Abstract
We examined the stage specificity and heterogeneity of 18p11 alterations in a series of tumors representing 96 microdissected samples. Significant loss of heterozygosity (LOH) (63%) was found, with 56% occurring early in ductal carcinoma in situ. Although most cases indicated LOH was clonally inherited, heterogeneity for 18p LOH occurred in 27% of tumors. When compared with other LOH data, 18p LOH was found in conjunction with allelic deletion on 3p, 9p, 17p and 17q, while 13q, 16q, and 11p were less frequently associated. These analyses suggest chromosome 18p11 alteration is a common and early event in breast disease.
Keywords: 18p, breast cancer, heterogeneity, loss of heterozygosity, progression
Synopsis
Introduction:
A complete understanding of the genetic events involved in breast tumorigenesis and their heterogeneity in relation to stage specificity will be critical to the development of successful treatments. This laboratory and other workers recently reported the presence of a novel LOH region in band 18p11.3 in non-small cell lung carcinomas, glioblastomas and invasive breast carcinomas [1,2]. This current study uses informative loci in 18p11.3 to investigate the stage specificity and extent of molecular heterogeneity exhibited by this locus in breast cancer progression. These results are analyzed in conjunction with other LOH data available for markers on 3p, 11p, 13q, 16p, 17p, and 17q in an effort to place chromosome 18p alterations on the breast cancer progression pathway.
Materials and methods:
The material used in this study, consisting of formalin-fixed, paraffin-embedded tissue from the archives of the Armed Forces Institute of Pathology, has been previously described [3]. Tumors were selected at random for those with the presence of lesions representing multiple stages of breast disease within individual patient samples. Ninety-six foci representing normal, intraductal and infiltrating tumor were microdissected from 30 tumors. Matching normal and tumor DNA were subjected to polymerase chain reaction (PCR)-based LOH analysis using two 18p11.3 markers (D18S59 and D18S481) and one 18p11.2 marker (D18S452). Forward primers were synthesized with either a fluorescent FAM tag (D18S481 and D18S452) or a fluorescent TET tag (D18S59) on the 5' end. PCR reactions were performed in 10 μl with aliquots of normal and tumor DNA using a PCR protocol consisting of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min for a total of 40 cycles. Horizontal ultrathin, high throughput fluorescence-based DNA fragment gel electrophoresis (GTI-9600; Genesys Technologies, Inc, Sauk City, WI, USA) was applied to the separation and analysis of PCR-generated alleles as previously described [4]. Allelic ratios were calculated and expressed as a percentage of loss of intensity for the tumor allele compared with the corresponding normal allele (D-value) after normalization.
Results:
A total of 96 foci were analyzed for LOH at several polymorphic markers in 18p11.3. The overall frequency of LOH in 18p11.3 was 63% (27/43). When analyzed by tumor stage, LOH on 18p was found in 56% (9/16) of ductal carcinoma in situ (DCIS) foci, 67% of invasive ductal carcinoma (IDC) foci (14/21), and 67% (4/6) of metastatic disease. A schematic diagram of these results appears in Figure 1.
Figure 1.
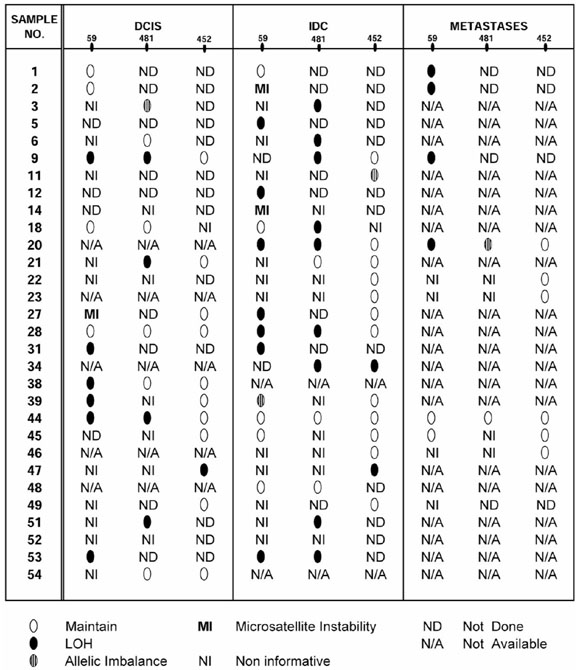
Schematic representation of 18p LOH in all stages of breast carcinoma. Tumor sample numbers are listed on the left. Analysis at markers D18S59, D18S481 and D18S452 are shown for all DCIS, IDC and metastases studied.
We also examined the extent of heterogeneity associated with chromosome 18p LOH (Fig. 2). In tumors where information was attainable for two or more disease stages, 73% (11/15) displayed a pattern of classic clonal progression. LOH first appeared in the DCIS stage for the majority. Four tumors (27%) were, however, interestingly identified with genetic heterogeneity for chromosome 18p events where DCIS-identified events were not found in associated invasive foci. Genetic events involving chromosome 18p sequences thus do not follow a classic progression paradigm in 27% of tumor cases analyzed as part of this study.
Figure 2.
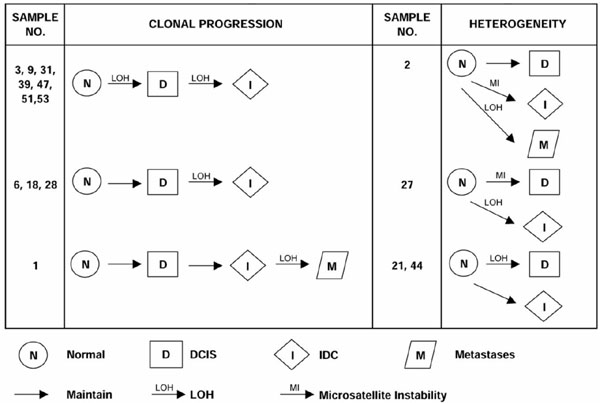
Heterogeneity for LOH on chromosome 18p in breast cancer progression. Schematic diagrams show the different patterns of progression based on this chromosome 18p study. Nine cases show clonal progression where 18p LOH occurs either in DCIS or IDC and is clonally inherited in related later stages of tumor progression. Two cases show LOH first appearing in the metastatic stage. In contrast, four cases show heterogeneity for alterations on 18p. Two specimens show LOH in the DCIS component but not in the associated IDC component. The other cases show the presence of microsatellite instability in earlier but not later stage disease.
Discussion:
Significant LOH on chromosome 18p11.3 (63%) suggests the presence of a putative tumor suppressor gene(s) with an important role in the pathogenesis of breast cancer. These measured allelic deletions represented alterations confined to 18p11.3 and not whole chromosome 18 loss based on results using marker D18S452, although the presence of simultaneous but independent LOH on 18q was not examined. It was discovered, by analyzing multiple foci occurring simultaneously within individual tumors, that chromosome 18p events occur at a significant frequency (56%) early in the tumorigenic process, making LOH at chromosome 18p one of the most common known events in DCIS tumors. A slightly higher frequency of LOH in IDC tumors (67%) as well as in the small number of metastatic samples studied further supports this early event hypothesis. The majority (73%) of these breast tumors progressed in a clonal fashion. Four cases (27%) were, however, representative of genetic heterogeneity. Such heterogeneity at multiple loci may account for treatment failure, and a combinatorial histological and genetic approach with sufficient information on survival and response profiles may provide more effective disease prevention and cure in the future.
Full article
Introduction:
The molecular pathogenetic pathway of breast tumor progression is not yet clearly defined. Although LOH at chromosomes 1, 3p, 6q, 7q, 8p, 9p, 10q, 11, 13q, 16q, 17, 18q, 22q, and X have all been reported [5,6], their relationship to known genes and, more specifically, to different stages of breast disease is still under investigation. Studies in benign fibroadenomas and fibrocystic disease showed no LOH on 1p, 3p, 7q, 11p, 17p, 17q or 18q [7]. However, 37% of ductal hyperplasias and 42% of atypical ductal hyperplasias exhibit LOH [8], suggesting that these foci are benign lesions resulting from the alteration of tumor suppressor genes. Furthermore, the discovery that morphologically normal terminal duct-lobule units adjacent to breast cancers exhibit LOH in up to 60% of cases when analyzed with chromosome 3p markers indicates these genetic events can be cryptically present before histopathological abnormalities are identifiable [9].
These aforementioned studies and other research support the idea that events affecting tumor suppressors can occur early and that preinvasive DCIS can be a direct precursor of invasive carcinoma, a paradigm supported pathologically by the frequency of DCIS lesions found in areas adjacent to invasive breast cancer [10]. These same analyses also confirm the complexity of this disease. Examination of multiple lesions from individual tumors, for example, reveals the presence of intratumoral heterogeneity [11,12,13]. Markers on chromosome 9p, in one study, demonstrate loss of opposite alleles in different ducts while examples of ducts retaining both alleles were also found [14]. An extensive analysis of markers on chromosome 11p15 showed LOH early in breast disease (DCIS), although examples of late stage LOH without concomittant DCIS involvement were also found [3]. This indicates that different components from an individual tumor can represent genetically divergent clones, even at the preinvasive stage of DCIS. The heterogeneous nature of breast cancer alterations could have important implications for the development of targeted therapies.
If LOH events define a localized region of increased risk for carcinoma, the presence of LOH may be clinically important. A complete understanding of all the genetic events involved in breast tumorigenesis and their heterogeneity in relation to stage specificity will therefore be critical to the development of successful treatments. This laboratory and other workers have reported the presence of a previously unidentified region of LOH in band 18p11.3 in non-small cell lung carcinomas, glioblastomas and a small series (14) of invasive breast carcinomas [1,2]. The current study uses informative loci in 18p11.3 to investigate, in microdissected material, the stage specificity and extent of molecular heterogeneity exhibited by chromosome 18p11 in breast cancer progression. A series of 30 tumors from which 96 multiple stages of disease have been microdissected, including normal, DCIS and IDC material, were studied. Associated metastases were also examined in some cases. These results were analyzed in conjunction with other LOH data available for markers on 3p, 11p, 13q, 16p, 17p, and 17q in an effort to place chromosome 18p alterations on the breast cancer progression pathway.
Materials and methods
Patient material
The material used in this study, consisting of formalin-fixed, paraffin-embedded tissue from the archives of the Armed Forces Institute of Pathology, has been previously described [3]. Tumors were selected at random for those with the presence of lesions representing multiple stages of breast disease within individual patient samples. Ninety-six foci representing normal, intraductal and infiltrating tumor were microdissected from 30 tumors. One-half of the patient samples contained both DCIS and IDC components. Seven patients had metastatic tumor tissue available in addition to both DCIS and IDC material, while five patients had invasive component material with corresponding metastatic disease. Aliquots of DNA from these microdissected lesions were used as the source template for PCR-based LOH analysis.
LOH analysis
Matching normal and tumor DNA were subjected to PCR-based LOH analysis using two 18p11.3 markers (D18S59 and D18S481) and one 18p11.2 marker (D18S452). These markers define a region on chromosome 18p previously shown to undergo LOH in invasive ductal breast carcinomas as well as non-small cell lung carcinomas and glioblastomas [1,2]. Forward primers were synthesized with either a fluorescent FAM tag (D18S481 and D18S452) or a fluorescent TET tag (D18S59) on the 5' end. PCR reactions were performed in 10 μl with aliquots of normal and tumor DNA using a PCR protocol consisting of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min for a total of 40 cycles.
Horizontal ultrathin gel electrophoresis
Horizontal ultrathin, high throughput fluorescence-based DNA fragment gel electrophoresis (GTI-9600; Genesys Technologies, Inc) was applied to the separation and analysis of PCR-generated alleles as previously described [4]. One microliter of amplified material was combined with 2 μl formamide loading dye (Amersham Inc, Piscat-away, NJ, USA) prior to electrophoresis. ROX 350 fluorescent size markers (0.7 μl; ABI, Foster City, CA, USA) were added to amplified tumor DNA to allow sizing of alleles. Samples were heated to 95°C, loaded on the 70 μm, 5% horizontal polyacrylamide gel and electrophoresed for 1 h and 15 min at 30 W in 1 × TBE using the GTI-9600. Data were collected while running the electrophoresis and stored using the accompanying Cartographer data analysis software program, which extracts the pixel intensity of individual peaks present in a lane trace, allowing for sizing of alleles and quantitation of fluorescent peak areas. In determining whether allelic deletion had occurred at individual markers, allelic ratios were calculated and expressed as a percentage of loss of intensity for the tumor allele compared with the corresponding normal allele (D-value) after normalization. When the allelic ratio in the tumor DNA was reduced by greater than 40% (DO.40) from that found in the normal DNA, the sample was denoted as having LOH at that locus.
Results
Chromosome 18p11 LOH in breast cancer progression
A total of 96 foci were individually microdissected from 30 breast tumor cases and analyzed for LOH at the polymorphic markers D18S59 and D18S481 spanning 18p11.3. These microdissections resulted in normal, DCIS and IDC material from most tumors. The overall frequency of LOH in 18p11.3, as measured for any stage of disease using D18S481 and/or D18S59, was 63% (27/43). This is significantly higher than the 21% frequency originally reported using non-microdissected IDC samples [1], suggesting that allelic deletion events may have been masked by contaminating normal stroma in earlier studies. A low frequency (3/96) of microsatellite instability was also detected at marker D18S59. These microsatellite instability results were confirmed in at least two independent experiments and correspond to previous published results for this marker [2].
LOH on 18p was found, when analyzed by tumor stage, in 56% (9/16) of DCIS foci, 67% of IDC foci (14/21), and 67% (4/6) of metastatic disease. A schematic representation of all tumor foci studied appears in Figure 1. The magnitude of LOH detected indicates that alterations in this region generally occur early in breast cancer progression. In four cases (cases 6, 18, 27, and 28), however, LOH was not detected until the invasive stage, and in two cases (cases 1 and 2) LOH was only detected in metastatic disease (Fig. 1). This suggests that allelic deletion in 18p11.3 may also arise late in progression, albeit with less frequency. Such heterogeneity in the stage specificity of genetic events has also been shown for other chromosomal regions harboring tumor suppressor loci [11,12,13].
The extent to which LOH is specific to band 18p11.3 was determined by analysis with marker D18S452. This marker resides at the border between bands 18p11.3 and 18p11.2, approximately 12 cM proximal to D18S481. Of the 15 tumor foci with LOH at 18p11.3 and for which D18S452 was informative, 73% (11/15) retained heterozygosity at this 18p11.2 marker. The majority of measured deletion events being detected are thus intrachromosomal events rather than loss of the entire chromosome 18.
Heterogeneity of 18p LOH in breast cancer progression
Previous genetic studies have shown that heterogeneous pathways exist for breast cancer progression [15,16]. We could examine, by studying multistage individually microdissected tumor components from the same specimen, the extent of heterogeneity associated with chromosome 18p LOH (Fig. 2). In tumors where allelic loss information was attainable for two or more disease stages, 73% (11/15) displayed a pattern of LOH indicative of a classic clonal progression. LOH first appeared in the DCIS stage for the majority of these cases (cases 3, 9, 31, 39, 47, 51 and 53). Four tumors were, however, interestingly identified with genetic heterogeneity for chromosome 18p events. LOH detected in the DCIS component was not found in the associated IDC component in two cases (tumors 21 and 44), suggesting that the invasive foci arose from cells other than those associated with the analyzed DCIS focus. Microsatellite instability and LOH distinguish multiple pathways of progression in the remaining two tumors (cases 2 and 27). Thus, although genetic events involving chromosome 18p sequences most often follow a classic progression paradigm, a significant proportion (27%) do not.
Correlations of allelic loss in chromosome 18p
Realizing that loss on 18p is only one part of the complex genetic profile for breast cancer, we sought to discover whether genetic alteration on this chromosome correlated with LOH in other regions known to undergo allelic deletion in cancer. These tumors had previously been shown to have undergone allelic deletion on chromosomes 3p (77%), 9p (75%), 11p (37%), 13q (39%), 16q (85%), 17p (100%) and 17q (71%). The LOH results specifically for those tumors also showing detectable chromosome 18p LOH are presented in Figure 3. In every instance where information was available, LOH on chromosomes 3p (region C13-CA373-D3S1300), 9p (region D9S171-D9S1748), 17p (region D17S1832-D17S1788 including p53) and 17q (region THRA1-D17S928 including nm23) was present when 18p LOH occurred. Chromosome 16q LOH (region D16S496-D16S513), in contrast, was only found in 81%, chromosome 13q (D13S260-D13S263) in 55%, and chromosome 11p (region D11S922-D11S988 including TH) in 48% of tumors also having 18p LOH. These frequencies suggest that alterations on chromosome 18p occur more often in the presence of alterations affecting regions containing such tumor suppressors as p16 (9p) [17] and p53 (17p) [18], while events on chromosomes 13q, 16q and 11p occur more independently.
Figure 3.
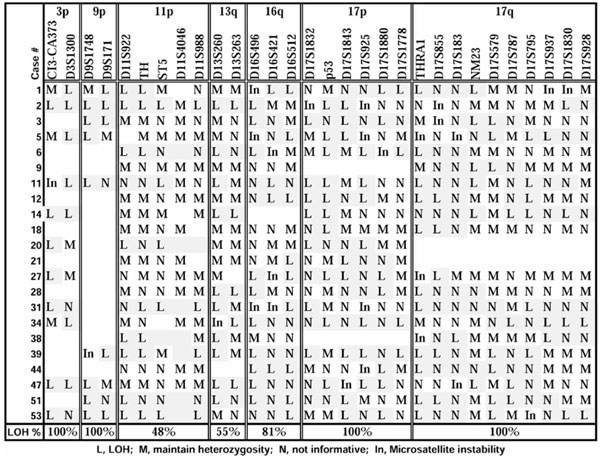
Other LOH in tumors with chromosome 18p allelic deletion.
Discussion
Current knowledge about the progression of breast cancer reveals that, by analogy to colon and brain tumors, genetic aberrations can occur in a disease stage or grade specific manner. Clonal evolution is, however, a dynamic process from which clones that are not necessarily committed to a malignant phenotype can also emerge, resulting in genetic heterogeneity in the tissue. Such genetic heterogeneity has been described for multiple loci in breast cancer [3,11,16,19,20], providing an understanding of the complex picture of breast cancer pathogenesis. Such an understanding is necessary if detection of LOH or other genetic abnormalities will ever allow for consistent and useful identification of preinvasive breast lesions at higher risk of progression.
The present study investigated stage specificity and level of genetic heterogeneity of LOH events in a new region (18p11.3) involved in breast cancer. Significant LOH for chromosome 18p11.3 (63%) was discovered using microdissected foci, suggesting the presence of a putative tumor suppressor gene(s) on 18p with an important role in the pathogenesis of breast cancer. The frequency reported in the present study (63%) is significantly higher than the original reported frequency of 21% [1] and most probably reflects the superior separation of tumor cells from normal contaminating tissue using a microdissection protocol. A series of unrelated microdissected DCIS tumors for which no invasive component was present also show this high LOH frequency (58%) when measured at 18p11.3 (data not shown), suggesting the 63% frequency reported in the present study is not a consequence of analyzing samples containing multiple stages of disease. These measured allelic deletions represent alterations specific to 18p11.3 and not entire chromosome 18 events based on results using marker D18S452, although the presence of simultaneous but independent LOH on 18q was not examined.
It was discovered, by analyzing multiple foci occurring simultaneously within individual tumors, that chromosome 18p events occur at a significant frequency (56%) early in the tumorigenic process, making LOH at chromosome 18p one of the most common known events in DCIS tumors. For 73% of the cases in which multiple stages of disease arose simultaneously and for which information on at least one marker was available, 18p11.3 LOH in the DCIS component could also be found in the invasive component isolated for analysis (Fig. 2). This suggests that the majority of breast tumors progress in a clonal fashion. Four cases (27%) were, however, consistent with genetic heterogeneity at this chromosomal region. These latter observations argue that invasive foci of an individual tumor can represent genetically divergent clones rather than progressive stages of the disease. Such conclusions are further supported by flow cytometric and comparative genomic hybridization analyses on primary and metastatic disease components [19,20]. In the comparative genomic hybridization study [20], 69% of metastatic lesions showed a high degree of clonal progression from the primary tumor, whereas 31% did not. These frequencies parallel the 70% clonal/30% heterogeneous progression frequencies reported for chromosome 18p events in the present study.
We are not yet at a stage in our understanding of breast disease such that genetic alterations can substitute for the histopathology of the lesion. A combinatorial histological and genetic approach with sufficient information on survival and response profiles may, however, make for more effective disease prevention and cure in the future. The power of genetic profiles in prognostic relevance has recently been shown by Emi et al [21]. Studying 15 loci in 264 women, Emi et al found that LOH at markers 1p34, 13q12, 17p13.3 and 17q21.1, as well as two pairs of markers (1p34/17p13.3 and 13q12/17p13.3), had significant prognostic value and carried significant relative risk of death. It would be interesting, given the high frequency of LOH discovered for chromosomal region 18p11.3 in this report, to include this marker along with these regions to determine its usefulness as a predictive risk marker in breast cancer.
To that extent, it was interesting to view a partial genetic profile for other regions of LOH in these tumors when chromosome 18 LOH was present. Chromosome 18p LOH always occurred in conjunction with LOH on 3p, 9p, 17p and 17q independent of tumor stage, suggesting this group of loci were critical to the development of breast cancer. Allelic deletion on chromosomes 16q, 13q and 11p occurred less consistently (81, 55 and 46%, respectively), suggesting these regional events are more varied depending on the stage and/or type of tumor foci analyzed. Of further interest is the putative tumor suppressor gene, DAL-1, which has recently been mapped to chromosome band 18p11.3 [22]. It will be important to learn more about the function of this gene and its potential mutational profile in breast cancer. Future studies will determine whether this or another tumor suppressor gene is the target of the high frequency of allelic deletion measured in this study for breast cancer.
Abbreviations
DCIS = ductal carcinoma in situ; IDC = invasive ductal carcinoma; LOH = loss of heterozygosity; PCR = polymerase chain reaction.
Acknowledgments
Acknowledgements
The authors would like to thank Dr Oliver Bogler for his critical reading of this manuscript. This work was supported in part by R29 CA77330 (IN). KK is the recipient of a Royal Thailand Scholar Fellowship.
References
- Tran Y, Benbatoul K, Gorse K, Rempel S, Futreal A, Green M, Newsham I. Novel regions of allelic deletion on chromosome 18p in tumors of the lung, brain and breast. Oncogene. 1998;17:3499–3505. doi: 10.1038/sj.onc.1202258. [DOI] [PubMed] [Google Scholar]
- Osborne RJ, Hamshere MG. A gemone-wide map showing common regions of loss of heterozygosity/allelic imbalance in breast cancer. Cancer Res. 2000;60:3706–3712. [PubMed] [Google Scholar]
- Lichy JH, Zavar M, Tsai MM, O'Leary TJ, Taubenberger JK. Loss of heterozygosity on chromosome 11p15 during histological progression in microdissected ductal carcinoma of the breast. Am J Pathol. 1998;153:271–278. doi: 10.1016/S0002-9440(10)65568-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsham IF, Gorse KM, Rempel SA, Luckey J, Golden JB, Bogler O. Use of horizontal ultrathin gel electrophoresis to analyze allelic deletions in chromsome band 11p15 in gliomas. Neuro-Oncol. 2000;2:1–5. doi: 10.1093/neuonc/2.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson S. Molecular genetics of breast cancer progression. Semin Cancer Biol. 1999;9:277–288. doi: 10.1006/scbi.1999.0124. [DOI] [PubMed] [Google Scholar]
- Beckmann MW, Niederacher D, Schnurch H-G, Gusterson BA, Bender HG. Multistep carcinogenesis of breast cancer and tumour heterogeneity. J Mol Med. 1997;75:429–439. doi: 10.1007/s001090050128. [DOI] [PubMed] [Google Scholar]
- Lizard-Nacol S, Lidereau R, Collin F, Arnal M, Hahnel L, Roignot P, Cuisenier J, Guerrin J. Benign breast disease: absence of genetic alterations at several loci implicated in breast cancer malignancy. Cancer Res. 1995;55:4416–4419. [PubMed] [Google Scholar]
- O'Connell P, Pekkel V, Fuqua SAW, Osborne CK, Clark GM, Allred DC. Analysis of loss of heterozygosity in 399 premalignant breast lesions at 15 genetic loci. J Natl Cancer Inst. 1998;90:697–703. doi: 10.1093/jnci/90.9.697. [DOI] [PubMed] [Google Scholar]
- Deng G, Lu Y, Zlotnikov G, Thor AD, Smith HS. Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science. 1996;274:2057–2059. doi: 10.1126/science.274.5295.2057. [DOI] [PubMed] [Google Scholar]
- Ottesen GL, Graversen HP, Blichert TM, Zedeler K, Andersen JA. Ductal carcinoma in situ of the female breast: short-term results of a prospective nationwide study. Am J Surg Pathol. 1992;16:1183–1196. doi: 10.1097/00000478-199212000-00005. [DOI] [PubMed] [Google Scholar]
- Lichy JH, Dalbegue F, Zavar M, Washington C, Tsai MM, Sheng ZM, Taubenberger JK. Genetic heterogeneity in ductal carcinoma of the breast. Lab Invest. 2000;80:291–301. doi: 10.1038/labinvest.3780034. [DOI] [PubMed] [Google Scholar]
- Bonsing BA, Devilee P, Cleton-Jansen A-M, Kuipers-Dijkshoorn N, Fleuren GJ, Cornelisse CJ. Evidence for limited molecular genetic heterogeneity as defined by allelotyping and clonal analysis in nine metastatic breast carcinomas. Cancer Res. 1993;53:3804–3811. [PubMed] [Google Scholar]
- Anbazhagan R, Hiroaki F, Gabrielson E. Allelic loss of chromosomal arm 8p in breast cancer progression. Am J Pathol. 1998;152:815–819. [PMC free article] [PubMed] [Google Scholar]
- Marsh KL, Varley JM. Loss of heterozygosity at chromosome 9p in ductal carcinoma in situ and invasive carcinoma of the breast. Br J Cancer. 1998;77:1439–1447. doi: 10.1038/bjc.1998.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsham IF. The long and short of chromosome 11 in breast cancer. Am J Pathol. 1998;153:5–9. doi: 10.1016/S0002-9440(10)65538-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Szumel R, Marsh C, Zhou W, Gabrielson E. Genetic progression, histological grade and allelic loss in ductal carcinoma in situ of the breast. Cancer Res. 1996;56:5260–5265. [PubMed] [Google Scholar]
- Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature (London) 1994;368:753–756. doi: 10.1038/368753a0. [DOI] [PubMed] [Google Scholar]
- Vos CB, ter Haar NT, Rosenberg C, Peterse JL, Cleton-Jansen AM, Cornelisse CJ, van de Vijver MJ. Genetic alterations on chromosome 16 and 17 are important features of ductal carcinoma in situ of the breast and are associated with histologic type. Br J Cancer. 1999;81:1410–1418. doi: 10.1038/sj.bjc.6693372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsing BA, Corver WE, Fleuren GJ, Cleton-Jansen AM, Devilee P, Cornelisse CJ. Allelotype analysis of flow-sorted breast cancer cells demonstrates genetically related diploid and aneuploid subpopulations in primary tumors and lymph node metastases. Genes Chrom Cancer. 2000;28:173–183. doi: 10.1002/(sici)1098-2264(200006)28:2<173::aid-gcc6>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Kuukasjärvi T, Karhu R, Tanner M, Kähkönen M, Schäffer A, Nupponen N, Pennanen S, Kallioniemi A, Kallioniemi OP, Isola J. Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res. 1997;57:1597–1604. [PubMed] [Google Scholar]
- Emi M, Yoshimoto M, Sato T, Matsumoto S, Utada Y, Ito I, Minobe K, Iwase T, Katagiri T, Bando K, Akiyama F, Harada Y, Fukino K, Sakamoto G, Matsushima M, Iida A, Tada T, Saito H, Miki Y, Kasumi F, Nakamura Y. Allelic loss at 1p34, 13q12, 17p13.3 and 17q21.1 correlated with poor postoperative prognosis in breast cancer. Genes Chrom Cancer. 1999;26:134–141. [PubMed] [Google Scholar]
- Tran YK, Bogler O, Gorse K, Wieland I, Green MR, Newsham IF. A novel member of the NF2.ERM/4.1 superfamily with growth suppressing properties in lung cancer. Cancer Res. 1999;59:35–43. [PubMed] [Google Scholar]


