Abstract
Subtype 2 Serotonin (5-Hydroxytryptamine, 5-HT) receptors are major drug targets for schizophrenia, feeding disorders, perception, depression, migraines, hypertension, anxiety, hallucinogens, and gastrointestinal dysfunctions.1 We report here the predicted structure of 5-HT2B and 5-HT2C receptor bound to highly potent and selective 5-HT2B antagonist PRX-08066 3, (pKi: 30 nM), including the key binding residues [V103 (2.53), L132 (3.29), V190 (4.60), and L347 (6.58)] determining the selectivity of binding to 5-HT2B over 5-HT2A. We also report structures of the endogenous agonist (5-HT) and a HT2B selective antagonist 2 (1-methyl-1-1,6,7,8-tetrahydropyrrolo[2,3-g]quinoline-5-carboxylic acid pyridine-3-ylamide). We examine the dynamics for the agonist-bound and the antagonist-bound HT2B receptors in explicit membrane and water finding dramatically different patterns of water migration into the NPxxY motif and the binding site that correlates with the stability of ionic locks in the D(E)RY region
Keywords: GPCR, Serotonin, Docking, Membrane protein structure, Molecular dynamics
INTRODUCTION
Three 5-HT2 receptors (2A, 2B, 2C) are major drug targets for schizophrenia, feeding disorders, perception, depression, migraines, hypertension, anxiety, hallucinogens, and gastrointestinal dysfunctions, but in many cases it is desirable to bind selectively to just one of these very similar receptors. 1 These 5-HT2 receptors are highly homologous with ~ 80% amino acid identity in the transmembrane (TM) domain, so that many 5-HT receptor antagonists (e.g., methylsergide, metergoline, mianserin, and ritanserin) have similar affinities for all three 5-HT2 receptor subtypes.2 Unfortunately, there is a paucity of antagonists selective for the 5-HT2B or 2C receptors, leading to cross-selectivity for drugs targeting either receptor.
Previously, we reported the predicted the 3-Dimensional (3D) structure for human 5-HT2C receptors (hHT2CR) using the MembStruk computational procedure.3 Based on this structure, we used the MSCDock computational procedure to predict the 3-D structures for bound ligand-protein complexes for agonists such as serotonin and antagonists such as ritanserin, metergoline, and methiothepin. The predicted structure-activity relationship (SAR) data for a series of psilocybin analogs, both agonists and antagonists shows a good agreement with the currently known experimental data.
Here, we report the new predicted structures of human 5-HT2B receptors (hHT2BR) and hHT2CR using newer methods, the MembEnsemb and the GenMSCDock techniques. To understand the subtype selectivity of hHT2BR and further drug development of the HT2B selective antagonist, we used the MembEnsemb (later version of the MembStruck) techniques to predict the 3D structure for the hHT2BR and hHT2CR and we used the GenMSCDock (later version of the MSCDock)3 techniques to predict the binding site for agonists (HT, SNF, RNF, desmethylNF, ethylNF), and antagonists (SB-206533 derivatives, PRX-08066), including some highly selective 5-HT2B antagonists known from the literature.
We report the predicted binding site and energies for five known agonists and nine antagonists (Fig. 1, Fig. 2, and Table 1), finding relative affinities that correlate well with experiment. We also report the key residues in the binding site that determine the selectivity of highly selective 5-HT2B ligands binding to hHT2BR over hHT2AR/hHT2CR. The different binding preference of agonists vs antagonists were studied through ensemble docking. Our molecular dynamics (MD) studies in explicit lipids and water show ligand-induced conformational changes, with the salt bridges in D(E)RY motif maintained in antagonist dynamics but broken in agonist dynamics. We observed that binding of the agonist induce water to flow into the NPxxY region which seems to be important in allowing the conformational transitions upon activation.
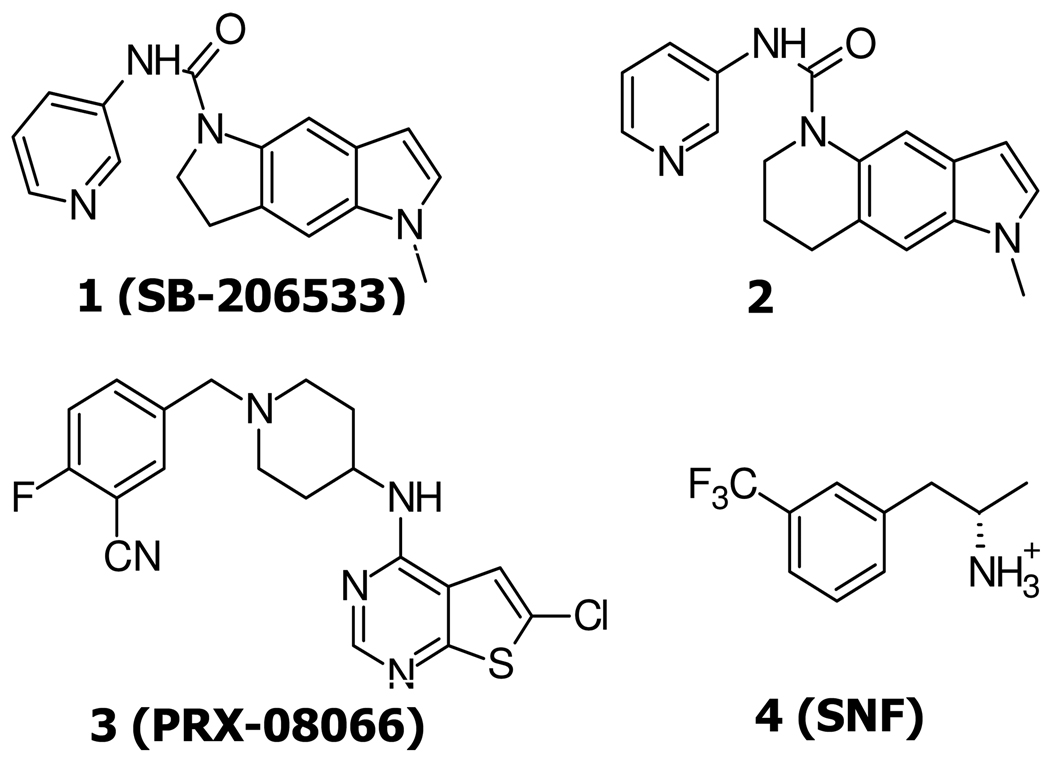
Fig. 1.
The chemical structures of 5-HT2B receptor antagonists, 1, 2, 3, and agonist 4.
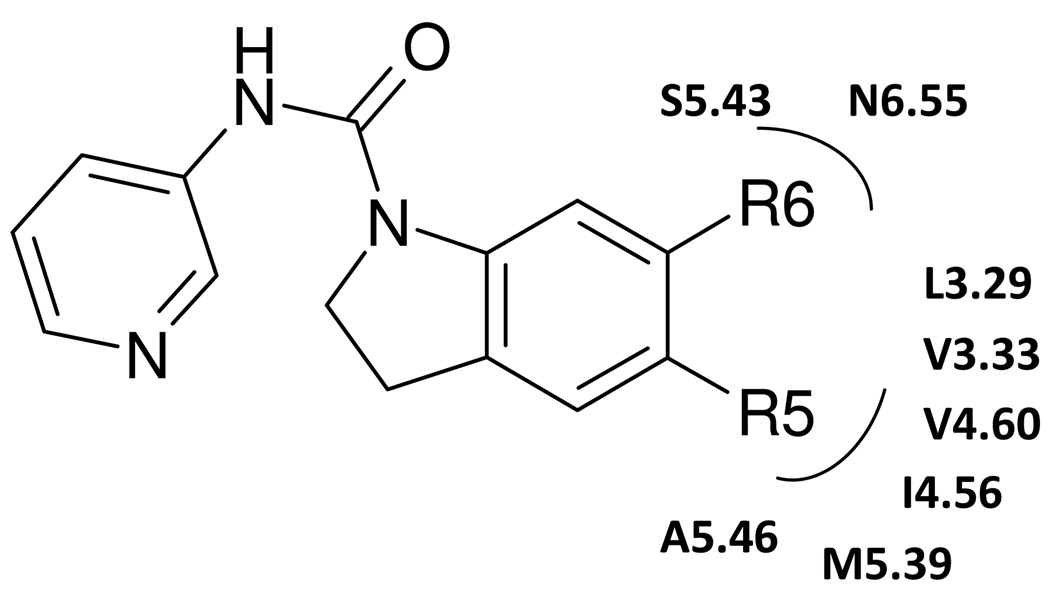
Fig. 2.
The chemical structures of several SB-206533 1 derivatives. R5 lipophilic substituent is surrounded by the aliphatic environments (V3.33, L3.29, I4.56, V4.60, M5.39, A5.46), while R6 electron-withdrawing group is in the proximity of L3.29, S5.43, and N6.55.
Table 1.
Cavity energy of several SB-206533 1 derivatives at human 5-HT2B and 2C receptors. The compound was ordered by binding energy for 5-HT2C receptors. Experimental data (Pki) were taken from reference30.
| 5-HT2B | 5-HT2C | |||||
|---|---|---|---|---|---|---|
| # | R5 | R6 | Pki | UnifiedCav | Pki | UnifiedCav |
| 46 | SMe | CF3 | 7.9 | −47.53 | 8.6 | −56.91 |
| 53 | OiPr | CF3 | 8.4 | −51.39 | 8.5 | −56.59 |
| 47 | SEt | CF3 | 8.0 | −50.43 | 8.5 | −55.19 |
| 48 | SnPr | CF3 | 7.8 | −46.51 | 8.2 | −54.57 |
| 56 | SMe | C2F5 | 7.5 | −48.49 | 8.4 | −54.09 |
| 36 | tBu | Cl | 6.8 | −41.67 | 7.7 | −43.38 |
pKi, 5-HT2B: Binding affinity (human cloned receptors, HEK 293 cells, [3H]-5-HT)
pKi, 5-HT2C: Binding affinity (human cloned receptors, HEK 293 cells, [3H]mesulergine)
UnifiedCav: Unified cavity E (unit: kcals/mol)
RESULTS
1. Structure predictions of the 5-HT2B structure
Over the last two years, structures for two family human G protein-coupled receptors (GPCRs) have been reported: human β2 adrenergic (hβ2AR)4–6 and human A2A adenosine receptors (hAA2AR)7. In addition the structures for turkey β1 (tβ1AR),8 bovine rhodopsin (bRho),9–13 and opsin14, 15 are available. Unfortunately, these structures include an inverse agonist or antagonist, providing little information about the structures involved upon activation by agonist binding.
The predicted seven TM regions for the three 5-HT2 receptors are shown in Fig. S1 in Supporting Information. Because the experimental structures for some GPCRs show α-helical extensions well beyond the surface of the membrane, we used a standard secondary structure prediction (PSIPRED)16 method to predict these extensions for the 5-HT2 receptors. These extensions lead to an additional 7 residues (EQRASKV) at the cytoplasmic N-terminus of TM6, as shown by underlines in Fig. S1. This includes the (D/E)RY motif expected to be important either in stabilizing the inactive state or facilitating activation.
1.1 TM predictions
The seven TM regions of the 5-HT2B receptor were first predicted by hydropathicity analysis and information from sequence alignments. At ExPaSy home page, a blast search on the query sequence (P41595, 5HT2B_HUMAN, 481 amino acids (a.a.) sequences) with NCBI BlastP 2.2.15 from UniProtKB Swiss-Prot DB (options; mammalian and no fragment) was performed. After multiple sequence alignment using the clustalW program, v.1.8.3, the TM regions were predicted using three ways depending on the input sequences; i) a variety of protein sequences from other family, ii) only 5-HT family receptors, iii) individual TM sequences. The following is the result of each case.
TM prediction 1 using a variety of protein sequences: A variety of 253 protein sequences from other GPCRs with sequence identities from 18 to 98% was first generated. The 253 sequences included HT (hydroxytryptammine, serotonin receptor), DR (dopamine receptor), AD (adrenergic receptor), HR (Histamine), TAA (Trace amine-associated receptor), CCKAR (Cholecystokinin receptor), GAST (Gastrin precursor), ACM (Muscarinic acetylcholine receptor), AA (Adenosine receptor), NMUR2 (Neuromedin-U receptor 2) family receptors, displaying gaps in TMs 1, 5, and 7. The second filtering generated total 118 sequences with identities from 18% to 98%, excluding HRH1 receptor and filtering the species of human, rat and mouse to avoid the redundancy of gene sequence. Total 118 sequences revealed gaps in TMs 1, 2, and 4. Three passes were run, eliminating gap sequences until the appropriate TM regions and hydrophobic centers were yielded. Removing the sequences of AD and TAA receptors, 91 sequences with 18 to 98% identity showed gaps in only TM2. Further removing the sequences which showed the bulge in TM2 (DRD1R, DRD5R, HT4R) and increasing the population of high or middle sequence identity, final 46 sequences with 20 to 98% identity successfully generated 7 TM regions without any gaps in TM regions.
TM prediction 2 using the 5-HT receptor family: After filtering out the non-5-HT receptor family from 46 sequences of TM prediction 1, total 29 sequences were used for predicting 7 TM regions. The 7 TM regions and the hydrophobic centers without any gaps were generated similarly.
TM prediction 3 using an individual TM as a query sequence and a variety of protein sequences from the other family as an input: Each TM regions were decided using an individual TM query sequence adding 4 to 6 a.a. at the end of TM region from the result of TM prediction 1 (40 a.a. for TM1, 47 a.a for TM2, 40 a.a. for TMs 3 and 4, 41 a.a. for TM5, 42 a.a. for TM6, and 43 a.a. for TM7). In each case except TMs 4 and 6, the sequences with the gaps were eliminated until the appropriate TM regions and hydrophobic centers were yielded.
The comparison of all three TM predictions in Table S1 in Supporting Information displayed that predicted TM 7 region was exactly same among three methods. The other TM regions had a good agreement within 1 up to 4 residue differences. The hydrophobic centers also showed within 1 or 2 a.a. deviation. However, the hydrophobic center of TM2 from TM prediction 2 revealed 10 residue differences.
To avoid the Pro problem in Helical Dynamics, four residues LVLC before Pro were added to the N-terminal of TM3. To study the ionic lock in DRY motif, two residues RA were considered as a helix part of the beginning of TM4. In addition, the 5-HT2B receptor was regenerated including EQRASKV at the cytoplasmic part of TM6. These helix extensions of TMs 3, 4, and 6 agreed well with the secondary structure prediction using PSIPRED server16 which was predicted those extensions as a helix part, as shown in Fig. S2. The final TM region of the 5-HT2B receptor was shown in Fig. S3.
1.2 The hydrophobic centers (Peak vs Area methods)
Initially we chose the center of each TM region from a combination of the middle of the predicted TM region modified by any maximum in the hydrophobicity17, 18 over this region. The peak method finds the stable window which does not deviate 5 or more from the value at window size 20 and average those stable values. We also use the area method, the centroid of the hydrophobicity area above the baseline, as an alternative method. The area method bisects the area in the hydrophobic curve using Metlab program.
In Table S2 in Supporting Information, hydrophobic centers of the 5-HT2B receptor were compared in TM prediction 1 to 3. In TM prediction 1, the hydrophobic center of TMs 2, 3, and 7 revealed around 5, 2, and 2 residue difference between two methods, respectively. In TM prediction 3, only TM2 displayed 3.5 residue difference and other TMs revealed similar centers.
Since the hydrophobic centers are important in the translational orientation of each individual helix, four structures of two different hydrophobic centers (the peak and the area methods) and energy scale (Eisenberg vs Octanol) from Cartesian-Neutral dynamics were generated for next step of PDB template generation and compared the protein packings.
1.3 PDB templates
For the generation of the 5-HT2B structure, three PDB templates, frog rhodopsin (fRho),19 mouse Mas-related gene (Mrg) C11 (mMrgC11),20 and human CCR1 (hCCR1) Chemokine receptor,21 were used. Each template displayed different sequence identity of whole sequence with the 5-HT2B receptor; 14% (17% in TM) for fRho, 13% (18% in TM) for mMrgC11, 17% (21% in TM) for hCCR121. Compared to the structure generated by the fRho template, the RMSD of the 5-HT2B structure generated by mMrgC11 and hCCR1 receptor templates showed 3.44 and 3.98 Å, respectively, as shown in Fig. S4 in Supporting Information. Major structural deviations are shown at the tilting of upper TM5 in mMrgC11and the tilting of lower TM2 and upper TM4 in hCCR1. Since the structure from mMrgC11 receptor displayed the similar structure with fRho, two structures of the 5-HT2B receptor from fRho and hCCR1 template were used for further steps.
Two minimized structures of the 5-HT2B receptors generated by two different templates, fRho and hCCR1 receptors were compared. The MPSim energy suggested the 5-HT2B receptor from fRho template displayed lower energy with ~60 kcal/mol than that from hCCR1 template at the same RMS force, 0.09 because of better van der Waals (vdW) interaction. Thus, the 5-HT2B receptor from fRho template was selected for the further study.
1.4 Secondary structure extension at TM6
To study the interaction of ionic locks in DRY motif, additional combinatorial set with the extension of TM6 as an α-helix structure was regenerated; −60° for TM1, 60° for TM2, 0° for TM3, 60° for TM4, −30 and −60° for TM5, 30 and 60° for TM6, 0 and 30° for TM7. Finally, Best 10 structures from 152 combinatorial sets were minimized and compared by Scream total energy (Etot = Tot-fm + Tot-ScSc), total energy of charged (MinEtot,) and neutral system (NeuEtot). Previous best structure (−60, −60, 0, 60, −30, 30, −30°) from new generated TM6 extended structure showed all winners.
To select which hydrophobic centers and energy scale to use, four structures of each receptor were analyzed for putative binding sites and H-bonding networks. When compared H-bonding networks of TMs 1-2-7 (N1.50, D2.50, N7.49) and TMs 2-3-4 (S2.45, H3.42, W4.50), the structure of the peak method and Octanol scale displayed the best H-bonding networks which are conserved in family A GPCRs, while the structure of the area method could not mimic the classical H-bonding networks of TMs 2-3-4 and revealed unrealistic kinks at TM7. In addition, when compared the orientation of the conserved residue in each TM, rotation by Octanol scale gave better orientation than rotation by Eisenberg scale except TM2. Thus, the structure of the peak method and Octanol scale was selected for further steps.
1.5 Helix scan
Hydrophobicity penalty decided the appropriate rotational range of hydrophobic scales within ~2 kcal/mol energy difference for further interhelical energy scan in Fig. S5 in Supporting Information. For example, in TM2, 0 to 90°, −90 to −150° in blue line were selected. Other TMs were also chosen by Penalty E; 90 to −180 degree for TM1, 0 to −120°/90 to 150° for TM3, all angles for TM4, 30 to −150° for TM5, 0 to −120° for TM6, −30 to 30°/180° for TM7. Interhelical energy scans within 2 kcal/mol hydrophobicity penalty energy difference using SCREAM22 yielded the optimal angles of rotation for each helix.
Hydrophobicity penalty and interhelical energy scans in Fig. S6 in Supporting Information yielded the optimal angles of rotation for each helix to produce a combinatorial set of hundreds of conformations. Three passes of combinations were produced, yielding 144 different conformations, one of which is to be selected as the best packing structure. For the first combinatorial set (total: 128), two energetically favorable angles for each helix were selected by Scream E1; 30 and −60 for TM1, 0 and −150 for TM2, 0 and 150 for TM3, 30 and 60 for TM4, 30 and −30 for TM5, 30 and 120 for TM6, 30 and 180 for TM7 as shown in red ball. For the second combinatorial set, 8 additional structures were generated; −60 for TM1, −30 for TM2, 0 and −30 for TM3, 30 and 60 for TM4, −30 for TM5, 30 for TM6, 0 and 30 for TM7. For the third combinatorial set, 8 additional structures were generated; −60 for TM1, −30 for TM2, 0 and −30 for TM3, 30 and 60 for TM4, −30 for TM5, 30 for TM6, 0 and 30 for TM7.
Total 144 combinatorial sets were sorted by polar screamed E, E1 (Totfm + Totscsc −Vscsc − Cfsc − Intern), E2 (Totscsc − Vscsc), and E3 (Totfm − Vtot − Intern). Since E3 best showed the best packing structure, −60, 60, 0, 60, −30, 30, −30 angles for each TM helix were chosen. However, E1 and E2 best structure (30, −150, 0, −60, −30, 120, 30°) didn’t show the classical H-bonding network of TMs 1-2-7, because the orientation of the side chains of D2.50 and N1.50 directed toward the membrane.
Finally, the hydrophobicity penalty and interhelical energy scans are to be re-examined for the best packing structure. Since two more energetically favorable angles for 120° in TM1 and 90° in TM2 were shown in the second MembScream, 4 more combinatorial sets were generated and reordered by E1 + P*9, E2 + P*9, and E3 + P*9. Current all 0 angles for all TM helixes were detected as an energetically favorable helix orientation. In addition, the final best structure revealed better H-bonding networks of TMs 1-2-7 and TMs 2-3-4 as well as ligand binding compared with the beginning structure before MembScream.
Fig. S6 in Supporting Information shows the energetic for the most stable 10 predicted conformations selected from a sequence of optimizations.
For self-consistency, the best packing structure from MembEnsemb was re-examined using the recently developed BiHelix method. BiHelix determines the optimum configuration by sampling all combinations of the rotations of each of the 7 helices through 30 ° rotations, leadings to (12)7 ~ 35 million conformations, for each of which the side chains are optimized using SCREAM.21 To make this practical we evaluate ESCREAM for each of the 144 helix pair combinations for each of the 12 nearest pairs of helices (1-2, 2-4, 4-5, 5-6, 6-7, 7-1, 3-1, 3-2, 3-4, 3-5, 3-6, 3-7) to estimate the total energy for the full 35 million 7-helix bundle conformational combinations. We use the CombiHelix step to build the 1,000 combinations with the lowest estimated energy into explicit 7-helix bundles and calculate the total energy after optimizing the side chains (SCREAM). Then we select the best 100 using ESCREAM and minimize for ~10 steps using the DREIDING 3 force field (FF)23. The best 20 of these bundles is then immersed in an implicit membrane using a Poisson-Boltzmann model with separate dielectric constants for the middle of the membrane, the surface regions of the membrane and the exterior. This provides membrane solvation effects that disfavor helix rotations exposing charged residues to inner part of the lipid bilayer.
1.6 The 5HT2C structure
To study the subtype selectivity, the hHT2CR structure was mutated from the final best structure of hHT2B receptor. All mutated side chains were reassigned by SCREAM method. The lowest neutral energy angles for the 5-HT2B receptors were 15, 15, 0, −15, 15, 15, 0 for TM 1 to 7, while the lowest energy angles for the 5-HT2C receptors were 0, 30, 0, −30, 0, 0, −30 for TM 1 to 7. The preferred angles for each TM between two subtypes revealed ± 15 angle differences among the subtypes. TM7 showed 30° angle deviation, while 0 is the best in TM3 in both cases.
Summarizing, we concluded that the best structure is the one with the lowest total energy (MPSim E) based on the hHT2BR (fRho template) using the peak hydrophobic center, the Eisenberg hydrophobicity scale, and the Cartesian-Neutral dynamics. The predicted best packing structure has the conformation {−60, 60, 0, 60, −60, 60, 0 °} with respect to the fRho template.
1.7 Bihelix/CombiHelix result
To validate the predicted structure, Bihelix/CombiHelix were performed for the best packing structure. The Table S3 in Supporting Information shows the top 10 structures out of ~35 million from the BiHelix analysis. The best packing structures from MembEnsemb (All 0 ° angles correspond to the angles of −60, 60, 0, 60, −60, 60, 0 ° from MembEnsemb) ranks as number 2 by total E. The best one differs by a −30° rotation of TM4. The top 10 from the 1000 structures of CombiHelix built explicitly are shown in Table 2. We find that the all 0 ° structure leads to the best total E. We also find low lying structures with ± 30° variations of TMs 4, 6, and 7.
Table 2.
The best 10 most stable packing structures of the human 5-HT2B receptor from CombiScream analysis.Total energy (kcal/mol) = ScreamTot + MembSolE.
| # | H1 | H2 | H3 | H4 | H5 | H6 | H7 | ScreamTot | MembSolE | Total E |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 605.0 | 97.6 | 507.4 |
| 2 | 0 | 0 | 0 | 330 | 0 | 0 | 0 | 679.5 | −101.0 | 578.5 |
| 3 | 0 | 0 | 0 | 0 | 0 | 30 | 0 | 693.9 | −102.9 | 591.0 |
| 4 | 0 | 0 | 0 | 0 | 0 | 30 | 330 | 694.3 | −96.7 | 597.6 |
| 5 | 0 | 0 | 0 | 0 | 0 | 0 | 330 | 708.0 | −99.8 | 608.2 |
| 6 | 210 | 0 | 0 | 0 | 0 | 0 | 0 | 737.4 | −96.8 | 640.6 |
| 7 | 0 | 0 | 0 | 0 | 0 | 30 | 300 | 746.3 | −89.6 | 656.7 |
| 8 | 0 | 0 | 0 | 30 | 0 | 0 | 0 | 755.3 | −93.5 | 661.8 |
| 9 | 0 | 0 | 0 | 120 | 0 | 0 | 0 | 739.9 | −76.1 | 663.8 |
| 10 | 0 | 0 | 270 | 0 | 0 | 0 | 0 | 744.6 | −76.1 | 668.5 |
This final predicted three-dimensional structure of the hHT2BR possesses the H-bonding network found in the other known class A GPCR structures.
In the middle of TM, D2.50 forms a hydrogen bond (HB) to N1.50.
H3.42 forms a HB with S2.45 and with W4.50.
In the expected ligand binding site of the apo-protein, we find a HB between D135 and Y370 (both are highly conserved within biogenic amine receptors). We find that the predicted hHT2BR structure leads to an ionic lock involving the (D/E)RY motif. Thus we find two salt-bridges between D152 (3.49) and R169 (4.39) and between R153 (3.50) and E253 (6.30) on the cytoplasmic end. These ionic bonds are consistent with two experiments;
The E6.30R mutation shows a highly constitutively active receptor with enhanced affinity for agonist through disruption of ionic interaction.24
R4.39E displays constitutively active arrestin mutant, stabilizing the agonist-high affinity state.25
2. Predicted structures for hHT2BR ligands
First, we docked the rigid SB-206533 1, 5-methyl-1-3,5-dihydro-2H-pyrrolo[2,3-f]indole-1-carboxylic acid pyridine-3-ylamide, (Fig. 1). This has >100 fold selectivity in binding to 5-HT2B over 5-HT2A and other receptors, which was based on the first selective pyridyl urea 5-HT2C/2B antagonist, SB-200646A.26 Then we studied another conformational restricted analogue 2 (1-methyl-1-1,6,7,8-tetrahydro-pyrrolo[2,3-g]quinoline-5-carboxylic acid pyridine-3-ylamide) through the introduction of a six membered ring which is clearly detrimental to 5-HT2C receptor affinity (pA2, 5-HT2B: 7.27 vs pKi, 5-HT2C: 5.39).27.
In addition, we predicted the binding site of PRX-08066 3 (Fig. 1) from EPIX pharmaceutical company known to be a highly potent (Ki ~ 1.7 nM) and selective 5-HT2B antagonist (Ki > 100 fold for more than 55 receptors tested).28 It (pKi, hHT2B: 30 nM) has good bioavailability, preclinical safety profile, and a low order of acute toxicity, which is under Phase II clinical trials for the treatment of pulmonary hypertension and hypoxia-induced pulmonary hypertension syndromes.
Receptor modeling studies of the 5-HT2C receptor suggested that the observed selectivity of 5-HT2B/2C receptors in SB-206533 1 was expected from two valines, V212 and V608, of which the corresponding amino acids were Leu in the 5-HT2A sequence, thus constricting the pocket.29 However, little is known about the selectivity between 5-HT2B and 2C receptors.
Here, we report the predicted binding site and energies for five known agonists and nine antagonists (Fig. 1, Fig. 2, and Table 1), finding relative affinities that correlate well with experiment. We also report the key residues in the binding site which determines the selectivity of the hHT2BR over the 5-HT2A/2C receptor through the docking study of highly selective 5-HT2B ligands.
2.1 Selective hHT2BR agonist 4 (SNF)
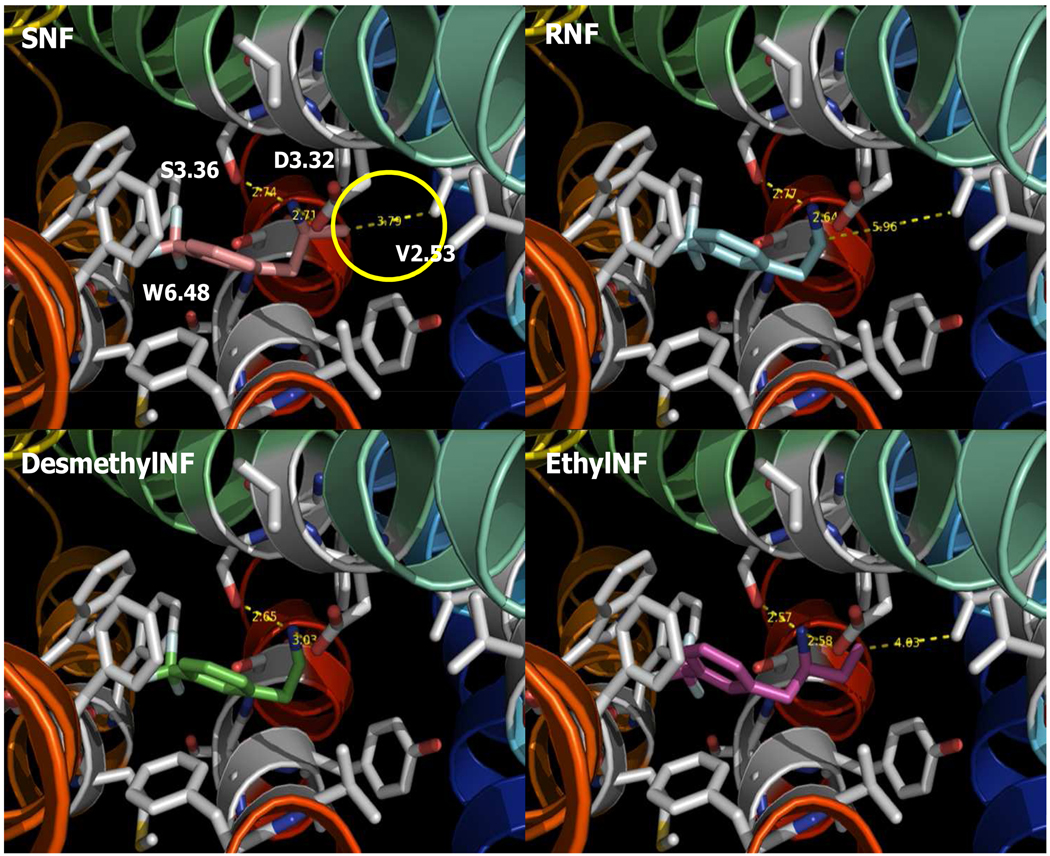
Site directed mutagenesis, binding studies, ligand docking, and MD simulations suggest that terminal methyl groups of V103 in the hHT2BR form stabilizing vdW interactions with the α-methyl group of SNF.1 The role of V103 in SNF binding to 5-HT2 receptors was subtype selective. Our predicted structure agrees with the experimental data in the reported literature.1 The protonated amine nitrogen of SNF 4 shows an ionic interaction at conserved D135 in the biogenic amine receptor. An additional vdW interaction between the α-methyl group carbon of SNF and the terminal γ-methyl carbon of V103 stabilizes the complex with 3.79 Å distances, as shown in Fig. 3.
Fig. 3.
The binding site of the 5-HT2B receptor agonist, SNF 4, RNF, DesMeNF, and EthylNF.
To validate our model, we studied the stereo selectivity using the other isomer. We find that the (R)-enantiomer of norfenfluramine binds in the same orientation as SNF, with a major anchoring interaction at D135. However, the α-methyl group of RNF is away from the terminal γ-methyl carbon of V103 by 5.96 Å, missing favorable vdW interactions in Fig. 3. We find that desmethylNF also has no vdW interaction at V103 because of the absence of the methyl group. We find that ethylNF has unfavorable vdW interaction with the β-methyl carbon of the terminal ethyl carbon. The V103L mutation has little effect on 5-HT affinity, while the V103L mutation markedly and uniquely affects SNF binding to hHT2BRs.1 Compared with SNF (−25.87 kcal/mol in neutral cavity E), the binding E indicates that RNF, DesMeNF, and EthylNF have a higher binding by 0.32, 0.45, 2.92 kcal/mol (see Table S2). Therefore, we predict that SNF is the strongest binder at 5-HT2BR, consistent with the experimental binding affinities (SNF > DesmethylNF > RNF > EthylNF).1
2.2 Selective hHT2BR antagonist 1 (SB-206553) and 2
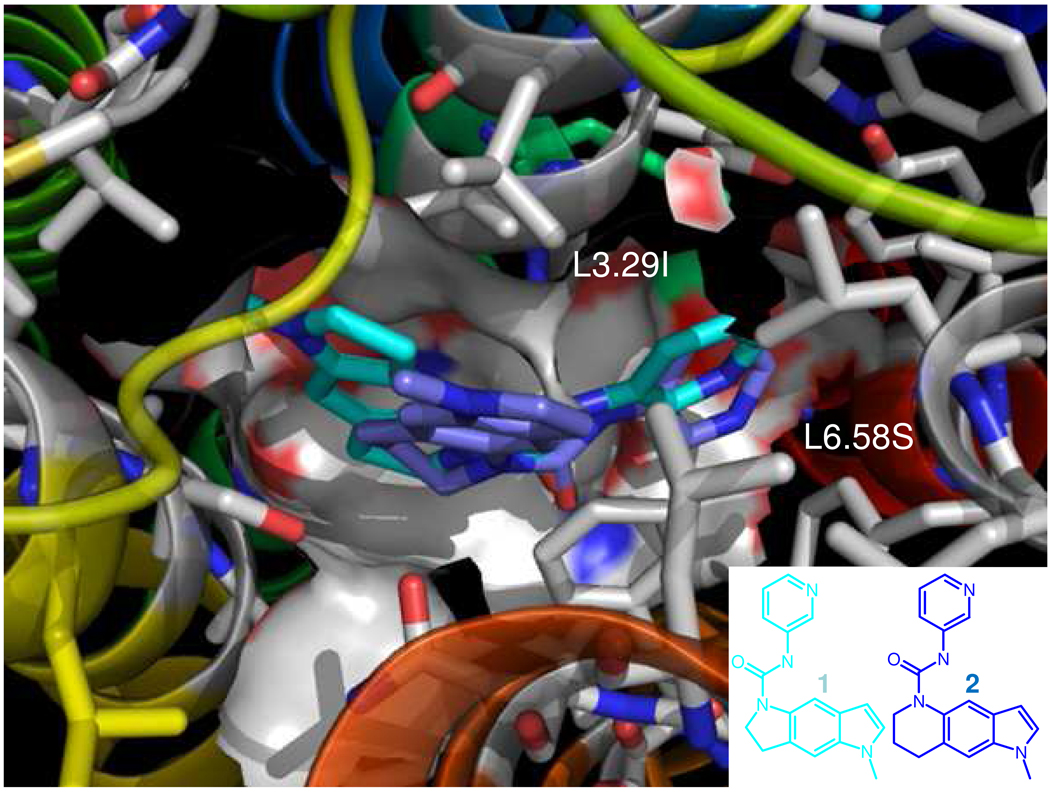
We docked the nonselective 5-HT2B/2C receptor antagonist, SB-206533 1, to both of the hHT2BR and hHT2CR, as shown in Fig. S7A and 7B in Supporting Information. Common HB interactions were found between the ureido-CO group and the Ser-OH side chain at conserved S139 among 5-HT2 family receptor. Comparing the cavity analyses, similar non-bonding E is shown at the hHT2BR and at the 5-HT2C receptor. Additional hydrophobic interactions stabilize the complex at conserved residues, V136, F274 (2B)/F272 (2C), and F299/F298. The N-methyl of the cyclopenta-indole ring interacts with the subtype variable residues, L132, I186, and V190 in the hHT2BRs with nonbond energies of −2.48, −0.10, and −0.76 kcal/mol, respectively. The corresponding amino acids at the 5-HT2C receptors, I132, V186, and I190, also have favorable interaction with nonbond energies. Thus, SB-206533 1 at the 5-HT2B and 2C receptor shows similar total cavity E, −42.48 and −42.28 kcal/mol, respectively. In addition, subtype-selective residues also reveal similar interaction in Table S4 in Supporting Information.
We predict that the 5-HT2B selective antagonist 2 in Fig. S7C/7D in Supporting Information. The ureido-CO group has a major HB interaction with Ser-OH at conserved S139, stabilizing hydrophobic interactions with surrounding hydrophobic residues at conserved residues, V136, F274, and F299. We predict that the major reason for reduced interaction at the HT2C receptor is bad contacts at I132, which has unfavorable vdw interaction (+6.24 kcal/mol), compared with the favorable vdW interaction at the hHT2BR (−2.32 kcal/mol), making binding at the 5-HT2C receptor unfavorable by +8.46 kcal/mol. Other subtype selective residues, V190/I190 in 2C, L281/S279 in 2C, showed similar binding energetic. Consistent with experiment, we find favorable interaction at HT2BR (−47.53 kcal/mol), while binding to the HT2CR is +11.47 kcal/mol less favorable (Table S4). Fig. 4 superimposes 1 and 2, showing that the N-methyl of the cyclohexa-indole ring of 2 is closer to the upper TM3 leading to unfavorable interactions with the bulkier Ile side chain in 5-HT2CR (I132), while the methyl of the cyclopenta-indole ring in 1 points toward the upper TM 4. In addition, the cyclohexa-indole ring is bordered by hydrophobic L281 in hHT2BR, while it is near the hydrophilic residue S279 in the 5-HT2CR.
Fig. 4.
Superimposition of the nonselective 5-HT2B/2C receptor antagonist (SB-206533 1) and the selective 5-HT2B receptor antagonist (2) at the human 5-HT2B receptor. The N-methyl of the cyclohexa indole ring of 2 is closer to the upper part of TM3 leading to unfavorable interactions with bulkier Ile side chains in the 5-HT2C receptors (I132), while the methyl of the cyclopenta-indole ring in SB-206533 1 is pointing toward the upper TM 4.
Further SAR studies were used to validate the current binding mode.30 Based on the experimental SAR in Table 1, the lipophilic group at R5 position and the electron-withdrawing group at the 6 position were optimized. Our predicted energies correlate well with the experimental binding affinity at 5-HT2BR/2CR, with correlation coefficient (r2 value) between experimental binding affinities (Pki) and the calculated cavity E of 0.83 and 0.91 at the human 5-HT2B and 2C receptors, respectively. We find that the R5 lipophilic substituent is bordered by an aliphatic environments (V3.33, L3.29, I4.56, V4.60, M5.39, A5.46), while R6 has electron-withdrawing group is in the proximity of L3.29, S5.46 and N6.55 (Fig. 2).
2.3 Highly selective hHT2BR antagonist, 3 (PRX-08066)
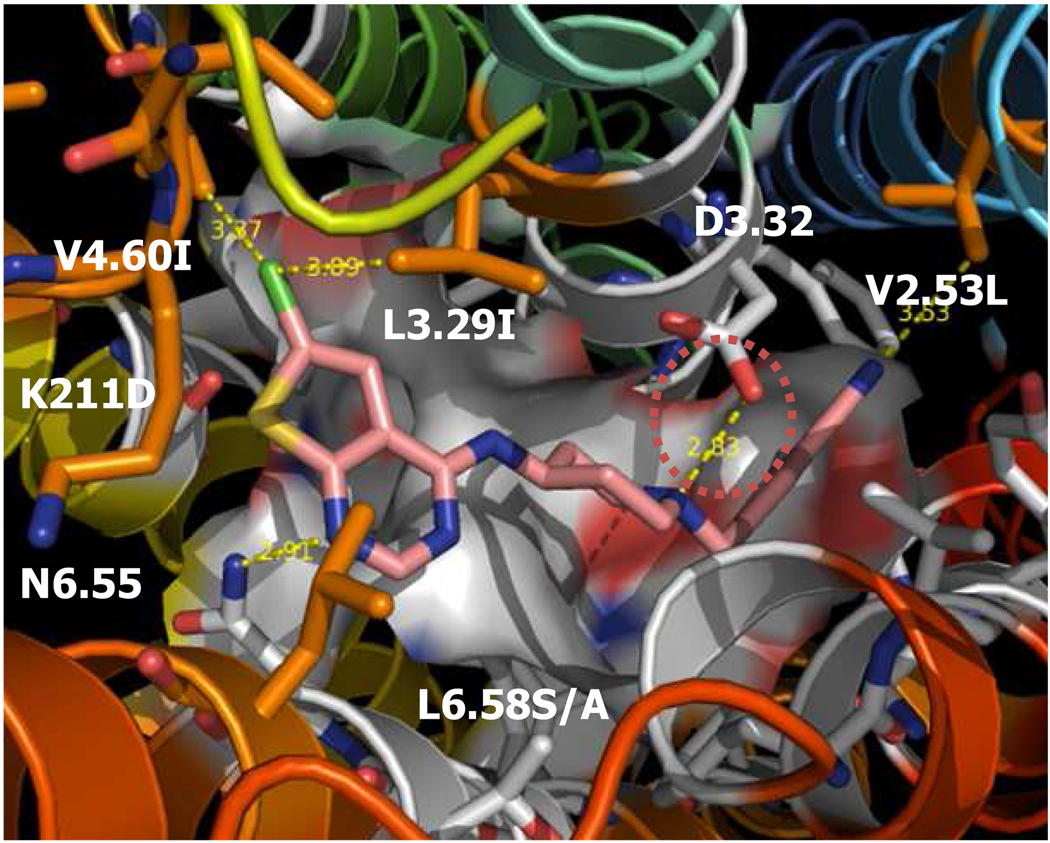
To obtain a better understanding of the molecular basis of the subtype selectivity of highly potent and selective hHT2BR antagonist 3, we dock this ligand (see Fig. 5 and Fig. S8). We find that D135/D134 in TM3 (2B/2C) have a common salt-bridge with a protonated amine in pyridine ring (−7.52 for 2B vs −7.70 for 2C). We find that the N atom of the thienopyrimidine ring makes an additional HB at with the side chain NH of N344/N311 in TM6. We find that several hydrophobic residues stabilized the complex through vdW interaction; the cyano group with V103/V102, the thienopyrimidine ring with F274/F272, L132/I132, and V190/I190 (Table S5 in Supporting Information). We predict that the binding is 24.27 kcal/mol more favorable interaction at 5HT2BR compared to 5HT2CR supports, consistent with experiment.
Fig. 5.
The predicted structure of the highly potent and selective 5-HT2B receptor antagonist PRX-08066 3.
To learn more about subtype selectivity, we examined the effect of mutations in the 5-HT2 receptors on binding of antagonist 3 based on the multiple alignment of 11 serotonin receptors in Fig. S9 in Supporting Information. Here we examined mutations at L132, L347, V103, M218, V190, S372, in the TM regions and K211, T210, E212 in extracellular loop (EL) 2. Each residue was mutated into all 20 amino acids and the side chains reoptimized. Then we match the ligand to the new binding site and minimized the E for each mutant. We found that all mutations decreased the affinity of 3. Thus the mutant L132I to mimic 5-HT2A/2C receptors revealed major decreased interactions by 6.8 kcal/mol compared with the wild type (see Fig. S10 Supporting Information), supporting the direct role of this residue in the selectivity of 3. Other mutants show that L347A (2A) or S (2C), L347A (2A), V103L (2A), M218V (2A/2C), V190I (2A/2C), and S372C (2C) also lead to small decreased interactions by 2.1, 3.4, 4.2, 0.3, 0.7, and 0.5 kcal/mol, respectively. Mutations in EL2, K211D (2A/2C), T210A (2A) or N (2C), E212D (2A) or F (2C) have a negative effect, decreasing binding by 1.1, 0.8, 0.8, 0.8 and 0.0 kcal/mol, but little contribution to the selectivity of 3). These results show a direct affect of mutations on affinity and selectivity. Our results are consistent with reported mutational studies. Mutations at V103 and A225 show that they play a role in the 2B/2C selectivity.31 We propose additional mutation studies at residues, L132, V190, and L347, to test our predictions that they are involved in 5-HT2B/2C selectivity. For the 5-HT2B selective ligands 2 and 3 studied, we found that the side chain at L132 directly determines relative ligand selectivity.
2.4 Docking of endogenous agonist, HT and selective antagonist, 2 to low lying structures of 5-HT2B
To determine whether other low lying structures of HT2B might play a role in activation, we docked the endogenous agonist, 5-HT, and 5-HT2B selective antagonist 2 into the five lowest-lying packing structures from the CombiHelix predictions in Table 2.
We find that agonist HT binds best to bind to the all 0 structures, but it binds unfavorably to the structure with TM6 rotated by 30° (clock-wise from the extracellular view). This agonist binding preference correlates with experimental data; suggesting that binding agonist causes TM6 to rotates anti-clockwise.32.
However, we find that antagonist 2 prefers a slightly different conformation, with TM7 rotated by − 30°, as shown in Table S6 in Supporting Information.
3. Dynamics
To study the effect of membrane and water on the structures of the ligand-GPCR complex, we inserted the predicted protein-ligand complexes into a periodic infinite membrane fully solvated with water and carried out 10 ns of MD at 300K.
3.1 Apo-hHT2BR dynamics
After 10 ns of MD, the major change in the structure observed is a ~ −20° rotation of TM 4. We find that TM6 exhibits large variations with fluctuations +/− 28°. However, the effective change of P6.50 position in TM6 compared to the starting structure is ~ −10° rotation.
The following stable HB networks among TMs in the simulation were shown in Apo-hHT2BR dynamics.
N72 (1.50) and D100 (2.50); the average heteroatom distance HB of 3.5 Å,
S142 (3.39) and S307 (7.46); the average heteroatom distance of 3.2 Å,
S142 (3.39) and N310 (7.49); the average heteroatom distance of 3.3 Å.
In the binding site, we find that three HBs are stable;
D135 (3.32) and Y304 (7.43); the average heteroatom distance of 3.7 Å,
D135 (3.32) and W131 (3.28); the average heteroatom distance of 3.6 Å.
S139 (3.36) and W271 (6.48); the average heteroatom distance of 2.9 Å.
In addition new HBs are formed in the cytoplasmic end;
S150 (3.47) and Y314 (7.53) in the NPxxY region; the heteroatom distance changes from 10.1 Å to 3.8 Å.
D152 (3.49) and R153 (3.28) in the D(E)RY region; the heteroatom distance changes from 11.9 Å to 3.8 Å.
3.2 Agonist (5-HT)-hHT2BR dynamics
After 10 ns of MD, the major change in the agonist (5-HT) bound structure is at TM 6 which rotates by ~ −80° (anti-clockwise), which is consistent with experiment.31 TM 6 also shows the largest fluctuations in the average η angle by +/− 55°. The change in η difference is 7 ° for N1.50 in TM1, −34 ° for D2.50 in TM2, 6 for D3.32 in TM3, 25 ° for W4.50 in TM4, −5 ° for P5.50 in TM5, −113 ° for P6.50 in TM6, and 15 ° for P7.50 in TM7.
Interestingly, we found that all of the salt-bridges in EL (E82-K84, E82-K385, D152-R169, R153-E253, E319-R321) and IL (K193-D216, K211-D351, R213-D351) in the agonist (HT2)-hHTBR are unstable, breaking by 8ns.
The following stable HB networks among TMs in the simulation were shown in Agonist (5-HT)-hHT2BR dynamics.
N72 (1.50) and D100 (2.50); the average heteroatom distance of 3.9 Å.
S95 (2.45) and H145 (3.42); the average heteroatom distance of 3.5 Å.
H145 (3.42) and W180 (4.50); the average heteroatom distance of 3.0 Å.
In the binding site, major anchoring interactions were stable;
D135 (3.32) and the protonated nitrogen; the average heteroatom distance of 3.5 Å,
S139 (3.36) and the OH group; the average heteroatom distance of 4.1 Å,
S222 (5.43) and the NH group; the average heteroatom distance of 3.4 Å.
3.3 Antagonist (2)-hHT2BR simulations
After 10 ns of MD, the major changes in the antagonist (2) bound structure a ~120° rotation (clockwise) at TMs 6, unlike the agonist (5-HT)-hHT2BR. However, the fluctuation of TM6 is +/−20 °. The average η difference of N1.50, D2.50, D3.32, W4.50, P5.50, P6.50, and P7.50 are 16, −27, −24, 15, 6, −20, and −1° angle. Compare to agonist (5-HT)-hHT2BR, the rotations of TMs 3, 5, and 7 are opposite. Although TM6 showed the same rotation, the change is much small (−20 °) in the antagonist (2)-hHT2BR simulations, compared to the agonist (HT)-hHT2BR model (−113°).
Major interactions in the binding site were stable; S139 and the ureido-CO; the average heteroatom distance of 3.0 Å. The ureido-CO group also revealed new weak H-bonding with 4.1 Å average heteroatom distance of H-bonds at W337 which also interacted with S139, resulting in final 2.9 Å heteroatom distance of H-bonds after 10 ns.
Unlike the agonist-bound structure, we found that two salt-bridges between K211-D351 and between K193 and D216 in EL relatively stable during the MD. Thus all three salt-bridges in IL (D152-R169, R153-E319, and E319-R321) are maintained.
With antagonist (2)-hHT2BR, we observed much less motion of the ligand in the binding site, and there were no big differences in the average backbone root mean square deviation (BRMSD) of TM helices (~ 2.0 Å for all three cases). After 10 ns the RMS change in antagonist 2 was 1.0 Å (the average RMSD of 0.6 Å), while the RMS change for agonist HT was 3.9 Å (the average RMSD of 3.1 Å). Thus binding of agonist induces motion in the active site while binding of antagonist leads to a more rigid structure. This increased rigidity of the antagonist (2)-hHT2BR system may block the conformational change for constitutive activation.
The following stable HB networks among TMs in the simulation were shown in Antagonist (2)-hHT2BR simulations.
N72 (1.50) and D100 (2.50); the average heteroatom distance of 2.8 Å
S95 (2.45) and H145 (3.42); the average heteroatom distance of 4.0 Å.
D135 (3.32) and Y370 (7.43); the average heteroatom distance of 3.1 Å
S139 (3.36) and W271 (6.48); the average heteroatom distance of 2.9 Å.
3.4 Water channel from the binding site into the NPxxY motif
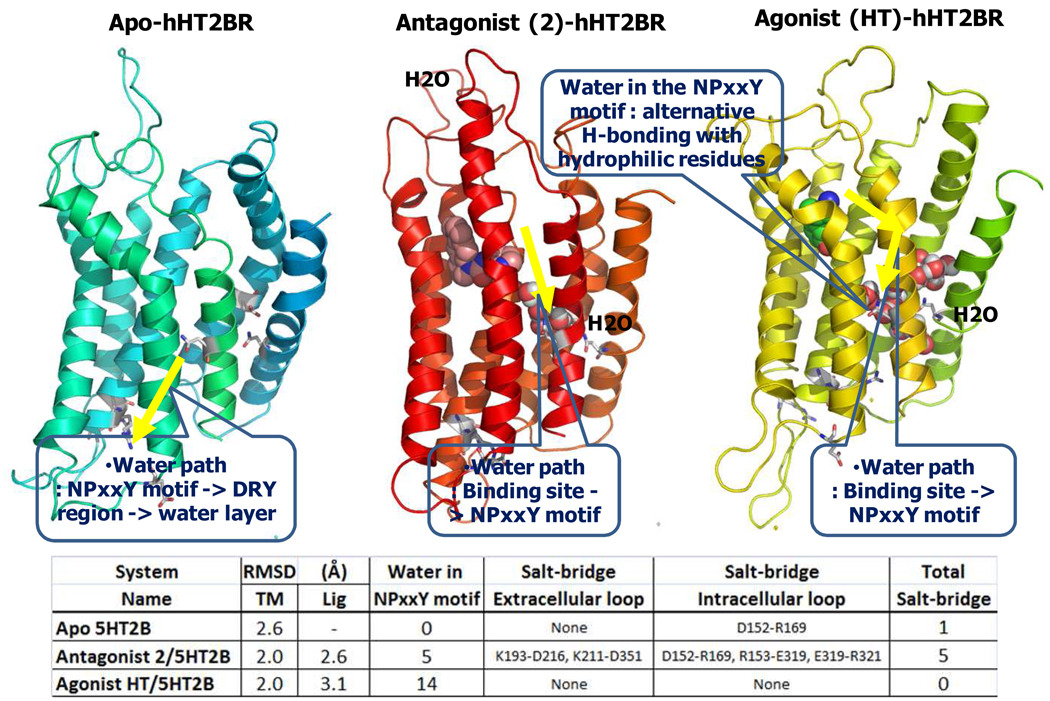
We observed water entering the conserved N1.50-D2.50-N7.49 area (NPxxY region) during the MD for all three cases. However just two waters moved into this region for the apo-hHT2BR, compared to 6 for the antagonist complex, and 14 for the agonist complex. We found a common water path in the ligand-bound system as shown in Fig. 6. Thus binding of agonist promotes migration of water inside the protein, which facilitates the conformational change upon activation compared with the antagonist.
Fig. 6.
Water path (yellow arrow) of apo-protein, antagonist (2) and agonist (HT) bound 5HT2B receptor complexes during 10 ns dynamics in explicit water and membrane. Ligands and water in the 5 Å proximity of N172, D100, and N310 (NPxxY region) are displayed using a space-filling model. The structure was taken from last trajectory of 10 ns dynamics.
In the apo-hHT2BR-membrane complex, two water in the NPxxY region (water within 5 Å of N1.50, D2.50, and N7.49 in VMD program). Thus by 1.7 ns, two water molecules in the NPxxY region were observed in the water layer, passing through the D(E)RY motif which was important for family A GPCR activation, as shown in Fig. 6.
Water in the proximity of D135 (3.32) which is the major anchoring point with the protonated nitrogen passed into S307 (7.46), S142 (3.39) into NPxxY region. This phenomenon suggests that ligand binding in the binding site might regulate the conformational change at the NPxxY region through water.
In the antagonist (2)-bound hHT2BR, water in the proximity of D135 or Y304 in the binding site enters into the NPxxY region, forming HBs with D100, N310, S142 and S307.
In contrast the dynamics of agonist (5-HT)-bound hHT2BR shows water in the proximity of N278 or Y304 in the binding site penetrate into the NPxxY region, forming HBs with D100, S142 and S307. More water in the NPxxY region are floating around in the water pocket through alternative HBs with neighboring hydrophilic residues, N72, D100, S142, S307 and N310. In particular, W271 which is involved in the water channel are thought to control the beginning of activation) as a rotamer switch. One water near R153 also can go into N72. This means the possibility of the cross-talk between the D(E)RY and the NPxxY regions.
The corresponding amino acids of S142 (3.39) and S307 (7.46) in bRho are the hydrophobic residues of Ala. Unlike bRho, hHT2BR displays a constitutive basal activity.33 The hHT2BR seems to provide more flexibility of the protein through the alternative H-bond interactions at two additional hydrophilic regions, S3.39 and S7.46, in the water channel. These additional hydrophilic residues in the water channel extending from the binding site to the NPxxY region correlate with the higher basal activity or protein instability compared to bRho.
Explicit water mediates the H-bond between S222 and the tryptophan NH-group of agonist (5-HT) after 1ns MD. After 9 ns, preserved interactions in the binding site between protonated nitrogen and D135 convert into water-mediated interactions.
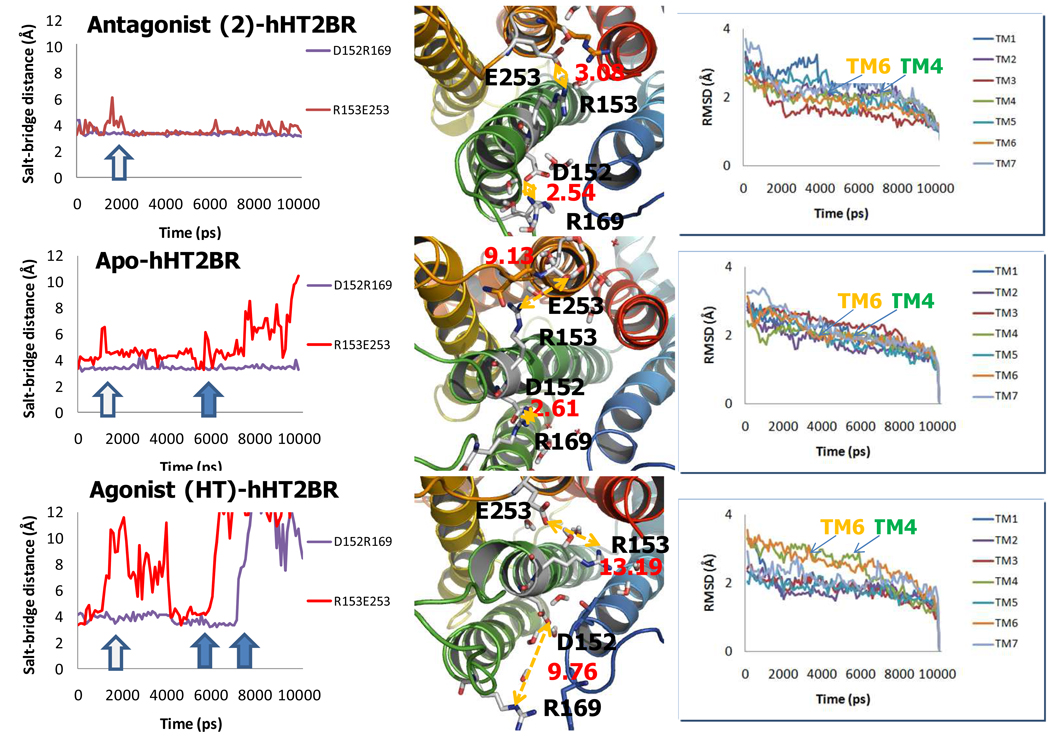
3.5 Ionic-lock stability in the D(E)RY motif
For the apo-protein structure of hHT2BR the ionic locks between R153 and E253 and between D152 and R169 in the D(E)RY region constrain the motions of helix 6 relative to 3.
In apo-protein dynamics, the interaction between TMs 3 and 4 is stable, while the other salt-bridge between TMs 3 and 6 is not stable, as shown in Fig. 7. The first break between R153 and E253 occurred at around 2 ns but the H-bond was reformed later. However after 6 ns, the distance between two hetero atoms of the counter charge increases with the final heteroatom distance of 9.1 Å, while the other final heteroatom distance is 2.6 Å.
In agonist (5-HT)-hHT2BR, both of the salt bridges are broken after 6 ns and 8 ns. The interaction between R153 and E253 is first destabilized. Final distances between R153 and E253 and between D152 and R169 were 13.2 and 9.8 Å, respectively. Supporting this, TMs 4 and 6 resulted in the high BRMSD in Fig. 7.
In the antagonist (2)-bound hHT2BR simulation, both interactions were stabilized. Final distances between R153 and E253 and between D152 and R169 were 3.1 and 2.5 Å, respectively.
These observation support the idea that the D(E)RY-motif plays an important role in protein activation.34 Thus, the motions observed with respect to helix 4 and 6 provide a strong indication that such changes are involved in protein activation.
Fig. 7.
Trajectory analysis of salt-bridge interactions in the D(E)RY region (Left), the final hetero-atom distance of human 5-HT2B receptor (hHT2BR) structures at 10 ns (Middle), and the root mean square deviation (RMSD) of backbone in each TM (Right). Top) antagonist (2)-hHT2BR, Middle) Apo-hHT2BR, Bottom) agonist (HT)-hHT2BR.
DISCUSSION AND CONCLUSIONS
The dynamics for the agonist-hHT2BR structure leading to substantial migration of water into the NPxxY region and the breaking of the ionic lock suggest that binding of the agonist might be able to cause activation of the GPCR. The second alternative is that there might be two or more stable states of the receptor (inactive state R and active state R* with the full agonists binding to R*, while inverse agonists binding to and stabilizing R). Indeed there is a growing body of experimental evidence for the existence of multiple conformational states.35 Our results indicate that several distinct conformations can bind to the agonist and the antagonist, supporting the idea of multiple conformations.
Our ensemble docking result with several lower-lying packing structures of hHT2BR from BiHelix/CombiHelix method revealed the different binding preference between an endogenous agonist (5-HT) and a HT2B selective antagonist 2. The antagonist 2 preferred to bind to the structures for which TM7 was rotated by −30 °. However, the agonist 5-HT revealed weak interactions in the structures for which TM6 was rotated by 30 ° in the clock-wise rotation from the extracellular view. The predicted agonist binding preference is consistent with the experimental data, the anti-clockwise rotation of TM 6 from the extracellular view.32.
Examination of the MD trajectory indicates that antagonist-hHT2BR simulations differ substantially from the dynamics of the apo-hHT2BR and of the agonist-hHT2BR systems. Both of the two salt-bridges in the D(E)RY regions of the antagonist (2)-bound hHT2BR simulation were relatively stable during the simulation, while both interactions were broken in agonist (5-HT)-hHT2BR after 6 ns and 8 ns, sequentially. Apo-dynamics display the intermediate stability of these interactions. The interaction of TMs 3 and 4 is stable, but the other salt-bridge between TMs 3 and 6 is not stable, as summarized in Fig. 7.
In addition, the comparison of the dynamics of agonist and antagonist-bound HT2BRs shows the high RMSD of agonist in the binding site. The more flexible agonist in the binding site allows more water inside the protein and forms a water channel from the binding site to the loosely packed NPxxY region. This region is believed to be important in allowing conformational transitions as there will be fewer steric restraints to side chain packing. Floating water in the NPxxY region is thought to act like a buffer to reduce a steric clash or an electrostatic repulsion between the same charges when the conformational change occurs.
As conclusions, the general agreement of the predicted structures with experimental mutation and binding data suggests that these methods protein structure prediction are reasonably accurate. Thus:
The final best structure of the hHT2BR from MembEnsemb and BiHelix leads to the ionic lock involving (D/E)RY motif and we expected TMs 1-2-7 and TMs 2-3-4 as networks of class A GPCRs.
SNF as an agonist is the strongest binder at the hHT2BR among four derivatives, consistent with their experimental binding affinity (SNF > DesmethylNF > RNF > EthylNF).1 The cavity energies of 6 antagonists paralleled with the experimental binding affinities at the hHT2BR and hHT2CR with the correlation coefficient (r2) values of 0.83 and 0.91.
Docking complex (2) suggests a novel binding interactions for these series of diaryl ureas, involving a hydrogen bonding interaction between the urea carbonyl oxygen of the ligand and D139 (3.36) which is unique at 5-HT2 family receptor.
In the docking models of the 5-HT2B selective antagonist 2 and 3 (PRX-08066), V103 (2.53), L132 (3.29), V190 (4.60), A225 (5.46), and L347 (6.58) lead to subtype selectivity among the 5-HT2 family receptors.
The ensemble docking study with several lower-lying packing structures reveals the different binding preference of a full agonist (5-HT) and an antagonist (2).
The simulations reveal that the salt bridges in D(E)RY motif are maintained in antagonist dynamics but broken in agonist dynamics. The NPxxY region filled with water from the ligand binding site is believed to be important in allowing the conformational transitions upon activation.
Thus, this modeling study will help to design more potent and more selective drugs for the hHT2BR without undesired side effects.
METHODS
1. Generation of the 5-HT2B structure using MembEnsemb program, v.4.30
First, the three-dimensional structures of the hHT2BR based only on its primary sequence were predicted using the MembEnsemb first principles method which was updated from the Membstruk first principles method.36, 37 All E and force calculations used the DREIDING FF23 for the ligand and CHARMM22 charges for the protein.38 The MembEnsemb procedure involves the following steps.
1.1 Predict TM region
Our method for predicting the seven TM domains, PredicTM which is developed from earlier version of TM2ndS,36 uses hydropathicity analysis combined with information from multiple sequence alignments. We start with about 50 to 200 sequences having sequence identities from the target structure varying uniformly from 20% to 90%.
The second step of TM2ndS is to calculate the consensus hydrophobicity for every residue position in the alignment using the average hydrophobicity of all the amino acids in that position over all the sequences in the multiple sequence alignment. Then, we calculate the average hydrophobicity over a WS of residues about every residue position, using WS ranging from 12 to 30. The average hydrophobicity value at each sequence position was plotted to yield the hydrophobic profile. MembStruk procedure used the Eisenberg hydrophobicity scale, but with MembEnsemb we now use 8 different hydrophobicity scales, 1) thermodynamic hydrophobic octanol as a default, 2) the interface of octanol, 3) the average of thermodynamic and biological hydrophobic scales, 4) biological hydrophobic scales, 5) hydrophobicity penalty with the solvent accessible surface area developed by Jenelle, 6) the Eisenberg hydrophobicity (the scale used by the original TMPred), 7) new Eisenberg, and 8) interface.
1.2 Create PDB template
The initial z position of each of the 7 helices and the initial orientation η, the initial x and y position is taken from the 7.5 Å electron density map of fRho,19 mMRgC11,20 and hCCR121 receptors which was generated using TMPred.
In addition, the tilt (θ) of each axis from the z axis and the azimuthal orientation (φ) is taken from this structure. This is the procedure used for previous applications of MembStruk. However, for MembEnsemb, we are now extracting the x, y, θ, and φ from those predicted structures of GPCRs that have been successful in predicting binding sites of known ligands and which have been subjected to MD optimization with full membrane and solvent. We will use one or more sets of x, y, θ. and φ from this data base as starting points for the GPCRs being studied. After optimization, the helix is reinserted in the bundle while preserving the major axis, which is followed by a full bundle optimization.
With the predicted TM regions for the 5-HT2B/2C receptors, the canonical helices were generated. The helical axes are positioned according to each template, with the hydrophobic centers on the same fitting plane.
1.3 Rotation by phobic face
The helices will be treated as canonical and have all C-alpha carbon positions (the middle 15 residues around the hydrophobic center) projected on to the plane that intersects the hydrophobic center and aligns the smallest moment of inertial along the Z-axis. Then all projected C-alpha positions will be assigned their hydrophobic scalar according to the hydrophobicity scale chosen and the program will determine the largest gap of the helix that does not face other helices and look the largest number summing the C-alpha numbers over that gap. The middle of the gap containing the largest sum of all C-alphas will become the hydrophobic moment and the helix will be rotated to move this position to 180 degrees from the center of the protein. The canonical helices for the predicted TM segments of the 5-HT2B/2C receptors were rotated by Octanol scale for better orientation of hydrophobic side chains facing toward the membrane.
1.4 Helix dynamics
The structures of the individual helices were optimized with E minimization followed by 100 ps Newton-Euler inverse mass operator (Neimo) torsional dynamics39, 40 and Cartesian dynamics with charged force field or neutral QeQ charge at 300K (NVT). This optimizes the bends and kinks in each helix due to the presence of Pro and Gly in some TM domains. This is an important step as it is believed that the individual helices interact with each other in the lipid bilayer during the folding process and by the start of this interhelical interaction they would have assumed their native bent or straight helix conformations. The presence of bent helices disallows many rotational combinations when the helices interact with each other.
The spatial orientation of the helices, helical bends and kinks were optimized by individual helix dynamics. The Neimo dynamics and the Cartesian-Charge dynamics produced severe kinking and unraveling of TM 2, eliminating them as a feasible option. The Cartesian-Neutral structure is feasible because there is little unraveling and minor kinking caused by the presence of Pro. All prior calculations were repeated using the different hydrophobic center of the area method.
1.5 Rotation by hydrophobic moments
The initial orientation (η) of the helix about this axis is determined by calculating the net hydrophobic moment of the middle one-third of the helix (centered at the hydrophobic center) but counting only the half circle that would be oriented toward the lipid. The calculation of the hydrophobic moments uses only the middle 15 residues around the hydrophobic center of each helix. Then these hydrophobic moments are projected onto the plane of intersection of the hydrophobic centers. The degree of rotation needed to point the end of hydrophobic vector 180 degrees away from the center of the protein is used for the individual rotation of the helices. This is pointed outward from the center of the xy positions of the seven helices. The optimized helices of the 5-HT2B receptor from Cartesian-Neutral Dynamics were rotated by Eisenberg or Octanol scale.
1.6 MembEnsemb
To select the initial orientation of the side chains we use a Monte Carlo procedure. Using SCREAM22 for selecting the side chain conformation considerably improves the accuracy in comparing different rotations, and we have now eliminated the ROTMIN step, replacing ROTSCAN with ROTSCREAM, but now the scan need only be in increments of 30°, rather than 5°. All helices are rotated by 30° increments while reassigning the side chain conformation by SCREAM method22. Hydrophobicity penalty and interhelical E scan are performed to maximize the best orientation of each helix and the side chain. For the optimization of rotational orientation of the helices, each helix is rotated through a grid of rotational angles while reassigning the side chain conformation by SCREAM method.
1.7 The combinatorial set using MembEnsemb
The combinatorial sets lead to an ensemble (3 to 10) of packed bundles, each of which we may use for docking candidate agonists and antagonists. Our working hypothesis is that GPCRs may sample 3 or 4 (or more) packings of the helices during their dynamics as they interact with agonists, antagonists, inverse agonists, or modulators. By determining the properties for each of the structures in this ensemble, we expect to obtain a better understanding of which structures might play a role in the various processes of the GPCRs (some might be more favorable for dimers). This systematic search over a grid of rotational angles and translational distances is important because there are likely to be substantial E barriers between some of the local minima, which molecular dynamics simulations may not find all of them.
1.8 BiHelix and CombiHelix
The top1 of the hHT2BR from MembEnsemb was used for running BiHelix and CombiHelix. To find a diverse ensemble of low-E packing of the TM helices and validate the current hHT2B structure, we started with the MembEnsemb structure and rotated independently 12 helix pairs (H1–H2, H1–H7, H2–H3, H2–H4, H2–H7, H3–H4, H3–H5, H3–H6, H3–H7, H4–H5, H5–H6, and H6–H7) in 30° degree increments through full a 360° (12 rotations in each helix), leading to explicit 1,728 (12×12×12) bundle configurations. For each of these theoretical 35 million TM bundle configurations, the pair E which was indicated in details in Table S1 was calculated.
In the combinatorial set, the best 100 structures were optimized all side chains using SCREAM, calculated the polar interhelical interaction E ignoring side chain-side chain EvdW or all vdW E, or internal E, total interhelical interaction E and the Membrane Solvation E (MembSolE), ordered by total E, total interhelical interaction E and MembSolE.
The CombiHelix results for best 10 are shown in Table 1. The current structure with all 0° angles for each helix ranked as Number 1. The second low-lying structure rotated −30° anti-clockwise way for TM4 from extracellular side. The third one showed a single rotation of TM6 by 30°. In addition to the rotation of TM6 by 30°, the fourth one showed an additional −30° rotation of TM7 which revealed in the fifth case. All other helices, TMs 1, 2, 3, and 5 showed 0° structure as the lowest-lying one. Thus the best angles within best 5 are: TM1= 0, TM2= 0, TM3= 0; TM4= 0, −30, TM5= 0, TM6= 0, 30, TM7: 0, −30.
The 0° case for each helix has the lowest E, validating that the current model is correctly predicted through MembEnsemb procedure. We believe the other low-lying packings of the helices may play a role in binding of agonists and/or in activation.
* The description of BiHelix energies.
The BiHelx pair energy (BiHelE) is the sum of the SCRAEM energies from all 12 pairs for each of the 127 7-helix conformations, properly corrected for overcounting of the intra helix interactions.
BiHelE = Sum_HiHj = Totintra-ii + Totintra-jj + Totinter-ij
where
Totintra-ii = Total intrahelical energy of Helix i in i-j pair.
Totintra-jj = Total intrahelical energy of Helix j in i-j pair.
Totinter-ij = Total interhelical energy between helices i and j.
TotinterH-ij = Interhelical hydrogen-bond energy between helices i and j.
Interhelical hydrogen-bond energy (InterHB) is the sum of all Interhelical hydrogen-bond energies between all 12 pairs of helices.
* The description of CombiHelix energies
For each of the 7-helix conformations selected from the BiHelix analysis we built full 7-helix bundle, scream all 7 helixes simultaneously and evaluated the energy, ScreamTot. In addition, we evaluated the membrane solvation free energy (MembSolE, Es) using a multi-dielectric (80|7|2|7|80) implicit membrane model of the total solvation free energy change associated with transfer of protein from implicit bulk water.
ScreamTot = Totfm + Totscsc + Intern
i) Totfm = Intern+Vfsc+VTotfm+CHTotfm
Intern = Valence energy terms of the SCREAMed residues.
Vfsc = vdW energy between SCREAMed residue side chains and fixed residue side chains.
VTotfm = Vfsc + vdW energy between SCREAMed residue side chains and all backbones.
CHTotfm = Coulomb plus Hydrogen bond (HB) energy between fixed residues and SCREAMed residue side chains.
ii) Totscsc = Vscsc+CHscsc
Vscsc = vdW energy among SCREAMed residue side chains.
CHscsc = Coulomb and HB energy among SCREAMed residue side chains.
MembSolE = Es = E1−E2+E3
E1=Polar solvation energy of the protein in a multi-dielectric (80|7|2|7|80) implicit membrane.
E2=Polar solvation energy of the protein in implicit bulk water.
E3=Non-polar solvation free energy change corresponding to transfer of protein from implicit bulk water to multi-dielectric (80|7|2|7|80) implicit membrane, approximated as γ × Lipid exposed solvent accessible surface area.
2. Generation of loops by CCBB loop builder
Loops were predicted by a new CCBB loop builder using the CCBB Monte Carlo method developed for predicting free energies of polymer chains combined with Restraint Generic Protein algorithm41 for handling the constraint of terminating the loop on the adjacent TM domains.
The disulfide bond between C128 and C207 was constructed. The final structure was optimized without any constraints. To save the simulation time in explicit membrane and water, truncated intracellular loop (IL) 3, from A247 to V313, which was far away from the binding site was used.
To provide initial loop structures for other low-lying packing structures, the loops of three 3 EL and 3 IL parts were connected into four other TM structure. Connection parts were minimized with trans-amide torsional constraints and backbone freeze atoms at the α-helix structure.
To check the quality of the model, the main chain parameters (Ramachandron plot quality assessment, peptide bond planarity, measure of bad non-bonded interactions, alpha carbon tetrahedral distortion, HB energies) and the side chain parameters (the standard deviation angle of Chi-a gauche minus, Chi-1 trans, Chi-1 gauche plus, Chi-2 trans angle) used Procheck program to reveal statistically better or reasonable results.
3. Docking Study
3.1 GenMSCDock
The putative binding sites for agonists and antagonists of the hHT2BRs were determined using GenMSCDock. GenMSCDock was the next generation of HierDock42and MSCDock,43 but many improvements have also been made. MSCDock was successfully applied to mMrgC1120 and hHT2C3 receptors, as described previously. This procedure solved the problem of allowing bulky residues and bulky ligands to accommodate each other. The validation of GenMSCDock has been described previously.44 We docked the antagonist, ZM241385, to the crystal structure of the hAA2AR (PDB ID: 3eml). Our best predicted structure (best cavity energy) is 0.80 Å RMSD from the x-ray structure. The largest deviation was at the phenoxy ring exposed toward EL 2 and 3 due to the three waters surrounding the phenoxy ring in the X-ray complexes.
GenMSCDock sampled a complete set of configurations for each conformation of the ligand. This leads to a hierarchy of families with a specified range of diversities, allowing the best to be selected at a coarse level by evaluating energies of family heads. This also leads to an ensemble of the best diversity families that are used for higher-level calculations. The steps in GenMSCDock include bulky residue alanination, diversity finder, Voronoi reclustering, side-chain refinement and neutralization.
Bulky residue alanination: To accommodate various orientations of the ligands, all bulky residues (Phe, Trp, Tyr, Val, Ile, and Leu) were replaced by Ala.
Diversity finder: A critical part of GenMSCDock is to sample all binding configurations in all potential binding sites and to group the results into families of related structures allowing whole family to be rejected when the parent of a family is weakly bound. The GenMSCDock method does two rounds, one to ensure completeness and the other to enrich the better families. Based on our experiences, a larger diversity parameter of 1.2 – 1.4 Å in the completeness step generates ~2500 families. To achieve an average of 6 children in each family, we resort to obtain 0.6 Å families in the enrichment step.
Voronoi re-clustering: As an enrichment step, Voronoi clustering fractionizes to ensure that each family has the closest children within it. We find that with a 0.6 Å diversity threshold all members of a family minimize to essentially the same structure.
Side chain refinement: For each of the 10% low E conformation from Voronoi clusters, we dealaninize the binding site with the ligand present using the SCREAM procedure22. SCREAM finds the optimum side chain conformations for the bulky residues compatible with each ligand configuration. After SCREAM, a short full complex minimization is performed. Total 50% subset from our hierarchy was selected for further steps.
Neutralization of charged groups: To remove the sensitivity of counter ions distant from the active site, charged residues and ligands are modified by protonating or deprotonated. This leads to a smoother electrostatic potential and smaller solvation E difference in the binding site.
Relaxation: For each of the 50% ligand-side chain combination, we now define a unified binding site and then minimize the atoms in the binding pocket. Then for another 50% subset, we minimize the structure for the full ligand-protein complex and pick a subset of 50% for annealing within 5 Å of the binding site.
Scoring E: GenMSCDock allows at each step for a percentage of the protein+ligand complexes to be eliminated based on E criteria including continuum solvent corrections. This improved procedure is now being automated to increase the number of ligands for practical considerations. In each step, the complex was scored using the following different E criteria.
* E scoring
-
-
UnifiedCav (Unified cavity): the E from cavity analysis where all residues in the unified cavity analysis contribute to the E.
-
-
LocalCav (Local cavity): the normal version of cavity analysis where only residues within the local 5.0Å binding cavity contribute to the E.
-
-
Total: the MPSim E of the total complex (all atoms movable).
-
-
FullSol (Full solvation E): the binding E with solvation for the complex, protein, and ligand.
BE = (complexvac + complexsolv)−[(proteinvac + proteinsolv) + (ligandvac + ligandsolv)]
-
-
PartialSol (Partial salvation E): the binding E with solvation only for the ligand.
BE = complexvac − [proteinvac + (ligandvac + ligandsolv)]
-
-
Interaction: the MPSim interaction E between the protein and ligand (protein fixed, ligand movable).
3.2 Ensemble docking
The best ligand conformation from GenMSCDock matched onto several low lying packing structures and relaxed the binding site to maximize the interaction through the following sequential steps, BindsiteScream (SCREAM the 5 Å binding site compatible with each ligand configuration), BindsiteMinimize (the 5 Å binding site minimization), iii) ComplexMinimize (full complex minimization), and BindsiteAnneal (binding site annealing), and Neutralize including full complex minimization.
4. MD simulations in explicit membrane and water
To construct the periodic cell (73×70×71 Å) for the MD simulations, the predicted structures of apohHT2BR, agonist (5-HT)-hHT2BR, and antagonist (2)-hHT2BR were each independently embedded in the center of a square periodic lipid bilayer consisting of 100 palmitoyloleoyl phosphatidylcholine (POPC) molecules and solvated with 6,689 water molecules. We used the CHARMM22 force field parameters for the protein, the TIP3 model for water,38 and the CHARMM27 force field parameters for the lipids45. Quantum charges from DFT/6311G** method were used for these ligands.
To compensate for the net charge of the protein and ligand, two waters for the apo-hHT2BR/the antagonist-bound hHT2BR, three waters for the agonist-bound hHT2BR, and seven waters for the antagonist-bound hHT2CR were replaced by chloride ions to attain a zero net charge for the entire system. These systems contains 38,051 atoms (apo-hHT2BR), 38,977 atoms (5-HT-hHT2BR), 38,672 atoms (antagonist 1-hHT2BR, 38,818 atoms (antagonist 2-hHT2BR), and 38,025 atoms (antagonist 1-hHT2CR and antagonist 2-hHT2CR).
The process was first to minimize the water, ion, and lipid bilayer while keeping the protein and ligand fixed. This was followed by an all-atom conjugate gradient minimization of the entire system for 1,000 steps. After this minimization, we carried out 500 ps of MD simulations for equilibration with 1-fs time steps, followed by 1,000 steps of minimization of the full system. Langevin dynamics was used for temperature control with the thermostat set at 310 K. The Nose´–Hoover Langevin piston pressure control was used to control fluctuations in the barostat, which was set at a pressure of 1 bar. Here the periodic cell was constrained to remain orthorhombic, but the cell parameters were allowed to vary. A dielectric constant of 1 was used for the electrostatic interactions, which were calculated by using the particle mesh Ewald method.46 The grid in the x, y, and z directions used for the particle mesh Ewald method was set at 72, 75, and 81 points, respectively. Every 10 ps a snapshot was written to the trajectory file for subsequent analysis.
Seven independent 1ns MD simulations were carried out by using the program NAMD 2.6 for all minimization and MD runs.47 Three further 10 ns simulations were done for apo, agonist-bound and antagonist 2-bound hHT2BR.
Supplementary Material
ACKNOWLEDGMENT
This work was funded partially by Boehringer-Ingelheim and by NIH. We thank Dr. Henry Levenson for helpful discussion.
Footnotes
Supporting Information Available: Predicted transmembrane (TM) region by three methods and hydrophobic centers by two methods (Table S1), The experiment binding affinity and predicted scoring energy (kcal/mol) of several 5-HT2B agonists at the human 5-HT2B receptor (Table S2), The 10 most stable 7-helix conformations for the human 5-HT2B receptor from the BiHelix analysis (Table S3), Cavity analysis of SB-206533 1 and 2 at human 5-HT2B and 2C receptors (Table S4). Cavity analysis of PRX-08066 3, at the human 5-HT2B and 2C receptors (Table S5), The ensemble docking results of the human 5-HT2B receptor for agonist, HT and antagonist, SB1, in charged and neutral system (Table S6) TMPredict multiple alignments of three 5-HT2 receptors, 2A, 2B and 2C (Fig. S1), The prediction of secondary structure for the human 5-HT2B receptor (Fig. S2), The sequence of seven transmembrane (TM) regions (Top) and its hydropathy plot (Bottom) of human 5-HT2B receptor predicted by TMPred program (Fig. S3), Superimposition of two different templates with the fRho19 template (Fig. S4) Interhelical interaction energies of MembScream (Fig. S5), Interhelical interaction energies of MembScream at the third round (Fig. S6), The complexes of the nonselective 5-HT2B/2C receptor antagonist (SB-206533 1) and the selective 5-HT2B receptor antagonist (2) at the human 5-HT2B and 2C receptors (Fig. S7), The binding mode of highly selective 5-HT2B receptor antagonist PRX-08066 3 at the human 5-HT2B (top) and 2C (bottom) receptors (Fig. S8), Multiple alignments of 11 serotonin receptors (Fig. S9), and The mutation study of subtype selective residue L132X (3.29) from the cavity analysis of the complex of PRX-08066 3/5-HT2B receptor in neutral system (Fig. S10). This material is available free of charge via the Internet at http://pubs.acs.org.
Contributor Information
Soo-Kyung Kim, Email: skkim@wag.caltech.edu.
Youyong Li, Email: youyong@gmail.com.
Ravinder Abrol, Email: abrol@wag.caltech.edu.
Jiyoung Heo, Email: jyheo@wag.caltech.edu.
William A. Goddard, III, Email: wag@wag.caltech.edu.
REFERENCES
- 1.Setola V, Dukat M, Glennon RA, Roth BL. Molecular determinants for the interaction of the Valvulopathic anorexigen norfenfluramine with the 5-HT2B receptor. Mol. Pharmacol. 2005;68:20–33. doi: 10.1124/mol.104.009266. [DOI] [PubMed] [Google Scholar]
- 2.Schmuck AG, Ullmer C, Engles P, Lübbert H. Cloning and Functional Characterization of The Human 5-HT2B Serotonin Receptor. FEBS Lett. 1994;342:85–90. doi: 10.1016/0014-5793(94)80590-3. [DOI] [PubMed] [Google Scholar]
- 3.Bray JK, Goddard WA., III The structure of human serotonin 2c G-protein-coupled receptor bound to agonists and antagonists. J. Mol. Graph. Model. 2008;27:66–81. doi: 10.1016/j.jmgm.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 5.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EY, Velasquez J, Kuhn P, Stevens RC. A specific cholesterol binding site is established by the 2.8 Å structure of the human beta2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi H-J, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Sci. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Sci. 2008;322:1211–1277. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AGW, Tate CG, Schertler GFX. Structure of a β1-adrenergic G-protein coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J. Mol. Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Edwards PC, Burghammer M, Villa C, Schertler GFX. Structure of bovine rhodopsin in a trigonal crystal form. J. Mol. Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 11.Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y. Fundamental role of internal water molecules in rhodopsin revealed by Xray crystallography. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teller DC, Okada T, Behnke CA, Palczewski K, Stenkamp RE. Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs) Biochem. 2001;40:7761–7762. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H. Crystal structure of rhodopsin: a G protein-coupled receptor. Sci. 2000:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 14.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 15.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 16.Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucl. Acids Res. 2005;33:36–38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayasinghe S, Hristova K, White SH. Energetics, stability, and prediction of transmembrane helices. J. Mol. Biol. 2001;312:927–934. doi: 10.1006/jmbi.2001.5008. [DOI] [PubMed] [Google Scholar]
- 18.Eisenberg D WRM, Terwilliger TC. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc. Natl. Acad. Sci. U.S.A. 1984;8:140–144. doi: 10.1073/pnas.81.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schertler GFX, Villa C, Henderson R. Structure of rhodopsin. Eye. 1998;12:504–510. doi: 10.1038/eye.1998.138. [DOI] [PubMed] [Google Scholar]
- 20.Heo J, Han S-K, Vaidehi N, Wendel J, Kekenes-Huskey P, Goddard WA., III Prediction of the 3D Structure of FMRF-amide Neutopeptides Bound to the Mouse MrgC11 GPCR and Experimental Validation. Chem. Bio. Chem. 2007;8:1527–1539. doi: 10.1002/cbic.200700188. [DOI] [PubMed] [Google Scholar]
- 21.Vaidehi N, Schlyer S, Trabanino RJ, Floriano WB, Abrol R, Sharma S, Kochanny M, Koovakat S, Dunning L, Liang M, Fox JM, de Mendonca FL, Pease JE, Goddard WA, III, Horuk R. Predictions of CCR1 Chemokine Receptor Structure and BX 471 Antagonist Binding Followed by Experimental Validation. J. Biol. Chem. 2006;281:27613–27620. doi: 10.1074/jbc.M601389200. [DOI] [PubMed] [Google Scholar]
- 22.Kam VWT, Goddard WA., III Flat-Bottom Strategy for Improved Accuracy in Protein Side-Chain Placements. J. Chem. Theo. & Comp. 2008;4:2160–2169. doi: 10.1021/ct800196k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayo SL, Olafson BD, Goddard WA., III DREIDING - a generic force field for molecular simulations. J. Phys. Chem. 1990;94:8897–8909. [Google Scholar]
- 24.Shapiro DA, Kristiansen K, Weiner DM, Kroeze WK, Roth BL. Evidence for a Model of Agonist-induced Activation of 5-Hydroxytryptamine 2A Serotonin Receptors That Involves the Disruption of a Strong Ionic Interaction between Helices 3 and 6. J. Biol. Chem. 2002;277:11441–11449. doi: 10.1074/jbc.M111675200. [DOI] [PubMed] [Google Scholar]
- 25.Gray JA, Bhatnagar A, Gurevich VV, Roth BL. The interaction of a constitutively active arrestin with the arrestin-insensitive 5-HT(2A) receptor induces agonist-independent internalization. Mol. Pharmacol. 2003;63:961–972. doi: 10.1124/mol.63.5.961. [DOI] [PubMed] [Google Scholar]
- 26.Forbes IT, Kennett GA, Gadre A, Ham P, Hayward CJ, Martin RT, Thompson M, Wood MD, Baxter GS, Glen A, Murphy OE, Stewart BA, Blackburn TP. N-(1-Methyl-5-indolyl)-N’-(3-pyridyl)urea Hydrochloride: The first selective 5-HT1C receptor antagonist. J. Med. Chem. 1993;36:1104–1107. doi: 10.1021/jm00060a019. [DOI] [PubMed] [Google Scholar]
- 27.Forbes IT, Ham P, Booth DH, Martin RT, Thompson M, Baxter GS, Blackburn TP, Glen A, Kennett GA, Wood MD. 5-Methyl-1-(3-pyridiiylcarbamoyl)-1,2,3,5-tetrahydropyrrolo[2,3-f]indole: A novel 5-HT2C/5-HT2B receptor antagonist with improved affinity, selectivity, and oral activity. J. Med. Chem. 1995;38:2524–2530. doi: 10.1021/jm00014a004. [DOI] [PubMed] [Google Scholar]
- 28.Tomillero A, Moral MA. Gateways to clinical trials. Methods Find. Exp. Clin. Pharmacol. 2008;30:543–588. [PubMed] [Google Scholar]
- 29.Forbes IT, Dabbs S, Duckworth DM, Ham P, Jones GE, King FD, Saunders DV, Blaney FE, Naylor CB, Baxter GS, Blackburn TP, Kennett GA, Wood MD. Synthesis, Biological activity, and molecular modeling studies of selective 5-HT2C/2B receptor antagonists. J. Med. Chem. 1996;39:4966–4977. doi: 10.1021/jm960571v. [DOI] [PubMed] [Google Scholar]
- 30.Bromidge SM, Dabbs S, Davies DT, Duckworth DM, Forbes IT, Ham P, Jones GE, King FD, Saunders DV, Starr S, Thewlis KM, Wyman PA, Blaney FE, Naylor CB, Bailey F, Blackburn TP, Holland V, Kennett GA, Riley GJ, Wood MD. Novel and selective 5-HT2C/2B receptor antagonists as potential anxiolytic agents: Synthesis, quantitative structure-activity relationships, and molecular modeling of substituted 1-(3-pyridylcarbamoyl)indulines. J. Med. Chem. 1998;41:1598–1612. doi: 10.1021/jm970741j. [DOI] [PubMed] [Google Scholar]
- 31.Almaula N, Ebersole BJ, Ballesteros JA, Weinstein H, Sealfon SC. Contribution of a helix 5 locus to selectivity of hallucinogenic and nonhallucinogenic ligands for the human 5-hydroxytryptamine2A and 5-hydroxytryptamine2C receptors: Direct and indirect effects on ligand affinity mediated by the same locus. Mol. Pharmacol. 1996;50:34–42. [PubMed] [Google Scholar]
- 32.Gether U, Lin S, Ghanouni P, Ballesteros JA, Weinstein H, Kobilka BK. Agonists induce conformational changes in transmembrane domains III and VI of the beta2 adrenoceptor. EMBO J. 1997;16:6737–6747. doi: 10.1093/emboj/16.22.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manivet P, Mouillet-Richard S, Callebert J, Nebiqil CG, Maroteaux L, Hosoda S, Kellermann O, Launay JM. PDZ-dependent activation of nitric-oxide synthetases by the serotonin 2B receptor. J. Biol. Chem. 2000;275:9324–9331. doi: 10.1074/jbc.275.13.9324. [DOI] [PubMed] [Google Scholar]
- 34.Kim S-K, Gao Z-G, Jeong LS, Jacobson KA. Docking studies of agonists and antagonists suggest an activation pathway of the A3 adenosine receptor. J. Mol. Graph Model. 2006;25:562–577. doi: 10.1016/j.jmgm.2006.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kenakin T. Ligand-selective receptor conformations revisited: the promise and the problem. Sci. 2003;24:346–354. doi: 10.1016/S0165-6147(03)00167-6. [DOI] [PubMed] [Google Scholar]
- 36.Trabanino RJ, Hall SE, Vaidehi N, Floriano WB, Kam V, Goddard WA., III First principles predictions of the structure and function of G-protein coupled receptors: validation for bovine rhodopsin. Biophys. J. 2004;86:1904–1921. doi: 10.1016/S0006-3495(04)74256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vaidehi N, Floriano WB, Trabanino R, Hall SE, Freddolino P, Choi EJ, Goddard WA., III Structure and Function of GPCRs. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12622–12627. doi: 10.1073/pnas.122357199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 39.Jain A, Vaidehi N, Rodriguez G. A fast recursive algorithm for molecular dynamics simulation. J. Comp. Phys. 1993;106:258–268. [Google Scholar]
- 40.Vaidehi N, Jain A, Goddard WA., III Constant temperature constrained molecular dynamics: The Newton-Euler inverse mass operator method. J. Phys. Chem. 1996;100:10508–10517. [Google Scholar]
- 41.Debe DA, Carlson MJ, Sadanobu J, Chan SI, Goddard WA., III Protein fold determination from sparse distance restraints: The Restrained Generic Protein Direct Monte Carlo method. J. Phys. Chem. B. 1999;103:3001–3008. [Google Scholar]
- 42.Floriano WB, Vaidehi N, Zamanakos G, Goddard WA., III HierVLS hierarchical docking protocol for virtual ligand screening of large-molecule databases. J. Med. Chem. 2004;47:56–71. doi: 10.1021/jm030271v. [DOI] [PubMed] [Google Scholar]
- 43.Cho A, Wendel JA, Vaidehi N, Kekenes-Huskey PM, Floriano WB, Maiti PK, Goddard WA., III The MPSim-Dock Hierarchical Docking Algorithm: Application to the eight trypsin Inhibitor co-crystals. J. Comp. Chem. 2005;26:48–71. doi: 10.1002/jcc.20118. [DOI] [PubMed] [Google Scholar]
- 44.Goddard WA, III, Kim S-K, Li Y, Trzaskowski B, Griffith AR, Abrol R. Predicted 3D structures for adenosine receptors bound to ligands: Comparison to the crystal structure. J. Struct. Biol. 2010;170:10–20. doi: 10.1016/j.jsb.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 45.Feller SE, MacKerell AD., Jr An improved empirical potential energy function for molecular simulations of phospholipids. J. Phys. Chem. B. 2000;104:7510–7151. [Google Scholar]
- 46.Bhandarkar M, Brunner R, Chipot C, Dalke A, Dixit S, Grayson P, Gullingsrud J, Gursoy A, Hardy D, Humphrey W, Hurwitz D, Krawetz N, Nelson M, Phillips J, Shinozaki A, Zheng G, Zhu F. PME parameters. In: Theoretical Biophysics Group, editor. NAMD User’s Guide. Chapter 5. Urbana: University of Illinois and Beckman Institute; 2010. pp. 49–50. Version 2.7. [Google Scholar]
- 47.Kalé L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. NAMD2: Greater scalability for parallel molecular dynamics. J. Comp. Phys. 1999;151:283–312. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.