Abstract
Peptidomimetic polymers consisting of poly-N-substituted glycine oligomers (polypeptoids) conjugated to biomimetic adhesive polypeptides were investigated as antifouling surface coatings. The polymers were immobilized onto TiO2 surfaces via an anchoring peptide consisting of alternating residues of 3,4-dihydroxyphenylalanine (DOPA) and lysine. Three polypeptoid side-chain compositions were investigated for antifouling performance and stability toward enzymatic degradation. Ellipsometry and XPS analysis confirmed that purified polymers adsorbed strongly to TiO2 surfaces, and the immobilized polymers were resistant to enzymatic degradation as demonstrated by mass spectrometry. All polypeptoid-modified surfaces exhibited significant reductions in adsorption of lysozyme, fibrinogen and serum proteins, and were resistant to 3T3 fibroblast cell attachment for up to seven days. Long-term in vitro cell attachment studies conducted for six weeks revealed the importance of polypeptoid side-chain composition, with a methoxyethyl side chain providing superior long-term fouling resistance compared to hydroxyethyl and hydroxypropyl side chains. Finally, attachment of both gram-positive and gram-negative bacteria for up to four days under continuous-flow conditions was significantly reduced on the polypeptoid-modified surfaces compared to unmodified TiO2 surfaces. The results reveal the influence of polypeptoid side-chain chemistry on short-term and long-term protein, cell and bacterial fouling resistance.
Introduction
Biofouling, in the form of protein, cell and bacterial attachment on medical devices, can alter device performance and lead to patient infection, device removal and increased healthcare costs. Some common examples of biofouling include accumulation of proteins and cells on biosensor surfaces,1 stents and cardiovascular implants,2–4 and the colonization of bacteria on catheters,5,6contact lenses7 and surgical tools.8 Fouling resistance of materials can be improved by surface-immobilization of self-assembled monolayers (SAMs) and antifouling polymers; a number of strategies and molecular compositions for accomplishing this have been reviewed by others.9–12 Among the more commonly employed systems are oligo(ethylene glycol)-terminated SAMs,10,13–15 grafted polymers based on poly(ethylene glycol) (PEG),16–23 zwitterionic polymers,24–26 and glycomimetic polymers.27 Although much progress has been made toward preventing short-term adhesion of proteins and cells, long-term biofouling resistance remains a challenge.
We recently reported the design and synthesis of a new class of antifouling polymers composed of N-substituted glycine (peptoid) chains.28 Through the addition of an adhesive peptide containing residues found in mussel-adhesive proteins,29,30 an N-methoxyethyl glycine peptoid adsorbed strongly to Ti surfaces and exhibited low protein adsorption and cell-fouling resistance for over five months in vitro. In addition to attractive features such as protease resistance and low immunogenicity,31,32 peptoid polymers also offer vast potential for chemical tailoring through the use of both natural and unnatural side-chain functional groups. In the present study we begin to explore this chemical versatility through synthesis and characterization of two new antifouling peptidomimetic polymers comprising different polypeptoid side-chain compositions. The antifouling performance of the polymers grafted on TiO2 was assessed by protein-, cell- and bacterial-adhesion measurements. All three peptidomimetic polymers provided good short-term resistance to protein, cell and bacterial fouling. However, long-term cell-attachment studies revealed significant differences in fouling performance, which can be attributed to peptoid side-chain composition.
Experimental
Materials
Methoxyethylamine, ethanolamine, triisopropylsilyl chloride, (±)-1-amino-2-propanol, triisopropylsilane (TIS), dimethylformamide (DMF), acetonitrile (ACN), N-morpholinopropanesufonic acid (MOPS) buffer salt, chicken egg-white lysozyme, fibrinogen, 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) buffer salt, trypsin, pronase, Tris buffer salt, sodium tetraborate, 2-propanol, sinapic acid matrix and α-cyano-4-hydroxycinnamic acid (CHCA) matrix were purchased from Aldrich (Milwaukee, WI, USA). Rink amide resin, Fmoc-Lys(Boc)-OH, and Fmoc-DOPA(acetonide)-OH were purchased from Novabiochem (San Diego, CA, USA). Acetic anhydride and N-methylpyrrolidone (NMP) were purchased from Applied Biosystems (Foster City, CA, USA). Trifluoroacetic acid (TFA) was obtained from Fisher Scientific (Pittsburgh, PA, USA). Silicon wafers were purchased from University Wafer (South Boston, MA, USA). Lyophilized whole human serum (Control Serum N) was purchased from Roche Diagnostics (Indianapolis, IN, USA). Immobilized TPCK trypsin was purchased from Pierce (Rockford, IL, USA). 1,1′-Dioctadecyl-3,3,3′,3′-tetramethyl-indocarbocyanine perchlorate (DiI), calcein-AM, and Syto 9 were purchased from Molecular Probes (Eugene, OR, USA). 3T3-Swiss albino fibroblasts, Dulbecco’s modified Eagle’s medium, fetal bovine serum, penicillin/streptomycin, trypsin-EDTA, Staphylococcus epidermidis RP62A (ATCC 12228), and Escherichia coli (ATCC 53323) were obtained from American Type Culture Collection (Manassas, VA, USA). Tryptic soy agar and broth and Luria-Bertani, Miller agar and broth were obtained from Becton, Dickinson and Co. (Sparks, MD, USA). The chemostat and modified Robbins device (MRD) were custom designed and constructed by Tyler Research (Edmonton, Alberta, Canada). Ultrapure water (U.P. H2O) used for all experiments was purified (resistivity ≥ 18.2 MΩ cm, total organic content ≤ 5 ppb) with a NANOpure Infinity System from Barnstead/Thermolyne Corp. (Dubuque, IA, USA).
Synthesis of peptidomimetic polymers
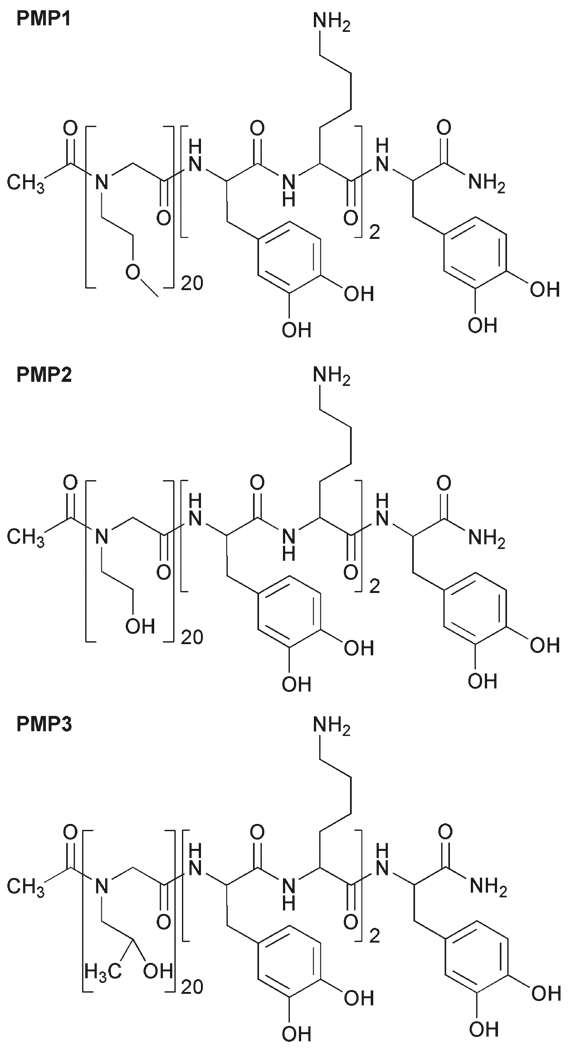
The peptidomimetic polymers were synthesized as described previously,28 using an ABI 433A (Applied Biosystems, Foster City, CA, USA) automated peptide synthesizer. Three polymers were synthesized with identical peptide anchors, but with different polypeptoid side chains as shown in Fig. 1. The C-terminal DOPA-Lys-DOPA-Lys-DOPA peptide anchor was synthesized on Rink amide resin using conventional Fmoc strategy of solid-phase peptide synthesis; the polypeptoid portion was then synthesized using a submonomer protocol33 and appropriate primary amine.34 The submonomer process was repeated until the desired number of monomers was added, after which acetic anhydride was used to acetylate the N-terminus of the polypeptoid.
Fig. 1.
Chemical structures of PMP1, PMP2 and PMP3.
Cleavage of the polymers from the resin and deprotection of the amino acid side chains was accomplished by treating the resin with 95% (v/v) TFA, 2.5% H2O and 2.5% TIS for 20 minutes, after which the cleaved polymer was removed by filtering and rinsing several times with TFA. Solvent was removed using a rotary evaporator; the oily product was dissolved in water–acetonitrile (1 : 1), frozen and lyophilized. The crude products were purified by preparative reversed-phase high performance liquid chromatography (RP-HPLC) (Waters, Milford, MA, USA) using a Vydac C18 column, and purified fractions were frozen and lyophilized. The purity of each final product was confirmed by RP-HPLC and matrix-assisted laser desorption–ionization mass spectrometry (MALDI-MS) (Voyager DE-Pro, Perspective Biosystem, MA, USA).
Surface modification
Silicon wafers were coated with a 20 nm thick layer of TiO2 (99.9% pure, West Cerac, Milwaukee, WI, USA) by electronbeam evaporation (Edwards Auto306, <10−5 Torr), and the coated wafers were cut into 8 by 8 mm pieces. The substrates were cleaned ultrasonically for ten minutes in 2-propanol and dried under N2. Surfaces were then exposed to O2 plasma (Harrick Scientific, Ossining, NY, USA) at ≤150 Torr and 100 W for three minutes. OWLS waveguide sensors were purchased from Micro Vacuum Ltd (Budapest, Hungary) and coated with a 10 nm thick layer of TiO2 by electron-beam evaporation as described above. Sensors were cleaned following the same procedure as TiO2 substrates. After use, OWLS waveguides were regenerated for subsequent use by sonication cycles (10 minutes) in 0.1 M HCl, U.P. H2O and 2-propanol followed by exposure to O2 plasma to remove adsorbates.
Clean substrates and sensors were immersed in a 1 mg mL−1 solution of peptidomimetic polymer in 3 M NaCl buffered with 0.1 M MOPS (pH = 6) at 50 °C for 24 hours. After modification, substrates were extensively rinsed with U.P. H2O to remove any unbound polymer, and then dried in a stream of filtered N2.
Surface characterization
Contact-angle measurements
The wettability of surfaces before and after modification was measured using a contact-angle goniometer with an auto pipetting system (Ramé-Hart, Mountain Lakes, NJ, USA). Advancing and receding contact angles were measured three times on each surface with U.P. H2O and the mean and standard deviation were reported.
Spectroscopic ellipsometry measurements
Prior to modification, substrates were cleaned as described above and measured using a M-2000 spectroscopic ellipsometer (J.A. Woollam, Lincoln, NE, USA). Measurements were made at 65°, 70° and 75° using wavelengths from 193–1000 nm. After modification, substrates were rinsed and dried as described above and measured again. The spectra were fit with multilayer models in the WVASE32 software (J.A. Woollam). Optical properties of the substrate were fit using a standard TiO2 model, while properties of the polymer layer were fit using a Cauchy model (An = 1.45, Bn = 0.01, Cn = 0).35 The obtained ellipsometric thicknesses represent the “dry” thickness of the polymer under ambient conditions. The average thickness and standard deviation of three or more substrates is reported for each polymer.
X-Ray photoelectron spectroscopy (XPS)
Survey and high resolution XPS spectra were collected on an Omicron ESCALAB (Omicron, Taunusstein, Germany) configured with a monochromated Al Kα (1486.8 eV) 300-W X-ray source, 1.5 mm circular spot size, a flood gun to counter charging effects, and an ultrahigh vacuum (<10−8 Torr). The takeoff angle was fixed at 45°. Substrates were mounted on standard sample studs using double-sided Cu adhesive tape. All binding energies were calibrated using the aliphatic hydrocarbon component of the C1s carbon signal (284.6 eV). Analysis consisted of a broad survey scan (70.0 eV pass energy) and high-resolution scans (26.0 eV pass energy) at 274–294 eV for C1s, 390–410 eV for N1s, 450–470 eV for Ti2p and 520–540 eV for O1s. Spectra were fitted using CasaXPS software; specifically a Shirley background subtraction and the sum of 90% Gaussian and 10% Lorentzian function were used. Atomic sensitivity factors were used to normalize peak areas from high-resolution spectra to intensity values, which were then used to calculate atomic compositions.36
Susceptibility to enzymatic degradation
Polymers in solution
In order to test the resistance of polypeptoid bonds to enzymatic degradation, a short polypeptoid consisting of 5 repeats of N-methoxyethyl glycine (NMEG5) was synthesized using the submonomer approach and purified using methods described previously. The purified NMEG5 was dissolved at a concentration of 1 mg mL−1 in 10 mM Tris buffer (pH = 7.5) and 1 mg of trypsin (~37 U) or pronase (~6 U) was added. Control samples with only enzyme or NMEG5 in buffer were prepared. Samples were incubated at 37 °C for 24 h with mild shaking. Solutions were filtered and analyzed by RP-HPLC using a 2–90% ACN gradient.
Additionally, peptidomimetic polymers (1.0 mg) were incubated with 0.25 mL immobilized TPCK trypsin (≥50 TAME units) in 0.75 mL of 0.1 M borate buffer (pH = 8.5) for 4 h at 37 °C. The trypsin was separated by centrifugation, and the remaining solution was dialyzed (MWCO 100) in U.P. H2O overnight to remove buffer salts and then lyophilized. Liquid chromatography-electrospray ionization mass spectrometry (LC-ESI-MS) (LCQ Advantage, Thermo Finnigan, Waltham, MA, USA) was used to analyze the degradation products for PMP1 solutions. For PMP2 and PMP3, the dialyzed solutions were purified by RP-HPLC (Waters, Milford, MA, USA) using a Vydac C18 column, and the appropriate fractions were collected, frozen and lyophilized. The fractions were analyzed using ESI-MS (LCQ Advantage).
Polymers adsorbed onto TiO2 substrates
TiO2 substrates were modified with PMP1, PMP2, or PMP3 as described previously, and incubated in 48 well plates with solutions of soluble trypsin or pronase (1 mg mL−1 in 10 mM Tris buffer solution, pH = 7.5). As controls, modified substrates were also incubated with buffer only and bare TiO2 substrates were incubated with the enzymes. Trypsin and pronase solutions were refreshed every 2 days; substrates were removed after 7 days, rinsed with U.P. water, and dried with N2. At the time of substrate removal, the solution from each well was collected, dialyzed (100 MWCO), lyophilized, and analyzed with ESI-MS. Substrates were spin-coated with CHCA matrix and analyzed using surface-MALDI-MS.
Protein-adsorption experiments
Proteins
The proteins investigated include whole human serum, lysozyme from chicken egg-white and fibrinogen from human plasma. Serum was reconstituted in water to reach the typical concentration in blood; lysozyme and fibrinogen were each dissolved at 3 mg mL−1 concentrations in HEPES buffer (10 mM HEPES, 150 mM NaCl, pH = 7.4).
Optical waveguide lightmode spectroscopy (OWLS)
OWLS is a technique that is well suited for monitoring of in situ protein adsorption because the sensitivity is near 0.5 ng cm−2, which correlates to approximately 0.5% of an average protein monolayer.37,38 For in situ protein-adsorption experiments, TiO2-coated waveguide sensors were modified with peptidomimetic polymers as described above. The sensors were then inserted into the measurement head of an OWLS110 (MicroVacuum Ltd) and exposed to HEPES buffer through the flow-through cell (16 µL volume) for at least 24 hours to allow for equilibration. The measurement head was mounted in the sample chamber and heated to 37 °C; the signal was recorded to ensure a stable baseline. A protein solution (1 mL total volume) was injected into the flow-through cell in stop-flow mode. The waveguide sensor was exposed to the protein solution for 20 minutes, subsequently rinsed with HEPES buffer (2 mL), and allowed to equilibrate for another 30 minutes. The measured incoupling angles, αTM and αTE were converted to refractive indices NTM and NTE by the Micro Vacuum software, and changes in refractive index at the sensor surface were converted to adsorbed mass using de Feijter’s formula.39
The refractive indices of solutions were measured using a refractometer (J157 automatic refractometer, Rudolph Research) under identical experimental conditions. A refractive index value of 1.33127 was used for the HEPES buffer, and a standard value of 0.182 cm3 g−1 was used for dn/dc in the protein-adsorption calculations.40 The large mass increase upon injection of protein solution reflects protein adsorption as well as the change in refractive index of the covering solution near the surface.
Mammalian cell-adhesion experiments
Cell culture
3T3-Swiss albino fibroblasts were maintained at 37 °C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) and 100 U ml−1 of penicillin/streptomycin. Immediately before use, fibroblasts of passage 12–16 were harvested using 0.25% trypsin-EDTA, resuspended in DMEM with 10% FBS and counted using a hemacytometer.
Cell-adhesion assays
Modified and unmodified TiO2 substrates were placed in a 12-well tissue-culture polystyrene (TCPS) plate and sterilized by exposure to UV light for 10 minutes, after which 1 ml of DMEM containing FBS was added to each well and incubated for 30 minutes at 37 °C and 5% CO2. The fibroblast cell suspension was diluted, and cells were seeded on each substrate at a density of 2.9 × 103 cells cm−2. For short-term studies, the substrates were maintained in DMEM with FBS at 37 °C and 5% CO2 for 4 hours, after which adherent cells were fixed in 3.7% paraformaldehyde for 5 minutes and stained with DiI for epifluorescent microscope counting. For long-term adhesion experiments substrates were reseeded with 3T3 fibroblasts at a density of 2.9 × 103 cells cm−2 twice per week. For live-cell staining, the medium was aspirated from each well to remove any non-adherent cells and PBS was used to rinse the substrates and wells. Fibroblasts were stained with 2.5 µM calcein-AM in complete PBS for 1 hour at 37 °C, transferred to new culture plates with fresh media and imaged weekly. After imaging, substrates were reseeded and placed back into the incubator; media was changed every three days.
Quantification of cell attachment and spreading
Quantitative cell-attachment data were obtained by acquiring nine images (10× magnification) from random locations on each substrate using a Leica epifluorescent microscope (W. Nuhsbaum Inc., McHenry, IL, USA) equipped with a SPOT RT digital camera (Diagnostics Instruments, Sterling Heights, MI, USA). Three identical substrates for each experiment were analyzed for statistical purposes, resulting in a total of 27 images per time point for each modification. The microscopy images were quantified using thresholding in Metamorph (Molecular Devices, Downingtown, PA, USA).
Bacterial cell-adhesion experiments
Bacterial culture and assay
Staphylococcus epidermidis (S. epidermidis) were streaked from frozen glycerol stocks onto tryptic soy agar and incubated overnight at 37 °C. A few colonies were then used to inoculate 25 mL of sterile tryptic soy broth (TSB) and grown overnight at 37 °C. The MRD, chemostat containing 500 mL TSB and a dewar containing TSB (diluted to 1/10 strength) were connected together and sterilized by autoclaving. Once the broth cooled, the chemostat was inoculated with 1 mL of the bacteria culture grown overnight. The chemostat was maintained at 37 °C with a dilution rate of 0.048 h−1 to allow the population of bacteria to stabilize as indicated by CFU count. The bacterial suspension was then pumped through the MRD containing unmodified and PMP1-, PMP2- and PMP3-modified TiO2 samples at a rate of 8 mL min−1. Half of the samples were removed from the device after 1 day, and half of the substrates were left in the device for 4 days to examine longer-term adhesion and stability of the coating under flow conditions. The same procedure was repeated for Escherichia coli (E. coli) using Luria-Bertani, Miller broth and agar.
Quantification of bacterial adhesion
Samples were removed from the MRD, placed into 24-well plates and gently rinsed with PBS to remove loosely adherent and planktonic bacteria. The attached bacteria were then stained with 2 µL mL−1 Syto 9 in PBS and visualized using a Leica epifluorescence microscope (40× magnification). Images were taken from 9 random places on each substrate and the bacterial cell area coverage was determined by threshold image analysis as explained previously for fibroblast cell adhesion.
Results and discussion
The chemical structures of the peptidomimetic polymers used in this study are shown in Fig. 1. Each polymer contains an identical peptide anchor consisting of a pentapeptide of alternating DOPA and lysine residues, which was previously shown to provide adequate attachment of peptoid polymers on Ti surfaces during extended exposure to biological fluids.28 The peptoid portions of the polymers were synthesized using the submonomer approach, which allows for easy alteration of the side-chain chemistry through the use of different primary amines.34 Following cleavage from the resin and standard work-up, the polymers were found to be pure and monodisperse as determined by RP-HPLC and MALDI-MS, respectively (ESI†, Fig. S1–S2).
In addition to the previously reported PMP1,28 which contains a methoxyethyl side chain reminiscent of the repeat unit of PEG, two additional polymers containing hydroxyl side chains were investigated (PMP2, PMP3). Hydroxyl-containing side chains were chosen for this study based on existing evidence that hydroxyl-terminated SAMs8,16 as well as hydroxylated polymers27,41 offer good biofouling resistance. The three different side chains employed also offered an opportunity to investigate the influence of peptoid wettability on fouling performance.
Surface modification and characterization
Titanium oxide was selected as the substrate material for this study because of its relevance to several titanium-based alloys that are used in medical devices,42 but also for the strong interaction that is known to exist between DOPA-containing peptides and TiO2 surfaces.43 The DOPA–lysine pentapeptide anchor was previously shown to provide robust anchorage to TiO2 surfaces over many months in vitro,28 thus allowing us to focus in this study on the effect of peptoid side-chain chemistry on fouling performance.
The purified polymers were grafted onto TiO2 substrates under marginal solvation or near ‘cloud point’ conditions, in which reduced chain repulsion leads to greater grafted polymer density.44,45 Temperature-dependent solubility behavior was expected for the peptidomimetic polymers because of the PEG-like peptoid side chains; a grafting temperature of 50 °C in 3 M NaCl buffered with 0.1 M MOPS (pH = 6) was selected based on temperature-solubility experiments (ESI†, Fig. S3).
Modification of TiO2 surfaces was confirmed by analyzing the wettability, thickness and chemical composition of the polymer-modified surfaces. Advancing and receding contact-angle results for water on unmodified TiO2 and polymer-modified substrates are shown in Table 1. PMP2-modified surfaces were considerably more hydrophilic than PMP1- and PMP3-modified surfaces. These results are consistent with published data for self-assembled monolayers on gold, which indicate that the hydrophilicity of terminal functional groups increases in the order methyl < methoxy ≪ hydroxy.46 Polymer thickness was measured using spectroscopic ellipsometry and the results are reported in Table 1. Thickness values for the polymer-modified surfaces are between 40 and 50 Å, with no statistically significant differences detected between the three polymers (ANOVA), suggesting that side-chain functional groups did not have a measurable effect on polymer thickness.
Table 1.
Wettability and polymer thickness for unmodified and polypeptoid-modified TiO2 substrates
| Contact angle/° ± SDa | Polymer | ||
|---|---|---|---|
| Substrate | Advancing | Receding | Thickness/Å ± SDa |
| TiO2 | 23.5 ± 6.5 | 15.3 ± 3.9 | — |
| PMP1 | 50.5 ± 0.6 | 35.3 ± 9.2 | 46.2 ± 6.1 |
| PMP2 | 31.3 ± 3.2 | 17.3 ± 4.2 | 40.7 ± 6.5 |
| PMP3 | 47.1 ± 4.0 | 29.9 ± 9.9 | 43.2 ± 5.8 |
SD = standard deviation.
Atomic compositions for the modified surfaces were determined from high-resolution XPS spectra and are shown in Table 2. The polymer-modified substrates exhibited peaks for the titanium oxide substrate, as well as oxygen, nitrogen and carbon contributions from the polymer layer. All polymer-modified surfaces exhibited significant increases in carbon and nitrogen concentration and decreases in titanium and oxygen concentration relative to unmodified TiO2. The nitrogen signal is attributed to the peptide/peptoid backbone and the lysine side chains; the oxygen signal is derived from both substrate and adsorbed polymer, and the carbon signal represents surface-adsorbed polymer and hydrocarbon contaminants. High-resolution C1s spectra of the polymer-modified substrates were indicative of differences in the polypeptoid side-chain compositions (ESI†, Fig. S4). Time-of-flight secondary ion mass spectrometry (TOF-SIMS) characterization of PMP1-modified Ti substrates published previously indicated the presence of numerous peptoid mass fragments and also fragments corresponding to catechol and DOPA residues complexed with Ti, suggesting interaction between the DOPA residues and substrate.28
Table 2.
Quantitative XPS analysis of atomic composition for unmodified and polypeptoid-modified TiO2 substrates
| Atomic composition (%) | ||||
|---|---|---|---|---|
| Substrate | Ti | O | N | C |
| TiO2 | 21.5 | 63.4 | 0.3 | 14.8 |
| PMP1 | 2.5 | 23.6 | 12.7 | 61.2 |
| PMP2 | 9.4 | 37.2 | 8.5 | 44.9 |
| PMP3 | 4.5 | 32.4 | 7.2 | 55.9 |
Resistance to enzymatic degradation
Enzymatic degradation of antifouling polymer coatings can be problematic for many applications, particularly those that require long-term fouling protection in the presence of biological fluids. As candidate antifouling polymers, N-substituted glycine peptoids are attractive due to their resistance to protease-enzyme degradation resulting from conjugation of the side chain to the amide nitrogen instead of the alpha carbon of the amino acid. For example, Miller et al. have previously demonstrated the resistance of sequence-specific polypeptoids to cleavage by four types of proteases.31 Here, we focused on the protease resistance of a model N-methoxyethyl glycine peptoid, as well as PMP1, PMP2 and PMP3 in solution and after adsorption on a substrate.
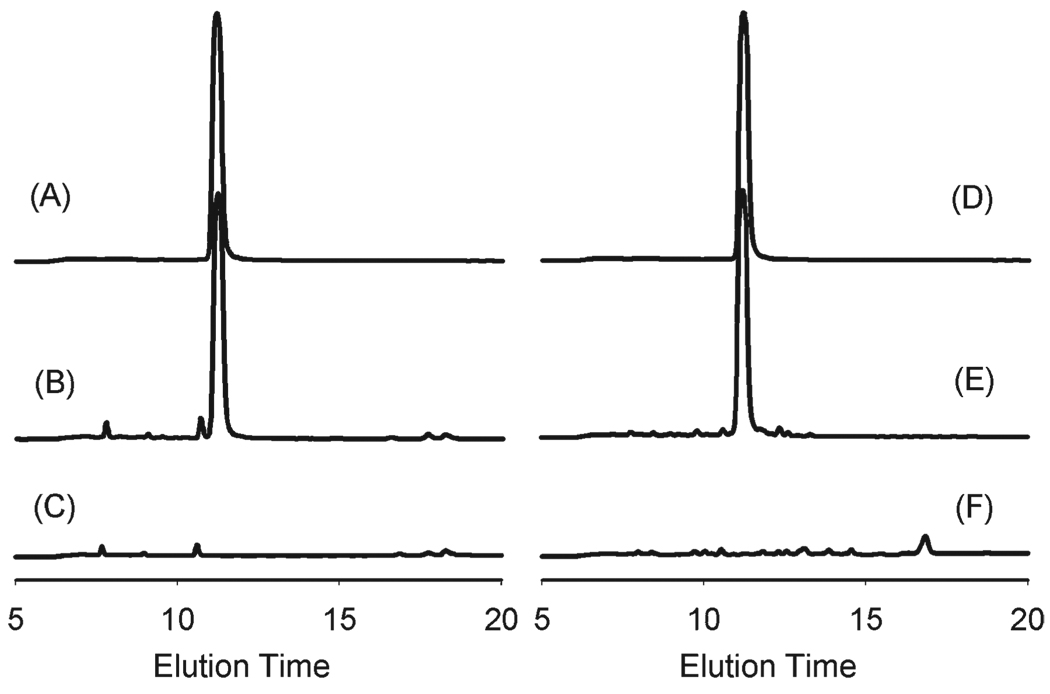
To evaluate the inherent protease stability of the N-substituted glycine oligomers, we synthesized a 5-mer N-methoxyethyl glycine (NMEG5) peptoid and incubated 1 mg of the purified molecule in solution with trypsin and pronase for 24 h. Analysis of the trypsin-incubated samples by RP-HPLC revealed no changes in elution time or peak intensity of the NMEG5 peak (Fig. 2), indicating no degradation by trypsin. Similar results were obtained with pronase, an enzyme with much broader activity. These results confirmed the resistance of N-methoxyethyl glycine backbones to degradation by common proteases.
Fig. 2.
RP-HPLC spectra for 5-mer N-methoxyethyl glycine peptoid (NMEG5). (A) NMEG5 incubated in buffer solution; (B) NMEG5 incubated with pronase in buffer solution; (C) pronase incubated in buffer solution. (D) NMEG5 incubated in buffer solution; (E) NMEG5 incubated with trypsin in buffer solution; and (F) trypsin incubated in buffer solution.
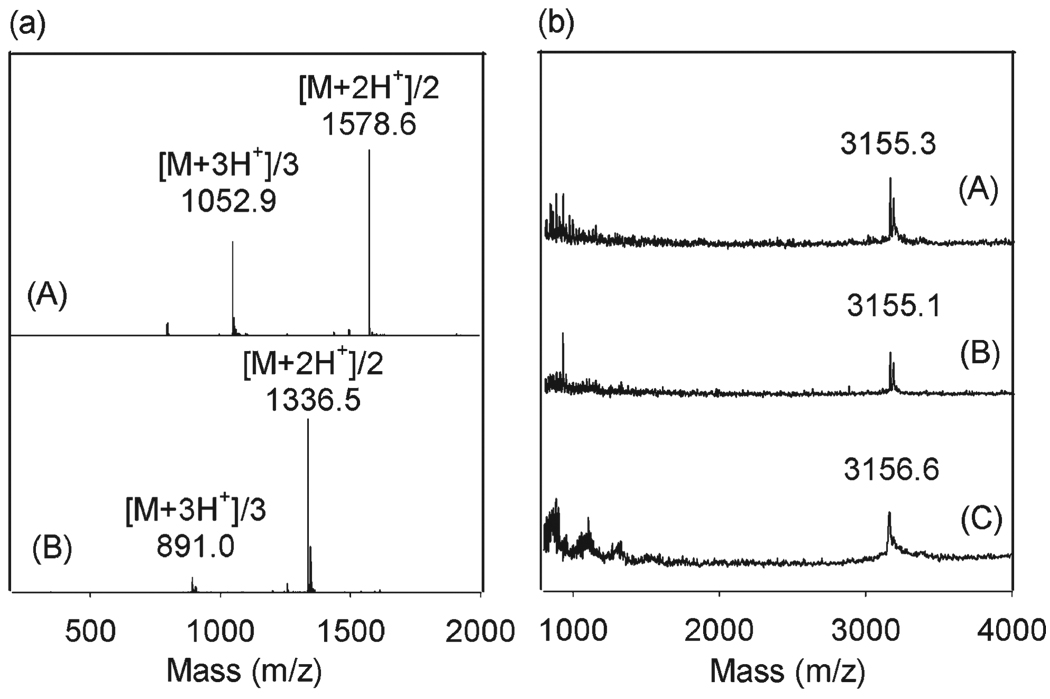
Enzyme resistance of the peptidomimetic polymers shown in Fig. 1 is complicated by their chimeric nature, as they contain both peptoid and peptide domains. In principle, trypsin is capable of cleaving the carboxy side of each lysine residue in the anchoring domain of PMP1-3. Incubation of PMP1 in solution with immobilized trypsin and analysis of the reaction mixture by mass spectroscopy indicated that PMP1 was cleaved by trypsin, yielding a fragment corresponding to PMP1 minus the three carboxy-terminal residues (i.e. two DOPA residues and one lysine residue) (Fig. 3a). Furthermore, no residual peaks corresponding to full length PMP1 (m/z = 3156) or PMP1 minus the terminal DOPA (m/z = 2976) were detected, suggesting that trypsin degradation of the terminal anchoring domain of PMP1 in solution was complete within 4 hours. Similar degradation patterns were observed for PMP2 and PMP3 in solution (ESI†, Fig. S5). Thus, we conclude that when the peptidomimetic polymers are mixed in solution with an enzyme, such as trypsin, the peptide bonds are accessible to the enzyme.
Fig. 3.
Enzymatic degradation of PMP1. (a) LC-MS spectra for: (A) PMP1 and (B) PMP1 sample after incubation with immobilized trypsin. (b) MALDI-MS spectra for PMP1-modified TiO2 after 1 week incubation in: (A) buffer only, (B) trypsin and (C) pronase.
However, enzymatic degradation may be impacted by adsorption of the polymer on a substrate surface. To gain a more accurate picture of the polymers in the form of surface coatings, we therefore also studied enzymatic degradation of PMP1-, PMP2- and PMP3-modified TiO2 substrates. Polymer-modified substrates were incubated in trypsin-containing solutions and directly analyzed in the adsorbed state by MALDI-MS. In addition, the enzyme solutions were subsequently analyzed by ESI-MS to detect any polymer or peptide fragments released into the medium as a result of enzyme degradation. Unlike the behavior of PMP1 in solution, MALDI-MS spectra for PMP1-modified TiO2 substrates were largely unchanged after 1 week of incubation with trypsin (Fig. 3b). Peaks were not detected either on the substrates or in the enzyme solutions for the expected fragments resulting from trypsin cleavage of the lysine bonds (ESI-MS spectra not shown), suggesting that the peptide-cleavage sites are inaccessible to the enzyme when the polymers are immobilized on surfaces. Similar results were observed for PMP1-modified TiO2 substrates exposed to pronase (Fig. 3b) and for PMP2- and PMP3-modified substrates exposed to trypsin and pronase (ESI†, Fig. S6).
Two factors could contribute to the observed long-term degradation resistance. The first is the strength of adhesion of the peptide to the surface. Incubation of a DOPA-Lys-DOPA-Lys-DOPA-modified TiO2 substrate in trypsin and analysis of the surface by MALDI-MS revealed intact peptide (ESI†, Fig. S7), suggesting that the adhesion of the peptide to the surface was sufficiently strong as to render the peptide backbone inaccessible to enzyme. In support of the role of DOPA in strong adhesion to the substrate, control adsorption experiments performed with Tyr-Lys-Tyr-Lys-Tyr showed that this peptide desorbed from the TiO2 surface during incubation in water (data not shown). Furthermore, a Tyr-Lys-Tyr-Lys-Tyr-(N-methoxyethyl glycine)10 polymer, analogous to PMP1 but without DOPA, adsorbed weakly to TiO2 (ESI†, Fig. S8) and exhibited poor protein and cell fouling resistance (data not shown). Collectively, the data suggest a very important role for DOPA in strong adhesion to the surface. A second contribution could be due to the inhibitory effect of the peptoid domain in preventing or slowing access of the enzyme to the surface-bound peptide. Further experiments will be necessary to fully elucidate these effects.
Resistance to protein adsorption
For the protein adsorption experiments we chose a exposure time of 20 minutes because the majority of protein adsorption to surfaces typically occurs during the first few minutes of exposure to physiological fluids and our previous study of PMP1 demonstrated that short-term protein adsorption was a good predictor of long-term fouling resistance.28 Mass-adsorption curves (data not shown) for all experiments appeared to reach saturation level within this time period. The proteins studied were lysozyme (14 kDa), fibrinogen (340 kDa), and human serum, which contains a mixture of proteins, the largest percentage being albumin (67 kDa).
Protein adsorption was determined for polymer-modified TiO2 and compared to unmodified TiO2 (Table 3). Protein-adsorption values on unmodified TiO2 waveguide sensors were comparable to published OWLS (optical waveguide lightmode spectroscopy) data for serum,20 fibrinogen,47 and lysozyme.48 While protein adsorption on all polymer-modified sensors was significantly lower (p < 0.0005) than unmodified TiO2, no statistically significant differences were observed for protein adsorption among polymer-modified surfaces with the exception of serum adsorption on PMP2-modified TiO2, which was greater than on PMP1-modified TiO2 (p < 0.02). Due to the multicomponent nature of serum it is impossible at this time to attribute the observed increase in serum protein adsorption on PMP2 to specific features of the peptoid side chain (e.g. length of side chain, hydroxy vs. methoxy terminal group), or to interactions with specific components of the protein mixture. Nevertheless, the amount of serum protein adsorption for all polymer-modified surfaces is similar to that adsorbed onto PEG coatings.17,19,20,23 Minimization of fibrinogen adsorption is important for blood-contacting applications where prevention of thrombosis is desired; a fibrinogen adsorption value of ~5 ng cm−2 has been claimed to be a value below which activation of the pathways for blood coagulation does not occur.20 The average values of fibrinogen adsorption on polymer-modified substrates approached this value, especially PMP1 (7 ± 3 ng cm−2), suggesting possible use of this polymer as a passivating coating for blood-contacting devices.
Table 3.
Average protein adsorption values with standard deviations for serum, fibrinogen, and lysozyme adsorption measured by OWLS
| Adsorbed protein mass/ng cm−2 | |||
|---|---|---|---|
| Substrate | Serum | Fibrinogen | Lysozyme |
| TiO2 | 342 ± 21 | 521 ± 61 | 209 ± 31 |
| PMP1 | 15 ± 16 | 7 ± 3 | 7 ± 5 |
| PMP2 | 83 ± 20 | 31 ± 21 | 39 ± 22 |
| PMP3 | 35 ± 25 | 18 ± 25 | 22 ± 11 |
The similarity of fibrinogen and lysozyme adsorption values on PMP1- and PMP2-modified sensors was not unexpected, as methoxyethyl- and hydroxyethyl-terminated alkanethiol SAMs exhibited nearly identical resistance to fibrinogen and lysozyme adsorption.8 Given the small size and positive charge of lysozyme, the significant decrease in lysozyme adsorption for all polymer-modified surfaces is worth noting and suggests that the adsorbed polymer density was sufficient to prevent lysozyme from penetrating through the peptoid layer and electrostatically interacting with the negatively charged oxide surface.
Resistance to mammalian cell adhesion
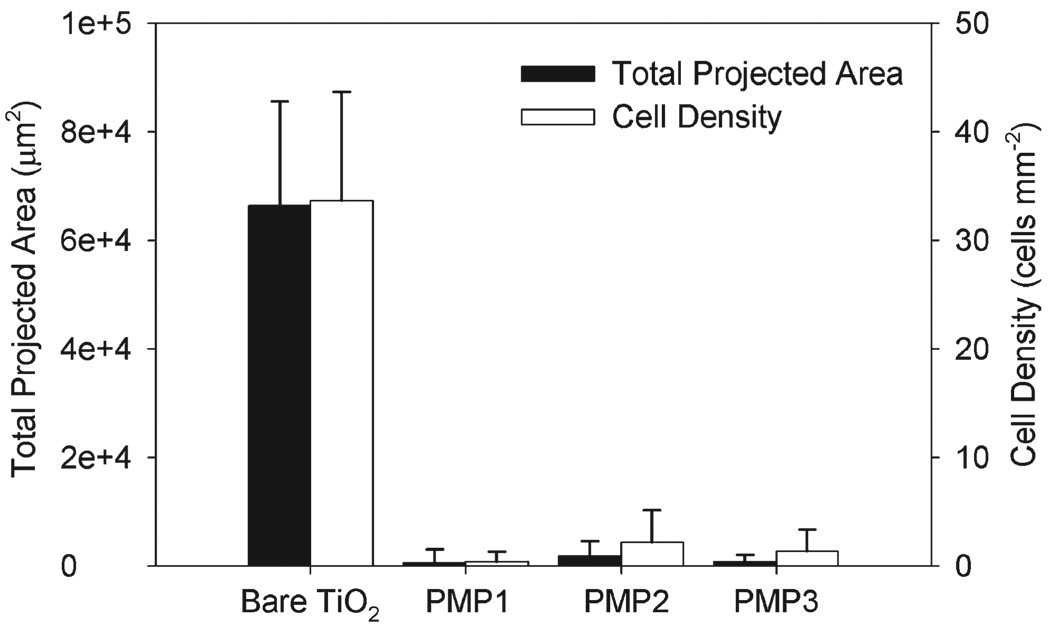
All peptidomimetic polymer-modified TiO2 substrates displayed excellent resistance to the adhesion of 3T3 fibroblast cells for a 4 h culture period (Fig. 4). Total projected area and cell density were statistically lower for all polymer-modified substrates compared to unmodified TiO2 substrates (p < 0.0005). There was no significant difference between any of the modified substrates, indicating that all polymers are equally suitable for repelling short-term cell adhesion.
Fig. 4.
Total projected area and cell density of 3T3 fibroblasts after 4 h culture. PMP1-, PMP2- and PMP3-modified TiO2 substrates all exhibited a significant reduction in adhesion and spreading compared to unmodified TiO2 substrates (p < 0.0005).
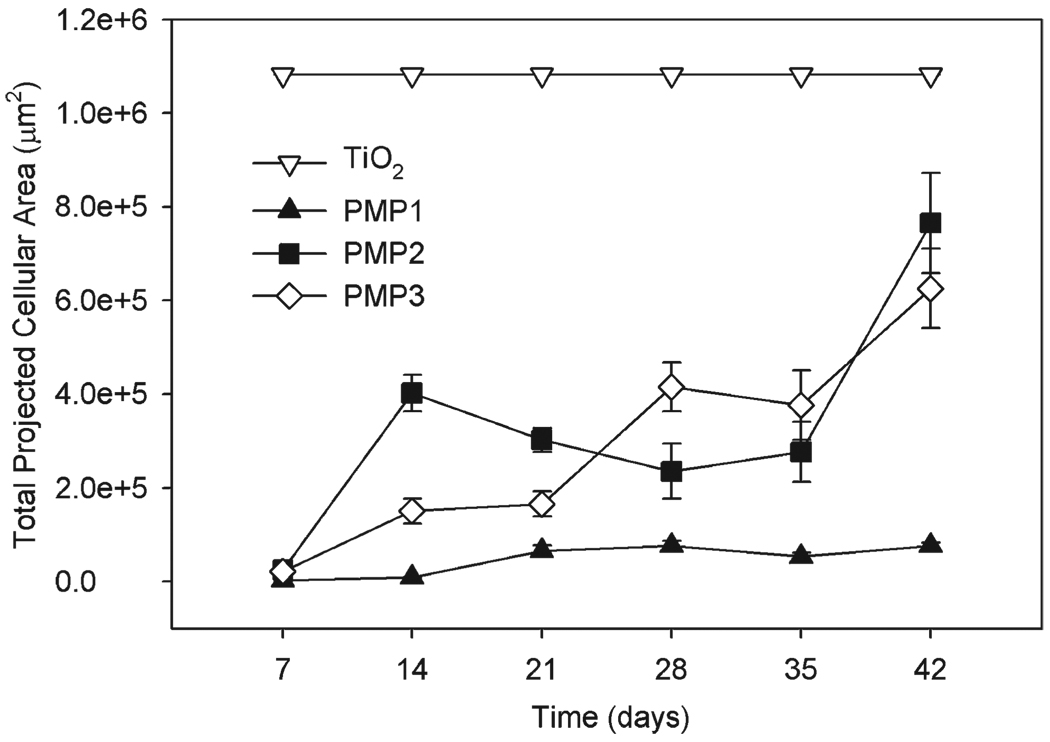
For long-term cell-attachment studies, substrates were seeded twice a week with fresh 3T3 fibroblasts suspended in serum-containing media. Cell attachment was quantified weekly for up to six weeks by live-cell staining, fluorescence microscopy and image analysis (Fig. 5). Fibroblasts formed a confluent monolayer on the unmodified TiO2 substrates within the first week, while the peptidomimetic polymer-modified surfaces all initially exhibited low levels of cell attachment in agreement with the short-term cell-attachment results. PMP1-modified substrates remained highly resistant throughout the experiment, which was expected based on our earlier 5 month study.28 Whereas cell attachment was minimal after one week on the PMP2 and PMP3 substrates, by week three some fibroblasts were attached in confluent patches as seen by visual inspection, and the number of attached cells grew consistently throughout the six-week experiment. However, these cells were apparently weakly adhered and easily removed by a gentle PBS rinse or simply by handling the substrate with tweezers for imaging. Large error bars for PMP2 and PMP3 reflect the spontaneous removal of some patches of cells and not others during handling. Although fibroblast adhesion to PMP2 and PMP3 substrates was clearly weaker than the adhesion to TiO2 control substrates, there was nevertheless stronger interaction between cells and PMP2 and PMP3 compared to PMP1.
Fig. 5.
Total projected area of 3T3 fibroblasts during long-term cell culture on unmodified TiO2, PMP1-, PMP2-, and PMP3-modified TiO2 substrates.
Long-term performance of PMP1 compares favorably to other antifouling systems in which multi-week experiments have been conducted. For example, surface-grafted PEG showed reduced cell-fouling resistance after two weeks in culture and loss of the PEG layer after 25 days,49 and OEG-terminated SAMs succumbed to cell fouling after seven days in serum-containing media.16 Surfaces modified with short- and long-chain PEGs were resistant to fouling by HUVEC cells for greater than 1 week in culture because of the high density of PEG chains.50A report of long-term antifouling performance of polymer brushes derived from surface-initiated polymerization of PEG-based monomers indicates that these systems can be cell resistant for many weeks,21 however another report showed that grafted PEG-based polymer brushes lose their efficacy after several weeks.51
The lack of major differences in protein adsorption on the three polymers is interesting in view of the superior long-term cell resistance of PMP1-modified substrates compared to PMP2- and PMP3-modified substrates. Given that the thickness of the grafted polymers and their resistance to protease degradation were similar, and the anchoring peptide was identical in each case, it is possible that the observed behavior can be attributed to differences in peptoid side-chain chemistry. For example, PMP2 and PMP3 have hydroxyl functional groups whereas PMP1 does not. Based on their studies of SAMs, Ostuni et al. made the observation that the presence of hydrogen-bond donors (e.g. hydroxyl and amine groups) generally increases protein adsorption8. However, it is unclear whether such trends would extend to the present system of grafted peptidomimetic polymers.
It may also be the case that short-term protein-adsorption experiments, which are often conducted for tens of minutes or hours at most, are not necessarily the most reliable predictors of biofouling events that play out over days, weeks or months. In fact, recent theoretical calculations by Szleifer and coworkers predicted that protein adsorption to some surfaces reaches maximum only at much longer time scales.52
Resistance to bacterial cell attachment
Bacterial colonization of medical device surfaces can lead to complications for patients and increased healthcare costs. There is evidence to suggest that adhesion and subsequent growth of bacteria on a device surface is preceded by protein adsorption,53 implying that surfaces resistant to the adhesion of proteins should exhibit bacterial-resistance properties as well. Other researchers have investigated bacterial attachment to low protein-fouling polymers, including polyamine thin films covalently attached to SAMs,53 PEG immobilized onto poly(ethylene terephthalate) (PET) surfaces,54 and PEO brushes covalently attached to glass surfaces.55
Bacterial adhesion to unmodified and polymer-modified TiO2 substrates was tested using a modified Robbins device, which is a parallel plate-flow chamber with multiple sample ports that is connected to a chemostat for continuous circulation of bacteria-containing media over the sample surfaces. This approach is favored over static experiments for relevance to bacterial attachment under conditions of fluid flow such as might exist at the surface of a catheter, cardiovascular stent or other device designed for transport of fluids, and has the additional advantage of allowing long-term attachment studies under conditions of approximately constant cell count over many hours.56
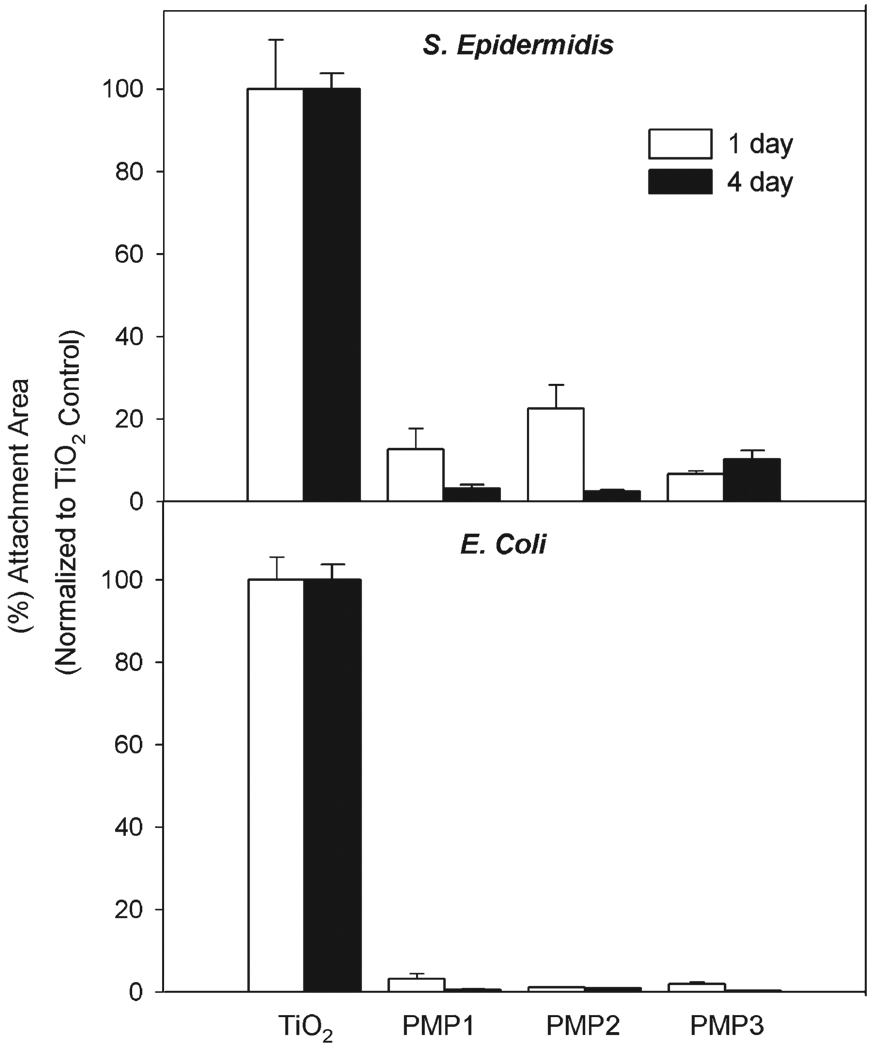
Peptidomimetic polymer-modified substrates exposed to a flowing solution of S. epidermidis for either 1 day or 4 days showed significant reduction in bacterial adhesion compared to unmodified TiO2 substrates (Fig. 6). Similar experiments were performed with E. coli, another common strain of bacteria found in surgically related infections;57 again polymer-modified surfaces showed a significant reduction in attachment for 1-day and 4-day time points (Fig. 6). There was no significant difference among the polymer-modified surfaces for either time period with S. epidermidis and E. coli. An apparent decrease in adhesion for the polymer-modified surfaces from 1-day to 4-day time points is due to normalizing of the control substrates, which had a large increase in adhesion between the two time points. Unfortunately it was not possible to conduct bacterial-adsorption experiments for longer than a few days due to the limited number of sample ports in our device and also due to instabilities of the bacterial-chemostat system, which is prone to clogging with biofilm material and contamination during sample exchange.58 Nevertheless, the low in vitro adhesion of both gram-positive and gram-negative bacteria over several days suggests that peptidomimetic polymer-modified surfaces may be capable of reducing biofilm formation over longer times in vivo.
Fig. 6.
S. epidermidis and E. coli adhesion to unmodified and polypeptoid-modified TiO2 substrates after 1 day (white bars) and 4 day (black bars) exposure.
Conclusions
Three peptidomimetic polymers, composed of a robust biomimetic peptide anchor coupled to N-substituted glycine peptoid oligomers, were determined to have significant protein, cell and bacterial antifouling properties when immobilized onto TiO2 substrates. The antifouling peptoid portions of the molecules were found to be resistant to enzymatic protease degradation. While the peptide anchor was susceptible to enzyme cleavage in solution, upon adsorption onto a surface the peptide remained intact presumably due to strong adsorption or inaccessibility of the enzyme to the peptide backbone. PMP1-modified substrates appeared to offer superior long-term cell-fouling resistance compared to PMP2- and PMP3-modified substrates, which possibly can be attributed to the differences in side-chain chemistry; further studies are necessary to determine the exact cause of the observed reduction in antifouling performance. Overall, the demonstrated resistance to the adsorption of several proteins, reduced attachment of fibroblast cells and decreased adhesion of E. coli and S. epidermidis are encouraging results for possible clinical applications of these peptidomimetic polymers.
Supplementary Material
Acknowledgements
This research was supported by NIH grant DE14193. A.E.B. acknowledges support from NIH – National Human Genome Research Institute (grant no. R01 HG002918-01). XPS surface analysis was performed at KeckII/NUANCE at Northwestern University; NUANCE Center is supported by NSF-NSEC, NSF-MRSEC, Keck Foundation, the State of Illinois, and Northwestern University. Mass spectrometry was completed at the Northwestern University Analytical Services Laboratory, supported by the NIH and NSF (grant no. DMR-0114235). Matthew O. Honaberger, Robert J. Meagher and Simonida Grubjesic are acknowledged for their assistance with synthesis.
Footnotes
Electronic supplementary information (ESI) available: HPLC and MALDI-MS spectra, cloud point UV-vis spectrum, XPS C1s spectra, and enzyme degradation spectra. See DOI: 10.1039/b711944e
References
- 1.Wisniewski N, Reichert M. Colloids Surf., B: Biointerfaces. 2000;18:197–219. doi: 10.1016/s0927-7765(99)00148-4. [DOI] [PubMed] [Google Scholar]
- 2.Brash JL. J. Biomater. Sci., Polym. Ed. 2000;11:1135–1146. doi: 10.1163/156856200744237. [DOI] [PubMed] [Google Scholar]
- 3.Horbett TA. Cardiovasc. Pathol. 1993;2:137S–148S. [Google Scholar]
- 4.Ratner BD. J. Biomed. Mater. Res. 1993;27:283–288. doi: 10.1002/jbm.820270302. [DOI] [PubMed] [Google Scholar]
- 5.Bryers JD. Colloids Surf., B: Biointerfaces. 1994;2:9–23. [Google Scholar]
- 6.Geesey GG, Bryers JD. In: Biofilms II. Bryersinc JD, editor. New York: Wiley-Liss, Inc.; 2000. pp. 237–279. [Google Scholar]
- 7.Bruinsma GM, van der Mei HC, Busscher HJ. Biomaterials. 2001;22:3217–3224. doi: 10.1016/s0142-9612(01)00159-4. [DOI] [PubMed] [Google Scholar]
- 8.Ostuni E, Chapman RG, Holmlin RE, Takayama S, Whitesides GM. Langmuir. 2001;17:5605–5620. doi: 10.1021/la0015258. [DOI] [PubMed] [Google Scholar]
- 9.Morra M. J. Biomater. Sci., Polym. Ed. 2000;11:547–569. doi: 10.1163/156856200743869. [DOI] [PubMed] [Google Scholar]
- 10.Nath N, Hyun J, Ma H, Chilkoti A. Surf. Sci. 2004;570:98–110. [Google Scholar]
- 11.Nuzzo RG. Nat. Mater. 2003;2:207–208. doi: 10.1038/nmat872. [DOI] [PubMed] [Google Scholar]
- 12.Senaratne W, Andruzzi L, Ober CK. Biomacromolecules. 2005;6:2427–2448. doi: 10.1021/bm050180a. [DOI] [PubMed] [Google Scholar]
- 13.Prime KL, Whitesides GM. Science. 1991;252:1164–1167. doi: 10.1126/science.252.5009.1164. [DOI] [PubMed] [Google Scholar]
- 14.Prime KL, Whitesides GM. J. Am. Chem. Soc. 1993;115:10714–10721. [Google Scholar]
- 15.Mrksich M, Whitesides GM. Annu. Rev. Biophys. Biomol. Struct. 1996;25:55–78. doi: 10.1146/annurev.bb.25.060196.000415. [DOI] [PubMed] [Google Scholar]
- 16.Luk Y-Y, Kato M, Mrksich M. Langmuir. 2000;16:9604–9608. [Google Scholar]
- 17.Xia N, Hu Y, Grainger DW, Castner DG. Langmuir. 2002;18:3255–3262. [Google Scholar]
- 18.Dalsin JL, Hu BH, Lee BP, Messersmith PB. J. Am. Chem. Soc. 2003;125:4253–4258. doi: 10.1021/ja0284963. [DOI] [PubMed] [Google Scholar]
- 19.Dalsin JL, Lin L, Tosatti S, Voros J, Textor M, Messersmith PB. Langmuir. 2005;21:640–646. doi: 10.1021/la048626g. [DOI] [PubMed] [Google Scholar]
- 20.Kenausis GL, Voros J, Elbert DL, Huang N, Hofer R, Ruiz-Taylor L, Textor M, Hubbell JA, Spencer ND. J. Phys. Chem. B. 2000;104:3298–3309. [Google Scholar]
- 21.Ma HW, Hyun JH, Stiller P, Chilkoti A. Adv. Mater. 2004;16:338. [Google Scholar]
- 22.Bearinger JP, Castner DG, Golledge SL, Hubchak S, Healy KE. Langmuir. 1997;13:5175–5183. [Google Scholar]
- 23.Bearinger JP, Terrettaz S, Michel R, Tirelli N, Vogel H, Textor M, Hubbell JA. Nat. Mater. 2003;2:259–264. doi: 10.1038/nmat851. [DOI] [PubMed] [Google Scholar]
- 24.Chen S, Zheng J, Li L, Jiang S. J. Am. Chem. Soc. 2005;127:14473–14478. doi: 10.1021/ja054169u. [DOI] [PubMed] [Google Scholar]
- 25.Ishihara K, Hanyuda H, Nakabayashi N. Biomaterials. 1995;16:873–879. doi: 10.1016/0142-9612(95)94150-j. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Chao T, Chen S, Jiang S. Langmuir. 2006;22:10072–10077. doi: 10.1021/la062175d. [DOI] [PubMed] [Google Scholar]
- 27.Holland NB, Qiu Y, Ruegsegger M, Marchant RE. Nature. 1998;392:799–801. doi: 10.1038/33894. [DOI] [PubMed] [Google Scholar]
- 28.Statz AR, Meagher RJ, Barron AE, Messersmith PB. J. Am. Chem. Soc. 2005;127:7972–7973. doi: 10.1021/ja0522534. [DOI] [PubMed] [Google Scholar]
- 29.Waite JH, Tanzer ML. Science. 1981;212:1038–1040. doi: 10.1126/science.212.4498.1038. [DOI] [PubMed] [Google Scholar]
- 30.Waite JH, Qin X. Biochemistry. 2001;40:2887–2893. doi: 10.1021/bi002718x. [DOI] [PubMed] [Google Scholar]
- 31.Miller SM, Simon RJ, Ng S, Zuckermann RN, Kerr JM, Moos WH. Drug Dev. Res. 1995;35:20–32. [Google Scholar]
- 32.Patch JA, Barron AE. Curr. Opin. Chem. Biol. 2002;6:872–877. doi: 10.1016/s1367-5931(02)00385-x. [DOI] [PubMed] [Google Scholar]
- 33.Zuckermann RN, Martin EJ, Spellmeyer DC, Stauber GB, Shoemaker KR, Kerr JM, Figliozzi GM, Goff DA, Siani MA, Simon RJ. J. Med. Chem. 1994;37:2678–2685. doi: 10.1021/jm00043a007. [DOI] [PubMed] [Google Scholar]
- 34.Zuckermann RN, Kerr JM, Kent SBH, Moos WH. J. Am. Chem. Soc. 1992;114:10646–10647. [Google Scholar]
- 35.Hilfiker JN, Synowicki RA. Solid State Technol. 1998;41:101. [Google Scholar]
- 36.Scofield JH. J. Electron Spectrosc. Relat. Phenom. 1976;8:129–137. [Google Scholar]
- 37.Kurrat R, Walivaara B, Marti A, Textor M, Tengvall P, Ramsden JJ, Spencer ND. Colloids Surf., B: Biointerfaces. 1998;11:187–201. [Google Scholar]
- 38.Voros J, Ramsden JJ, Csucs G, Szendro I, De Paul SM, Textor M, Spencer ND. Biomaterials. 2002;23:3699–3710. doi: 10.1016/s0142-9612(02)00103-5. [DOI] [PubMed] [Google Scholar]
- 39.de Feijter JA, Benjamins J, Veer FA. Biopolymers. 1978;17:1759–1772. [Google Scholar]
- 40.Pasche S, De Paul SM, Voros J, Spencer ND, Textor M. Langmuir. 2003;19:9216–9225. [Google Scholar]
- 41.Scales CW, Vasilieva YA, Convertine AJ, Lowe AB, McCormick CL. Biomacromolecules. 2005;6:1846–1850. doi: 10.1021/bm0503017. [DOI] [PubMed] [Google Scholar]
- 42.Brunette DM, Tengvall P, Textor M, Thomsen P. Titanium in Medicine. Berlin: Springer-Verlag; 2001. [Google Scholar]
- 43.Lee H, Scherer NF, Messersmith PB. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12999–13003. doi: 10.1073/pnas.0605552103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karlström G, Engkvist O. In: Poly(ethylene glycol): chemistry and biological applications. Harris JM, Zalipsky S, editors. Vol. 680. Washington, DC: American Chemical Society; 1997. pp. 16–30. [Google Scholar]
- 45.Kingshott P, Thissen H, Griesser HJ. Biomaterials. 2002;23:2043–2056. doi: 10.1016/s0142-9612(01)00334-9. [DOI] [PubMed] [Google Scholar]
- 46.Lee S-W, Laibinis PE. Biomaterials. 1998;19:1669–1675. doi: 10.1016/s0142-9612(98)00044-1. [DOI] [PubMed] [Google Scholar]
- 47.Hook F, Voros J, Rodahl M, Kurrat R, Boni P, Ramsden JJ, Textor M, Spencer ND, Tengvall P, Gold J, Kasemo B. Colloids Surf., B: Biointerfaces. 2002;24:155–170. [Google Scholar]
- 48.Pasche S, Voros J, Griesser HJ, Spencer ND, Textor M. J. Phys. Chem. B. 2005;109:17545–17552. doi: 10.1021/jp050431+. [DOI] [PubMed] [Google Scholar]
- 49.Branch DW, Wheeler BC, Brewer GJ, Leckband DE. Biomaterials. 2001;22:1035–1047. doi: 10.1016/s0142-9612(00)00343-4. [DOI] [PubMed] [Google Scholar]
- 50.Satomi T, Nagasaki Y, Kobayashi H, Otsuka H, Kataoka K. Langmuir. 2007;23:6698–6703. doi: 10.1021/la0624384. [DOI] [PubMed] [Google Scholar]
- 51.Fan X, Lin L, Messersmith PB. Biomacromolecules. 2006;7:2443–2448. doi: 10.1021/bm060276k. [DOI] [PubMed] [Google Scholar]
- 52.Fang F, Satulovsky J, Szleifer I. Biophys. J. 2005;89:1516–1533. doi: 10.1529/biophysj.104.055079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chapman RG, Ostuni E, Liang MN, Meluleni G, Kim E, Yan L, Pier G, Warren HS, Whitesides GM. Langmuir. 2001;17:1225–1233. [Google Scholar]
- 54.Kingshott P, Wei J, Bagge-Ravn D, Gadegaard N, Gram L. Langmuir. 2003;19:6912–6921. [Google Scholar]
- 55.Roosjen A, Kaper HJ, van der Mei HC, Norde W, Busscher HJ. Microbiology. 2003;149:3239–3246. doi: 10.1099/mic.0.26519-0. [DOI] [PubMed] [Google Scholar]
- 56.Lappin-Scott HM, Jass J, Costerton JW. In: Microbial Biofilms: formation & control. Denyer SP, Gorman SP, Sussman M, editors. Oxford: Blackwell Scientific Publications; 1993. pp. 1–12. [Google Scholar]
- 57.Jose B, Antoci V, Jr, Zeiger AR, Wickstrom E, Hickok NJ. Chem. Biol. 2005;12:1041–1048. doi: 10.1016/j.chembiol.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 58.Millar MR, Linton CJ, Sherriff A. Methods Enzymol. 2001;337:43–62. doi: 10.1016/s0076-6879(01)37005-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.