Abstract
Pyrrolysine, the 22nd amino acid to be found in the natural genetic code1–4, is necessary for all known pathways of methane formation from methylamines5,6. The residue is comprised of a methylated pyrroline carboxylate in amide linkage to the ε-amino group of L-lysine2,7,8. The three different methyltransferases that initiate methanogenesis from different methylamines9–11 have genes with an in-frame amber codon12,13 translated as pyrrolysine2,7,8. E. coli transformed with pylTSBCD from methanogenic Archaea can incorporate endogenously biosynthesized pyrrolysine into protein14. The decoding of UAG as pyrrolysine requires pylT1,6 which produces tRNAPyl (also called tRNACUA), and pylS1 encoding a pyrrolysyl-tRNA synthetase4,15,16. The pylBCD genes1 are each required for tRNA-independent pyrrolysine synthesis14. Pyrrolysine has been the last remaining genetically encoded amino acid with an unknown biosynthetic pathway. Here, we provide genetic and mass spectroscopic evidence for a pylBCD-dependent pathway in which pyrrolysine arises from two lysines. We show that a new UAG encoded residue, desmethylpyrrolysine, is made from lysine and exogenous D-ornithine in a pylC, then a pylD, dependent process, but is not further converted to pyrrolysine. These results indicate that the radical S-adenosyl-methionine (SAM) protein PylB mediates a lysine mutase reaction producing 3-methylornithine, which is then ligated to a second molecule of lysine by PylC before oxidation by PylD results in pyrrolysine. The discovery of lysine as sole precursor to pyrrolysine will further inform discussions of the evolution the genetic code and amino acid biosynthetic pathways, while intermediates of the pathway may provide new avenues by which the pyl system may be exploited for production of recombinant proteins with useful modified residues.
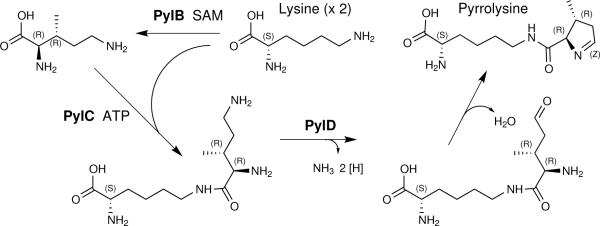
Previously hypothesized pathways of pyrrolysine biosynthesis have suggested the ligation of lysine to a ring precursor derived from other cellular metabolites such as ornithine, glutamate, proline, or isoleucine5,14,17. The results presented here instead support the pathway seen in Fig. 1. We employed E. coli transformed with Methanosarcina acetivorans pylTSBCD and Methanosarcina barkeri mtmB116 to demonstrate that lysine is the only precursor to pyrrolysine. The mtmB1 gene encodes pyrrolysyl-protein MtmB, the predominant monomethylamine methyltransferase of M. barkeri13 in which pyrrolysine is proposed to bind and activate methylamines for methyl group transfer to a cognate corrinoid protein2,5. With no known pathway of lysine degradation to central metabolites, E. coli BL21 primarily incorporates lysine into protein, or decarboxylates it to cadaverine18. This allows analysis of lysine incorporation into pyrrolysyl-peptides without extensive timecourses, as peptide ions without lysine are unlabeled.
Fig. 1.
Proposed pathway of pyrrolysine biosynthesis by the products of pylB, pylC, and pylD from two molecules of lysine. Lysine is first used to make (2R, 3R) 3-methylornithine. After ligation to a second molecule of lysine, the terminal amine of the methylornithyl-εN-lysine intermediate is oxidized to an aldehyde. Spontaneous elimination of water forms pyrrolysine. Supporting data is described throughout the text.
Chymotryptic digests of MtmB labeled using 13C615N2-lysine at 99% enrichment were analyzed by mass spectrometry (MS) using an LTQ-Orbitrap and methods for accurate peptide mass determination, as well as tandem MS for peptide sequencing and residue masses (Table 1, Supplementary Tables 1–3). The MtmB peptide 194AGRPGMGVO202GPETSL208 (O is pyrrolysine) studied previously8 was readily observed as multiple ions in unlabeled controls. In contrast, digests of labeled MtmB, contained no detectable unlabeled pyrrolysyl-peptide, but a single labeled peptide was observed by multiple ions with m/z values within 2.5 to 5.4 ppm of the theoretical value for the pyrrolysyl-peptide with a 15 Da (twelve 13C and three 15N atoms) mass shift. Tandem spectra revealed the increased mass was due to the pyrrolysyl-residue uniformly labeled with 13C and 15N.
Table 1.
Effect of labeled lysine and D-ornithine on MtmB pyrrolysyl-peptide ionsa
| K isotopeb | D-orn | Observed m/zc | UAG Residue Formula Assignedb | m/z error (ppm)d | UAG-Residue Mass (Da)e | Residue Identity (mass shift Da) |
|---|---|---|---|---|---|---|
| C6N2 | − | 783.40802+ | C12H19N3O2 | 0.77 | 237.05 | O |
| 13C6 15N2 | − | 790.92252+ | 13C12H19 15N3O2 | 2.53 | 252.05 | O (+15) |
| α–l5N | − | 792.40312+f | C12H19 15N2NO2 | 0.25 | 239.14 | O (+2) |
| ε-15N | − | 783.90842+ | C12H19 15NN2O2 | 1.27 | 238.16 | O (+1) |
| C6N2 | + | 783.40652+ | C12H19N3O2 | 3.06 | 237.15 | O |
| 776.39932+ | C11H17N3O2 | 2.19 | 223.12 | O* | ||
| 13C6 15N2 | + | 790.92242+ | 13C12H19 15N3O2 | 0.12 | 252.19 | O (+15) |
| 780.40992+ | 13C6C5H17 15N2N1O2 | 0.26 | 231.16 | O* (+8) | ||
| C6N2g | + | 776.40252+ | C11H17N3O2 | 1.93 | 223.13 | O* |
194AGRPGMGV(O or O*)202GPETSL208 from chymotryptic digestion of MtmB produced in E. coli transformed with pylTSBCD (except where indicated) and mtmB1 supplemented with the listed isotopic form of lysine in the presence and absence of D-ornithine.
Isotopic composition is most abundant natural isotope, unless otherwise indicated.
Accurate mass measured with LTQ-Orbitrap.
Difference between observed and theoretical m/z calculated with indicated elemental formula for UAG-encoded residue.
Average calculated mass (Da) using y- and b-series ions from MS/MS analysis.
Mox replaces M.
Only pylTSCD present.
As the experiment described above shows the pyrrolysyl-residue (C12H19N3O2) is made only from lysine, one amine must be eliminated from one of two lysines forming the lysyl chain and ring. To identify which, incorporation of 95%-enriched α-15N-lysine or ε-15N-lysine into the pyrrolysyl-residue was ascertained. With the former label, both the accurate peptide mass and residue mass indicated incorporation of two 15N nuclei into pyrrolysine, whereas with ε-15N-lysine the data supported incorporation of a single 15N atom into pyrrolysine (Table 1, Supplementary Tables S4 and S5). Different levels of label incorporation into the pyrrolysyl-peptide were not observed with either label in directed searches. Therefore, the ε-amine group from one lysine must be eliminated to make pyrrolysine, presumably to form the ring with its single imine bond.
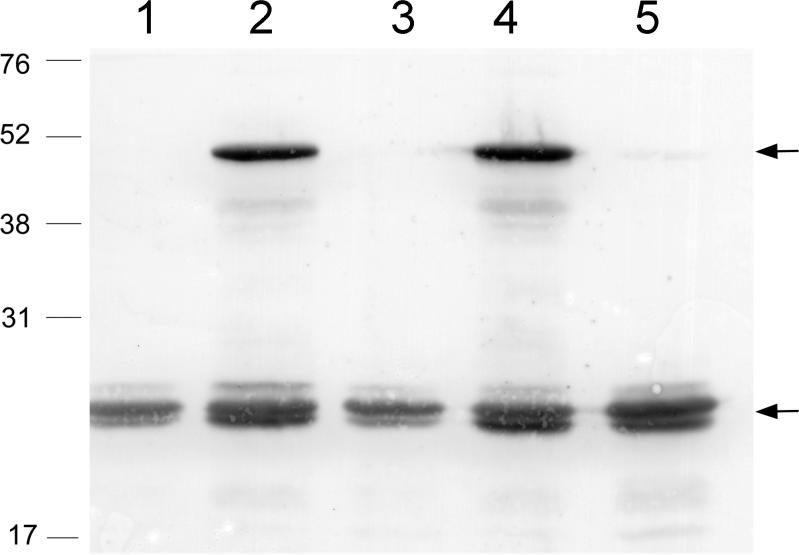
Pyrrolysine biosynthesis from lysine as sole precursor led us to examine a report that D-ornithine stimulated UAG readthrough in E. coli bearing pylTSBCD and was proposed as an intermediate to pyrrolysine19. D- but not L-ornithine (Supplementary Fig. S1) increased UAG readthrough from mtmB1 in the presence of pylTSBCD, indicating that formation of D-ornithine or a derivative might limit UAG translation, possibly via limiting UAG-encoded residue biosynthesis. Strains lacking any one of pylBCD are incapable of pyrrolysine synthesis14, but this proved untrue for D-ornithine stimulation of UAG translation. Strains that possessed only pylC and pylD were capable of significant D-ornithine dependent synthesis of full-length MtmB (Fig. 2), indicating that lack of pylB could be overcome by addition of D-ornithine for formation of the UAG-encoded residue.
Fig. 2.
D-ornithine-dependent UAG translation requires cells with pylTSCD, but not pylB. E. coli bearing the indicated plasmids was induced for expression of mtmB1 and pyl genes then analyzed by immunoblot for full-length 50 kDa mtmB1 product (upper arrow), or 23-kDa amber-terminated mtmB1 product (lower arrow). E. coli transformed with pDLBADHis and: pK parent vector pACYC-Duet1 (lane 1), pK13 bearing pylB, pylC, pylD (lane2), pK14 bearing pylB, pylD (lane 3), pK15 bearing pylC, pylD (lane 4), or pK16 pylB, pylC (lane 5) were grown in the presence of D-ornithine.
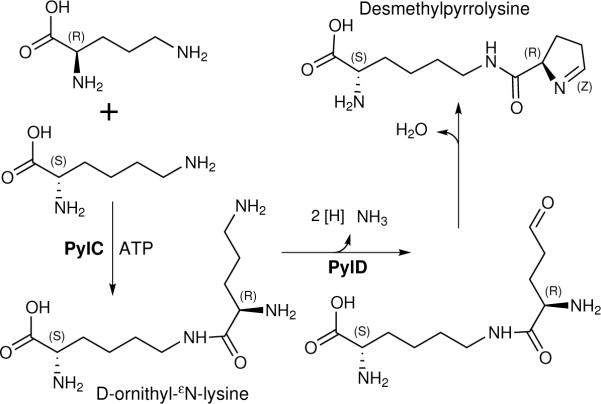
Chymotryptic digests of MtmB, made in E. coli bearing pylTSBCD and supplemented with D-ornithine, contained ions of 194AGRPGM(or Mox)GVO202GPETSL208 with O having the pyrrolysine residue mass of 237 Da (Table 1, Supplementary Tables S6, S7). However, new ions corresponding to 194AGRPGMGVX202GPETSL208 were also observed (Table 1, Supplementary Table S6 and Fig. S2), where X corresponded to a new UAG-encoded residue. The observed peptide masses averaged 14.0185 +/− 0.0027 (s.d.) Da less than the theoretical mass of corresponding pyrrolysyl-peptides, and tandem MS corroborated a UAG-encoded residue mass of 14 Da less than pyrrolysine (Table 1, Supplementary Table S6 and Fig. S2). This mass difference is equivalent to the substitution of a proton for a methyl group, indicating the residue is desmethylpyrrolsyine (Fig. 3). Using the desmethylpyrrolysyl-residue elemental formula (C11H17N3O2) yields a peptide theoretical m/z value of 776.40102+, within 1.8 to 3.7 ppm from observed m/z values. This assignment explains the significant charging of tRNAPyl with desmethylpyrrolysine in vivo, as lysylated ring compounds are a necessity for significant PylS function20–22.
Fig. 3.
Pathway of desmethylpyrrolysine biosynthesis by the products of the pylC and pylD genes from D-ornithine and L-lysine.
Ions corresponding to 194AGRPGMGVO*202GPETSL208 (where O* is desmethylpyrrolysine) were readily detected in digests of MtmB made in E. coli bearing pylTSCD (lacking pylB) and the residue mass of desmethylpyrrolysine observed upon tandem MS of this ion (Table 1, Supplementary Table S6). However, a pyrrolysyl-peptide ion could not be detected in digests of MtmB from cells lacking pylB, confirming that pylB is required for producing pyrrolysine, but not desmethylpyrrolysine.
If D-ornithine and desmethylpyrrolysine are intermediates to pyrrolysine, both desmethylpyrrolysine and pyrrolysine should incorporate D-ornithine. Unfortunately, isotopically labeled DL- or D-ornithine could not be obtained; therefore, isotopic dilution experiments were undertaken with 13C6, 15N2-lysine and D-ornithine (Table 1, Supplementary Table S8, Fig. S2). Recombinant MtmB made in the presence of heavy lysine and D-ornithine was subjected to chymotryptic digestion and MS analysis. The labeled pyrrolysyl-peptide was readily identified with a mass shift of 15 Da due to complete biosynthesis of the pyrrolysyl-residue from lysine. Ions indicating pyrrolysine was unlabeled, or labeled with less than 15 heavy nuclei, were sought, but could not be observed. The 8 Da mass shift of the desmethylpyrrolysyl-peptide ion, and of the desmethylpyrrolysyl-residue in tandem MS revealed incorporation of the equivalent of an intact lysine 13C615N2 skeleton. The observed m/z values for the peptide fits the theoretical value for this assignment with an error of less than 0.8 ppm. No desmethylpyrrolysyl-peptides with different contributions from labeled lysine could be identified. Thus, unlike pyrrolysine, desmethylpyrrolysine is synthesized with the equivalent of only one lysine, consistent with the formation of the amide linkage between lysine and the ring precursor made from D-ornithine.
As PylC is a member of the carbamoyl phosphate synthetase family of proteins forming amides between carbon dioxide (or carboxyl groups), and ammonia (or amines)1,23, we hypothesized that PylC ligates the εN of lysine to the carboxyl of D-ornithine as a first step in forming desmethylpyrrolysine (Fig. 3). To test this idea, extracts of E. coli transformed with pK18 (bearing only pylC) were examined and an m/z=261.1915 ion was detected. The corresponding mass is 2.3 ppm removed from the theoretical mass predicted for C11H24N4O3, i.e., the empirical formula of D-ornithyl-εN-lysine (Fig. S3). This signal was undetectable in pylC-containing cells not given D-ornithine, and was further dependent on the presence of pylC in the recombinant strain as shown by vector-only controls. Similar experiments with E. coli bearing only pylD yielded no evidence of D-ornithine oxidation (data not shown). However, PylD has a region highly similar to NAD binding folds of several dehydrogenases23, and reiterative PSI-BLAST using the tools at the National Center for Biotechnology Information reveal significant similarity of PylD to several amino acid dehydrogenases yielding alignments with expect values as low as 10−95. Given that pylD is required for desmethylpyrrolysine synthesis, the PylD relationship to amino acid dehydrogenases is thus consistent with function in the oxidation of the δ–amine of D-ornithyl residue of D-ornithyl-εN-lysine, leading to spontaneous dehydration to form desmethylpyrrolysine (Fig. 3).
If desmethylpyrrolysine represents an intermediate to pyrrolysine, methylation of the ring would be a final step of synthesis. As pyrrolysine can be formed entirely from lysine, this possibility would imply that lysine would first be used to make D-ornithine to form the pyrroline ring which is lysylated to form desmethylpyrrolysine, which is then methylated with a methyl group also derived from lysine. This is unprecedented and unlikely biochemistry, and no data supported D-ornithine as a precursor to pyrrolysine. Further, desmethylpyrrolysine as an intermediate would act as a competitor for pyrrolysine ligation to tRNAPyl, and a mixture of O and O* would result that is not found in native MtmB8. Desmethylpyrrolysine is thus likely to be a pyrrolysine analog made with an analog (D-ornithine) of an actual intermediate in the pathway.
A much more tenable model for lysine conversion to the methylated pyrroline ring of pyrrolysine is rearrangement of lysine to (R, R)-3-methylornithine via a lysine mutase reaction, thus generating the D-ornithine derivative that is a substrate for ligation to lysine by PylC (Fig. 1). PylB is a radical SAM protein23 and such proteins are known to function as lyases, reductases, and mutases24. As the PylB requirement for a UAG-translatable residue can be overcome with D-ornithine, PylB is proposed to function in vivo as the lysine mutase. Certain B12 dependent enzymes carry out amino acid mutase reactions with inversion of configuration of chiral centers25, and PylB as a lysine mutase could potentially could give rise to the R chiral centers of the pyrroline ring using even L-lysine as substrate in a radical mechanism dependent on SAM. The pathway explains the loss of the εN amine from one lysine during pyrrolysine biosynthesis, as the εN of one lysine would become the δ-amino group of the methyl-ornithine intermediate oxidized to form the imine bond of the ring.
Hypotheses of how the genetic code evolved are sometimes interwoven with the evolution of the biosynthetic pathways. For example, the coevolution theory considers families of amino acids arising from a common precursor as having non-randomly similar codon assignments26. The finding that pyrrolysine is entirely a derivative of lysine firmly identifies it as part of the aspartate family in Bacteria and Archaea, and its codon within those of that family of amino acids26. The identification of the precursor and intermediate analogs of the pyrrolysine and desmethylpyrrolysine pathway may have a more applied outcome. Recent years have seen the pylS and pylT genes serving to introduce useful modified residues into recombinant proteins22,27–29. Synthetic analogs of pyrrolysine intermediates, such as D-ornithine derivatives, may provide a route by which wild type or modified PylC and PylD can make endogenously biosynthesized pyrrolysine derivatives with desired properties for incorporation into tailored recombinant proteins.
Methods Summary
E. coli BL21 Tuner (DE3) (Novagen, Inc., Madison, WI) transformed with pK13, pK14, pK15, pK16, or with parent vector pACYCDuet-1 (Novagen), and pDLBADHis14 was used for production of Methanosarcina barkeri MtmB with a C-terminal hexahistidine sequence. For labeling, exponential phase cultures in defined medium with the twenty common amino acids were resuspended in fresh medium with or without 5 mM D-ornithine and with isotopic forms of lysine present prior to induction of pylTSBCD and mtmB1, and MtmB detected by immunoblotting14. Urea-solubilized MtmB was purified by affinity chromatography prior to in-gel chymotryptic digestion4,8. Capillary liquid chromatography-tandem MS employed a Thermo Finnigan LTQ orbitrap mass spectrometer. Tandem MS was acquired with a microspray source (Michrom Bioresources Inc, Auburn, CA) with a spray voltage of 2 kV and a capillary temperature of 175 °C. The scan sequence of the mass spectrometer was based on the TopTen™ method with a MS scan between 300 – 2000 Da followed by 10 consecutive MS/MS scans of the ten most abundant peaks to generate product ion spectra. The resolution of full scan on the orbitrap was set at 30,000 for high mass accuracy determinations. MS/MS scans were then performed in ion trap mode to obtain higher signal intensity for better peptide sequencing, albeit with lowered mass accuracy. The RAW data files collected on the mass spectrometer were converted to mzXML and MGF files using MassMatrix30 tools (http://www.massmatrix.net/download). Data was later searched with MassMatrix30 for peptides containing pyrrolysine or derivatives. For detection of D-orn-εN-lys, E. coli bearing pK18 (derived by EagI digestion of pK16 and religation to delete pylB) with 10 mM D-ornithine and lysine was grown to exponential phase and induced with IPTG14 prior to extraction with hot aqueous methanol at 70°C. The clarified extract was acidified before microspray ionization and introduction into the LTQ-Orbitrap.
Methods
Plasmid and strain construction
BL21 Tuner (DE3) (Novagen, Inc., Madison, WI) transformed with pK13, pK14, pK15, pK1614, or parent vector pACYCDuet-1 (Novagen Inc., Madison, WI) was also transformed with pDLBADHis for recombinant expression of MtmB with a C-terminal GGHHHHHH tag. The pDLBADHis plasmid was constructed from pDLBAD14. The M. barkeri mtmB1 gene was amplified from pDLBAD using primers MtmBNdeIF, (CATATGACATTTAGAAAATCATTTG) and MtmBCHisXhoI (CTCGAGTTATTATTATTATTAGTGGTGGTGGTGGTGGTGTCCTCCGAATACAAGTCC CAGGTCTTCGAGCTTCTTCCT). The altered mtmB1 PCR fragment and pDLBAD were digested with NdeI and XhoI so as to replace the unmodified mtmB1 gene, creating pDLBADHis. Plasmids pK17 and pK18 were respectively created from pK14 and pK16 by excision of the EagI fragment from each plasmid. Following re-ligation of the plasmid backbone, pK17 contained only pylD, and pK18 only pylC.
Production of recombinant MtmB in pyl transformed E. coli
Overnight cultures of E. coli bearing pDLBADHis and pK13 (or pK14, pK15, pK16, as appropriate) were grown in M9 minimal media supplemented with 0.2% glucose, 1 mM each of the canonical 20 amino acids, 2 mM MgSO4, 80 μM CaCl2, 36 μM FeSO4, 100 μg/ml ampicillin and 34 μg/ml chloramphenicol. These cultures were then used to inoculate 1 L of the same medium. At OD600 = 0.4–0.6, cells were pelleted then resuspended in 1 L of the same medium with 80 μM IPTG for induction of pylTSBCD. For labeling experiments, 1 mM isotopically labeled lysine (Cambridge Isotope Labs, Inc., Andover MA) substituted for unlabeled lysine. When noted, the cultures were further supplemented with 5 mM D-ornithine. After cells were grown for 1 hour at 37°C with shaking, mtmb1 expression was induced with 0.02% L-arabinose. After 2 hours MtmB1 production was assessed by immunoblotting with polyclonal anti-MtmB antibody14.
Purification of recombinant MtmB
Cells suspended in 50 mM MOPS pH 7.0 were lysed by French pressure cell, then centrifuged at 1000 ×g for 5 minutes. The supernatant was centrifuged at 25,000 ×g for 25 min. The pellet was resuspended in 3.5 M urea, 50 mM MOPS, pH 7.0, centrifuged, and the pellet was resuspended in 7 M urea, 50 mM MOPS, pH 7.0 before application to a Ni-activated HiTrap Chelating HP column (GE Healthcare BioSciences, Piscataway, NJ) equilibrated in 10 mM imidazole, 500 mM NaCl, 7 M urea in 20 mM sodium phosphate buffer, pH 7.2. MtmB then eluted with an imidazole gradient to 500 mM in the same buffer. Aliquots of fractions were assessed by immunoblotting with anti-MtmB antibody for MtmB, which eluted at approximately 220 mM imidazole.
Mass spectrometric analysis of peptides
MtmB-containing fractions were subjected to SDS-PAGE. The 50-kDa MtmB protein was excised and subjected to in-gel chymotrypsin digestion prior to analysis4,8. Capillary-liquid chromatography- tandem mass spectrometry (Capillary-LC/MS/MS) for protein/peptide identification was performed on a Thermo Finnigan LTQ orbitrap mass spectrometer operated in positive ion mode. Samples were separated on a capillary column (0.2 × 150 mm Magic C18AQ 3μ 200A, Michrom Bioresources Inc, Auburn, CA) using an UltiMate™ 3000 HPLC system (LC-Packings, Sunnyvale, CA). Each sample was injected into the trapping column (LC-Packings), and desalted with 50 mM acetic acid for 10 minutes before column injection. Mobile phase A was 0.1% formic acid in water and 0.1% formic acid in acetonitrile was used as mobile phase B. Flow rate was set at 2μl/min. Mobile phase B was increased from 2% to 15% in 30 min and again from 30–50% in 45 min. Mobile B was then increased from 50%–90% in 5 min and then kept at 90% for another 5 min before being brought back quickly to 2% in 1 min. The column was equilibrated at 2% of mobile phase B (or 98% A) for 25 min before the next sample injection. The MS/MS was acquired with a microspray source (Michrom Bioresources Inc, Auburn, CA) operated with a spray voltage of 2 kV and a capillary temperature of 175 °C. The scan sequence of the mass spectrometer was based on the data dependant TopTen™ method: the analysis was programmed for a full MS scan recorded between 300 – 2000 Da followed by 10 consecutive MS/MS scans of the ten most abundant peaks in the spectrum to generate product ion spectra for the determination of the amino acid sequence. The resolution of full scan on the orbitrap was set at 30000 to achieve high mass accuracy mass determination. MS/MS scans were then performed in ion trap mode to obtain higher signal intensity for better peptide sequencing, albeit with lowered mass accuracy. The CID fragmentation energy was set to 35%. Multiple MS/MS detection of the same peptide was excluded after detecting it three times. The RAW data files collected on the mass spectrometer were converted to mzXML and MGF files by use of MassMatrix data conversion tools (version 1.3, http://www.massmatrix.net/download). Data was later searched with MassMatrix30 for peptides containing pyrrolysine or its derivatives. The mass accuracy of the precursor ions was set to 1.2 Da to accommodate accidental selection of the natural abundance 13C ion, and the fragment mass accuracy was set to 0.8 Da. Possible hits were manually verified.
D-ornithine metabolite analysis
E. coli transformed with pK17 (bearing pylD), pK18 (bearing pylC), or the parent vector pACYC-Duet1 were grown in 500 mL Luria-Bertani medium supplemented with 10 mM lysine in the presence or absence of 10 mM D-ornithine. At OD600= 0.6, 80 μM IPTG was added to induce pylC or pylD. After 2 hours, cells were pelleted by centrifugation, washed twice in 50 mM MOPS, pH 7.2, resuspended in 3 mL ddH2O, then extracted with 9 mL of 66% methanol at 70°C for 30 minutes. The extract was then centrifuged at 16,100 ×g for 10 minutes before the supernatant was concentrated to 250 μL by vacuum centrifugation. The sample was then diluted with 50% methanol in 2% acetic acid and infused into the electrospray source set as described above. The data was recorded between 100–300 Da and the resolution of full scan was set at 30000 to achieve high mass accuracy MS determination. Data were acquired in continuum mode until well-averaged data were obtained.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health to KG-C, and grants from the Department of Energy and the National Institute of Health to JAK. The authors are grateful to David Longstaff for preparing the tagged mtmB1 version of pDLBAD.
Footnotes
Reprints and permissions information is available at www.nature.com/reprints.
Supplementary Information accompanies the paper on www.nature.com/nature.
The authors declare that they have no competing financial interests.
References
- 1.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 2.Hao B, et al. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science. 2002;296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 3.Atkins JF, Gesteland R. The 22nd amino acid. Science. 2002;296:1409–1411. doi: 10.1126/science.1073339. [DOI] [PubMed] [Google Scholar]
- 4.Blight SK, et al. Direct charging of tRNACUA with pyrrolysine in vitro and in vivo. Nature. 2004;431:333–335. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]
- 5.Krzycki JA. Function of genetically encoded pyrrolysine in corrinoid-dependent methylamine methyltransferases. Curr. Opin. Chem. Biol. 2004;8:484–491. doi: 10.1016/j.cbpa.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Mahapatra A, et al. Characterization of a Methanosarcina acetivorans mutant unable to translate UAG as pyrrolysine. Mol. Microbiol. 2006;59:56–66. doi: 10.1111/j.1365-2958.2005.04927.x. [DOI] [PubMed] [Google Scholar]
- 7.Hao B, et al. Reactivity and chemical synthesis of L-pyrrolysine— the 22nd genetically encoded amino acid. Chemistry and Biology. 2004;11:1317–1324. doi: 10.1016/j.chembiol.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 8.Soares JA, et al. The residue mass of L-pyrrolysine in three distinct methylamine methyltransferases. J. Biol. Chem. 2005;280:36962–36969. doi: 10.1074/jbc.M506402200. [DOI] [PubMed] [Google Scholar]
- 9.Burke SA, Krzycki JA. Reconstitution of monomethylamine:coenzyme M methyl transfer with a corrinoid protein and two methyltransferases purified from Methanosarcina barkeri. J. Biol. Chem. 1997;272:16570–16577. doi: 10.1074/jbc.272.26.16570. [DOI] [PubMed] [Google Scholar]
- 10.Ferguson DJ, Jr., Gorlatova N, Grahame DA, Krzycki JA. Reconstitution of dimethylamine:coenzyme M methyl transfer with a discrete corrinoid protein and two methyltransferases purified from Methanosarcina barkeri. J. Biol. Chem. 2000;275:29053–29060. doi: 10.1074/jbc.M910218199. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson DJ, Jr., Krzycki JA. Reconstitution of trimethylamine-dependent coenzyme M methylation with the trimethylamine corrinoid protein and the isozymes of methyltransferase II from Methanosarcina barkeri. J. Bacteriol. 1997;179:846–852. doi: 10.1128/jb.179.3.846-852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burke SA, Lo SL, Krzycki JA. Clustered genes encoding the methyltransferases of methanogenesis from monomethylamine. J.Bacteriol. 1998;180:3432–3440. doi: 10.1128/jb.180.13.3432-3440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paul L, Ferguson DJ, Krzycki JA. The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons. J. Bacteriol. 2000;182:2520–2529. doi: 10.1128/jb.182.9.2520-2529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Longstaff DG, et al. A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine. Proc. Natl. Acad. Sci. USA. 2007;104:1021–1026. doi: 10.1073/pnas.0610294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polycarpo C, et al. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12450–12454. doi: 10.1073/pnas.0405362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krzycki JA. The direct genetic encoding of pyrrolysine. Curr. Opin. Microbiol. 2005;8:706–712. doi: 10.1016/j.mib.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Ambrogelly A, Palioura S, Soll D. Natural expansion of the genetic code. Nat. Chem. Biol. 2007;3:29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 18.Jeong H, et al. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3) J. Mol. Biol. 2009;394:644–652. doi: 10.1016/j.jmb.2009.09.052. [DOI] [PubMed] [Google Scholar]
- 19.Namy O, et al. Adding pyrrolysine to the Escherichia coli genetic code. FEBS Lett. 2007;581:5282–5288. doi: 10.1016/j.febslet.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 20.Li WT, et al. Specificity of pyrrolysyl-tRNA synthetase for pyrrolysine and pyrrolysine analogs. J. Mol. Biol. 2009;385:1156–1164. doi: 10.1016/j.jmb.2008.11.032. [DOI] [PubMed] [Google Scholar]
- 21.Polycarpo CR, et al. Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase. FEBS Letts. 2006;580:6695–6700. doi: 10.1016/j.febslet.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yanagisawa T, et al. Multistep engineering of pyrrolysyl-tRNA synthetase to genetically encode N(epsilon)-(o-azidobenzyloxycarbonyl) lysine for site-specific protein modification. Chem. Biol. 2008;15:1187–1197. doi: 10.1016/j.chembiol.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Rother M, Krzycki JA. Selenocysteine, pyrrolysine, and the unique energy metabolism of methanogenic archaea. Archaea. 2010;2010 doi: 10.1155/2010/453642. doi:10.1155/2010/453642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frey PA, Hegeman AD, Ruzicka FJ. The radical SAM Superfamily. Crit. Rev. Biochem. Mol. Biol. 2008;43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 25.Banerjee R. Radical carbon skeleton rearrangements: catalysis by coenzyme B12-dependent mutases. Chem. Rev. 2003;103:2083–2094. doi: 10.1021/cr0204395. [DOI] [PubMed] [Google Scholar]
- 26.Wong JT. Coevolution theory of the genetic code at age thirty. Bioessays. 2005;27:416–425. doi: 10.1002/bies.20208. [DOI] [PubMed] [Google Scholar]
- 27.Fekner T, Li X, Lee MM, Chan MK. A pyrrolysine analogue for protein click chemistry. Angew. Chem. Int. Ed. Engl. 2009;48:1633–1635. doi: 10.1002/anie.200805420. [DOI] [PubMed] [Google Scholar]
- 28.Chen PR, et al. A facile system for encoding unnatural amino acids in mammalian cells. Angew. Chem. Int. Ed. Engl. 2009;48:4052–4055. doi: 10.1002/anie.200900683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding Nε-acetyllysine in recombinant proteins. Nat. Chem. Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 30.Xu H, Freitas MA. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics. 2007;8:133. doi: 10.1186/1471-2105-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.