Abstract
Purine nucleoside phosphorylase (PNP) catalyzes the reversible phosphorolysis of 6-oxypurine (2′-deoxy)ribonucleosides, generating (2-deoxy)ribose 1-phosphate and the purine base. Transition-state models for inosine cleavage have been proposed with bovine, human, and malarial PNPs using arsenate as the nucleophile, since kinetic isotope effects (KIEs) are obscured on phosphorolysis due to high commitment factors. The Phe200Gly mutant of human PNP has a low forward and reverse commitment factors in the phosphorolytic reaction, permitting the measurement of competitive intrinsic KIEs on both arsenolysis and phosphorolysis of inosine. The intrinsic 1′-14C, 1′-3H, 2′-2H, 9-15N, and 5′-3H2 KIEs for inosine were measured for arsenolysis and phosphorolysis. Except for the remote 5′-3H2, and some slight difference between the 2′-2H KIEs, all isotope effects originating in the reaction coordinate are the same within experimental error. Hence, arsenolysis and phosphorolysis proceed through closely related transition states. Although electrostatically similar, the volume of arsenate is greater than phosphate and supports a steric influence to explain the differences in the 5′-3H2 KIEs. Density functional theory calculations provide quantitative models of the transition states for Phe200Gly human PNP-catalyzed arsenolysis and phosphorolysis, selected upon matching calculated and experimental KIEs. The models confirm the striking resemblance between the transition states for the two reactions.
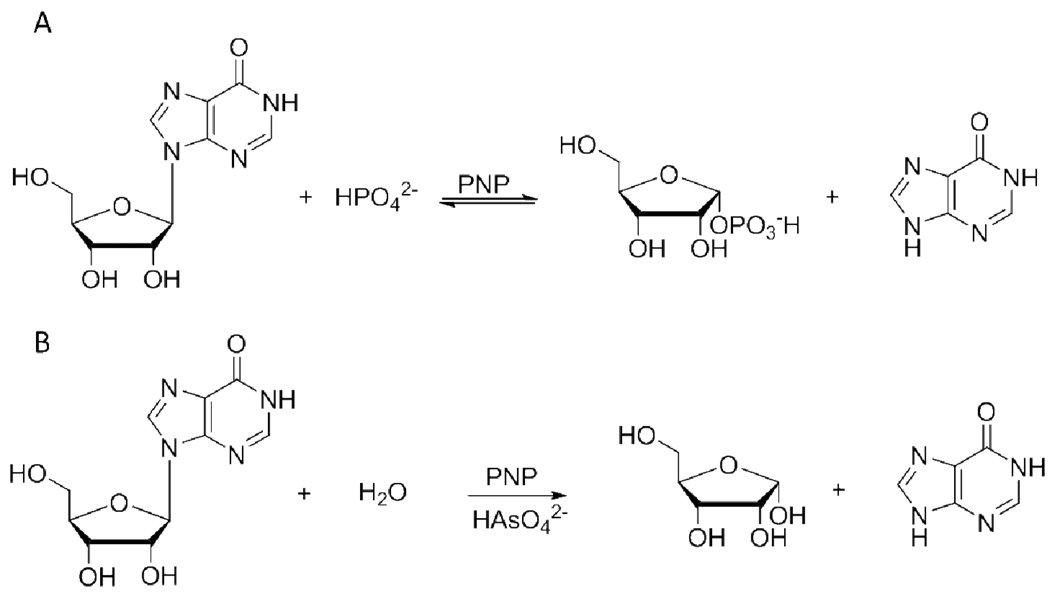
Purine nucleoside phosphorylase (PNP)1 (EC 2.4.2.1) catalyzes the reversible phosphorolysis of 6-oxopurine (2′-deoxy)ribonucleosides to generate ribose 1-phosphate (R-1-P) and the corresponding purine base, a key step of the purine salvage pathway and an essential step in formation of uric acid in humans (1). The reaction causes inversion of configuration from β-nucleosides to α-D-(2-deoxy)ribose 1-phosphate (2) (Scheme 1A). The genetic deficiency of PNP activity in humans impairs T-cell-based immunological response, while other tissues are unaltered (3). This led to the recognition of PNP as a target for inhibitor design, aiming at the treatment of autoimmune diseases, T-cell lymphomas and gout (4). Four generations of transition state analogues designed for human PNP (HsPNP) are exemplified by immucillin-H, DADMe-immucillin-H, DATMe-immucillin-H, and SerMe-immucillin-H, with overall dissociation constants (Ki*) in the picomolar range (5).
Scheme 1.
PNP-catalyzed (A) phosphorolysis and (B) arsenolysis of inosine.
Kinetic isotope effects (KIEs) arise from differences in bond vibrational modes as a molecule goes from the ground state to the transition state. Consequently, their measurement can provide information on the nature of the transition states of chemical reactions. However, enzyme-catalyzed reactions are often affected by kinetically complicating commitment factors that obscure the expression of intrinsic KIEs (6). In the case of HsPNP, KIEs measured on phosphorolysis are near unity due to forward and reverse commitment factors. To overcome commitment factors, phosphate has been replaced by arsenate in the bovine PNP (BtPNP) reaction (7) (Scheme 1B). The chemical similarity between arsenate and phosphate makes it reasonable to use the former as a mimic of the latter. Accordingly, this strategy has been employed in several biochemical systems (8, 9). The use of arsenate in the BtPNP reaction decreased the commitment factor for the nucleoside and rendered the reaction irreversible, since α-D-ribose 1-arsenate is readily hydrolyzed. Use of arsenate allowed the measurement of intrinsic KIEs and transition state analysis indicated a dissociative, ribooxocarbenium ion-like transition state with significant bond order to the leaving group and no bond order to the approaching nucleophile (7). The same approach was also employed with HsPNP and Plasmodium falciparum PNP (PfPNP). Their isotope effects reported on the formation of a ribooxocarbenium transition state with no bond order to either the leaving group or the nucleophile (10). These studies provided the foundation for design of the powerful PNP transition state analogues mentioned above (11, 12). More recently, these properties of arsenate were used to probe the effect of remote mutations on the geometry of the transition states for Lys22Glu-His104Arg HsPNP (13) and Val39Thr-Asn123Lys-Arg210Gln BtPNP (14).
In spite of the apparent success of the method, the validity of using arsenate as a mimic of phosphate in ribosyl-transfer reactions has been questioned with the suggestion that chemical differences in the two nucleophiles would lead to distinct transition states (15). Here we test this hypothesis by taking advantage of the Phe200Gly mutant of HsPNP, which possesses low commitment factors for inosine in both arsenolysis and phosphorolysis. Phenylalanine 200 is an important catalytic site amino acid for substrate binding and catalysis in HsPNP (16). The Phe200Gly mutant has ~30 fold decreased kcat and ~900 fold increased KM resulting in a 27,000-fold decrease in the specificity constant (kcat/KM) for inosine relative to the native enzyme (17). The Phe200Gly HsPNP was used in both phosphorolysis and arsenolysis reactions and transition-state models were built based on intrinsic KIE values and gas-phase density functional theory calculations using both arsenate and phosphate. The results indicate that the transition states are remarkably similar with either nucleophile. Furthermore, the structure and geometry of the transition state for Phe200Gly HsPNP differs from those found for native or other mutant HsPNPs.
EXPERIMENTAL PROCEDURES
Materials
D-[1-3H]Ribose, D-[1-14C]ribose, D-[6-3H2]glucose, D-[6-14C]glucose, and D-[5-3H]glucose were purchased from American Radiolabeled Chemicals, Inc. Pyruvate kinase (PK) myokinase (MK), hexokinase (HK), glucose-6-phosphate dehydrogenase (G6PDH), glutamic acid dehydrogenase (GDH), 6-phosphogluconic acid dehydrogenase (6PGDH), adenosine deaminase (ADA), and phosphoriboisomerase (PRI) were from Sigma. Alkaline phosphatase (AP) was from Roche. Ribokinase (RK) and phosphoribosyl-α-1-pyrophosphate synthetase (PRPPase) were prepared as previously described (18, 19). Hypoxanthine-guanine phophoribosyltransferase (HGPRT) and wild-type HsPNP were kind gifts from Keith Hazleton and Dr. Achelle Edwards of this laboratory. All other chemicals and reagents were obtained from commercially available sources and were used without further purification.
Expression, purification, and activity assay of Phe200Gly HsPNP
The N-terminal His-tagged mutant enzyme was expressed and purified to homogeneity, as judged by SDS-PAGE (20), according to previously described methods (17). Protein concentration was determined spectrophotometrically at 280 nm using the theoretical extinction coefficient (ε280) of 29,800 M−1 cm−1 (ExPASy). Phe200Gly HsPNP activity was coupled to xanthine oxidase and measured spectrophotometricaly as previously described (11, 17), at 25 °C, in 50 mM Tris-HCl (pH 7.5), with 100 nM Phe200Gly HsPNP and 60 mU bovine xanthine oxidase. Saturation curves for Phe200Gly HsPNP with inosine were obtained by measuring initial rates at varying concentration of inosine (10 – 50 mM), and fixed, saturating concentrations of either arsenate (50 mM) or phosphate (50 mM), and fitted to the Michealis-Menten equation.
Synthesis and purification of isotopically labeled inosines
[1′-3H]IMP and [1′-14C]IMP were enzymatically synthesized at 37 °C from [1-3H]ribose and [1-14C]ribose, respectively (Scheme S1, available as Supporting Information). Each reaction mixture contained 1 mM ribose, 20 mM phosphoenolpyruvate (PEP), 0.2 mM ATP, 20 mM MgCl2, 1.2 mM hypoxanthine, 100 mM KH2PO4 (pH 7.5), 50 mM KCl, 1 mM DTT, 0.2 U mL−1 of PRPPase, 2 U mL−1 of PK, 2 U mL−1 of MK, 0.2 U mL−1 of HGPRT, and 1 U mL−1 of RK.
For the synthesis of [5′-3H2]-, [4′-3H]-, and [5′-14C]IMP, D-[6-3H2]-, D-[5-3H]-, and D-[6-14C]glucose were, respectively, utilized as precursors (Scheme S1, available as Supporting Information). Each reaction mixture contained 2 mM glucose, 20 mM PEP, 0.2 mM ATP, 20 mM MgCl2, 2.5 mM hypoxanthine, 140 mM KH2PO4 (pH 7.5), 50 mM KCl, 2.5 mM DTT, 0.2 mM NADP+, 20 mM α-ketoglutarate (α-KG), 5.5 mM NH4Cl, 0.2 U mL−1 of PRPPase, 2 U mL−1 of PK, 2 U mL−1 of MK, 0.2 U mL−1 of HGPRT, 1 U mL−1 of G6PDH, 1 U mL−1 of 6PGDH, 0.5 U mL−1 of PRI, 1 U mL−1 of GDH, and 0.2 U mL−1 of HK. To synthesize [5′-14C,2′-2H]IMP, the reaction mix with D-[6-14C]glucose was prepared in D2O. The deuterium was incorporated by the reaction catalyzed by PRI, and the deuterium content in the final product was determined by electrospray ionization mass spectrometry (ESI-MS).
The labeled IMPs were purified by reverse-phase HPLC (50 mM triethylamine:acetic acid, pH 5.0) using an analytical C18 Deltapak column (Waters). The solvent was evaporated in a speedvac, and the pellet was dissolved in water. The labeled IMPs were converted to labeled inosines in the AP-catalyzed reaction, and the labeled inosines were purified with the same protocol utilized for IMP purification.
The synthesis of [5′-14C,9-15N]inosine employed [5′-14C,9-15N]ATP, synthesized as previously described (13), as initial material (Scheme S2, available as Supporting Information). A solution of 10 mM glucose, 1 mM ATP, 100 mM KH2PO4 (pH 7.5), 20 mM MgCl2, 2.5 mM DTT, 10 U mL−1 of MK, and 6 U mL−1 of HK was allowed to react overnight, after which alkaline buffer and 0.05 U mL−1 of AP were added, and the reaction proceeded for a few additional hours at 37 °C. Labeled adenosine was isolated as described above for IMP, and subsequently converted to labeled inosine by incubation with 50 mM Tris-HCl (pH 7.5) and 0.5 U mL−1 of ADA.
Determination of forward commitment factors
The forward commitments (Cf) for inosine with Phe200Gly HsPNP in arsenolysis and phosphorolysis reactions were measured by the isotope trapping method (21). Pulse incubation mixtures (20 µL) containing 200 µM Phe200Gly HsPNP, 5 mM [5′-14C]inosine, and 50 mM Tris-HCl (pH 7.5), at 25 °C, were chased with a solution (480 µL) consisting of 20 mM inosine, 50 mM Tris-HCl (pH 7.5), and either 0 to 30 mM arsenate or 0 to 10 mM phosphate. After 5 s, the reactions were quenched with 50 µL of 1 N HCl and loaded onto charcoal columns (14, 19). [5-14C]Ribose or [5-14C]ribose 1-phosphate was eluted with 3 mL of 10 mM ribose in 10% ethanol (v/v), dried by speedvac, and dissolved in 100 µL of water and 10 mL of scintillation fluid (Perkin-Elmer). Radioactivity was counted for 10 min in a Wallac 1414 LSC scintillation counter (Perkin-Elmer). The values obtained in the absence of arsenate and phosphate were used as background to correct the others. Forward commitments were calculated with eq 1, where Cf is the forward commitment, and Y is the asymptotic value of a hyperbolic fit of the data.
| eq 1 |
Measurement of KIEs
All KIEs were measured by the competitive radiolabel method (19), at 25 °C. A typical reaction mixture for arsenolysis contained 30 mM NaH2AsO4 (pH 7.5), 7 µM Phe200Gly HsPNP, 50 mM Tris-HCl (pH 7.5), and 1 mM inosine (3H-labeled, 14C-labeled, and cold carrier), while a reaction mixture for phosphorolysis consisted of 10 mM NaH2PO4 (pH 7.5), 50 µM Phe200Gly HsPNP, 50 mM Tris-HCl (pH 7.5), and 50 µM inosine (3H-labeled, 14C-labeled, and cold carrier). The arsenolysis was allowed to proceed to 20 to 25% completion and the phosphorolysis reaction to 10 to 15% completion, when half of each reaction mixture was loaded onto charcoal columns (14, 19). The remainder of each reaction mixture was completely converted to products by addition of 10 µM of native HsPNP and in the case of the phosphorolysis reaction, 5 mM of NaH2AsO4 (pH 7.5). After both reactions were complete, they were also loaded onto charcoal columns. Ribose or ribose 1-phosphate was separated from inosine and prepared for scintillation counting by the same procedures described for the commitment experiment. A sample with only [14C]inosine was also counted as a standard.
The samples were counted in dual-channel fashion, with the 3H signal appearing only in channel 1, and 14C signal, in both channels. The total 3H signal was assessed by eq 2, and the total 14C signal, by eq 3, in which 3H is the total number of counts per minute (cpm) for this isotope, 14C is the total number of cpm for this isotope, channel 1 and channel 2 are the number of cpm in each channel, and r is the channel 1 to channel 2 ratio of 14C standard (19).
| eq 2 |
| eq 3 |
Experimental KIEs were calculated and extrapolated to 0% reaction using eq 4, where Rf and R0 are the ratios of heavy to light isotopes at partial and complete conversions, respectively, and f is the fraction of substrate conversion.
| eq 4 |
Computational methods
The reaction of arsenate and phosphate with inosine in mutant PNP was studied in B3LYP using a 6-31G* basis set as implemented in Gaussian 03 and 09 (22, 23). Final transition structures without geometric constraints were optimized in B3LYP using a 6-31+G** basis set. All starting material structures were located as global minima and frequency calculations performed on the optimized structures contained no imaginary frequencies. Almost all transition structures that were located, with and without geometric constraints, had only one calculated imaginary frequency (structures with a second imaginary frequency were only used in the analysis if the second frequency was less than 10% of the first). The errors associated with isotope effect predictions for geometries with more than one imaginary frequency are discussed elsewhere (24). The isotope effects for each of the theoretical structures were calculated from conventional transition state theory by the method of Bigeleisen and Mayer using ISOEFF98 (25–27). from the scaled theoretical vibrational frequencies. Tunneling corrections were applied using a one-dimensional infinite parabolic barrier model (28).
RESULTS AND DISCUSSION
Steady-state apparent kinetic parameters
Phe200Gly HsPNP follows Michaelis-Menten kinetics for both inosine arsenolysis and phosphorolysis (data not shown). Fitting the data to the Michaelis-Menten equation yielded apparent values of kcat = 0.90 ± 0.01 s−1, Kinosine = 18 ± 2 mM, and kcat/Kinosine = 50 M−1 s−1 with phosphate, in agreement with previously reported values (17), and kcat = 0.30 ± 0.02 s−1, Kinosine = 18 ± 3 mM, and kcat/Kinosine = 16 M−1 s−1 with arsenate.
Comparison between arsenate and phosphate ions
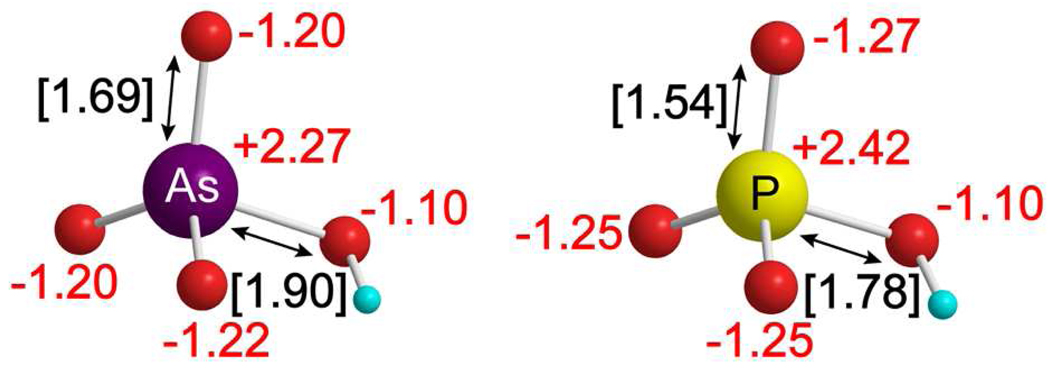
A comparison of the electronic and structural properties of phosphate and arsenate ions shows small differences in NBO charges for the nucleophilic oxygens (0.07), indicating similar electronic structure of the ions (Figure 1). The pKa values for arsenate (pKa1 = 2.21, pKa2 = 6.95, pKa3 = 11.49) and phosphate (pKa1 = 2.16, pKa2 = 7.21, pKa3 = 12.32) are also similar, supporting closely related nucleophilicity. A notable difference between the ions is the longer bond lengths of the arsenate ion (greater than phosphate by 0.15 Å for the As–O bond and by 0.12 Å for the O–H bond). One possible effect of the increased size of arsenate relative to phosphate could be a change in the conformation of the enzymatic ion-binding pocket to accommodate the larger ion. However, similarities in the electronic structure and pKa values of arsenate and phosphate support arsenate as a reasonable mimic for phosphate in the reaction of PNP.
Figure 1.
Comparison between arsenate and phosphate dianions. NBO charges for phosphate and arsenate are shown in red text. Bond distances for the P–O and As–O bonds are shown in black text with brackets. Oxygen atoms are shown in red, hydrogen in cyan, arsenic in purple, and phosphorus in yellow.
Potential energy surfaces for arsenolysis versus phosphorolysis
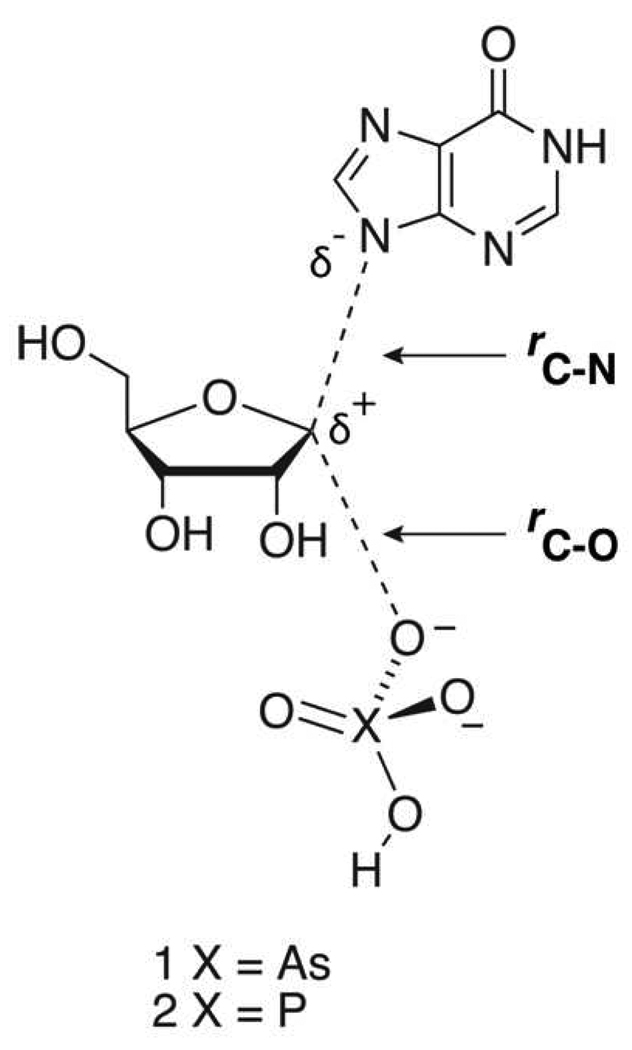
The potential energy surfaces for the reactions of arsenate and phosphate with inosine were analyzed to compare the relative landscapes for conversion of reactants to products. Scheme 2 shows general transition structures for the substitution reactions of arsenate (1) and phosphate (2) with inosine. The leaving-group bond distance between C-1′ and N-9 is defined as rC-N and the attacking nucleophile bond distance between C-1′ and the O (of the ion) is defined as rC-O. Energy grids were generated by varying rC-N and rC-O of transition structures 1 and 2 along the reaction coordinate by 0.2 Å increments. Structures for the grids were conformationally similar to the lowest energy structure of the inosine starting material and all structures were optimized using B3LYP/6-31G* calculations in the gas phase.
Scheme 2.
General transition state for arsenolysis and phosphorolysis of inosine.
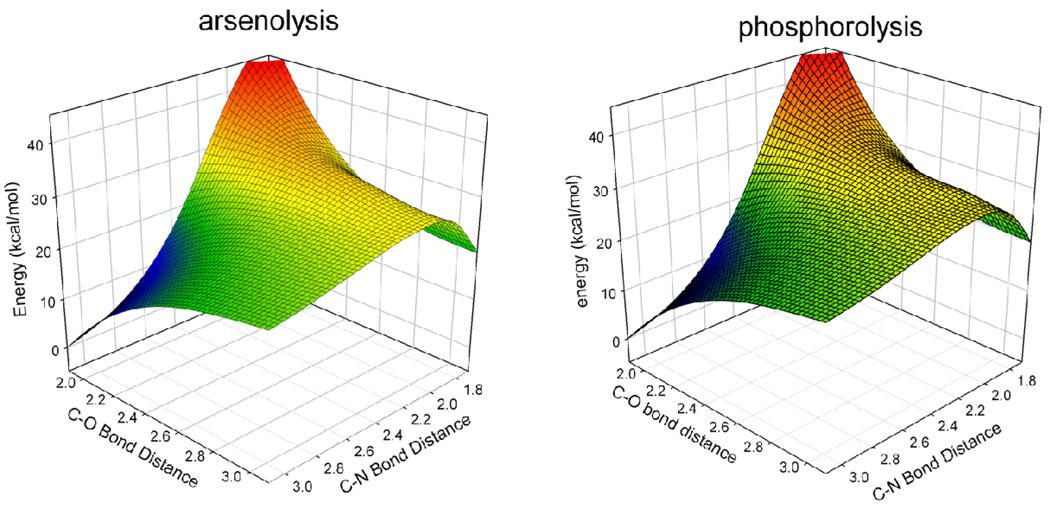
The potential energy surfaces, including zero-point energy (ZPE), for the reactions of arsenate and phosphate are virtually identical with an RMSD2 of 0.982 (Figure 2). The potential energy surfaces display a flat area in the transition state region and the range is similar for both arsenolysis and phosphorolysis reactions. In the enzymatic reaction, stabilizing interactions of the substrates with the enzyme environment are known to considerably alter the position of the transition state, causing the enzymatic transition state to lie at a non-stationary point on the gas-phase surface (29). Consequently, a transition structure located for the enzymatic reaction at a non-stationary position on the analogous surfaces involving arsenate or phosphate should be similar in both reactions unless the catalytic site environment is disturbed by arsenate interactions. The similarity between the surfaces of arsenolysis and phosphorolysis provides further evidence to support the ability of arsenate to mimic phosphate in the reaction with inosine without altering the reaction coordinate.
Figure 2.
Potential energy surfaces for the reaction of arsenate with inosine and phosphate with inosine are shown above. The similarity in the surfaces (RMSD = 0.982) supports the ability of arsenate to mimic phosphate in the PNP reaction.
Transition structures of stationary points
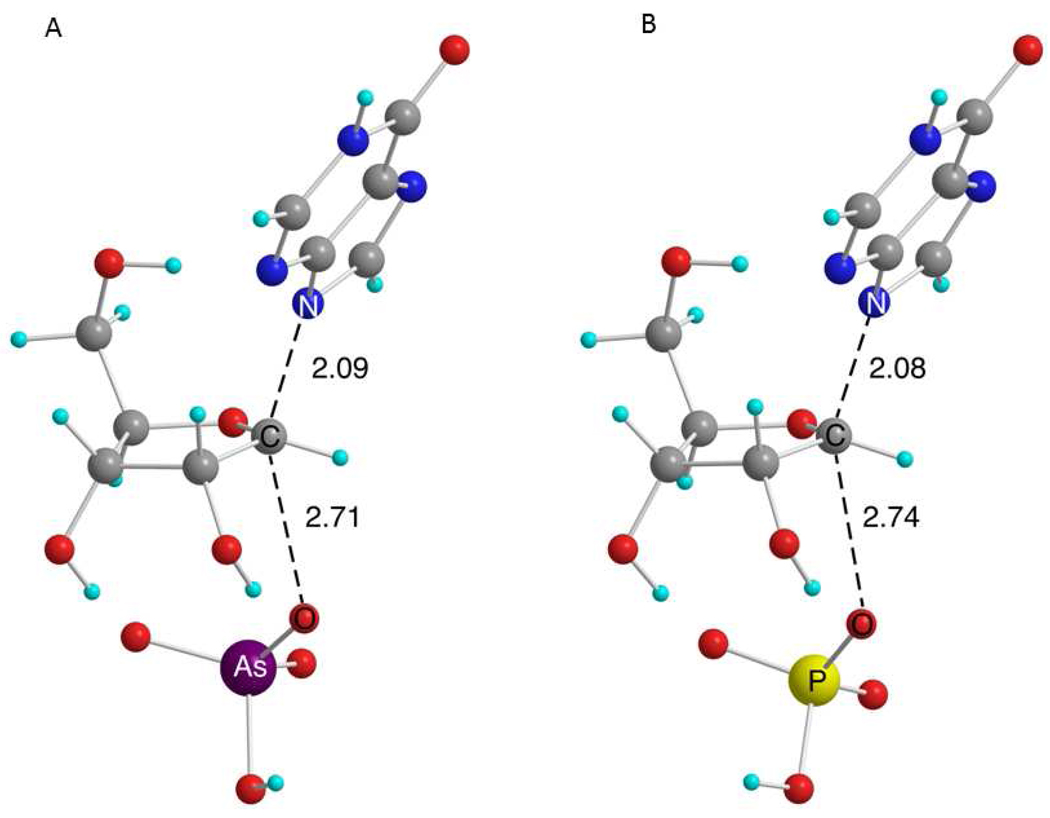
A comparison of transition structures for the reactions of arsenolysis (1) and phosphorolysis (2), without imposing any geometric constraints upon the structure, will give an unbiased picture of the differences and similarities in the substitution reactions of the two ions with inosine. From a systematic search of the reactions of arsenate and phosphate with inosine, a total of four different transition structures were located in B3LYP calculations using a 6-31G* basis set. Variations in the transition structures primarily involve changes in the conformation of the catalytically remote 5′-OH group. Corresponding conformations of the transition structures for arsenolysis (1) and phosphorolysis (2) displayed similar KIE predictions at every position and nearly identical rC-N and rC-O (coordinates for all geometries are available in the Supporting Information). The lowest energy transition structures of 1 and 2 were calculated in B3LYP using a 6-31+G** basis set (Figure 3). The theoretical structures for both arsenolysis and phosphorolysis are similar with a deviation in rC-N and rC-O of 0.01 Å and 0.03 Å, respectively. Furthermore, the predicted KIEs are similar with the largest deviation being 0.6% at the 5′-3H2 (Table 1). The resemblance in both the geometry and predicted KIEs of corresponding arsenolysis and phosphorolysis transition structures is striking and provides strong evidence for the similarity in reactivity of arsenate and phosphate in substitution reactions of nucleosides.
Figure 3.
Transition structures (B3LYP/6-31+G**) for the substitution reaction of (A) arsenate and (B) phosphate on the substrate inosine in gas phase without imposing geometric constraints. The numbers are measures of rC-N and rC-O. Carbon atoms are shown in gray, oxygen in red, hydrogen in cyan, nitrogen in blue, arsenic in purple, and phosphorus in yellow.
Table 1.
Predicted KIEs for stationary point transition states for arsenolysis and phosphorolysis of inosine in gas phase.
| KIE position | Type of KIE | Predicted KIE for arsenolysis |
Predicted KIE for phosphorolysis |
Difference in predicted KIEs |
|---|---|---|---|---|
| 1′-3H | α-Secondary | 1.081 | 1.082 | 0.001 |
| 1′-14C | Primary | 1.026 | 1.027 | 0.001 |
| 2′-2H | β-Secondary | 1.097 | 1.101 | 0.004 |
| 9-15N | Primary | 0.971 | 0.974 | 0.003 |
| 5′-3H2 | δ-Secondary | 1.134 | 1.141 | 0.007 |
Commitments to catalysis
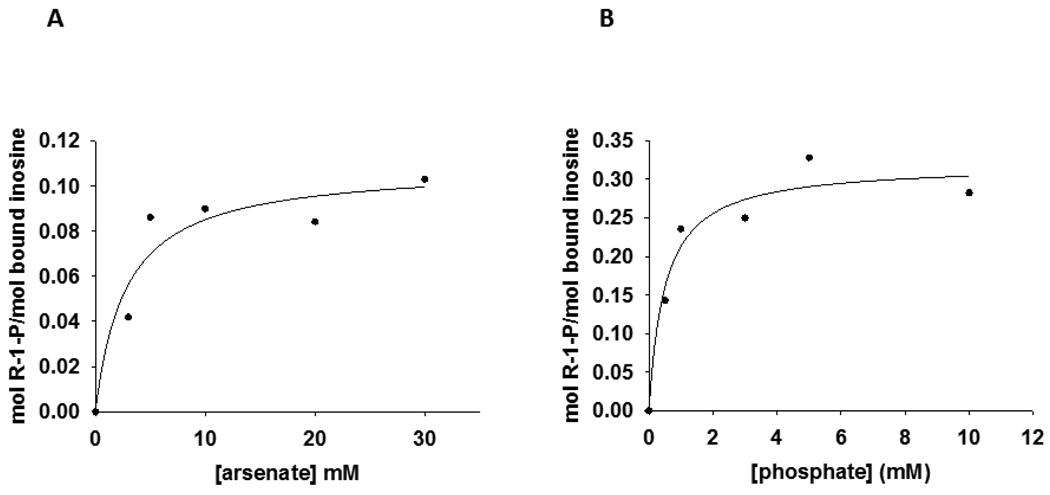
A forward commitment factor (Cf) represents the tendency of a substrate to undergo catalysis, once the Michaelis complex is formed, as opposed to dissociating to free substrate. The commitment factor is defined as the ratio of the rate constant for the isotope-sensitive step to the net rate constant of dissociation for the substrate from the catalytically competent complex (6). In the internal competition method of KIE measurement, Cf always refers to the substrate that carries the isotopic label (30), in the present case, inosine. Significant Cf masks the full expression of isotope effects in enzymatic reactions, and its value must be known to permit calculation of intrinsic KIEs (6). Accordingly, Cf upper limits were estimated for inosine with Phe200Gly HsPNP, and values of 0.115 ± 0.019 and 0.471 ± 0.058 were obtained under arsenolytic and phosphorolytic conditions, respectively (Figure 4). The Cf for Phe200Gly HsPNP-catalyzed inosine arsenolysis is the range found for native HsPNP (0.147), PfPNP (0.105) (10), and BtPNP (0.190) (7), but is smaller than the value reported for Lys22Glu-His104Arg HsPNP (0.239) (13). The Cf in the phosphorolysis reaction is in the range to permit calculation of intrinsic KIEs for the Phe200Gly HsPNP-catalyzed phosphorolysis of inosine.
Figure 4.
Isotope trapping-estimated forward commitment to catalysis for inosine with (A) arsenate or (B) phosphate as nucleophiles.
For native PNP, the Cf for inosine with phosphate is 0.687. However, the isotope effects are near unity (eg. 1.00 for [1′-14C]inosine) because of on-enzyme equilibration (reverse commitment). Enzyme-bound products on native PNP (hypoxanthine and α-D-ribose 1-phosphate) return to enzyme-bound reactants several times before product release, further reducing experimental KIE values toward equilibrium isotope effects. Both of these effects are reduced in the Phe200Gly PNP used here.
Experimental and intrinsic KIEs
Competitive KIEs were measured for Phe200Gly HsPNP-catalyzed arsenolysis and phosphorolysis reactions of inosine. Intrinsic KIEs were determined by correcting for Cf, according to eq 5, where LV/K is the observed KIE on V/K, and Lk is the intrinsic KIE (6). Terms other than the Cf are small since the values found for the primary 15N KIEs, after correction for Cf, are equal, within experimental error, to the maximum values found computationally for these isotope effects in inosine arsenolysis and phosphorolysis, as discussed below.
| eq 5 |
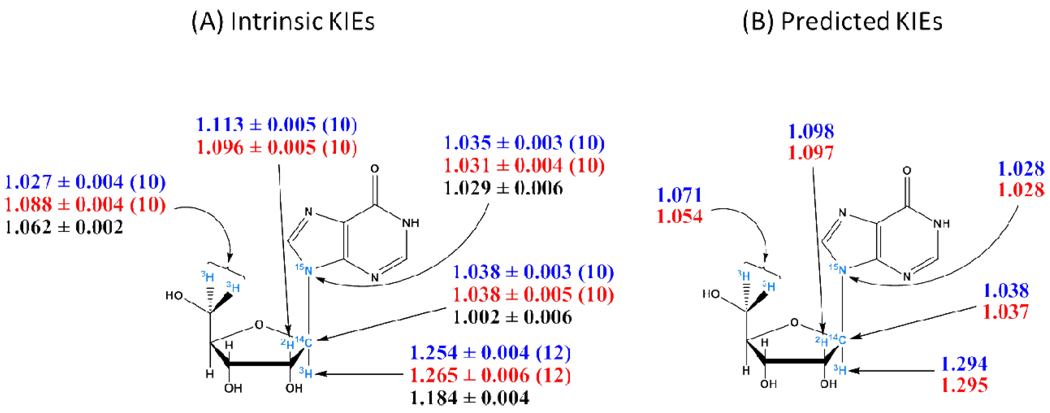
Except for the remote 5′-3H2 KIEs and, to a much lesser extent, the 2′-2H KIEs, intrinsic KIEs for arsenolysis are the same, within experimental errors, as those for phosphorolysis, indicating that Phe200Gly HsPNP-catalyzed arsenolysis and phosphorolysis of inosine proceed through closely related transition states (Table 2) (Figure 5A). The large primary 9-15N KIEs suggest substantial C1′-N9 bond loss, and the large α-secondary 1′-3H and β-secondary 2′-2H KIEs establish a ribooxocarbenium ion-like transition state. These characteristics are also present in the transition states for native bovine, human, and malarial PNP-catalyzed reactions (7, 10). However, the primary 1′-14C KIEs are too large to support a fully formed oxocarbenium transition state as found with HsPNP (10). Thus, significant bond order between the 1′-carbon and the attacking nucleophile must exist at the transition state. Nucleophile participation in the transition state has also been reported for human and bovine PNPs carrying mutations remote from the active site (13, 14).
Table 2.
KIEs and their standard errors on arsenolysis and phosphorolysis of inosine catalyzed by Phe200Gly HsPNP.
| Heavy inosine |
Light inosine |
Experimental KIE with AsO4 |
Experimental KIE with PO4 |
Intrinsic KIE with AsO4 |
Intrinsic KIE with PO4 |
|---|---|---|---|---|---|
| 1′-3H | 5′-14Ca | 1.228 ± 0.003 | 1.180 ± 0.003 | 1.254 ± 0.004 | 1.265 ± 0.006 |
| 1′-14C | 4′-3H | 1.034 ± 0.002b | 1.026 ± 0.003b | 1.038 ± 0.003 | 1.038 ± 0.005 |
| 5′-14C,2′-2H | 4′-3H | 1.101 ± 0.004c,d | 1.065 ± 0.004c,d | 1.113 ± 0.005 | 1.096 ± 0.005 |
| 5′-14C,9-15N | 4′-3H | 1.031 ± 0.002c | 1.021 ± 0.003c | 1.035 ± 0.003 | 1.031 ± 0.004 |
| 5′-3H2 | 5′-14C | 1.024 ± 0.003 | 1.060 ± 0.003 | 1.027 ± 0.004 | 1.088 ± 0.004 |
| 4′-3H | 5′-14C | 1.022 ± 0.002 | 0.986 ± 0.002 | - | - |
The remote 5′-14C KIE is assumed to be unity.
Experimental values corrected for remote 4′-3H KIE according to the equation KIE = KIEobs × 4′-3H KIE.
Experimental values corrected for remote 4′-3H KIE according to the equation KIE = KIEobs / 4′-3H KIE.
Experimental values corrected for 2′-2H content according to the equation KIE = (KIEobs − 1 + F)/F, where F is the fraction of 2H in the substrate.
Figure 5.
(A) Intrinsic KIEs and their standard errors for Phe200Gly HsPNP-catalyzed arsenolysis (blue) and phosphorolysis (red) of inosine, and WT-HsPNP catalyzed (black) arsenolysis of inosine (10). Between parentheses is the number of replicates. (B) Predicted KIEs for the best match non-stationary point TS for arsenolysis (blue) and phosphorolysis (red) of inosine. The labels and their respective positions are shown in cyan.
Enzymatic transition structures
A match of intrinsic KIEs to theoretical predictions is a powerful technique in providing detailed information about the nature of enzymatic transition states (31). Stabilization by the enzyme alters the position of the transition state relative to the gas phase potential energy surface, necessitating the restriction of geometries and predictions from non-stationary points. This method of calculating enzymatic transition structures has provided excellent predictions in previously modeled systems resulting in powerful transition state mimics (7, 11). A detailed chemical and theoretical test of this method of transition state analysis has shown that errors are minimal (24). Transition structures for the reaction of arsenate (1) and phosphate (2) with inosine were explored from the calculated grid of structures (Figure 2). Predicted KIEs for 112 non-stationary point structures were obtained from the Gaussian 03-calculated frequencies for the heavy atom isotope effects, 1′-14C and 9-15N, along the reaction coordinate (a table of predicted KIEs is included in the Supporting Information). Several theoretical structures exhibit 1′-14C and 9-15N predicted KIEs that fit within experimental error of the measured values. Theoretical structures with a sum of rC-N and rC-O between 5.4–5.6 Å provided the best fit to the experimental data. The full set of KIEs, including 1′-3H, 2′-2H, and 5′-3H2, were then predicted for this subset of grid structures. The 2′-2H position provides an accurate gauge of the electron density at the reactive center of the substitution because of the hyperconjugation effect at the 2′ position. Theoretical structures with longer rC-N more accurately predict the measured KIEs at the 2′-2H position. Importantly, the predicted KIEs were virtually the same in geometrically similar theoretical transition state structures involving arsenate (1) and phosphate (2) (see the predicted KIEs for the grid geometries in the Supporting Information). Predictions for the 5′-3H2 KIEs were inverse in all structures derived from the grid. To aid in the prediction of accurate KIEs and to avoid errors associated with fixed parameters, a more complete computational model was attempted incorporating several residues from the active site of HsPNP.
Dynamic studies have reported that the coordination of residue His257 to the 5′-OH is involved in the reaction coordinate of HsPNP (32). A pyrrole mimic of the histidine residue was added at the 5′ position of the sugar moiety in the second analysis. The pyrrole mimic was used rather than imidazole to avoid unwanted coordination of the second imidazole nitrogen. Residues Glu201 and Asn243 are also catalytically important (16) and were added to the model to mimic enzymatic stabilization of the nucleobase. New structures were explored with this more complete model containing the side chains of residues Glu201, Asn243, and the His257 mimic. All structures were calculated in B3LYP using a 6-31G* basis set. Bond distances rC-N and rC-O were necessarily fixed to predict the enzymatic transition structures. Several conformations of the transition structure were explored, including the position of the base, conformations of the sugar moiety, and the position of the attacking phosphate; however, the overall nucleobase and residue orientations derived from previous molecular dynamic studies were maintained (32, 33).
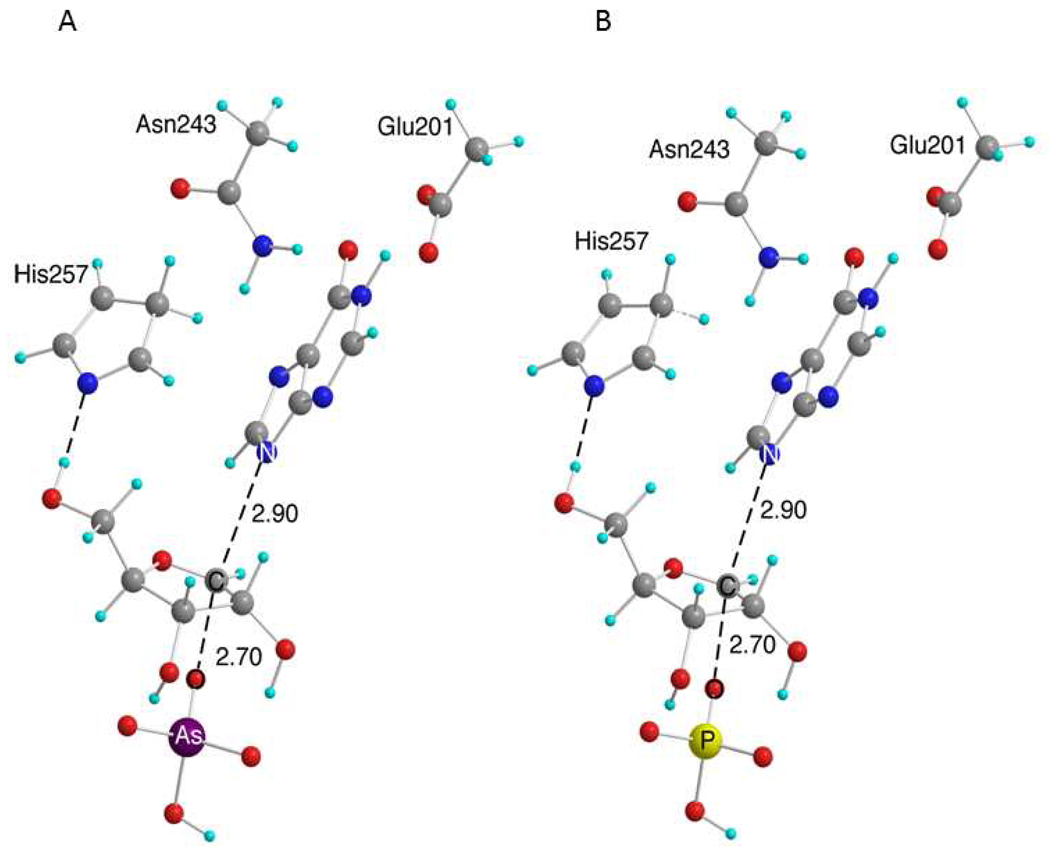
A comparison of experimental intrinsic and theoretically predicted KIEs was used to gauge the best theoretical structure to represent the enzymatic transition states for arsenolysis (1) and phosphorolysis (2) reactions. Several structures were explored with fixed rC-N in the range of 3.0–2.8 Å and fixed rC-O in the range of 2.7–2.5 Å (ranges were based upon previously explored structures). The transition structures for both arsenolysis and phosphorolysis exhibit rC-N of 2.9 Å and rC-O of 2.7 Å; however, structures within approximately 0.1 Å of each of the bond lengths give predicted values within the range of experimental error (Figure 6).
Figure 6.
Transition structures for the reaction of (A) arsenate and (B) phosphate with inosine in the reaction of Phe200Gly HsPNP. The numbers by the dashed lines are measures of rC-N and rC-O. Carbon atoms are shown in gray, oxygen in red, hydrogen in cyan, nitrogen in blue, arsenic in purple, and phosphorus in yellow.
The transition structures for arsenolysis and phosphorolysis represent dissociative ANDN transition states, exhibiting significant oxocarbenium ion character with the breaking of the rC-N, the leaving nucleobase being advanced and with small but significant formation of the rC-O bond with the nucleophile. The calculated transition structures exhibit remarkable similarity with either arsenate or phosphate as nucleophiles. Thus, the two ions lead to virtually the same TS in the reaction catalyzed by Phe200Gly HsPNP.
Predicted KIEs
The agreement between experimental and predicted KIEs is impressive for both arsenolysis and phosphorolysis given the simplicity of the model (Figure 5). Heavy atom predictions (1′-14C and 9-15N) are within the uncertainty of the experimental measurements, indicating that the model accurately represents the reaction coordinate. Excellent predictions of the 2′-2H KIEs support the accuracy of the ribose ring conformation of the calculated structures. The ribose ring of the inosine lies nearly flat at the transition state, allowing for strong hyperconjugation of the 2′-H into the oxocarbenium center. The 1′-3H KIEs are slightly over predicted by approximately 3–4% for both reactions; however, considering the magnitude of the effect and the difficulty in predicting 1′-3H KIEs in related reactions, the agreement is reasonable. The 9-15N KIEs are slightly under predicted in all structures explored for both reactions, and this is probably due to the sensitivity of nitrogen to the electronic environment in the active site, which differs from the gas phase calculations.
The 5′-3H2 KIEs are predicted to be normal for both transition structures; however, the 5′-3H2 KIEs are over predicted for arsenolysis and under predicted for phosphorolysis. The slight mismatch in predictions at this position is probably caused by differences in the coordination of the His257 mimic to the 5′-OH in the enzyme pocket as compared to the gas phase calculations (Figure 6). A dynamic study of HsPNP using transition path sampling suggests compression of the 5′-O with His257 at the transition state initiates N-ribosidic bond loss (32). A full enzymatic model involving the mode of compression at the 5′-hydroxyl group is beyond the scope of the theory used here. Nonetheless, the assumption that the 5′-3H2 KIEs increase under this mode of compression is reasonable and explains the predicted KIEs for the reaction with phosphate. Conversely, the arsenate ion is larger than the phosphate one, and the ion-binding pocket of PNP must necessarily open to accommodate the larger ion. Molecular dynamic studies on HsPNP show that the phosphate-binding pocket interacts with the residues surrounding the 5′-OH of inosine (33). A reasonable hypothesis is that the pocket at the 5′-OH position is forced to open up in the reaction involving arsenate causing a decreased interaction between the His257 and the 5′-OH. To further examine this idea, the 5′-3H2 KIEs were modeled as a function of the distance between the His257 mimic and the 5′-OH. The magnitude of the 5′-3H2 KIEs were inversely proportional to the bond distance between the 5′-OH and the N of the His257 mimic. The explanation above is consistent with the reduced intrinsic KIE at the 5′-H2 positions for the arsenolysis reaction relative to phosphorolysis. In summary, theoretical analysis of the experimentally measured KIEs for the arsenolysis and phosphorolysis reactions with mutant HsPNP has confirmed the strong similarity between the transition state structures of both reactions.
Alternative mechanisms
Only results involving an ANDN mechanism have been shown. However, other mechanistic possibilities were also considered. Previous studies with native HsPNP have shown that the N7 position of the nucleobase is protonated at the transition state (10). All attempts to locate a transition structure for a concerted ANDN mechanism with protonation at the N7 position of inosine proved to be inconsistent with the results for Phe200Gly HsPNP. The possibility of a DN*AN‡ mechanism involving first the departure of the nucleobase (with or without protonation at N7) followed by rate-limiting attack of the ion, as reported for Lys22Glu-His104Arg HsPNP (13), was also explored. In that case, the isotope effect at 9-15N should represent an EIE in the case of a DN*AN‡ mechanism. The isotope effect was predicted to be 1.024 and 1.028 for hypoxanthine with and without protonation at N7, respectively. The second step of the DN*AN‡ mechanism, involving attack of the ion (arsenate or phosphate) to the oxocarbenium, was explored by including only the ion and the oxacarbenium (no nucleobase) and varying rC-O. The mismatch between the predicted and intrinsic values of 1′-14C, 1′-3H, and 2′-2H KIEs at all distances of rC-O excluded the DN*AN‡ as a mechanistic possibility (all geometries and KIE predictions for the attack of arsenate and phosphate on the oxocarbenium are included in the Supporting Information).
A final mechanistic possibility is a DN‡*AN mechanism involving rate-limiting dissociation of the nucleobase, followed by the attack of the ion (arsenate or phosphate) to the oxocarbenium, as reported for BtPNP (7). The DN‡*AN mechanism was explored by varying rC-N for various structures involving only the inosine (excluding the arsenate and phosphate). The mismatch between the predicted and intrinsic values of 1′-14C, 1′-3H, and 2′-2H KIEs at all distances of rC-N, specifically the under prediction of the 2′-2H KIE, excluded the DN‡*AN as a mechanistic possibility (all geometries and KIE predictions for rate-liming dissociation of the nucleobase is included in the Supporting Information). Transition structures for the dissociation of a nucleobase protonated at N7 could not be located3. Both the DN‡*AN and DN*AN‡ mechanisms can be excluded as mechanistic possibilities for both the arsenolysis and phosphorolysis of Phe200Gly HsPNP based on a mismatch of the predicted and experimental KIEs. The maximum predicted isotope effect at 9-15N for all investigated mechanisms was 1.035, which is equal, within experimental error, to the intrinsic values obtained for arsenolysis and phosphorolysis after correction for Cf. Thus, the reaction displays no significant terms other than Cf to influence the intrinsic KIEs.
The ability of arsenate to mimic phosphate in the reaction of Phe200Gly HsPNP has been shown using a combination of experimental and theoretical techniques. The ions of arsenate and phosphate exhibit similar electronic properties, and calculated transition structures for the reaction of arsenate and phosphate with inosine without geometric restriction are nearly identical. The strongest evidence justifying previous studies that necessarily involve the substitution of arsenate for phosphate is the similar KIE measurements at multiple positions. The results indicate the remarkable similarity between the TS reached with either arsenate or phosphate in the Phe200Gly HsPNP-catalyzed inosine cleavage, and also suggest the same is true for other nucleophilic substitution reactions of nucleosides.
Supplementary Material
ACKNOWLEDGEMENT
The authors thank Keith Z. Hazleton and Dr. Achelle Edwards for their kind gifts of hypoxanthine-guanine phosphoribosyltransferase and wild-type human purine nucleoside phosphorylase, respectively. The authors acknowledge Yong Zhang for insightful discussions regarding the computational modeling.
Footnotes
This work was supported by NIH grant GM41964 to V. L. S.
Abbreviations used: PNP, purine nucleoside phosphorylase; R-1-P, ribose-1-phosphate; HsPNP, human PNP; BtPNP, bovine PNP; PfPNP, Plasmodium falciparum PNP; KIE, kinetic isotope effect; EIE, equilibrium isotope effect; Cf, forward commitment; ZPE, zero-point energy; PK, pyruvate kinase; MK, myokinase; HK, hexokinase; G6PDH, glucose-6-phosphate dehydrogenase; GDH, glutamic acid dehydrogenase; 6PGDH, 6-phosphogluconic acid dehydrogenase; ADA, adenosine deaminase; PRI, phosphoriboisomerase; AP, alkaline phosphatase; RK, ribokinase; PRPPase, phosphoribosyl-α-1-pyrophosphate synthetase; HGPRT, hypoxanthine-guanine phosphoribosyltransferase; PEP, phosphoenolpyruvate
The RMSD for the two curves were determined using the curve-fitting package in the MATLAB software. The surfaces were fit to a 5th degree polynomial using the Lowess interpolation method.
Theoretical structures were located with rC-N distances of 2.5, 2.7, and 2.9 Å. These structures had no, or very small, imaginary frequency associated with the geometry, and therefore represent structures with a fully dissociated nucleobase. The heavy atom equilibrium isotope effects for these structures are much too low in comparison with experimental values.
SUPPORTING INFORMATION AVAILABLE: Synthetic schemes for labeled IMPs (Scheme S1 and S2), and full geometries and tables of KIEs for all calculated structures. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Kalckar HM. Differential spectrophotometry of purine compounds by means of specific enzymes; determination of hydroxypurine compounds. J Biol Chem. 1947;167:429–443. [PubMed] [Google Scholar]
- 2.Parks RE, Jr, Agarwal RP. Purine nucleoside phosphorylase. In: Boyer PD, editor. The Enzymes. New York: Academic Press; 1972. pp. 483–514. [Google Scholar]
- 3.Giblett ER, Ammann AJ, Wara DW, Sandman R, Diamond LK. Nucleoside-phosphorylase deficiency in a child with severely defective T-cell immunity and normal B-cell immunity. Lancet. 1975;1:1010–1013. doi: 10.1016/s0140-6736(75)91950-9. [DOI] [PubMed] [Google Scholar]
- 4.Stoeckler JD. Purine nucleoside phosphorylase: a target for chemotherapy. In: Glazer RI, editor. Developments in Cancer Chemotherapy. Boca Raton: CRC Press Inc.; 1984. pp. 35–60. [Google Scholar]
- 5.Ho MC, Shi W, Rinaldo-Matthis A, Tyler PC, Evans GB, Clinch K, Almo SC, Schramm VL. Four generations of transition-state analogues for human purine nucleoside phosphorylase. Proc Natl Acad Sci U S A. 2010;107:4805–4812. doi: 10.1073/pnas.0913439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Northrop DB. Steady-state analysis of kinetic isotope effects in enzymic reactions. Biochemistry. 1975;14:2644–2651. doi: 10.1021/bi00683a013. [DOI] [PubMed] [Google Scholar]
- 7.Kline PC, Schramm VL. Purine nucleoside phosphorylase. Catalytic mechanism and transition-state analysis of the arsenolysis reaction. Biochemistry. 1993;32:13212–13219. doi: 10.1021/bi00211a033. [DOI] [PubMed] [Google Scholar]
- 8.Krebs HA, Eggleston LV, Knivett VA. Arsenolysis and phosphorolysis of citrulline in mammalian liver. Biochem J. 1955;59:185–193. doi: 10.1042/bj0590185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nemeti B, Gregus Z. Mechanism of thiol-supported arsenate reduction mediated by phosphorolytic-arsenolytic enzymes: I. The role of arsenolysis. Toxicol Sci. 2009;110:270–281. doi: 10.1093/toxsci/kfp112. [DOI] [PubMed] [Google Scholar]
- 10.Lewandowicz A, Schramm VL. Transition state analysis for human and Plasmodium falciparum purine nucleoside phosphorylases. Biochemistry. 2004;43:1458–1468. doi: 10.1021/bi0359123. [DOI] [PubMed] [Google Scholar]
- 11.Miles RW, Tyler PC, Furneaux RH, Bagdassarian CK, Schramm VL. One-third-the-sites transition-state inhibitors for purine nucleoside phosphorylase. Biochemistry. 1998;37:8615–8621. doi: 10.1021/bi980658d. [DOI] [PubMed] [Google Scholar]
- 12.Evans GB, Furneaux RH, Lewandowicz A, Schramm VL, Tyler PC. Synthesis of second-generation transition state analogues of human purine nucleoside phosphorylase. J Med Chem. 2003;46:5271–5276. doi: 10.1021/jm030305z. [DOI] [PubMed] [Google Scholar]
- 13.Luo M, Li L, Schramm VL. Remote mutations alter transition-state structure of human purine nucleoside phosphorylase. Biochemistry. 2008;47:2565–2576. doi: 10.1021/bi702133x. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Luo M, Ghanem M, Taylor EA, Schramm VL. Second-sphere amino acids contribute to transition-state structure in bovine purine nucleoside phosphorylase. Biochemistry. 2008;47:2577–2583. doi: 10.1021/bi7021365. [DOI] [PubMed] [Google Scholar]
- 15.Edwards PN. A kinetic, modeling and mechanistic re-analysis of thymidine phosphorylase and some related enzymes. J Enzyme Inhib Med Chem. 2006;21:483–499. doi: 10.1080/14756360600721075. [DOI] [PubMed] [Google Scholar]
- 16.Erion MD, Takabayashi K, Smith HB, Kessi J, Wagner S, Honger S, Shames SL, Ealick SE. Purine nucleoside phosphorylase. 1. Structure-function studies. Biochemistry. 1997;36:11725–11734. doi: 10.1021/bi961969w. [DOI] [PubMed] [Google Scholar]
- 17.Ghanem M, Murkin AS, Schramm VL. Ribocation transition state capture and rebound in human purine nucleoside phosphorylase. Chem Biol. 2009;16:971–979. doi: 10.1016/j.chembiol.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh V, Lee JE, Nunez S, Howell PL, Schramm VL. Transition state structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Escherichia coli and its similarity to transition state analogues. Biochemistry. 2005;44:11647–11659. doi: 10.1021/bi050863a. [DOI] [PubMed] [Google Scholar]
- 19.Parkin DW, Leung HB, Schramm VL. Synthesis of nucleotides with specific radiolabels in ribose. Primary 14C and secondary 3H kinetic isotope effects on acid-catalyzed glycosidic bond hydrolysis of AMP, dAMP, and inosine. J Biol Chem. 1984;259:9411–9417. [PubMed] [Google Scholar]
- 20.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Rose IA. The isotope trapping method: desorption rates of productive E.S complexes. Methods Enzymol. 1980;64:47–59. doi: 10.1016/s0076-6879(80)64004-x. [DOI] [PubMed] [Google Scholar]
- 22.Frisch MJ, et al. Gaussian 03, Revision E.01. Wallingford, CT: Gaussian, Inc.; 2004. [Google Scholar]
- 23.Frisch MJ, et al. Gaussian 09, Revision A.02. Wallingford CT: Gaussian, Inc.; 2009. [Google Scholar]
- 24.Hirschi JS, Takeya T, Hang C, Singleton DA. Transition-state geometry measurements from (13)c isotope effects. The experimental transition state for the epoxidation of alkenes with oxaziridines. J Am Chem Soc. 2009;131:2397–2403. doi: 10.1021/ja8088636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bigeleisen J, Mayer MG. Calculation of Equilibrium Constants for Isotopic Exchange Reactions. J. Chem. Phys. 1947;15:261–267. [Google Scholar]
- 26.Wolfsberg M. Theoretical evaluation of experimentally observed isotope effects. Acc. Chem. Res. 1972;5:225–233. [Google Scholar]
- 27.Anisimov V, Paneth P. ISOEFF98. A program for studies of isotope effects using Hessian modifications. J. Math. Chem. 1999;26:75–86. [Google Scholar]
- 28.Bell RP. The tunnel effect in chemistry. London: Chapman and Hall; 1980. [Google Scholar]
- 29.Berti PJ, Tanaka KSE. Transition State Analysis Using Multiple Kinetic Isotope Effects: Mechanisms of Enzymatic and Non-enzymatic Glycoside Hydrolysis and Transfer. Adv. Phys. Org. Chem. 2002;37:239–314. [Google Scholar]
- 30.Cleland WW. The use of isotope effects to determine enzyme mechanisms. Arch Biochem Biophys. 2005;433:2–12. doi: 10.1016/j.abb.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 31.Schramm VL. Enzymatic transition-state analysis and transition-state analogs. Methods Enzymol. 1999;308:301–355. doi: 10.1016/s0076-6879(99)08015-5. [DOI] [PubMed] [Google Scholar]
- 32.Saen-Oon S, Quaytman-Machleder S, Schramm VL, Schwartz SD. Atomic detail of chemical transformation at the transition state of an enzymatic reaction. Proc Natl Acad Sci U S A. 2008;105:16543–16548. doi: 10.1073/pnas.0808413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirschi JS, Arora K, Brooks CL, 3rd, Schramm VL. Conformational dynamics in human purine nucleoside phosphorylase with reactants and transition-state analogues. J Phys Chem B. 2010;114:16263–16272. doi: 10.1021/jp108056s. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.