Supplementary Figure 2. The Principal Components Analysis (PCA) completely separated Tourette Syndrome (TS) patients (purple on the right) from the Healthy Control (HC) (blue on the left) using the 1403 differentially expressed exon probesets identified by human Affymetrix Exon array (P<0.01, fold change > |1.2|) The three principal components accounted for 37.3% of the total variance.