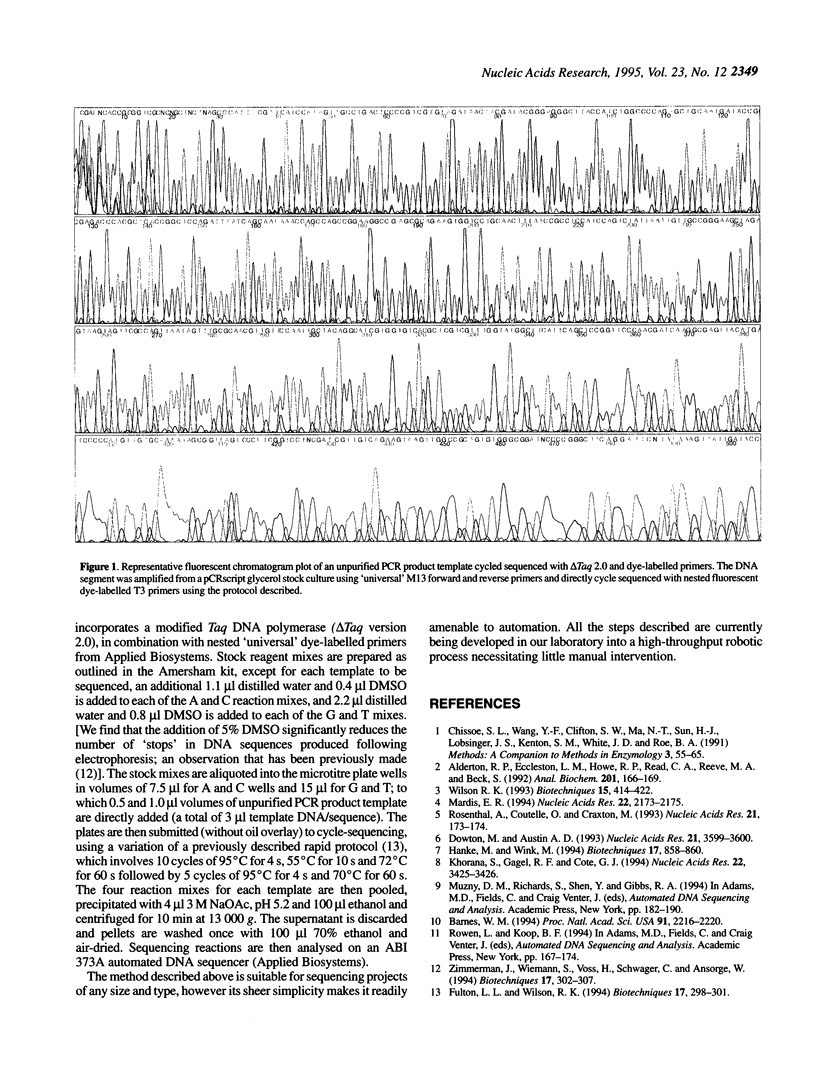
Abstract
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alderton R. P., Eccleston L. M., Howe R. P., Read C. A., Reeve M. A., Beck S. Magnetic bead purification of M13 DNA sequencing templates. Anal Biochem. 1992 Feb 14;201(1):166–169. doi: 10.1016/0003-2697(92)90190-i. [DOI] [PubMed] [Google Scholar]
- Barnes W. M. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc Natl Acad Sci U S A. 1994 Mar 15;91(6):2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowton M., Austin A. D. Direct sequencing of double-stranded PCR products without intermediate fragment purification; digestion with mung bean nuclease. Nucleic Acids Res. 1993 Jul 25;21(15):3599–3600. doi: 10.1093/nar/21.15.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton L. L., Wilson R. K. Variations on cycle sequencing. Biotechniques. 1994 Aug;17(2):298–301. [PubMed] [Google Scholar]
- Hanke M., Wink M. Direct DNA sequencing of PCR-amplified vector inserts following enzymatic degradation of primer and dNTPs. Biotechniques. 1994 Nov;17(5):858–860. [PubMed] [Google Scholar]
- Khorana S., Gagel R. F., Cote G. J. Direct sequencing of PCR products in agarose gel slices. Nucleic Acids Res. 1994 Aug 25;22(16):3425–3426. doi: 10.1093/nar/22.16.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis E. R. High-throughput detergent extraction of M13 subclones for fluorescent DNA sequencing. Nucleic Acids Res. 1994 Jun 11;22(11):2173–2175. doi: 10.1093/nar/22.11.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal A., Coutelle O., Craxton M. Large-scale production of DNA sequencing templates by microtitre format PCR. Nucleic Acids Res. 1993 Jan 11;21(1):173–174. doi: 10.1093/nar/21.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. K. High-throughput purification of M13 templates for DNA sequencing. Biotechniques. 1993 Sep;15(3):414-6, 418-20, 422. [PubMed] [Google Scholar]
- Zimmermann J., Wiemann S., Voss H., Schwager C., Ansorge W. Improved fluorescent cycle sequencing protocol allows reading nearly 1000 bases. Biotechniques. 1994 Aug;17(2):302–passim. [PubMed] [Google Scholar]