Abstract
Repetitive DNA is often packaged into heterochromatin structures that prevent illicit recombination events that cause genomic instability. A recent study by Chiolo et al. (2011) published in Cell finds that DNA double strand breaks formed within heterochromatin are shuttled to adjacent sites that are “safe” to complete repair by recombination.
Defined by early cytological studies as the portion of the eukaryotic genome that remains visibly condensed throughout the cell cycle, heterochromatin is typically associated with centromeres and telomeres, domains that contain a high percentage of repetitive DNA. One key function for heterochromatin is to repress aberrant recombination between densely packed arrays of DNA repeats, thereby preserving genome stability (Peng and Karpen, 2008). Indeed, disruption of heterochromatin increases the occurrence of spontaneous DNA double strand breaks (DSBs) in these regions, leads to the expansion and contraction of DNA repeat arrays, and can cause chromosomal translocations and other types of genomic rearrangements (Peng and Karpen, 2009).
Seemingly in contradiction to this repressive role for heterochromatin, significant mitotic recombination is detected within heterochromatic loci. It has even been suggested that these domains may be recombination hot spots (Jaco et al., 2008). Thus, mechanisms must exist that regulate recombination within such repetitive regions of the genome to ensure that deleterious genomic rearrangement events do not occur. In a recent issue of Cell, Chiolo and colleagues (2011) make the remarkable discovery that although radiation-induced DSBs formed within heterochromatin are initially processed for homologous recombination (HR), further repair is blocked until the recombination intermediates are mobilized to loci adjacent to the heterochromatin domain. HR is then completed at these peripheral locations without the complications associated with a high density of repetitive DNA sequences.
Recombination events are initiated by formation of a DSB, which can be caused by DNA damaging agents or replication fork collapse (for review see Ciccia and Elledge, 2010). The early response to DSB formation includes phosphorylation of the histone variant H2AX (H2Av in Drosophila) within a large domain of chromatin that surrounds each DSB (often visualized by indirect immunofluorescence as foci of H2AX-phos, also called γ-H2AX). Phosphorylation of H2AX requires the ATM or ATR checkpoint kinases. This histone mark provides interaction surfaces for other checkpoint and repair factors. If the DSB is to be repaired by HR, the DNA ends are processed to generate long single strand DNA (ssDNA) tails required to recruit the ATRIP complex for triggering the DNA damage checkpoint. Subsequently, the Rad51 recombinase is loaded onto the processed DSB, forming a Rad51-DNA filament that searches for a homologous DNA duplex and catalyzes strand invasion, a precursor to the final steps of HR. Although the Rad51 filament typically locates the homologous sister chromatid, the high density of DNA repeats within heterochromatin creates numerous opportunities for inappropriate invasion events that compromise genome stability.
Surprisingly, previous studies have shown that very few foci of γ-H2AX occur within heterochromatin when DSBs are induced with ionizing radiation (IR) (Cowell et al., 2007), suggesting that heterochromatin structures may prevent DSB formation – certainly a simple way to prevent extensive recombination. Chiolo et al. (2011) revisited this phenomenon in their study and monitored DSB and γ-H2Av formation in cultured Drosophila cells exposed to ionizing radiation. While they do not detect DSBs and γ-H2Av foci within heterochromatin domains at 60 minutes after IR treatment, consistent with previous studies, they do detect DSBs and γ-H2Av foci at earlier timepoints (•10 min 10 min) at levels equal to that of non-heterochromatic sites. Furthermore, heterochromatic DSBs form ATRIP foci at these early timepoints, implying successful DSB processing and checkpoint activation. However, Rad51 foci are rarely observed. Thus, heterochromatin appears to block assembly of the complete HR machinery at a DSB. Why do DSBs seem to disappear at late timepoints? Live cell imaging demonstrated that the DSBs do not actually disappear, but instead they move from within heterochromatin to adjacent regions. Strikingly, the authors find that DSB re-localization also correlates with the appearance of Rad51 foci at these peripheral sites, indicating that the heterochromatin block has been alleviated and that assembly of the HR machinery has resumed.
What induces the movement of DSBs from inside to outside of a heterochromatin domain? The authors find that treatment of cells with the ATR kinase inhibitor, caffeine, partially blocks DSB movements, as does RNAi-mediated depletion of ATR. Likewise, depletion of DSB processing enzymes eliminates DSB mobilization. Interestingly, ATR and a processed DSB are also required for a rapid, IR-dependent expansion of the heterochromatin domain and the appearance of dynamic protrusions. Heterochromatin expansion occurs with timing similar to DSB movement and re-localized DSBs, visualized by Rad51 foci, appear at the tips of heterochromatic protrusions. Thus, a processed DSB signals a change in the organization of the heterochromatin domain that correlates with DSB re-localization to peripheral locations. As many chromatin remodeling enzymes are known to be recruited to DSBs, one compelling possibility is that one or more of these enzymes displace heterochromatin factors (Sinha et al., 2009), altering the global structure of the domain and enhancing the nuclear mobility of chromatin that contains a DSB.
The dynamic movements of heterochromatin DSBs share striking similarities to the behavior of DSBs induced within the repetitive rDNA locus of budding yeast (Torres-Rosell et al., 2007). This cluster of 100-200 rDNA copies is not found within a typical heterochromatic domain, but is compartmentalized in the nucleolus. Similar to the situation with DSBs in Drosophila heterochromatin, a DSB within the rDNA cluster is formed and processed within the nucleolus before being re-localized to the exterior of this compartment where recombinational repair is completed. In budding yeast, the Smc5/6 SUMO ligase complex is required to block recombinational repair within the nucleolus, and loss of this complex leads to persistent DSBs within the nucleolus as well as aberrant recombination events. Strikingly, Chiolo and colleagues find that the Drosophila homologs of Smc5/6 are abundant components of heterochromatin, and that depletion of Smc5/6 prevents re-localization of IR-induced, heterochromatic DSBs. These aberrant, heterochromatic DSBs also recruit Rad51, indicating that Smc5/6 not only blocks the re-localization of DSBs but also recombinational repair. Inhibition of ectopic recombination appears to be a major function for Smc5/6, as depletion of this complex in cells not treated with IR leads to aberrant recombination products within heterochromatin that produce heterochromatic DNA bridges after mitosis.
How does Smc5/6 prevent Rad51 foci formation and how does this complex control DSB movement? It does not appear that Smc5/6 functions by contributing to some novel heterochromatin structure, as depletion of Smc5/6 has no impact on the size of heterochromatin domains nor does it impact recruitment of the key heterochromatin protein, HP1α. A more likely model evokes the enzymatic activity of Smc5/6 as a SUMO ligase (Figure 1). Smc5/6 could catalyze sumoylation of one or more components of the recombination machinery and block further assembly of the HR machinery. How might sumolyation also induce DSB re-localization? Again, a parallel story in budding yeast seems highly relevant. In budding yeast, unrepairable DSBs are re-localized from the nuclear interior to peripheral sites close to nuclear pores (Nagai et al., 2008; Oza et al., 2009). In this process, it appears that the DSB becomes trapped at the nuclear periphery through association between several proteins bound to the processed DSB, including the telomerase complex, and a resident nuclear envelope protein. By analogy, the mobilized, heterochromatic DSB could be bound by a euchromatic SUMO-binding protein, trapping the DSB outside the heterochromatic domain. Completion of HR may be triggered by subsequent removal of the SUMO group. Alternatively, the assembly of Rad51 foci that follows loss of the SUMO group may be the event that traps the DSB outside heterochromatin (Figure 1). This latter version of the trapping model is consistent with the observation that depletion of Rad51 blocks the re-localization of DSBs, even though Rad51 foci are not observed within heterochromatin (Chiolo et al., 2011). In either case, DSB trapping outside the heterochromatin domain would allow HR to be completed in an environment that lacks a high density of repetitive elements.
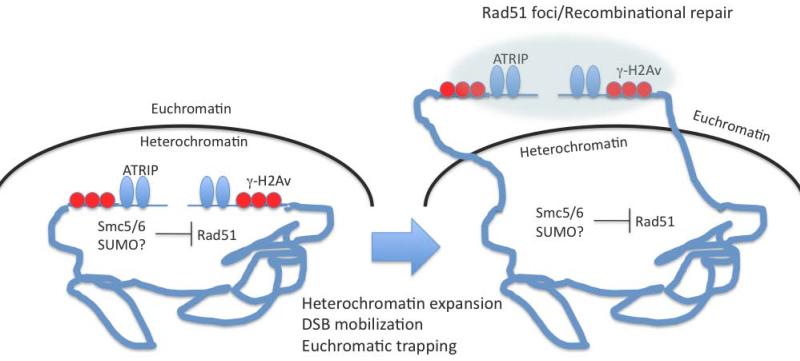
Figure 1. Model for re-localization of DSBs induced in a heterochromatic domain.
Initial DSB recognition and processing occurs within heterochromatin (left). The Smc5/6 SUMO ligase complex could then inhibit recruitment of the Rad51 recombinase. This allows DSB processing to induce heterochromatin expansion and DSB mobilization to a euchromatic site where the DSB is trapped outside the heterochromatin domain for completion of repair (right).
Of course, all of these models remain highly speculative with many features yet untested. For instance, what actually causes IR-induced heterochromatin expansion? Is the SUMO ligase activity of Smc5/6 required for heterochromatin function? Does DSB re-localization involve “trapping” or does some other mechanism operate? How does the repaired chromatin region become reunited with its heterochromatin domain? Luckily, the experimental tools appear to be in place to resolve each of these issues, hopefully in the near future.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Cell. 2011;144 doi: 10.1016/j.cell.2011.02.012. xxx-xxx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell IG, Sunter NJ, Singh PB, Austin CA, Durkacz BW, Tilby MJ. PLoS ONE. 2007;2:e1057. doi: 10.1371/journal.pone.0001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaco I, Canela A, Vera E, Blasco MA. J Cell Biol. 2008;181:885–892. doi: 10.1083/jcb.200803042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. Genes Dev. 2009;23:912–927. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. Curr Opin Genet Dev. 2008;18:204–211. doi: 10.1016/j.gde.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. PLoS Genet. 2009;5:e1000435. doi: 10.1371/journal.pgen.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha M, Watanabe S, Johnson A, Moazed D, Peterson CL. Cell. 2009;138:1109–1121. doi: 10.1016/j.cell.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragon L, Lisby M. Nat Cell Biol. 2007;9:923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]