SUMMARY
Coenzyme Q is an isoprenylated benzoquinone lipid that functions in respiratory electron transport and as a lipid antioxidant. Dietary supplementation with Q is increasingly used as a therapeutic for treatment of mitochondrial and neurodegenerative diseases, yet little is known regarding the mechanism of its uptake. As opposed to other yeast backgrounds, EG103 strains are unable to import exogenous Q6 to the mitochondria. Furthermore, the distribution of exogenous Q6 among endomembranes suggests an impairment of the membrane traffic at the level of the endocytic pathway. This fact was confirmed after the detection of defects in the incorporation of FM4-64 marker and CPY delivery to the vacuole. A similar effect was demonstrated in double mutant strains in Q6 synthesis and several steps of endocytic process; those cells are unable to uptake exogenous Q6 to the mitochondria and restore the growth on non-fermentable carbon sources. Additional data about the positive effect of peptone presence for exogenous Q6 uptake support the hypothesis that Q6 is transported to mitochondria through an endocytic-based system.
INTRODUCTION
Coenzyme Q (ubiquinone or Q) serves as a redox active lipid in the respiratory chain of prokaryotes and eukaryotes [1, 2], and there is an increasing appreciation of its use in therapeutic, nutritional and cosmetic applications. Coenzyme Q10 (the subscript designates the number of isoprene units in the polyisoprene tail) has been used successfully to aid patient recovery following ischemia post reperfusion in heart transplantation [3] and cardiac surgery [4], and also shows promise as a therapeutic agent in other cardiovascular diseases including atherosclerosis [5], and chronic heart failure [6].
Dietary supplementation with Q10 helps slow the progression of symptoms in patients with Parkinsons disease [7, 8], and may have similar benefits in Alzheimer [9] and Huntington diseases [10]. There is intense interest in mitochondrial diseases based on their apparent relationship with neurodegenerative diseases. A subset of mitochondrial diseases associated with decreased levels of Q10 such as cerebellar ataxia [11, 12] Q-deficient mitochondrial diseases [13, 14] can be alleviated by Q10 administration. Dietary supplementation with Q has been proposed to extend lifespan of rats fed diets enriched with polyunsaturated fatty acids [15]. Several studies demonstrate a decline in Q content during ageing [16], however dietary supplementation with Q10 did not affect life span of mice fed standard chow diets [17]. There are also studies that indicate a decrease in Q biosynthesis may actually extend lifespan of nematodes and mice [18]. Thus, there is a complex relationship between the beneficial functions of Q (as an antioxidant and in energy metabolism) and its potential to act as a source of oxidative stress (with the protein complexes of the respiratory electron transport chain).
The efficacy of Q supplementation is determined by its bioavailability to tissues and cells. Studies measuring uptake of Q10 have shown an extremely diverse distribution [19]. Despite the prevalent use of Q10 as a dietary supplement, little is known about the mechanisms responsible for its uptake. It is possible that both cellular uptake of exogenously supplied Q and intracellular transport of endogenously synthesized Q share a common pathway. Studies in human cells by cell fractionation indicate that Q10 is synthesized within the mitochondria and distributed to other cell membranes via the endomembrane system [20]. This system also participates in the uptake of exogenous Q10 suggesting that the endo-exocytic pathways play a role in distributing Q among membranes. In eukaryotes, Q is not only present in mitochondria, but is also located in a wide variety of cellular membranes and organelles, including the Golgi and endoplasmic reticulum [21]. The distribution of Q among cell membranes suggests that it may perform other functions. The reduced form of coenzyme Q, ubiquinol (QH2), acts as an antioxidant and may prevent lipid peroxidation in membranes [22, 23]. QH2 acts directly to scavenge lipid peroxyl radicals and also acts as a co-antioxidant to quench α-tocopheryl radicals and regenerate α-tocopherol [24, 25]. In addition, Q functions in the plasma membrane redox system in Saccharomyces cerevisiae [26] and mammalian cells [27, 28], and may also participate in the electron transport chain of lysosomal membranes [29].
Despite the widespread distribution of Q, the biosynthesis of this molecule appears to be restricted. Studies in mammalian cells suggested that Q was synthesized in mitochondria, Golgi apparatus and endoplasmic reticulum [21], and it was proposed that the high level of Q synthesis in Golgi apparatus serves as a reservoir for its subsequent distribution among cell membranes. However, more recent studies in yeast, nematodes, and mammals indicate that the enzymes of Q biosynthesis are located in the matrix side of the inner mitochondrial membrane. In the yeast Saccharomyces cerevisiae the sub-mitochondrial localization of each of the Coq1p–Coq9p polypeptides was determined by subcellular fractionation and mitochondrial in vitro import experiments [30]. Furthermore, human, nematode and rat gene homologues rescue the Q biosynthetic defects of the coq3, coq5 and coq7 yeast mutants [31–35]. Recent studies have demonstrated that homologues of yeast Coq7p in Caenorhabditis elegans and mouse are located in mitochondria [36–38]. Further, human Coq4 is also located in mitochondria [39].
Results described above indicate that Q biosynthesis occurs within mitochondria, at least in yeast but the fact that Q is located in every cell membrane component suggests the existence of a mechanism for its distribution. Lipid transport, including phospholipids, has been extensively studied in Saccharomyces cerevisiae [40, 41], however, very little is known about the transport of Q. Some evidence for mobilization of Q is indicated by studies with the HL-60 cell line. HL-60 cells that have lost mitochondrial DNA by ethidium bromide treatment or have been grown in a serum-deprived media contain increased amounts of Q at the plasma membrane [42]. This observation suggests a specific Q transport mechanism that may regulate its distribution among cell membranes in response to stress or metabolic demands.
In this study, we have tested the role of the membrane traffic pathway in cellular uptake of exogenous Q. In the first approach we used two genetic backgrounds of the yeast Saccharomyces cerevisiae, EG103 and CEN.PK2-1C. In Q6 defficient mutants of both backgrounds was determined the distribution of Q6 after it exogenous supplementation. While EG103 is known to have defects in the import of exogenous Q6 to the mitochondria, CEN.PK2-1C is competent for such import. This behavior was associated with defects in typical traffic membrane markers. Therefore, in the second approach we analyzed the distribution of exogenous Q6 in endocytic-deficient strains with defects in Q6 biosynthesis. Both approaches delineate a general map of Q6 transport that explains the bioavailability of exogenous Q in eukaryotic cells.
MATERIAL AND METHODS
Yeast strains and growth media
The yeast strains used in this study are described in Table I. Yeast were grown in rich YPD medium (1% yeast extract, 2% peptone, and 2% dextrose) and in YPG (1% yeast extract, 2% peptone, and 3% glycerol). Defined media contained 0.18% yeast nitrogen base without amino acids, 2% dextrose, 0.14% NaH2PO4, 0.5% (NH4)2SO4, and either a complete amino acid supplement (SDc) or without uracil (SDc-ura). Yeast liquid cultures were grown at 30°C with shaking at 200 rpm.
Table I.
Genotype and sources of S. cerevisiae strains used in this work
| Strain | Genotype | Source |
|---|---|---|
| EG103 | α his3Δ1, leu2-3,112 trp1-289, ura3-52, gal2 | [62] |
| EG103 coq7 | EG103Δcat5::HIS3 | [50] |
| EG103 coq3 | EG103Δcoq3::LEU2 | This study |
| CEN.PK2-1C | A, his3-Δ1, leu2–3,112, trp1–289, ura3–52, MAL2–8c, MAL3, SUC3 | [63] |
| CEN coq7 | CEN PK2-1C Δcat5::HIS3 | [63] |
| CEN coq3 | CEN PK2-1C Δcoq3::LEU2 | This study |
| BY4741 | A, his3Δ1, leu2Δ0,met15Δ0, ura3Δ0 | Euroscarf |
| BY4741 coq3 | BY4741Δcoq3::LEU2 | This study |
| Y01709 (tlg2) | BY4741 YOL018c::KanMX4 | Euroscarf |
| Y01709 coq3 | Y01709Δcoq3::LEU2 | This study |
| Y04462 (vps45) | BY4741 YGL095c::KanMX4 | Euroscarf |
| Y04462 coq3 | Y04462Δcoq3::LEU2 | This study |
| Y00788 (pep12) | BY4741 YMR202w::KanMX4 | Euroscarf |
| Y00788 coq3 | Y00788Δcoq3::LEU2 | This study |
| Y01812 (erg2) | BY4741 YOR036w::KanMX4 | Euroscarf |
| Y01812 coq3 | Y01812Δcoq3::LEU2 | This study |
| NY431 | MAT a ura3-52 sec18-1 | [64] |
In situ disruption of the coq3 gene in yeast mutants harboring defects in endocytic pathways
A coq3::LEU2 disruption cassette was obtained from the plasmid pCleu12 [43]. In this plasmid a 0.7-kb BglII DNA segment of the yeast COQ3 coding region has been replaced with a 2.85-kb BglII fragment of YEp13 containing the LEU2 gene. Digestion of pCleu12 with PstI and BamHI generates a linear fragment of 4.65-kb that was used to disrupt the COQ3 gene in each of the designated mutants of the endocytic pathway. All strains were transformed with 1 µg of the PstI/BamHI linear fragment from pCleu12 by the lithium acetate/PEG method [44]. In the strain NY431 (sec18-1) the coq3 disruption required a coq3::URA3 cassette. The URA3 gene cassette was obtained by PCR from the plasmid pRS316 using specific primers containing flanking sequences complementaries with the gene COQ3 (COQ3-URA3-F 5’-AAC GAG ATG TAA GAG CAC AGA TGC GGA ATA AGG GCG ACA C-3’ and COQ3-URA3-R 5’-AAG AGT ATA CCT TTT TCG GGA TTT GAC AGC TTA TCA TCG A-3’). The amplicon was purified from the agarose gel and used to transforms the NY431 strain by the lithium acetate/PEG method [44].
The presence of the coq3 gene deletion in the different yeast strains was ascertained by HPLC-ECD detection of Q6 (Figure S2, Panel A) and by PCR with specific primers that amplify a COQ3 DNA segment internal to the fragment excised by BglII digestion (Figure S2, Panel B). Primers and experimental conditions were designed with Primer Premier 5 software (Premier Biosoft International, USA). PCR was performed with Touch-Down PCR (annealing temperature 62°C to 50°C at 0.5°C/cycle) with the sense primer coq3U96 (5’-AACGAGATGTAAGAGCACAGATGC-3’) and the antisense primer coq3L660 (5`-AAGAGTATACCTTTTTCGGGATTT-3’). Because this is a negative test, each PCR reaction contained a second set of primers amplifying an internal sequence of the gene ACT1 (ACT1F 5’-TTCTGAGGTTGCTGCTTTGG-3’ and ACT1R 5’-GATCCACATTTGTTGGAAGGTGGTAGTC-3’). ACT1 primers were designed to avoid non-desired interactions with COQ3 primers and first were tested in a separate reaction.
Purification of yeast subcellular fractions
Yeast plasma membranes were purified as described with some modifications [45]. Yeast cells were lysed by vortexing with glass beads and centrifuged (10 min at 700 × g) to remove debris. The supernatant was centrifuged (30 min at 20,000 × g) to obtain a crude membrane pellet. Crude membranes were resuspended in sucrose buffer (20% w/w sucrose, 10 mM Tris-HCl, pH 7.6, 1 mM EDTA and 1 mM dithiothreitol) and applied to a sucrose step gradient comprised of 4 ml 43% (w/w) sucrose and 2 ml 53% (w/w) sucrose in the same buffer. After centrifugation (4 h at 100,000 × g), plasma membranes were recovered at the 43/53 interface and reapplied to a second sucrose step gradient as before. To remove non-intrinsic plasma membrane proteins, the samples were treated with 100 mM Na2CO3, pH 11.5 [46], and suspended in the same buffer with 0.33 M sucrose.
Detergent-insoluble glycolipid-enriched complexes (DIGs) were isolated as described previously [47]. Cells (10–20 OD660 units) were broken with glass beads in TNE buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA) and centrifuged (10 min at 700 × g) to remove debris. The cleared lysate (1.5 ml) was incubated with TX100 (1% final concentration) for 30 min on ice. After the extraction with TX100, the lysate was adjusted to 40% Optiprep (Sigma, Spain) by adding 3 ml 60% Optiprep solution and overlaid with 7.2 ml of 30% Optiprep in TXNE (TNE, 0.1% TX100) and 1.2 ml of TXNE. The samples were subjected to centrifugation (140,000 × g, 10 h), and six fractions of 1.4 ml were collected from the top. The second fraction was subjected to a second incubation with TX100 and reapplied to a second Optiprep (Sigma, Spain) gradient as before to obtain a fraction of purified DIG´s by collecting the second 1.4 ml fraction from the top.
Other membrane fractions required the preparation of yeast spheroplasts, obtained by treatment with zymolyase in buffer B (1.2 M sorbitol, 20 mM potassium phosphate, pH 7.4) [48]. Spheroplasts were resuspended with C buffer (0.6 M manitol, 20 mM potassium phosphate, pH 7.4), lysed with a Dounce homogenizer and centrifuged (10 min at 700 × g) to remove debris. The supernatant was centrifuged 10 min at 12,000 × g to obtain a pellet of crude mitochondria. The resulting supernatant was centrifuged 30 min at 20,000 × g to remove a microsomal pellet fraction. The supernatant was centrifuged 45 min at 32,500 × g to obtain rough endoplasmic reticulum pellet, and then 1 hour at 100,000 × g to obtain smooth endoplasmic reticulum pellet. Crude mitochondria were applied to a Nycodenz step gradient of 4.5 ml 14.5% Nycodenz and 4.5 ml 20% Nycodenz in C buffer. After centrifugation (1 hour at 100,000 × g), mitochondria were recovered from the 14.5/20 interface. The upper fraction of the Nycodenz gradient was used to purify MAM (mitochondria associated microsomes); this fraction was applied to a sucrose step gradient of 5 ml 50% sucrose and 5 ml 22.5% sucrose in C buffer. After centrifugation (1 hour at 100,000 × g), the MAM fraction was recovered from the top of the gradient.
Vacuole and Golgi apparatus fractions were purified as described by Wu et al. [49]. Spheroplasts were obtained as above, resuspended in 0.6 M Sorbitol, 20 mM K+-MES, pH 6.0 and lysed with a Dounce homogenizer. After cell lysis, the homogenate was centrifuged first at 700 × g to remove debris and 12,000 × g to remove an enriched mitochondria fraction. The supernatant was centrifuged 30 min at 30,000 × g and the pellet obtained was suspended in the same buffer and applied to a sorbitol step gradient of 5 ml 80% (w/v) sorbitol and 5 ml 25% (w/v) sorbitol in 10 mM triethanolamine, pH 7.4. The interface between the 25 and 80% sorbitol layers was collected and adjusted to 43% sorbitol and layered on a gradient prepared in 40, 43, 60, 70 and 80% increments of 2.3 ml each. Following centrifugation (120,000 × g, 48 h), fractions were collected from the top (0.6 ml) and monitored by immunoblot with antibodies that recognized vacuole and Golgi apparatus specific proteins.
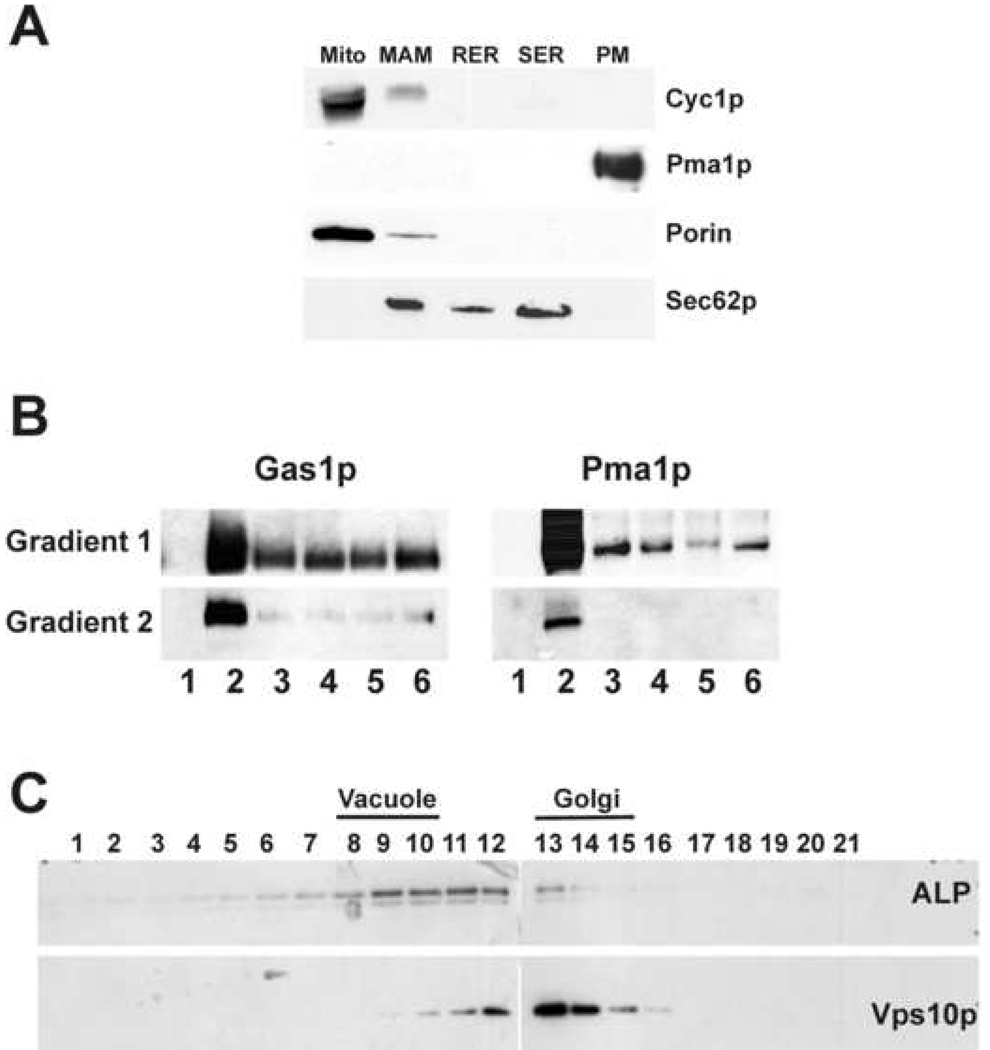
To check the purity of membrane fractions, SDS-PAGE and Western blotting were performed using standard methods (Figure 1). Primary antibodies that recognized specific proteins of membrane fractions were used at the following dilutions: 1:50,000 Pma1p (plasma membrane); 1:20,000 Gas1p (DIGs); 1:5000 porin (mitochondria outer membrane); 1:10,000 cytochrome c1 (mitochondria inner membrane); 1:10,000 Sec62p (endoplasmic reticulum); 4 µg/ml Vps10p (Golgi) and 2 µg/ml Alkaline phosphatase (vacuole) from Molecular Probes. Horseradish peroxidase-linked secondary antibodies to rabbit and mouse IgG were used in 1:10,000 (rabbit) and 1:5,000 (mouse) dilutions.
Figure 1.
Assessment of purity of yeast membrane fractions from the CEN.PK2-1C strain. In Supplementary Material was shown the corresponding analysis for the EG103 strain. Panel A: The extent of cross-contamination in endomembrane samples (30 µg of protein) was determined by SDS-PAGE and Western blot with specific antibodies to marker proteins. Panel B: DIGs were purified by an Optiprep gradient (see Experimental Procedures). Fraction 2 of the first gradient (enriched in both Pma1p and Gas1p marker proteins) was applied to a second gradient. Fraction 2 from the second gradient was considered as the pure DIGs fraction. Pma1p and Gas1p are considered as specific marker proteins of DIGs membranes. Panel C: Vacuole and Golgi membrane fractions were purified from a sorbitol step gradient. Fractions most enriched with vacuole and Golgi marker proteins were identified; ALP for vacuole, and Vps10p for Golgi.
Lipid extraction and determination of Q6
Lipid extractions and quantification of Q6 from purified membrane samples were performed as described previously [50]. Aliquots of purified membrane fractions (500 µl, 0.5–1 mg of protein) were mixed with an equal volume of 2% SDS and vortexed 1 min. Two ml of 5% isopropyl alcohol in ethanol was added, and samples were vortexed again for 1 min. To recover quinones, 5 ml of hexane were added, and the mixture was vortexed at top speed for 1 min and centrifuged at 1000 × g for 5 min. The upper phases recovered from three extractions were pooled and dried in a rotatory evaporator. Lipid extracts were suspended in 1 ml of ethanol dried in a speed-vac and kept at −20°C. Samples were suspended in a suitable volume of ethanol prior to HPLC injection. Q6 and Q9 were separated by reverse-phase high performance liquid chromatography with a C18 column and quantified with an ESA Coulochem III electrochemical detector and a 5010 analytical cell (E1, −500 mV; E2, +500 mV). A separate precolumn guard cell was set to an oxidizing mode (E, +500 mV) to convert all hydroquinones to quinones. The mobile phase was adjusted to a flow rate of 1 ml/min and was composed of methanol/ethanol/2-propanol (88/24/10) and 13.4 mM lithium perchlorate. Q6 and Q9 were quantified from the electrochemical detector results with Q6 and Q9 as external standards. The use of Q9 as an internal standard (10 µl of a 2 mM stock) indicated a recovery of 90–100% of total Q9 added to samples.
CPY secretion plate assay
Yeast cultures were freshly grown to logaritmic phase and washed with deionized water. 50 × 106 yeast cells were resuspended in 1 ml of sterile water and three ten fold dilutions were made. All dilutions were spotted on a nitrocellulose membrane which was placed on the surface of YPD plates and incubated at 30 °C for 24 h. The membranes were lifted and washed with deionized water to remove all the cells. Proteins adsorbed on the membrane were detected by inmunoblotting using monoclonal CPY antibody (Molecular Probes; 1:5,000 dilution).
Vacuolar staining with FM4-64
Yeast cells were grown in YPD media to logarithmic phase. Aliquotes of 1 ml were harvested by centrifugation and washed twice with 1 ml of PBS. Then, cells were resuspended in 1 ml of PBS and incubated with 2 µM FM4-64 at 30°C for 30 min. After incubation, yeast cells were washed twice with PBS, resuspended again in fresh PBS and observed under the fluorescence microscope (λexc = 515 nm, λem = 640 nm).
RESULTS
Exogenous coenzyme Q6 is differentially distributed among cell membranes in CEN.PK2-1C and EG103 yeast
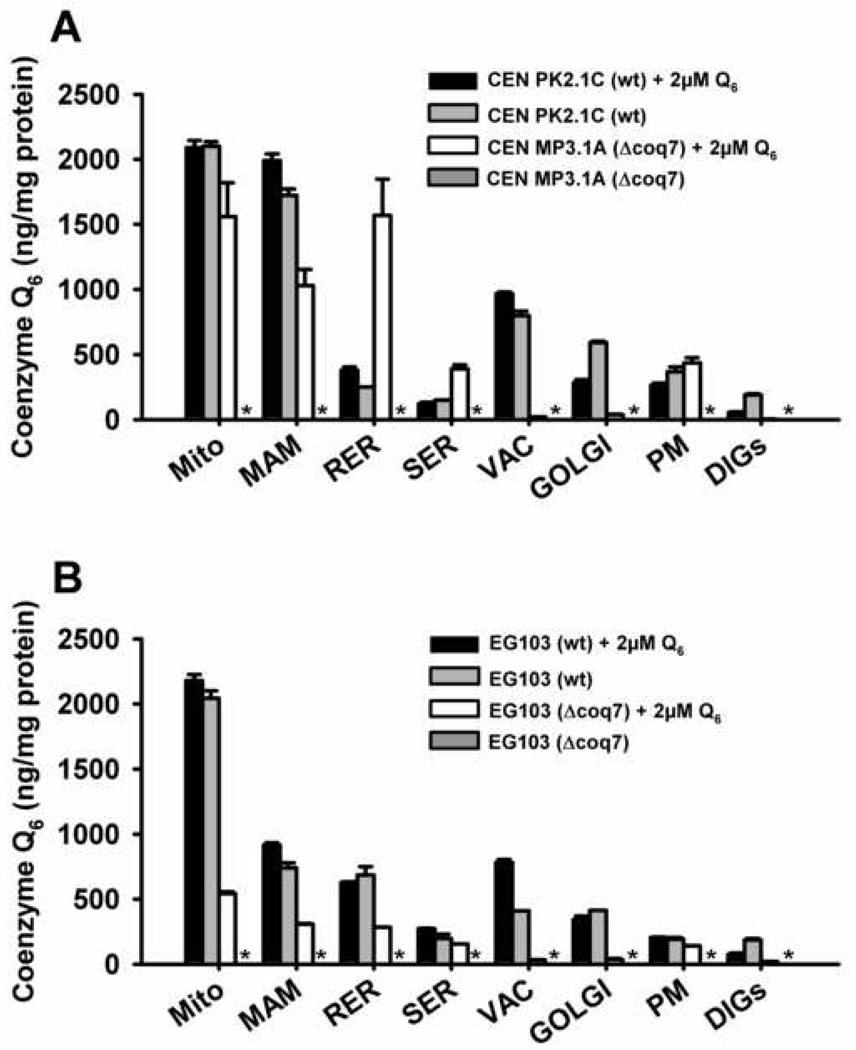
In order to delineate the influence of yeast genetic background on the distribution of Q6 among yeast cellular membranes, the Q6 content was analyzed in membranes of wild-type and coq mutant yeast strains derived from the CEN.PK2-1C and EG103 genetic backgrounds. Wild-type strains were grown in rich medium containing glucose as a fermentable carbon source (YPD). Null coq7 mutants were cultured in the same media supplemented with 2 µM exogenous Q6. All coq mutants are unable to synthesize Q6 and the Q6 detected come from the exogenous supplementation The content of Q6 was determined in several membrane fractions including mitochondria, mitochondria-associated microsomes (MAM), smooth and rough endoplasmic reticulum (SER and RER), Golgi apparatus, vacuole, plasma membrane and detergent-insoluble glycolipid-enriched complexes (DIGs) (Figure 1 and S1). The subcellular fractionation was not significantly affected in the different mutants analyzed. Lipid extracts were prepared from each membrane fraction, and the content of Q6 was determined by HPLC-ECD (Figure 2, Panel A and B). In wild-type cells (black bars) Q6 content was highest in wild-type mitochondria and vacuole, being lowest in DIGs, smooth ER and plasma membrane. The MAM fraction of CEN.PK2-1C yeast was dramatically enriched in Q6 (Figure 2, Panel A), but this enrichment was not evident in the EG103 yeast.
Figure 2.
Coenzyme Q6 content in yeast membrane fractions. Lipid extracts were obtained from purified membrane fractions and the content of Q6 was determined with HPLC-ECD as described in Experimental Procedures. Results shown are the average of three injections ± SD of two independent extractions. Panel A: CEN.PK2-1C (wt) and CENcoq7 strains cultured in absence or presence of 2 µM. Panel B: EG103 (wt) and EG103 coq7 strains cultured in absence or presence of 2 µM Q6. All cells were cultured in YPD. The asterisk (*) indicates that Q6 was not detected.
Addition of exogenous Q6 restores respiration in the coq7 null mutant derived from CEN.PK2-1C but not EG103 yeast parental strains [50]. Thus it was of interest to compare the relative amounts of exogenously added Q6 contained in membrane fractions prepared from these coq7 mutants. The presence of exogenously added Q6 led to dramatic differences in the distribution of Q6 among membrane fractions, as evidenced by the much higher Q6 content in mitochondria, MAM and RER in the coq7 mutant derived from CEN.PK2-1C as compared to EG103 (Figure 2, Panels A and B, white bars). The level of Q6 in EG103coq7 mitochondria was only 22% of wild-type, while the Q6 content of the CEN coq7 mutant was 75% of wild-type. Q6 levels in wild type strains are not affected significantly after the exogenous Q6 supplementation.
Two phenotypes associated to membrane transport defects are present in EG103 genetic background
Q6 distribution in coq7 null mutant from EG103 genetic background suggest that membrane traffic might be involved in the uptake and transport of this lipid to the mitochondria. With the above in mind, we decided to further investigate a potential membrane trafficking defect in EG103 yeast cells.
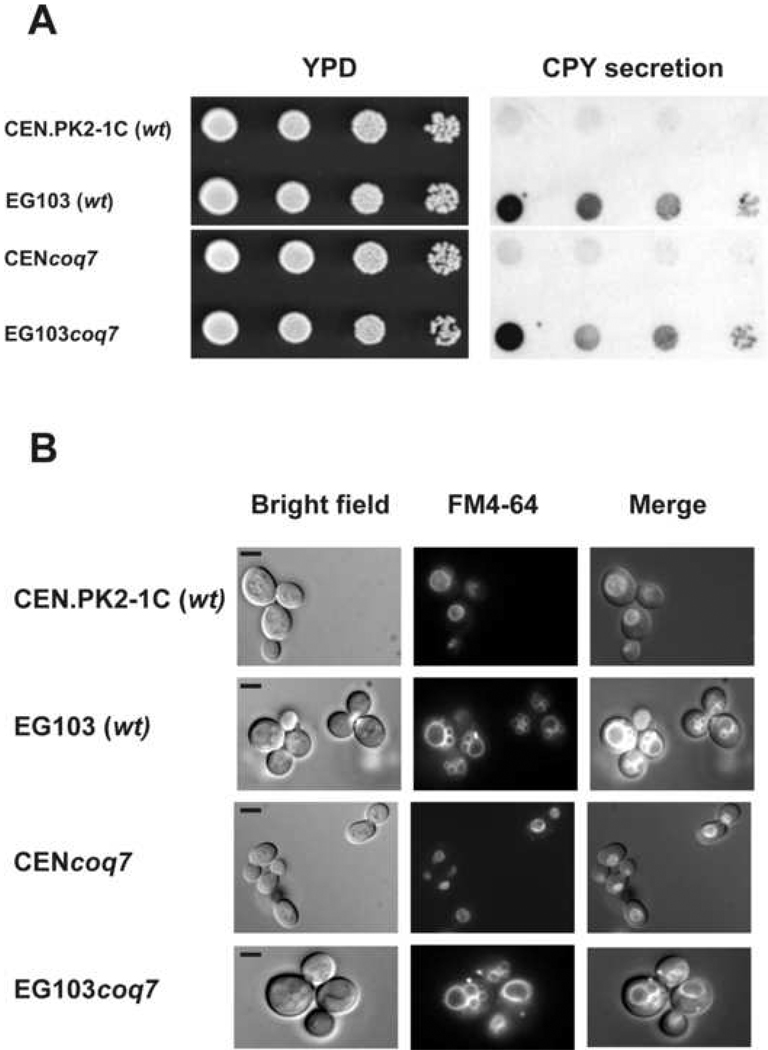
Carboxypeptidase Y (CPY) is synthesized as a prepro form and is transported across the endoplasmic reticulum membrane. Following signal peptidase cleavage in the ER to produce a 67 kDa precursor (p1) form, it is transported through the Golgi where it acquires sugar modifications to become a 69 kDa (p2) form. Wild type cells are able to correctly deliver CPY to its final destination within the vacuolar lumen, where it is processed to a 61 kDa mature form. However, yeast mutants with defects in protein/membrane trafficking to the vacuole secrete a portion of the p2 form of CPY [51]. To investigate whether EG103 cells present this phenotype, a plate assay was performed as described in Experimental Procedures. We did no detect any CPY secretion associated to the parental and coq7 null mutant strains derived from CEN.PK2-1C genetic background. Interestingly, a mild CPY secretion was detected in yeast strains from EG103 (Figure 3, Panel A), indicating a potential membrane trafficking defect associated to this genetic background.
Figure 3.
Analysis of traffic membrane markers in CEN.PK2-1C and EG103 cells. Panel A: CPY secretion. Equal number of cells from freshly grown yeast cultures were deposited as undiluted, 1:10, 1:100 or 1:1000 dilutions (left to right) on a nitrocellulose filter overlaid on YPD plates. The plates were incubated for 24 hours at 30 °C and washed as described under Experimental Procedures. Extracellular CPY secreted from colonies was detected by inmunostaining with anti-CPY antibody (Molecular Probes). Panel B: FM4-64 vacuolar staining. Yeast cell grown to logarithmic phase in YPD were incubated with 2 µM FM4-64 at 30°C during 30 min. After incubation, cells were washed with PBS and observed under the fluorescence microscope. Micrographs are obtained with 1000× magnification. Bar, 5 µm. Results are representative of a set of three experiments.
It should be noted that whereas yeast mutants with defects in membrane traffic to the vacuole secrete CPY, there are many others mutants that secrete low levels of this protein [52]. However, it is well known that defects in trafficking to the vacuole lead to alterations of the vacuolar morphology [53]. Therefore, to further confirm the above results, we decided to analyze the vacuolar morphology of the yeast cells in both genetic backgrounds studied performing FM4-64 staining. Yeast cells grown to logarithmic phase in YPD media were harvested and incubated with the dye as described in Experimental Procedures. The results (Figure 3, Panel B) evidenced that either parental or coq7 null cells from CEN.PK2-1C have generally one well defined vacuolar organelle. However, a number of small vacuoles were observed in yeast cells from EG103 background, which supports above CPY secretion results and strongly suggests an altered membrane trafficking pathway in EG103 cells.
The above results support the hypothesis that altered Q6 distribution among cell membranes observed in EG103 cells may be a consequence of a partial impairment in the normal membrane trafficking processes that occur within the cell.
The endocytic pathway is involved in Q6 uptake in yeast
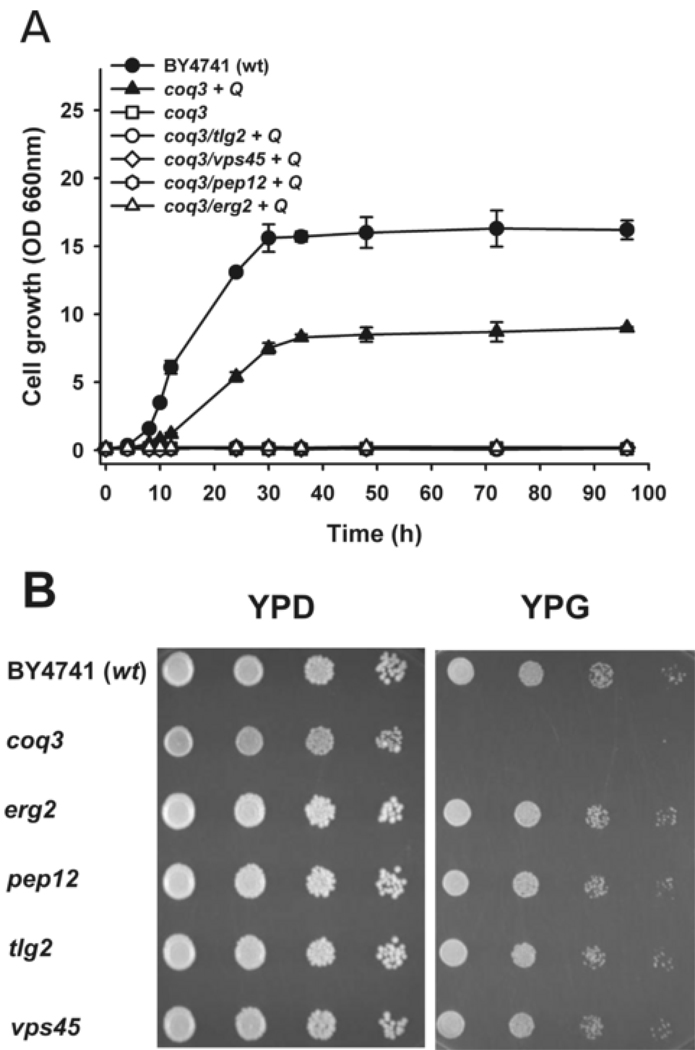
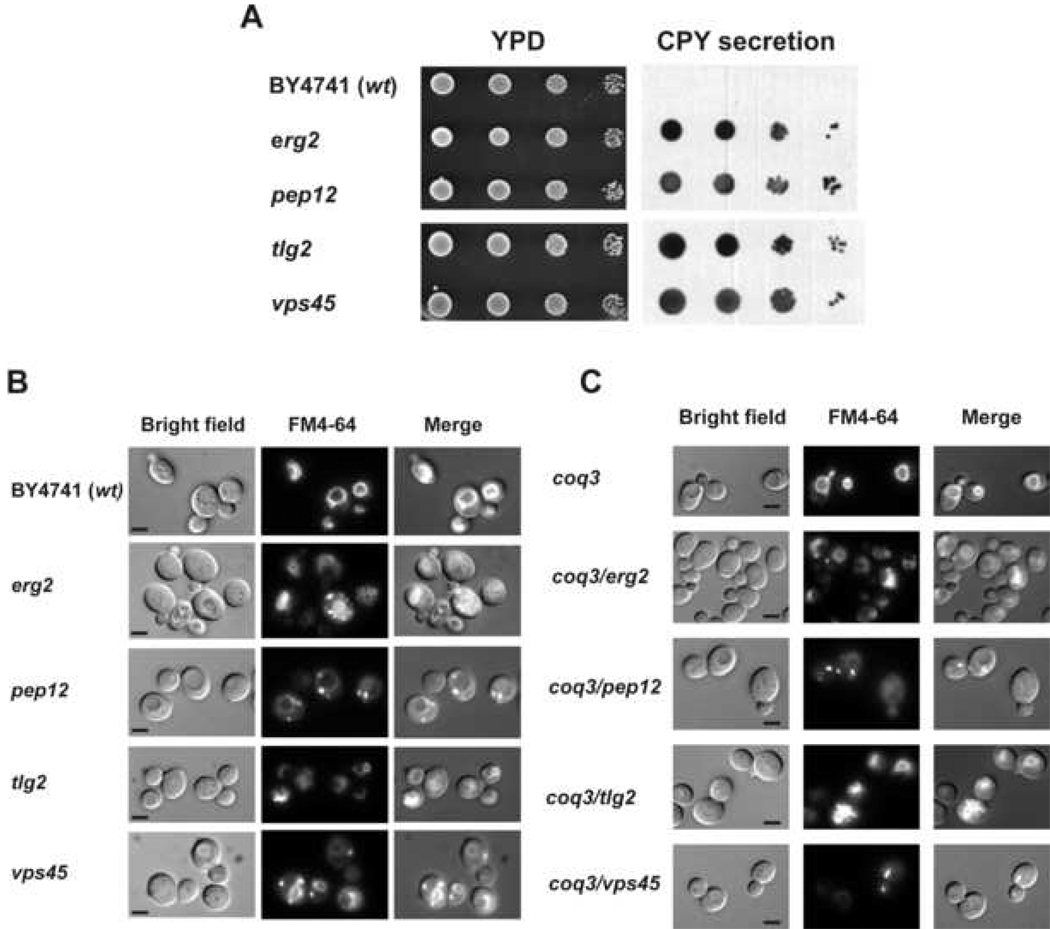
To confirm whether either an impaired endocytosis or a defective membrane traffic lead to an alteration of coenzyme Q uptake and its transport to the mitochondria, we analyzed the uptake and distribution of exogenous Q6 in mutant strains with gene defects in several endocytic transport steps such as ERG2, PEP12, TLG2 or VPS45 genes. Erg2p catalyzes a step in ergosterol biosynthesis and this deficiency induces the accumulation of sterol intermediates that support growth and exocytosis but fail to allow endocytic transport [54]. Pep12p is a syntaxin controlling the vesicular traffic in the prevacuolar compartment [55]. Tlg2p is a t-SNARE (target soluble N-ethyl maleimide-sensitive factor attachment protein receptor) protein required for the fusion of trans-Golgi network vesicles (TGN vesicles) with endocytic vesicles to produce early endosomes [56]. Vps45p is a SM protein (Sec1p-like/Munc-18) required for Tlg2p function [57]. These single mutant yeast strains (in the BY4741 genetic background) were obtained from the Euroscarf repository, and each retained the ability to grow on media containing glycerol as a nonfermentable carbon source (Figure 5, Panel B). Membrane traffic impairment in those mutants was confirmed by analyzing CPY secretion (Figure 4, Panel A) and performing a vacuolar staining with FM4-64 (Figure 4, Panel B). As expected, all the mutant strains showed a high level of CPY secretion and different kind of alterations in the vacuolar morphology. Using these strains and the parental BY4741 strain, mutant strains unable to synthesize Q6 were obtained by in situ gene disruption of the COQ3 gene. Null coq3 mutants show the same behavior as null coq7 mutants in terms of having defects in Q6 biosynthesis, inability to grow on nonfermentable carbon sources, and defects in respiration. Each of the double mutants was shown to have a defect in producing Q6 (Figure S2, Panel A) and to lack the wild-type COQ3 gene (Figure 5, Panel B). The transformation of double mutants with the plasmid pRS12A-2.5SB (containing the COQ3 gene) restores the growth on YPG plates (Figure S2, Panel C). Moreover, the additional COQ3 deletion in double mutant strains did not alter either CPY secretion or FM4-64 staining phenotypes observed in BY4741 parental and mutant strains (Figure 4, panel C).
Figure 5.
Exogenous Q6 fails to rescue yeast coq3 mutants containing additional deletions in genes required for endocytosis. Panel A: YPG media with 2 µM Q6 was inoculated with 0.1 OD660nm units/ml of the designated yeast strains and incubated at 30°C with shaking. Data correspond to the average ± SD of five measures of the same culture. Experiment is representative of a set of two independent experiments. Panel B: Yeast strains bearing defects in membrane trafficking are able to grow in non fermentable carbon source. Equal number of cells from freshly grown yeast cultures were deposited as undiluted, 1:10, 1:100 or 1:1000 dilutions (left to right) on YPD and YPG plates. The plates were incubated for 48 hours at 30 °C.
Figure 4.
Analysis of traffic membrane markers. Panel A: CPY secretion in single membrane traffic mutants. Equal number of cells from freshly grown yeast cultures were deposited as undiluted, 1:10, 1:100 or 1:1000 dilutions (left to right) on a nitrocellulose filter overlaid on YPD plates. The plates were incubated for 24 hours at 30 °C and washed as described under Experimental Procedures. Extracellular CPY secreted from colonies was detected by inmunostaining with anti-CPY antibody (Molecular Probes). Panel B: FM4-64 vacuolar staining. Yeast cell grown to logarithmic phase in YPD were incubated with 2 µM FM4-64 at 30°C during 30 min. After incubation, cells were washed with PBS and observed under the fluorescence microscope. Results are representative of a set of three experiments. Micrographs are obtained with 1000x magnification. Bar, 5 µm. Panel C: FM4-64 vacuolar staining in membrane traffic/coenzyme Q biosynthesis double mutants. Yeast cell grown to logarithmic phase in YPD were incubated with 2 µM FM4-64 at 30°C during 30 min. After incubation, cells were washed with PBS and observed under the fluorescence microscope. Results are representative of a set of three experiments. Micrographs are obtained with 1000× magnification. Bar, 5 µm.
To test exogenous Q6 uptake, each double mutant yeast strain was cultured in YPG medium supplemented with 2 µM Q6. None of the double mutants were able to grow in this medium (Figure 5). However, the coq3 single mutant prepared in the BY4741 genetic background showed growth rescue in YPG medium supplemented with Q6 (Figure 6). These results suggest that endocytosis and membrane trafficking are required for Q6 to reach mitochondria in these cells.
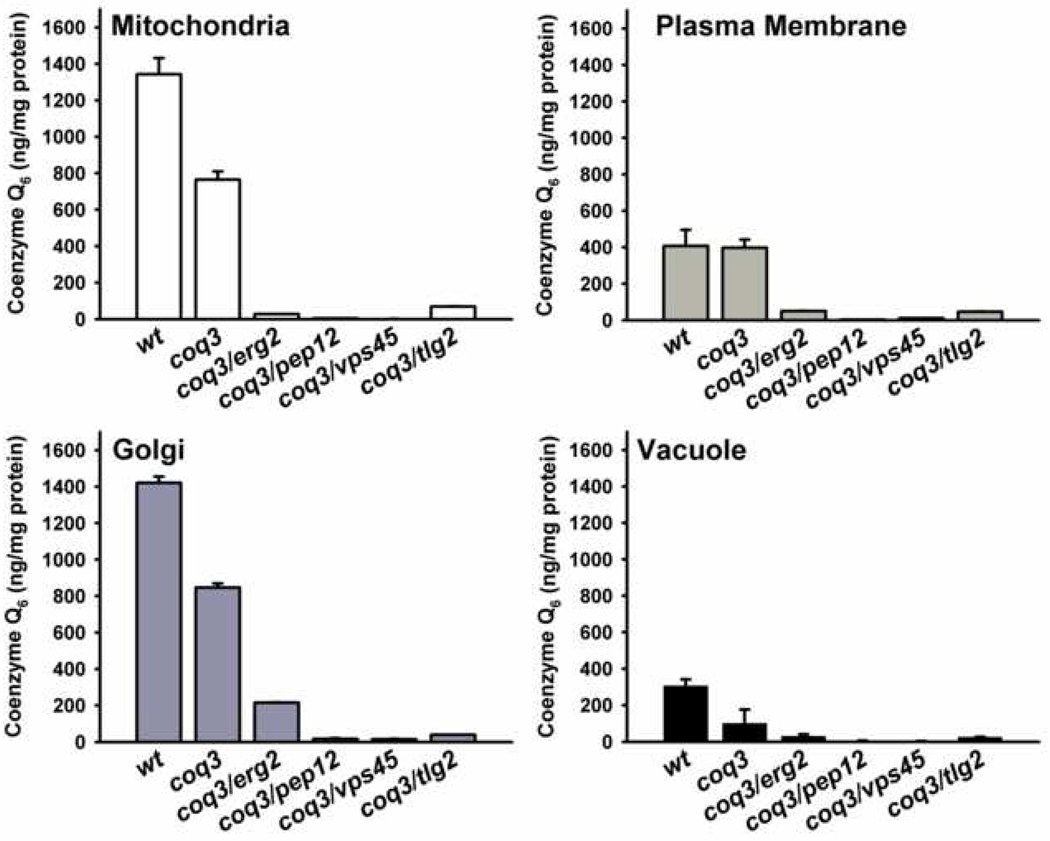
Figure 6.
Coenzyme Q6 content in endomembranes of coq3/endocytic double mutants. Lipid extracts were obtained from mitochondria, plasma membrane, Golgi and vacuole fractions purified from parental (wt), coq3 single mutant, and coq3/endocytic double mutants cultured in YPD with 2 µM Q6. The content of Q6 was determined by HPLC-ECD and results are expressed as the average ± SD of three injections. Data are representative of two independent experiments.
To determine whether the exogenously supplied Q6 was able to reach endomembranes, the Q6 content was determined in purified fractions of plasma membrane, Golgi apparatus, vacuole and mitochondria from the parental, the coq3 single mutant, and each of the double mutant strains cultured in Q6-supplemented YPD media (Figure 7, Panel A). The content of Q6 in plasma membrane of wild-type and coq3 mutant strains contained a similar amount of Q6, while Golgi, mitochondria, and vacuoles isolated from the coq3 mutant contained 59%, 57% and 32% of the wild-type Q6 content. However, each of the coq3-double mutant strains lacking endocytosis related genes showed dramatically decreased Q6 content in all membrane fractions analyzed. The low Q6 content in plasma membranes of the coq3-double mutants is particularly intriguing, as it suggests that most of the exogenously supplied Q6 cannot be directly incorporated into the plasma membrane. A direct insertion of Q6 into the plasma membrane seems to be difficult due the extremely low solubility in water. Given that another lipids such as sterols are transported to the cell using protein–lipid complexes through the endocytic pathway [41], we analyzed the possibility that peptone plays the function of a vehicle for exogenous Q6.
Figure 7.
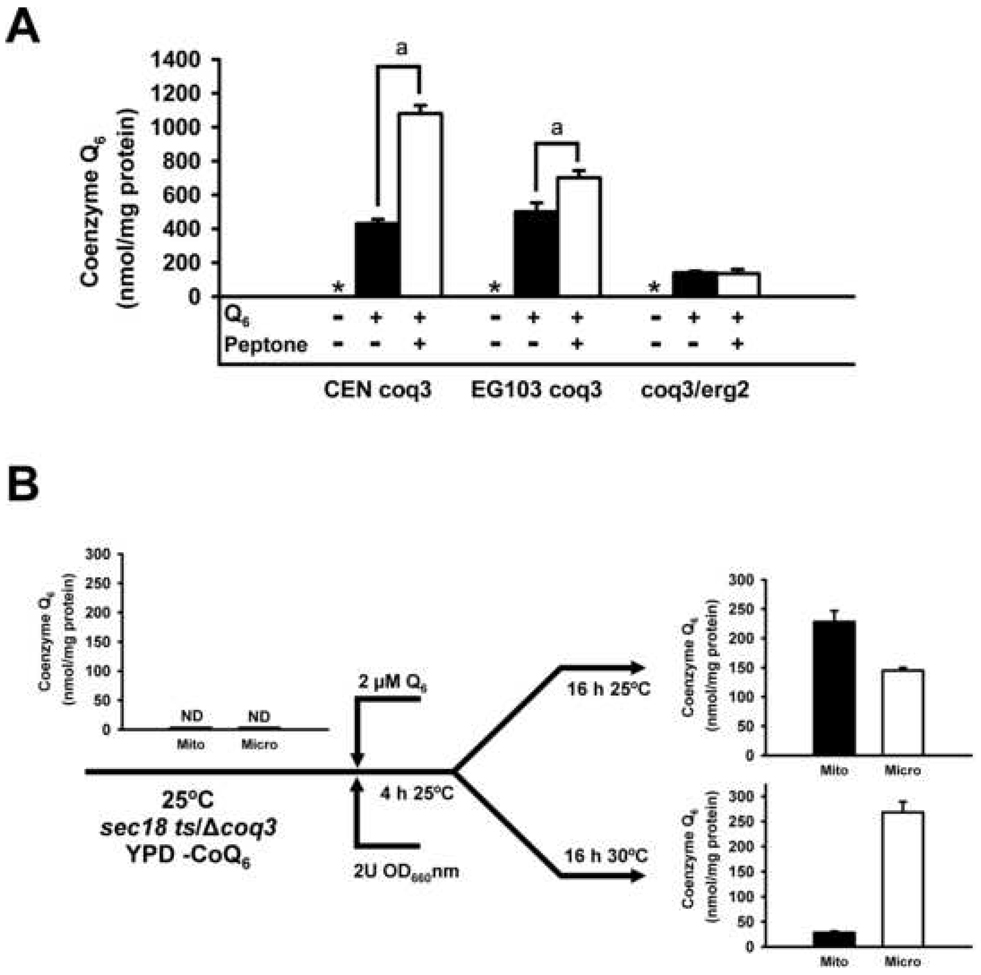
Coenzyme Q6 uptake is favored by the presence of peptone in culture media and is affected in termosensitive endocytic mutants at non-permisive temperature. Panel A: Cells from several Q-deficient strains (CEN coq3, EG103 coq3 and coq3/erg2) were cultured in SD medium at 0.1 OD660nm/ml for 4 hours and then exogenous Q6 (2 µM) was added with or without 2% peptone. Cells were cultured for 48 hours, harvested, and the cell wall removed by Zymolyase digestion. Lipid extracts of protoplasts were prepared and Q6 quantified as described in Material and Methods. Data correspond to the average ± SD of three Q6 determinations from the same experiment. Data are representative of a set of two independent experiments. a The addition of peptone increase significantly the levels of Q6 (p< 0.01). * Indicates that Q6 was not detected in those cells and culture conditions. Panel B: Double mutants cells (sec18-ts/coq3) were cultured at permisive temperature (25°C) in YPD without the presence of Q6 until reach 2 OD units at 660 nm. At this point exogenous Q6 (2 µM) was added and cells were cultured during 4 hours a the same conditions. The initial culture was splitted in two cultures; one was cultured 16 hours at permisive temperature (25°C) and the second at non-permisive temperature (30°C). Cultures were used to purify mitochondria (Mito) and microsome (Micro). Mitochondrial fractions were purified according to Material and Methods and the microsomal fractions were purified after the centrifugation of postmitochondrial supernatant at 100,000 × g 1 h at 4°C. Lipid extracts of both samples were prepared and Q6 quantified as described in Material and Methods. Data correspond to the average ± SD of three Q6 determinations from the same experiment. Data are representative of a set of two independent experiments.
However, the use of constitutive deleted mutants could introduce in the analysis already affected transport pathways or putative transporters. To solve this question, has been analyzed the Q6 exogenous transport in a double mutant NY431 coq3 (sec18-1/coq3) under nonpermisive temperature (Figure 7, Panel B). Sec18p is an AAA-ATPase, the orthologue of NSF in mammalian cells. In the yeast expressing the sec18-1 mutant protein, the transport ceases after shifting the cells to the nonpermissive growth temperature [58]. NY431 coq3 is unable to synthesize Q6 given that harbors the coq3 mutation. Before the addition of exogenous Q6, this molecule is not detected in mitochodria or microsomes. After the addition of exogenous Q6, at permisive temperature (25°C) the Q6 is readily transported to mitochondria while at non-permisive temperature (30°C) is accumulated mainly in microsomes.
Q6 uptake by cells requires soluble proteins
Result suggests that exogenous Q6 could utilize the hydrophilic phase in the lumen of endocytic vesicles to be transported into the yeast cell. Since Q6 is a hydrophobic molecule, this process would require the binding to a water soluble factor that would carry this lipid into the cell. Q6 uptake assays were carried out with cells cultured in a rich medium containing 2% peptone, an enzymatic digest of animal protein. The digested proteins seem to be a good candidate to bind exogenous Q6 to facilitate its transport into the cell. To test this possibility, a Q6 uptake assay was performed with cells cultured in SD medium, which does not contain digested proteins as nitrogen source. Thus, the coq3 null mutant strains CENcoq3 and EG103coq3 and the double mutant strain coq3/erg2 (Y01812coq3) were cultured until stationary phase in the presence of 2 µM Q6 with or without 2% peptone. Cells were collected after 48 hours, spheroplasts prepared, and the content of Q6 determined (Figure 7). EG103coq3 and CENcoq3 strains are unable to produce Q6 and also show a moderate amount of Q6 when were cultured with exogenous Q6 in synthetic media. However, the amount of Q6 was increased after the peptone addition but only in CENcoq3 strain, that does not show defects on membrane traffic.
DISCUSSION
Several studies performed in mammalian systems have shown that certain steps in the Q biosynthetic pathway take place in both mitochondrial and ER membranes [21, 59, 60]. Studies in yeast, however, suggest that Q6 biosynthesis is carried out solely within mitochondria [30]. Here we show a higher Q6 content in mitochondria as compared with other cell membranes in wild-type yeast strains derived from different genetic backgrounds, CEN.PK2-1C and EG103. Although Q6 content is highest in mitochondria, it is detected in all yeast membranes indicating a Q6 transport process from mitochondria to other cellular membranes. In this study we employed the CENcoq7 null mutant, previously shown to be capable of taking up exogenously supplied Q6 and respiratory electron transport is restored [50]. Exogenous Q6 uptake by the CENcoq7 null mutant shows a distribution profile similar in many aspects to its wild-type parent. This finding suggests that mitochondrial Q6 uptake in the coq7 mutant may mimic the transport of Q6 from mitochondria to cell membranes in wild type strains.
Several authors have indicated that the ER-Golgi system participates in the coenzyme Q secretion to the blood plasma in mammalian cells [21]. Other studies suggest that the MAM participates in the secretory pathway as a component that supplies lipids for the final assembly into very low-density lipoproteins [61]. Thus there is a precedent for the hypothesis that Q6 transported to ER or MAM from mitochondria may use the secretory pathway to reach other cell membranes. Similarly, endocytic vesicles could also transport exogenous Q6 to internal cell membranes. Our studies show a low but significant uptake of exogenous Q6 by the EG103coq7 strain, while the corresponding coq7 mutant in the CEN.PK2-1C background exhibited nearly wild-type Q6 content in mitochondria and plasma membrane, and higher than wild-type content of exogenous Q6 in ER and MAM. Interestingly, the analysis of two well defined phenotypes for defective membrane trafficking (CPY secretion and anormal vacuolar morphology) showed that EG103 strains may have a mild defect in those processes, which suggest that Q6 transport among membranes is dependent of normal endocytic and membrane transport within the cell. Furthermore, the findings reported here that yeast mutants with defects in either endocytosis (erg2) or several steps in membrane traffic (pep12, tlg2 and vps45) fail to transport exogenous Q6 to mitochondria strongly support this hypothesis. Indeed, not only there was no delivery of Q6 to mitochondria, but none of the endocytosis mutants were able to incorporate exogenous Q6 into other cellular membranes at significant levels, including the plasma membrane. This last observation is perhaps most surprising, as it indicates that uptake of exogenously supplied coenzyme Q6 by the plasma membrane requires an intact endocytosis membrane trafficking system. A time course analysis of radiolabeled CoQ10 in human cells demonstrated that it is first incorporated in mitochondria and then delivered to other membranes including plasma membrane [20].
Even though Q6 is a hydrophobic molecule, it is possible that when it is added exogenously in aqueous media, it is bound to soluble polypeptides, and taken up by cells via endocytic vesicles. Here we demonstrated that exogenous Q6 uptake by cells is facilitated by peptone, indicating that Q6 binding to the soluble proteins and peptides present in peptone partitions this lipid into the aqueous phase and allows its incorporation into the lumen of endocytic vesicles. Mutant strains defective in endocytosis (erg2) fail to take up exogenous Q6 even with soluble proteins in the medium, supporting this hypothesis. These data agree with the general scheme of cholesterol trafficking in mammalian cells [62]. Uptake of CoQ10 by HL60 cells requires specifically lipoproteins [20]. Therefore, we propose a mechanism for uptake, in which exogenous Q6 binds to soluble proteins, is taken up via endocytosis and travels via endocytic vesicles to the vacuole, where it must be retrieved in order to be delivered to the plasma membrane and mitochondria. The Q6 uptake detected in synthetic media (Figure 7, Panel A) can represent a non receptor-mediated endocytic uptake (fluid-phase endocytosis) or a direct insertion of the Q6 molecule in the plasma membrane. Also, is possible that the selected mutants do not show a total lack of endocytosis, a crucial function in the cells, and maintains a minimal activity that does not support the Q6 uptake to mitochondria. Receptor-mediated or fluid-phase endocytosis pathways share some components. In a recent and extensive study has been found 14 genes in yeast that are related to fluid-phase endocytosis [63]. One of those genes, TLG2, has been used in this work and probably a tlg2-deleted mutant strain accumulates defects on both pathways. This interplay between both procceses may also explain the low amount of Q6 detected in endomembranes from double mutants of endocytosis and Q6 biosynthesis pathway. Each of the membrane traffic mutants used in this study were unable to take up exogenous Q6 and have defects at various stages of endocytosis including the initial point of uptake (erg2), late endosome formation (tlg2, vps45), or vacuole maturation (pep12).
A point of criticism may be the use of deleted-mutants of the endocytic pathway. However, when was analyzed the Q6 uptake in a temperature restrictive mutant such as sec18-1/coq3 was found that at permisive temperature was produced a typical Q6 uptake to mitochondria. This uptake was blocked at non-permisive conditions. That allow us discard that the lack of Q6 uptake was produced by the absence of transport proteins or others factors related with the mutated genes that could not be expressed in permanent endocytic mutants. However, this experiment does not demonstrate completely that endocytic process was required for a proper Q6 uptake. Better, it demonstrates that at least part of the process, the transport from the endomembrane system to the mitochondria is supported by the membrane traffic machinery.
Hence, this model accounts for the absence of significant amounts of exogenous Q6 in the plasma membrane and vacuoles in each of these mutants. However, it is curious that there is very little exogenous Q6 detected in vacuoles isolated from either the single coq7 or coq3 mutants (Figures 2 and 6, respectively). Perhaps the relatively high content of Q6 in the vacuole of the wild-type strain represents the trafficking bottleneck for endogenously synthesized Q6 originating from the mitochondria, while the trafficking bottleneck for Q6 supplied exogenously may reside in the ER or MAM. It is interesting that other studies of exogenous Q10 uptake by human cells indicate that the Q10 accumulates mainly in a vesicular endocytic compartment [20], and only a small but significant amount of Q10 was assimilated by mitochondria. While the uptake of exogenous Q6 and its delivery to mitochondria by coq mutant yeast is quite high, uptake into mitochondria of Q6-replete yeast is quite low [50]. Thus, the amount of uptake of exogenous Q and its delivery to mitochondria in both human and yeast cells may reflect mechanisms that regulate mitochondrial Q content. In fact, exogenous Q10 uptake by human cells induces a decrease of endogenous biosynthesis maintaining Q10 homeostasis [20].
The vesicle traffic system for transport of exogenous Q6 would allow transport of Q6 to plasma membrane, and also to mitochondria. The trafficking of Q6, whether synthesized de novo or supplied exogenously, is likely to share mechanisms in common with higher eukaryotes. An understanding of the processes involved in yeast uptake and assimilation of exogenously supplied Q6 will provide insights into the process by which mammalian cells assimilate Q10.
Supplementary Material
ACKNOWLEDGMENTS
We thank Seasson Phillips Vitellio from the School of Medecine, University of Rochester, USA for helping us with the FM4-64 staining procedure and also the Dr. Peter Novick from the Department of Cell Biology, Yale University School of Medicine, USA for gift us the strain NY431. This work was supported by the Spanish Ministerio de Ciencia y Tecnología, Grant BFU2005-03017/BMC, and by NIH GM45952 to CFC.
The abbreviations used are
- DIGs
detergent-insoluble glycolipid enriched complexes
- CPY
carboxypeptidase Y
- MAM
mitochondria associated microsomes
- Q
coenzyme Q or ubiquinone
- RER
rough ER
- SM
Sec1p-like/Munc-18
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- TGN
trans-Golgi network
- t-SNARE
target SNARE
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Brandt U, Trumpower B. The protonmotive Q cycle in mitochondria and bacteria. Crit. Rev. Biochem. Mol. Biol. 1994;29:165–197. doi: 10.3109/10409239409086800. [DOI] [PubMed] [Google Scholar]
- 2.Trumpower BL. The protonmotive Q cycle. Energy transduction by coupling of proton translocation to electron transfer by the cytochrome bc1 complex. J Biol Chem. 1990;265:11409–11412. [PubMed] [Google Scholar]
- 3.Langsjoen PH, Langsjoen AM. Overview of the use of CoQ10 in cardiovascular disease. BioFactors (Oxford, England) 1999;9:273–284. doi: 10.1002/biof.5520090224. [DOI] [PubMed] [Google Scholar]
- 4.Rosenfeldt F, Marasco S, Lyon W, Wowk M, Sheeran F, Bailey M, Esmore D, Davis B, Pick A, Rabinov M, Smith J, Nagley P, Pepe S. Coenzyme Q10 therapy before cardiac surgery improves mitochondrial function and in vitro contractility of myocardial tissue. J Thorac Cardiovasc Surg. 2005;129:25–32. doi: 10.1016/j.jtcvs.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 5.Thomas SR, Neuzil J, Stocker R. Cosupplementation with coenzyme Q prevents the prooxidant effect of alpha-tocopherol and increases the resistance of LDL to transition metal-dependent oxidation initiation. Arteriosclerosis, Thrombosis, and Vascular Biology. 1996;16:687–696. doi: 10.1161/01.atv.16.5.687. [DOI] [PubMed] [Google Scholar]
- 6.Strey CH, Young JM, Molyneux SL, George PM, Florkowski CM, Scott RS, Frampton CM. Endothelium-ameliorating effects of statin therapy and coenzyme Q10 reductions in chronic heart failure. Atherosclerosis. 2005;179:201–206. doi: 10.1016/j.atherosclerosis.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Sharma S, Kheradpezhou M, Shavali S, El Refaey H, Eken J, Hagen C, Ebadi M. Neuroprotective actions of coenzyme Q10 in Parkinson's disease. Methods Enzymol. 2004;382:488–509. doi: 10.1016/S0076-6879(04)82027-5. [DOI] [PubMed] [Google Scholar]
- 8.Winkler-Stuck K, Wiedemann FR, Wallesch CW, Kunz WS. Effect of coenzyme Q10 on the mitochondrial function of skin fibroblasts from Parkinson patients. J Neurol Sci. 2004;220:41–48. doi: 10.1016/j.jns.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Beal MF. Mitochondrial dysfunction and oxidative damage in Alzheimer's and Parkinson's diseases and coenzyme Q10 as a potential treatment. J Bioenerg Biomembr. 2004;36:381–386. doi: 10.1023/B:JOBB.0000041772.74810.92. [DOI] [PubMed] [Google Scholar]
- 10.Beal MF, Shults CW. Effects of Coenzyme Q10 in Huntington's disease and early Parkinson's disease. BioFactors (Oxford, England) 2003;18:153–161. doi: 10.1002/biof.5520180218. [DOI] [PubMed] [Google Scholar]
- 11.Lagier-Tourenne C, Tazir M, López LC, Quinzii CM, Assoum M, Drouot N, Busso C, Makri S, Ali-Pacha L, Benhassine T, Anheim M, Lynch DR, Thibault C, Plewniak F, Bianchetti L, Tranchant C, Poch O, DiMauro S, Mandel J-L, Barros MH, Hirano M, Koenig M. ADCK3, an Ancestral Kinase, Is Mutated in a Form of Recessive Ataxia Associated with Coenzyme Q10 Deficiency. Am. J. Hum. Gen. 2008;82:661–672. doi: 10.1016/j.ajhg.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quinzii CM, Kattah AG, Naini A, Akman HO, Mootha VK, DiMauro S, Hirano M. Coenzyme Q deficiency and cerebellar ataxia associated with an aprataxin mutation. Neurology. 2005;64:539–541. doi: 10.1212/01.WNL.0000150588.75281.58. [DOI] [PubMed] [Google Scholar]
- 13.Cooper JM, Schapira AH. Friedreich's Ataxia: disease mechanisms, antioxidant and Coenzyme Q10 therapy. BioFactors (Oxford, England) 2003;18:163–171. doi: 10.1002/biof.5520180219. [DOI] [PubMed] [Google Scholar]
- 14.Hart PE, Lodi R, Rajagopalan B, Bradley JL, Crilley JG, Turner C, Blamire AM, Manners D, Styles P, Schapira AH, Cooper JM. Antioxidant treatment of patients with Friedreich ataxia: four-year follow-up. Arch Neurol. 2005;62:621–626. doi: 10.1001/archneur.62.4.621. [DOI] [PubMed] [Google Scholar]
- 15.Quiles JL, Ochoa JJ, Huertas JR, Mataix J. Coenzyme Q supplementation protects from age-related DNA double-strand breaks and increases lifespan in rats fed on a PUFA-rich diet. Exp Gerontol. 2004;39:189–194. doi: 10.1016/j.exger.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Kamzalov S, Sohal RS. Effect of age and caloric restriction on oenzyme Q and alpha-tocopherol levels in the rat. Exp Gerontol. 2004;39:1199–1205. doi: 10.1016/j.exger.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 17.Sohal RS, Kamzalov S, Sumien N, Ferguson M, Rebrin I, Heinrich KR, Forster MJ. Effect of coenzyme Q10 intake on endogenous coenzyme Q content, mitochondrial electron transport chain, antioxidative defenses, and life span of mice. Free Radic Biol Med. 2006;40:480–487. doi: 10.1016/j.freeradbiomed.2005.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, Hekimi S. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 2005;19:2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bentinger M, Dallner G, Chojnacki T, Swiezewska E. Distribution and breakdown of labeled coenzyme Q10 in rat. Free Radic Biol Med. 2003;34:563–575. doi: 10.1016/s0891-5849(02)01357-6. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez-Ayala DJ, Brea-Calvo G, Lopez-Lluch G, Navas P. Coenzyme Q distribution in HL-60 human cells depends on the endomembrane system. Biochim Biophys Acta. 2005;1713:129–137. doi: 10.1016/j.bbamem.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Kalen A, Norling B, Appelkvist EL, Dallner GCQsyf. Ubiquinone biosynthesis by the microsomal fraction from rat liver. Biochimica Et Biophysica Acta. 1987;926:70–78. doi: 10.1016/0304-4165(87)90183-8. [DOI] [PubMed] [Google Scholar]
- 22.Niki E. Mechanisms and dynamics of antioxidant action of ubiquinol. Mol Aspects Med. 1997;18:S63–S70. doi: 10.1016/s0098-2997(97)00035-6. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto Y, Komuro E, Niki E. Antioxidant activity of ubiquinol in solution and phosphatidylcholine liposome. J. Nutr. Sci. Vitaminol. 1990;36:505–511. doi: 10.3177/jnsv.36.505. [DOI] [PubMed] [Google Scholar]
- 24.Kagan VE, Tyurina YY, Witt E. Role of coenzyme Q and superoxide in vitamin E cycling. Subcell Biochem. 1998;30:491–507. doi: 10.1007/978-1-4899-1789-8_20. [DOI] [PubMed] [Google Scholar]
- 25.Kagan VE, Nohl H, Quinn PJ. In: Coenzyme Q: Its role in scavenging and generation of radicals in membranes. Cadenas E, Packer L, editors. vol. 1. New York: Marcel Decker Inc.; 1996. pp. 157–201. [Google Scholar]
- 26.Santos-Ocaña C, Villalba JM, Córdoba F, Padilla S, Crane FL, Clarke CF, Navas P. Genetic evidence for coenzyme Q requirement in plasma membrane electron transport. J. Bioenerg. Biomembr. 1998;30:465–475. doi: 10.1023/a:1020542230308. [DOI] [PubMed] [Google Scholar]
- 27.Sun IL, Sun EE, Crane FL, Morré DJ, Lindgren A, Löw H. Requirements for coenzyme Q in plasma membrane electron transport. Proc. Natl. Acad. Sci. USA. 1992;89:11126–11130. doi: 10.1073/pnas.89.23.11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villalba JM, Navarro F, Córdoba F, Serrano A, Arroyo A, Crane FL, Navas P. Coenzyme Q reductase from liver plasma membrane: Purification and role in trans-plasma-membrane electron transport. Proc. Natl. Acad. Sci. USA. 1995;92:4887–4891. doi: 10.1073/pnas.92.11.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gille L, Nohl H. The existence of a lysosomal redox chain and the role of ubiquinone. Arch Biochem Biophys. 2000;375:347–354. doi: 10.1006/abbi.1999.1649. [DOI] [PubMed] [Google Scholar]
- 30.Tran UC, Clarke CF. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion. 2007;7 Suppl:S62–S71. doi: 10.1016/j.mito.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonassen T, Clarke CF. Isolation and functional expression of human COQ3, a gene encoding a methyltransferase required for ubiquinone biosynthesis. The Journal of biological chemistry. 2000;275:12381–12387. doi: 10.1074/jbc.275.17.12381. [DOI] [PubMed] [Google Scholar]
- 32.Jonassen T, Marbois BN, Kim L, Chin A, Xia Y-R, Lusis AJ, Clarke CF. Isolation and sequencing of the rat Coq7 gene and the mapping of mouse Coq7 to chromosome 7. Arch. Biochem. Biophys. 1996;330:285–289. doi: 10.1006/abbi.1996.0255. [DOI] [PubMed] [Google Scholar]
- 33.Marbois BN, Hsu A, Pillai R, Colicelli J, Clarke CF. Cloning of a rat cDNA encoding dihydroxypolyprenylbenzoate methyltransferase by functional complementation of a Saccharomyces cerevisiae mutant deficient in ubiquinone biosynthesis. Gene. 1994;138:213–217. doi: 10.1016/0378-1119(94)90810-9. [DOI] [PubMed] [Google Scholar]
- 34.Vajo Z, King LM, Jonassen T, Wilkin DJ, Ho N, Munnich A, Clarke CF, Francomano CA. Conservation of the Caenorhabditis elegans timing gene clk-1 from yeast to human: a gene required for ubiquinone biosynthesis with potential implications for aging. Mamm Genome. 1999;10:1000–1004. doi: 10.1007/s003359901147. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez-Aguilera JC, Asencio C, Ruiz-Ferrer M, Vela J, Navas P. Caenorhabditis elegans ubiquinone biosynthesis genes. BioFactors (Oxford, England) 2003;18:237–244. doi: 10.1002/biof.5520180226. [DOI] [PubMed] [Google Scholar]
- 36.Felkai S, Ewbank JJ, Lemieux J, Labbe JC, Brown GG, Hekimi S. CLK-1 controls respiration, behavior and aging in the nematode Caenorhabditis elegans. EMBO J. 1999;18:1783–1792. doi: 10.1093/emboj/18.7.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang N, Levavasseur F, McCright B, Shoubridge EA, Hekimi S. Mouse CLK-1 is imported into mitochondria by an unusual process that requires a leader sequence but no membrane potential. The Journal of biological chemistry. 2001;276:29218–29225. doi: 10.1074/jbc.M103686200. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi M, Asaumi S, Honda S, Suzuki Y, Nakai D, Kuroyanagi H, Shimizu T, Honda Y, Shirasawa T. Mouse coq7/clk-1 orthologue rescued slowed rhythmic behavior and extended life span of clk-1 longevity mutant in Caenorhabditis elegans. Biochem. Biophys. Res. Comm. 2001;286:534–540. doi: 10.1006/bbrc.2001.5439. [DOI] [PubMed] [Google Scholar]
- 39.Casarin A, Jimenez-Ortega JC, Trevisson E, Pertegato V, Doimo M, Ferrero-Gomez ML, Abbadi S, Artuch R, Quinzii C, Hirano M, Basso G, Ocana CS, Navas P, Salviati L. Functional characterization of human COQ4, a gene required for Coenzyme Q10 biosynthesis. Biochem. Biophys. Res. Comm. 2008;372:35–39. doi: 10.1016/j.bbrc.2008.04.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast (Chichester, England) 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 41.van Meer G. Lipids of the Golgi membrane. Trends Cell Biol. 1998;8:29–33. doi: 10.1016/s0962-8924(97)01196-3. [DOI] [PubMed] [Google Scholar]
- 42.Gómez-Díaz C, Rodríguez-Aguilera JC, Barroso MP, Villalba JM, Navarro F, Crane FL, Navas P. Antioxidant ascorbate is stabilized by NADH-coenzyme Q10 reductase in the plasma membrane. J. Bioenerg. Biomembr. 1997;29:251–257. doi: 10.1023/a:1022410127104. [DOI] [PubMed] [Google Scholar]
- 43.Clarke CF, Williams W, Teruya JH. Ubiquinone biosynthesis in Saccharomyces cerevisiae. Isolation and sequence of COQ3, the 3,4-dihydroxy-5-hexaprenylbenzoate methyltransferase gene. J. Biol. Chem. 1991;266:16636–16644. [PubMed] [Google Scholar]
- 44.Agatep R, Kirkpatrick RD, Parchaulik RA, Wood RA, Gietz RD. Transformation of Saccharomyces cerevisiae by the lithium acetate/single-stranded carrier DNA/polyethylene glycol (LiAc/ss-DNA/PEG) protocol. Technical Tips Online. 1999 [Google Scholar]
- 45.Serrano R. H+ATPase from plasma membranes of Saccharomyces cerevisiae and Avena sativa roots: purification and reconstitution. Methods Enzymol. 1988;157:533–544. doi: 10.1016/0076-6879(88)57102-1. [DOI] [PubMed] [Google Scholar]
- 46.Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: Application to endoplasmic reticulum. J. Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bagnat M, Keränen S, Shevchenko A, Shevchenko A, Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glick BS, Pon LA. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods In Enzymology. 1995;260:213–223. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- 49.Wu W-I, Routt S, Bankaitis VA, Voelker DR. A New Gene Involved in the Transport-dependent Metabolism of Phosphatidylserine, PSTB2/PDR17, Shares Sequence Similarity with the Gene Encoding the Phosphatidylinositol/Phosphatidylcholine Transfer Protein, SEC14. J. Biol. Chem. 2000;275:14446–14456. doi: 10.1074/jbc.275.19.14446. [DOI] [PubMed] [Google Scholar]
- 50.Santos-Ocana C, Do TQ, Padilla S, Navas P, Clarke CF. Uptake of Exogenous Coenzyme Q and Transport to Mitochondria Is Required for bc1 Complex Stability in Yeast coq Mutants. J. Biol. Chem. 2002;277:10973–10981. doi: 10.1074/jbc.M112222200. [DOI] [PubMed] [Google Scholar]
- 51.Bowers K, Stevens TH. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2005;1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 52.Bonangelino CJ, Chavez EM, Bonifacino JS. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2486–2501. doi: 10.1091/mbc.02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Munn AL, Heese-Peck A, Stevenson BJ, Pichler H, Riezman H. Specific sterols required for the internalization step of endocytosis in yeast. Mol. Biol. Cell. 1999;10:3943–3957. doi: 10.1091/mbc.10.11.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerrard SR, Levi BP, Stevens TH. Pep12p is a multifunctional yeast syntaxin that controls entry of biosynthetic, endocytic and retrograde traffic into the prevacuolar compartment. Traffic. 2000;1:259–269. doi: 10.1034/j.1600-0854.2000.010308.x. [DOI] [PubMed] [Google Scholar]
- 56.Abeliovich H, Grote E, Novick P, Ferro-Novick S. Tlg2p, a yeast syntaxin homolog that resides on the Golgi and endocytic structures. J. Biol Chem. 1998;273:11719–11727. doi: 10.1074/jbc.273.19.11719. [DOI] [PubMed] [Google Scholar]
- 57.Bryant NJ, James DE. Vps45p stabilizes the syntaxin homologue Tlg2p and positively regulates SNARE complex formation. EMBO J. 2001;20:3380–3388. doi: 10.1093/emboj/20.13.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Juschke C, Zachter A, Schwappach B, Seedorf M. SEC18/NSF-independent, protein-sorting pathway from the yeast cortical ER to the plasma membrane. J. Cell Biol. 2005;169:613–622. doi: 10.1083/jcb.200503033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dallner G, Sindelar PJ. Regulation of ubiquinone metabolism. Free Radic Biol Med. 2000;29:285–294. doi: 10.1016/s0891-5849(00)00307-5. [DOI] [PubMed] [Google Scholar]
- 60.Teclebrhan H, Olsson J, Swiezewska E, Dallner G. Biosynthesis of the side chain of ubiquinone:trans-prenyltransferase in rat liver microsomes. J Biol. Chem. 1993;268:23081–23086. [PubMed] [Google Scholar]
- 61.Rusinol AE, Cui Z, Chen MH, Vance JE. A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins. J. Biol. Chem. 1994;269:27494–27502. [PubMed] [Google Scholar]
- 62.Simons K, Ikonen E. How cells handle cholesterol. Science. 2000;290:1721–1726. doi: 10.1126/science.290.5497.1721. [DOI] [PubMed] [Google Scholar]
- 63.Wiederkehr A, Meier KD, Riezman H. Identification and characterization of Saccharomyces cerevisiae mutants defective in fluid-phase endocytosis. Yeast (Chichester, England) 2001;18:759–773. doi: 10.1002/yea.726. [DOI] [PubMed] [Google Scholar]
- 64.Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.