Abstract
The proteins that mediate the analgesic and other effects of opioid drugs and endogenous opioid peptides are known as opioid receptors. Opioid receptors consist of a family of four closely-related proteins belonging to the large superfamily of G-protein coupled receptors. The three types of opioid receptors shown unequivocally to mediate analgesia in animal models are the mu (MOR), delta (DOR), and kappa (KOR) opioid receptor proteins. The role of the fourth member of the opioid receptor family, the nociceptin or orphanin FQ receptor (ORL), is not as clear as hyperalgesia, analgesia, and no effect was reported after administration of ORL agonists. There are now cDNA sequences for all four types of opioid receptors that are expressed in the brain of six species from three different classes of vertebrates. This review presents a comparative analysis of vertebrate opioid receptors using bioinformatics and data from recent human genome studies. Results indicate that opioid receptors arose by gene duplication, that there is a vector of opioid receptor divergence, and that MOR shows evidence of rapid evolution.
Keywords: Opioid Receptor, Analgesia, Evolution, Bioinformatics, Review
2. INTRODUCTION AND HYPOTHESES
G-protein coupled receptors (GPCR) represent the largest group of membrane proteins encoded in the human genome. There are at least 2,000 GPCRs within the rhodopsin-like superfamily recognized in the human genome, with more than a third of these receptors classified as ‘orphan GPCRs’ in that the endogenous ligand that binds to them is not known (1–3). GPCR genes represent about 5% of the coding genome, are the target for more than 60% of all prescription drugs, and the object of intense scrutiny for new drug development in the pharmaceutical industry (4). GPCRs are classified by their sequence homology which parallels classification based solely on ligand binding selectivity (5, 6). This correlation between receptor sequence and ligand specificity, while obvious from first principles (sequence determines structure determines activity), is at the crux of fully understanding receptor selectivity and ensuing pharmacological action.
The opioid receptors are membrane proteins of the Type A or rhodopsin-like GPCRs that mediate the analgesic effects of opioid agents like morphine. Opioid receptors also enable substance-abuse with drugs like heroin. The natural ligands for opioid receptors are endogenous opioid peptides; most notably beta-endorphin, met- and leu-enkephalin and dynorphin. The role of endogenous opioid receptors in physiological processes is vast; besides an obvious role in decreasing nociceptive transmission, nearly any Pubmed search with the Boolean parameters of ‘any physiological process and opioid’ brings up pages of references. This is true for terms like reproduction, growth, development, respiration, blood pressure, renal function, thermoregulation, endocrinology, seizures, stress, immunology, pregnancy and aging. It is also true when considering behaviors using the terms like eating, drinking, sex, learning, memory, locomotion and driving (I challenge the reader to enter any term plus opioid and not get a hit). A compendium of the literature on various opioid effects entitled ‘Endogenous opiates (sic) and behavior’ is published each year in the journal Peptides (and has been for the last 30 years) largely due to the efforts of Kastin and Bodnar (7). While there appears to be no limit to the effects of opioids, both endogenous and exogenous, on any system in the brain and body, underlying all opioid effects is the fundamental event of an opioid agonist binding and activating an opioid receptor.
Given the utmost importance of the opioid receptor system for producing analgesia and the myriad of effects on other regulated processes mentioned above, it comes as no surprise that opioid receptors are found in the nervous tissue of all vertebrate species examined. However, there are few papers analyzing the different types of vertebrate opioid receptors as a group or providing an explanation of how they may have arisen. This review posits the hypothesis that the family of four opioid receptor genes in extant vertebrates arose by gene duplication events from a single, ancestral opioid unireceptor gene. Furthermore, that the opioid receptor proteins encoded by these genes show an evolutionary vector of increased pharmacological type-selectivity. Finally, that one gene product of this family of duplicate genes, the mu opioid receptor (MOR) protein, is unique among opioid receptor types in showing evidence of rapid protein evolution and underlying adaptive evolution.
3. A SHORT HISTORY OF OPIOID RECEPTORS
There was a time when receptors were solely defined by the structure-activity relationships of various ligands that acted upon them. In this way, early studies by W. R. Martin initially proposed multiple opioid receptors based on the differential action of morphine and ketocyclazocine in a spinal dog model (8). Thus, Martin proposed the initial idea of mu (Greek ‘m’ for morphine; MOR) and kappa (for ketocyclazocine; KOR) opioid receptors. Later, Hans Kosterlitz and colleagues, working with the newly discovered enkephalin peptides in the mouse vas deferens preparation, proposed a delta (for deferens; DOR) opioid receptor type (9). Much work towards the end of the last century solidified the structure-activity relationship of these primary opioid receptor proteins, aided in large part by the development of highly-selective (type-specific) opioid agonists and antagonists. The development of highly-selective opioid antagonists by Takemori and Portoghese ushered in a new era of opioid pharmacology (10–12). These selective opioid antagonists show 100- to 1000-fold or greater selectivity for their respective type of opioid receptor. Common highly-selective opioid antagonists used are beta-funaltrexamine (beta-FNA, for MOR) (10), naltrindole (NTI, for DOR) (11), and nor-binaltorphimine (nor-BNI, for KOR) (12). Pharmacological evidence of three separate opioid receptors mediating analgesia in mammalian models was confirmed using these antagonists.
Although the existence of opiate receptors was postulated as early as the mid-1950s (13), definitive evidence of stereospecific opioid binding sites from radioligand binding studies using rodent brain homogenates did not emerge until 1973 from the labs of Snyder, Simon, and Terenius (14–16). Ongoing refinements of such ‘grind and bind’ studies clearly established three main opioid receptor binding profiles (17, 18) and a number of pharmacological subtypes for each of the main receptors (19, 20). The ultimate discovery and isolation of the cDNA for the mouse delta opioid receptor (mDOR1) from the labs of Evans on this continent and Kieffer abroad in 1992 (21, 22) for the first time linked an opioid receptor sequence to a pharmacological type of opioid receptor. Both groups screened a cDNA library derived from NG108–15, a species-hybrid cell line derived from mouse neuroblastoma x rat glioma cells using radiolabeled delta opioid peptides to identify positive clones. Individual cDNA clones were re-transfected into heterologous cells and characterized using selective opioid ligands, revealing the presence of a delta opioid receptor. It is interesting that both groups identified the mouse DOR from NG108-15 cells as these hybridomas contain a varying polyploidy of both mouse and rat chromosomes, although mouse chromosomes maintain predominance over rat chromosomes in stable NG108-15 clones (23).
Once the mDOR cDNA sequence was known, homology cloning led to the identification of receptor sequences for numerous rodent and monkey species and the human opioid receptors, hMOR (24), hDOR (25, 26), and hKOR (27–29). About this time, cloning studies revealed another ‘opioid receptor-like’ (ORL) sequence from numerous labs that was similar to the known MOR, DOR, and KOR sequences but differed in its ligand binding characteristics (30–36). This opioid receptor-like protein remained an ‘orphan’ receptor until the endogenous ligand for ORL was simultaneously identified as the neuropeptide named nociceptin by the lab of Meunier (37) or named orphanin FQ from the team assembled by Civelli (38). Thus the nociceptin (or orphanin FQ) receptor (ORL) is the fourth member of the opioid receptor family expressed in the nervous system of mammals (5, 39). However, the results of nociceptin or orphanin FQ binding to the ORL protein is not clear as behavioral studies show hyperalgesia (the origin of the name nociceptin) and analgesia or sometimes no effect (40). Additionally, the vast majority of opioids do not bind at all to ORL receptors, nor do the opioid antagonists, naloxone or naltrexone. For this reason, MOR, DOR, and KOR are considered the classical opioid receptor types of the family and ORL a distant cousin. In vivo studies using antisense oligodeoxynucleotides designed from MOR, DOR, and KOR cDNA sequences confirm that each one of the three opioid receptor proteins can independently produce analgesia in mammalian models (41–43).
It would be an error not to redress the confusing terminology in the literature describing the members of the opioid receptor family as types or subtypes. As suggested by Avram Goldstein2, an early leader in the opioid research field and discover of the endogenous opioid peptide dynorphin, the different receptor proteins coded by individual genes should be called ‘types’ of opioid receptors. ‘Types’ of receptors as each opioid receptor protein arises from its own gene (see below) and belongs to the family of opioid receptors, so it is a type of opioid receptor. The term ‘subtype’ should be reserved for pharmacologically defined differences in receptor binding as shown by structure-activity relationships. Each classical opioid receptor type mediates analgesia in mammals as shown by basic and clinical studies using type-selective opioid agonists and antagonists. However this use of type and subtype is not universal. In the adrenergic field, so-called ‘subtypes’ of receptors (e.g. alpha1 or alpha2 receptors) are actually different gene products. Likewise in the muscarinic receptor field. The logical convention used here for opioid receptors is calling the four receptor proteins from different genes as ‘types’ and variations in binding defined by pharmacological studies as ‘subtypes’.
There is also much evidence of opioid receptor isoforms in mammalian species resulting from alternative splicing of receptor mRNA (44–46). Interestingly, it appears that many more splice variants are noted for MOR than for the other types of vertebrate opioid receptors. Driven by the prodigious efforts of Pan and Pasternak, there are now at least 20 alternative splice variants of mMOR (47) and at least 6 variants of hMOR (48). Many of these splice variants show different ligand binding or signaling pathways (49). In contrast, only a few alternative splice variants are reported for KOR (50, 51), and even less for DOR (50) and ORL (52). This finding may reflect sampling bias as MOR transcripts have been examined to a greater degree, but more likely supports the notion that MOR is very different than the other types of vertebrate opioid receptors, a theme developed further throughout this review.
4. OPIOID RECEPTORS IN NON-MAMMALIAN VERTEBRATES
Early studies employing a hot plate test and electrified floor grid in amphibians did not detect an antinociceptive effect of morphine (53, 54). It is likely that this was due to inappropriate experimental design, as previously noted (55). A behavioral assay using acetic acid as the noxious stimulus and the wiping response as the nocifensive behavior was developed using the Northern grass frog, Rana pipiens, and is discussed in greater detail in the next section, along with a handful of studies in other non-mammalian species. Although not reviewed here, Dores and colleagues have gathered an impressive body of work describing endogenous opioid peptides in non-mammalian species (56–64).
4.1. Analgesic effects of opioids in non-mammalian models
Initial studies of the analgesic or antinociceptive effects of opioids in amphibians (Rana pipiens) were conducted using non-selective opioid agonists, endogenous opioid peptides, and antagonists (65–68). The assay used in the amphibian model is called the acetic acid test (55). The acetic acid test activates nociceptors and nociceptive primary afferent fibers exclusively (69–71). Tolerance to the analgesic effects of daily morphine administration was documented (72) and stress-induced release of endogenous opioids produced analgesia which was potentiated by enkephalinase inhibitors and blocked by naltrexone (73, 74). These studies showed that both exogenous opioid agonists and endogenous opioid peptides could raise the nociceptive threshold in amphibians by an action at opioid receptors. Other behavioral studies in amphibians include an investigation of the effects of opioids on noxious and non-noxious sensory modalities (69, 75), an examination of agents acting on alpha2 adrenergic receptors after systemic and spinal administration (76, 77), and recent reports of analgesia produced by remifentanil (78) and xendorphin (79).
Results of systematic studies examining the antinociception of selective mu, delta, or kappa opioid agonists administered by different routes in amphibians yielded an important finding: The relative antinociceptive potency of mu, delta, or kappa opioid agonists after systemic, intraspinal, or intracerebroventricular administration in amphibians was highly correlated to that observed in typical mammalian models and to the relative analgesic potency of opioid analgesics in human clinical studies (80–82). These data established the amphibian model as a robust and predictive adjunct model for the testing of opioid analgesics (83–86). Other studies in amphibians used the acetic acid test to demonstrate that regional hypothermia produces antinociceptive effects that is mediated in part by endogenous opioid peptides (87). A tail-flick test was also used to assess the antinociceptive effects of opioids in the Japanese firebelly newt, Cynops pyrrhogaster. (88). In this study, intraperitoneal injection of met-enkephalin and the putative opioid peptide, RFamide, produced a mild antinociceptive effect that was blocked by the concurrent administration of the opioid antagonist, naloxone. In the one study examining the behavioral effects of nociceptin in a non-mammalian model, nociceptin and other ORL agonists produced antinociceptive effects following spinal administration in Rana pipiens which was blocked by ORL but not opioid antagonists (Stevens et al., submitted).
Studies of the antinociceptive effects of opioids in species from the class of Pisces are not easily accomplished for obvious reasons. A study aimed primarily at the metabolism of morphine in goldfish (Carasius auratus) also described a behavioral assay using an electric prod applied caudal to the dorsal fin as the noxious stimulus (89). The nocifensive behavior observed was called the agitated swim response. The voltage threshold for eliciting the swim response was consistent for each animal and with repeated testing. Morphine sulfate added to the tank water produced a dose-dependent increase in the electrical voltage need to elicit the agitated swim response. These authors also showed that goldfish became tolerant to the antinociceptive effects of morphine with repeated administration (89). There was no attempt to attain a measure of morphine’s potency (i.e. no ED50 value of morphine was reported) nor was opioid receptor involvement verified by pretreatment with an opioid antagonist such as naloxone. Further studies using the same model in goldfish showed that morphine administered via the intracranial route (into a space above the optic tectum) produced a dose-dependent increase in the voltage needed to elicit the swim response (90). In this study, opioid receptor involvement was shown by naloxone antagonism of morphine’s effect. The antinociceptive effects of dermorphin, a potent and selective MOR opioid peptide originally isolated from the skin of Phyllomedusa frogs (91), was tested using electrodes implanted in the caudal region of Derjugin codfish, Gadus morhuamarisalbi, as the noxious stimulus (92). The nocifensive response was similar to the agitated swim response mentioned above but the fish was held in a flow-through chamber and the swim response was quantified by a force transducer. Dermorphin was administered into the intranasal passages (olfactory sacs) of the codfish and produced a dose-dependent antinociceptive effect. Further studies using rainbow trout, Oncorhynchus mykiss, with the same sophisticated behavioral apparatus, mapped the most sensitive areas of noxious stimulation on the trout and confirmed the antinociceptive effect of intranasal dermorphin in this species (93). In codfish, intraperitoneal and intramuscular administration of the MOR selective opioid peptide, beta-casomorphin, produced antinociceptive effects (94). The opioid analgesic, tramadol, a combined MOR agonist and norepinephrine/serotonin re-uptake inhibitor, produced dose-dependent antinociception in the carp, Cyprinus carpio (95). In this last study, pretreatment with the opioid antagonist naloxone blocked the antinociception produced by tramadol. A chronic pain model was developed in fish by injecting acetic acid or bee venom into the lip region of trout (96, 97). These animals develop pain-related behaviors that can be quantified and antinociception determined by the reduction in pain-related behaviors. Morphine had inhibiting effects on nociceptive behaviors per se and in the presence of novel objects (98, 99).
To test the antinociceptive effects of opioids in reptiles, a tail-flick apparatus was used in the green anole lizard, Anolis carolinensis (100). Intraperitoneal injection of morphine (5 mg/kg) produced a weak, but significant increase in the latency of the lizard to remove its tail from a noxious heat stimulus. A number of nociceptive assays were characterized in the Nile crocodile, Crocodylus niloticus (101). Crocodiles placed on a hot plate attempt to guard their foot pads by lifting them alternately as well as attempting to escape the heated surface. A dilute capsaicin solution instilled in the eye of the crocodile produces behavioral responses including blinking, rubbing and head shaking. Injection of formalin in the crocodile forepaw results in the development of paw guarding behaviors. Morphine or meperidine (pethidine) produced antinociception as measured by the increased latency for crocodiles to lift their leg or escape on the hot plate test, which was the most suitable of three assays (102). However, these studies did not include dose-response analysis, nor assess the opioid nature of the antinociceptive effect by pretreatment with an opioid antagonist, such as naloxone or naltrexone. As mentioned in a clinical review of analgesia for exotic species (103), there is preliminary data that the partial opioid agonist, butorphanol, produced antinociception in the green iguana lizard, Iguana iguana, using a thermal tail-flick test.
An early attempt to measure the antinociceptive effect of opioids in chicks (Gallus gallus) using toe-pinch as the noxious stimulus and limb withdrawal as the nocifensive behavior did not find any antinociceptive effect of morphine up to a dose of 200 mg/kg (104). Using an electroshock to the underside of the wing as the noxious stimulus and an elicited flight response as the nocifensive behavior in chickens, morphine at a single dose of 30 mg/kg produced measurable antinociception in this model (105). Using a heated floor grid, other studies found that morphine produced a decrease in the latency for chickens to jump (106). This hyperalgesic effect was dependent on the age of the chicken; morphine administered to young cockerels less than 14 days old did not produce hyperalgesia (107). The hyperalgesic effect of morphine was dose-dependent and exhibited a U-shaped time course curve (108). Using selective opioid antagonists, the hyperalgesic effect of morphine in chickens was shown to be predominantly mediated by mu opioid receptors (109). Codeine produced both hypoalgesia (antinociception) and hyperalgesia in this model (110). A formalin algesiometric assay was developed in chickens, quantified by the pain-related behaviors observed after injection of formalin in to the foot (111). There was a strain difference in the hyperalgesic effect of morphine, perhaps due to differences in descending monoamine pathways (112). Carageenan, a chemical irritant like formalin, provided the noxious stimulus in a chronic inflammatory model using the Hargreaves method (113). In this assay, chickens are measured for the latency to withdrawal an inflamed foot from a lamp placed under the floor grid. Morphine produced a dose-dependent antinociception but had no effect on the edema produced by carageenan. A chronic arthritis model was developed in chickens using using intra-articular injection of urate crystals, which models gouty arthritis (114, 115). However, intra-articular injection of morphine, fentanyl, or buprenorphine did not have any effect in reducing pain-related behaviors due to the experimentally-induced arthritis (116).
In all the above behavioral studies using non-mammalian models, it can be summarized that opioids have similar antinociceptive effects as observed in mammals. However, the types of opioid receptor mediating these effects in non-mammalian vertebrates are not as clear as in mammals. There has been only one study in amphibians examining opioid agonist effects with co-administration of highly-selective MOR, DOR, and KOR antagonists. The most common highly-selective opioid antagonists used in mammalian studies are beta-funaltrexamine (beta-FNA, for MOR) (10), naltrindole (NTI, for DOR) (11), and nor-binaltorphimine (nor-BNI, for KOR) (12). Beta-FNA, NTI, and nor-BNI were tested against the spinal antinociceptive effects of mu, delta, and kappa opioid agonists in the amphibian model (117). Each of the highly-selective opioid antagonists blocked the antinociceptive effects produced by all three types of selective opioid agonists in amphibians. For example, beta-FNA, a highly-selective MOR antagonist in mammalian models, blocked the effects of selective MOR, DOR, and KOR agonists. Similar results were noted for NTI and nor-BNI. The most parsimonious interpretation, following Occam’s razor3, was that there was one type of receptor mediating opioid analgesia in amphibians; the short-lived but beautiful opioid unireceptor hypothesis (117, 118).
4.2. Radioligand binding studies using non-mammalian vertebrates
There were a number of radioligand binding studies in brain homogenates from non-mammalian species including fish (119–122), reptiles (123–125) and birds (126–129). However, the largest effort came from the study of opioid binding sites in amphibian brain membranes. The first studies of opioid receptor purification used amphibian brain tissue, as it was recognized to be a rich source of opioid receptors (130). Biochemical and receptor isolation studies using amphibian brain tissues demonstrated a high proportion of opioid receptors compared to mammals (131–134). These reports and many others examining opioid binding sites using the European water frog, Rana esculenta, were produced from the Hungarian research group headed by Wolleman, Simon, Borsodi, and Benyhe (135–142). One of the salient findings from this body of work is that amphibian brain expresses kappa-like opioid binding sites, as its competitive binding profile was most correlated to the selectivity profile of the mammalian kappa opioid receptor. The main difference observed was a greater affinity of mu and delta selective opioids for the amphibian kappa-like site, and a lesser affinity for kappa-selective opioids, compared to mammalian kappa opioid receptors (143, 132). These results provided the first hint from radioligand studies that opioid receptors in non-mammalian species may be less selective than their mammalian counterparts.
The non-selective opioid ligand, diprenorphine, bound to Rana pipiens brain tissue homogenates with high affinity (144). Using the non-selective opioid antagonist, naloxone, binding studies using brain and spinal cord membranes from Rana pipiens showed a single high-affinity site which was displaced by mu, delta, and kappa selective opioid agonists with apparent affinities ranging from 1.8.6 nM to 31 μM. Surprisingly, the highly-selective opioid antagonists (beta-FNA, NTI and nor-BNI) displaced (3H)-naloxone binding with equal affinity to opioid receptors in brain and spinal cord tissue, each with an apparent affinity of about 3.0. nM (145, 146). This finding was consistent with behavioral studies showing non-selectivity of these selective antagonists and also supports the hypothesis that opioid receptors from earlier-evolved vertebrates are less selective than mammalian receptors (see below). However, using the selective opioid agonist radioligands, (3H)-DAMGO (MOR), (3H)-DPDPE (DOR), and (3H)-U65953 (KOR), three distinct opioid binding sites were identified based on different binding densities and selective competitive displacement of agonist radioligand by mu, delta, and kappa opioid ligands (147). With agonist radioligand binding, selectivity was observed such that cognate ligands were potent displacers of the selective agonist radioligand and the rank order of mu, delta, and kappa selectivity was similar to that observed in mammals. As opposed to their equipotent displacement of (3H)-naloxone binding, the highly-selective opioid antagonists (beta-FNA, nor-BNI, and NTI) were highly-selective in displacing the binding of their respective mu, delta, and kappa opioid agonist radioligands (147).
The summary of the above studies is that brain tissue of non-mammalian vertebrates contains the three types of classical opioid binding sites, similar to those characterized in mammalian tissues, but with significant differences in the binding of selective opioid ligands. There is one study characterizing ORL sites by the Hungarian group using Rana esculenta showing high-affinity, saturable binding of labeled nociceptin to brain homogenates (135). They went on to show that nociceptin initiated signal transduction through an ORL receptor by observing an increase in GTP binding to G-proteins that was blocked by ORL antagonists but not opioid antagonists.
4.3. Cloned opioid receptors in non-mammalian vertebrates
Prior to the identification of any non-mammalian, full-length opioid receptor sequences, a study utilizing a PCR strategy and degenerate primers was able to isolate opioid receptor-like fragments from genomic DNA obtained from each of the major vertebrate classes except Reptilia (148, 149). Two different sets of oligonucleotides (one having greater degeneracy than the other) were used to amplify 162 bp fragments spanning a region highly conserved in opioid receptors; the first intracellular loop to the third transmembrane domain. With this approach, partial opioid receptor sequences were identified in human (Homo sapiens), rat (Rattus norvegicus), mouse (Mus musculus), cow (Bos taurus), chicken (Gallus domesticus), bullfrog (Rana castesbeiana), shark (Alopias vulpinus), striped bass (Morone saxatilis), and hagfish (Eptatretus stoutii), but not in any of the invertebrates tested.
The first full-length clone sequenced from a non-mammalian species was MOR from the brain of the white suckerfish, Catostomus commersoni (150). Other non-mammalian species with full-length opioid receptors cloned are from the zebrafish, Danio rerio, by Rodriguez and colleagues (151–154) and from the rough-skinned newt, Taricha granulosa, in the laboratory of Moore (at the other OSU out west) (155–157). Using the same sets of primers that Evans group used in the first phylogenetic study of opioid receptors (148), the author’s research group was able to clone all four opioid receptor types expressed in Rana pipiens brain tissue; rpMOR, rpDOR, rpKOR, and rpORL (158). These were the first opioid receptors cloned from the class of Amphibia, and, on an amino acid level, showed 70 – 84% identity with their orthologous mammalian counterparts.
As noted above, the concept of an opioid unireceptor arose as the most parsimonious interpretation of the unusual pharmacology of highly-selective, type-specific opioid antagonists following spinal administration in amphibians (117, 118). Not yet having information on the types of opioid receptor proteins expressed in amphibians, it was posited that a single type of receptor mediates opioid action. The slaying of this beautiful hypothesis by the ugly fact4 of four types of opioid receptors cloned and expressed in amphibian brain and spinal cord (158) led quickly to a rebirth of the opioid unireceptor in a much earlier context, as resurrected in the last section of this review. As shown in Table 1, at present there is a dataset of six vertebrates from three different classes that have all four opioid receptor sequences from brain tissue cDNA deposited in Genbank.
Table 1.
Existing vertebrate species with cloned cDNA sequences for MOR, DOR, KOR, and ORL receptors. Access numbers are given for the NCBI protein database
| Vertebrate Class | Common name | Genus species | Access #s MOR | DOR | KOR | ORL |
|---|---|---|---|---|---|---|
| Pisces | zebrafish | Danio rerio | AAK01143 | CAA04862 | AAG60607 | AAN46747 |
| Amphibia | leopard frog | Rana pipiens | AAQ09991 | AAQ09992 | ABY82593 | AAR08905 |
| Amphibia | newt | Taricha granulosa | AAV28689 | AAV28690 | AAU15126 | AAU26067 |
| Mammalia | rat | Rattus norvigecus | AAA41630 | AAA19939 | AAA18261 | AAA50827 |
| Mammalia | mouse | Mus muscularis | AAA86878 | AAA37522 | AAA39363 | CAA62922 |
| Mammalia | human | Homo sapiens | AAA20580 | AAA83426 | AAC50158 | AAA84913 |
5. THE VERTEBRATE OPIOID RECEPTOR FAMILY
The vertebrate dataset of opioid receptor cDNA sequences yields 24 nucleotide and 24 protein sequences for phylogenetic analysis. As the focus of this review is the relationship between amino acid sequence and pharmacological action, the analyses were done using the protein data.
5.1. Alignment of protein sequences
As shown below (Figure 1), a ClustalW alignment of the present dataset of vertebrate opioid receptors shows a high degree of sequence similarity both within types of opioid receptors and among all 24 sequences. Key residues thought to be important for the function of GPCRs are present, such as the DRY motif at alignment positions 189–191 (159, 160) and the conserved Cys-Cys bridge between EL1 and EL2 provided by residues at 165 and 245 (161, 162). Na+ is known to regulate the agonist binding of GPCRs and is dependent on an allosteric binding site at the conserved Asp (134) in the TM2 domain (163, 164). There is high overall homology among existing vertebrate MOR, DOR, KOR, and ORL proteins, with nearly identical sequences among the seven transmembrane helical regions. Intracellular loop domains were also highly conserved and the N-terminus regions were most divergent, followed by the C-terminus and extracellular loop domains.
Figure 1.
Alignment of protein sequences of available sets of four opioid receptor types in six vertebrates. Protein alignment was done using ClustalW with default values (MEGA v. 4.0. at http://www.megasoftware.net). Identical sites are indicated by white text on black background, conservative substitutions noted by gray background (using Boxshade at http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py?form=boxshade). Boxed-in domains indicate the seven transmembrane regions of the receptor proteins and numbers below the sequences are the alignment positions. Key: humans, Homo sapiens (h); mouse, Mus musculus (m); rat, Rattus norvegicus (r); leopard frog, Rana pipiens (rp); rough-skinned newt, Taricha granulosa (tg); and the zebrafish, Danio rerio (dr).
There are also obvious differences between mammalian and non-mammalian sequences. For example, mammalian DOR contains an extended C-terminus sequence compared to the non-mammalian species. More striking is the eleven amino acid C-terminus extension of mammalian MOR (C-terminus, alignment positions 419–429) compared to the shorter sequence in earlier-evolved vertebrates. Phosphorylation sites along this extended sequence include Thr (position 425) which is critical for internalization and desensitization of rat mu opioid receptors (165). Interestingly, this ‘add-on’ piece of protein represents translation of exon 4, which is lacking in some hMOR and mMOR splice variants (47, 48). The differences observed in amphibians with regard to selective opioid ligands in behavioral and radioligand binding studies may result from the other substitutions in the primary sequence. The mu opioid antagonist, beta-FNA, is dependent on Lys (233) in EL2 for covalent attachment and selective mu opioid blockade in rMOR (166). The amphibian sequences show conservation of this residue in all three types of classical opioid receptors. The action of the kappa-selective opioid antagonist, nor-BNI, is dependent on the kappa-specific Glu (335) in EL3 (167) and the amphibian sequence contains a Val in this position.
5.2. Phylogenetic analysis of vertebrate opioid receptors
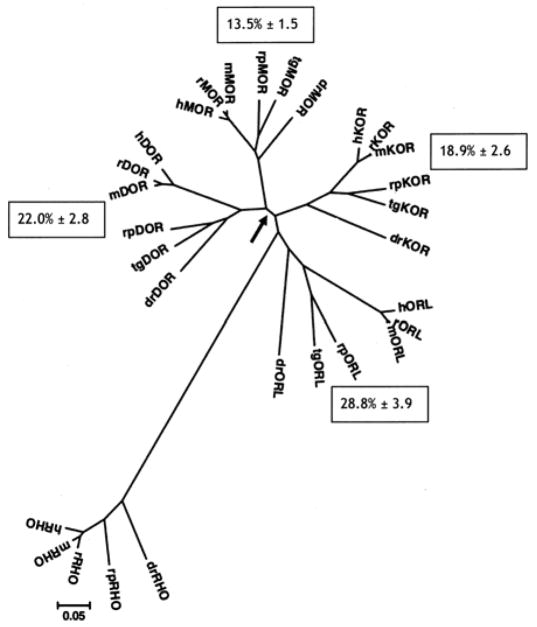
The analyses below are based on the sequence of the canonical MOR, DOR, KOR, and ORL proteins for each species listed in Table 1. Each type of opioid receptor (each set of orthologs) provided a pattern of vertebrate evolution consistent with established fossil evidence and phenotypic characteristics (see Figure 2). The radial tree was rooted with the sequences of rhodopsin (RHO, lower left) from each species with sequence available in Genbank. The ‘tree’ was rooted with an outlier group of sequences, those of the rhodopsin (RHO) protein for each species where available, a base model protein for Type A GPCRs. Overall, the four groups of opioid receptor sequences formed a dyad, with MOR and DOR sequences sharing a common ancestor (node) and KOR and ORL sharing a different common node. A similar, but unrooted tree was generated after all four types of opioid receptors were cloned and sequenced in the newt (155).
Figure 2.
Phylogenetic analysis of MOR, DOR KOR and ORL sequences in six vertebrates. Bioinformatics software (MEGA v. 4.0.) was used to generate a radial phylogenetic tree using the neighbor-joining method, rooted with the available matching sequences of rhodopsin (RHO). Protein sequences were from species listed in Table 1, where access numbers may also be found. Key: humans, Homo sapiens (h); mouse, Mus musculus (m); rat, Rattus norvegicus (r); leopard frog, Rana pipiens (rp); rough-skinned newt, Taricha granulosa (tg); and the zebrafish, Danio rerio (dr). The arrow shows the bifurcation of MOR and DOR sequences from KOR and ORL. Values in boxes by each opioid receptor type are the mean ± SEM of the pairwise distance (% divergence) among members of each type. Branch length is equal to the proportional difference among the sequences (scale bar = 0.0.5 or 5% difference in amino acid sequence).
The graphical view of MOR and DOR sharing one fork of the tree and KOR and ORL sharing the other is supported by an early study comparing a mixture of mouse and rat opioid receptor protein sequences by Chaturvedi and colleagues (168). They noted that the pairings of MOR and DOR, and KOR and ORL, were the most closely related among the rodent sequences. (168). Using the present vertebrate dataset and simple bioinformatics, these findings were confirmed, with MOR most identical to DOR (and vice versa) and KOR most identical to ORL (169).
5.3. Divergence and convergence of opioid receptor types
As pharmacological selectivity is correlated to similarity of amino acid sequences at the GPCR family level (e.g. opioid receptors vs. muscarinic receptors) type-selectivity within family members (e.g. MOR, DOR, or KOR) is also correlated with percent identity or similarity. The results of pair-wise Blast analysis (170) for the three classical types of opioid receptor sequences within each species yielded a rank order of divergence such that in earlier-evolved vertebrates, MOR, DOR, and KOR proteins were more closely related to each other than in humans and other mammals (see Table 2). For the percent similarity comparison, these values reached statistical difference such that the mean values of similarity of MOR, DOR, and KOR proteins in non-mammals were greater than the similarity of MOR, DOR, and KOR proteins in mammals. Insofar as divergence of molecular sequence is related to the greater type selectivity of opioid receptors, this finding gave rise to the hypothesis that opioid receptors are more type-selective in mammals than in non-mammalian species.
Table 2.
Comparison of vertebrate MOR, DOR, and KOR protein sequences within vertebrate species and by group using BLAST1
| Group/Species | MOR vs. DOR | MOR vs. KOR | DOR vs. KOR | Species mean (sem) | Group mean (sem) |
|---|---|---|---|---|---|
| Percent amino acid identity | |||||
| Non-mammals | |||||
| Danio rerio | 70 | 62 | 65 | 65.7. (2.3) | |
| Rana pipiens | 73 | 65 | 63 | 67.0. (3.1) | |
| Taricha granulosa | 70 | 66 | 68 | 68.0. (1.2) | 66.9. (1.2)3 |
| Mammals | |||||
| Rattus norvegicus | 66 | 61 | 61 | 62.7. (1.7) | |
| Mus musculus | 61 | 60 | 61 | 60.7. (0.3) | |
| Homo sapiens | 62 | 60 | 59 | 60.3. (0.9) | 61.2. (0.7) |
| Percent amino acid similarity | |||||
| Non-mammals | |||||
| Danio rerio | 82 | 79 | 79 | 80.0. (1.0)2 | |
| Rana pipiens | 85 | 81 | 77 | 81.0. (2.3)2 | |
| Taricha granulosa | 82 | 80 | 82 | 81.3. (0.7)2 | 80.8. (0.7)3 |
| Mammals | |||||
| Rattus norvegicus | 77 | 75 | 75 | 75.6. (0.7) | |
| Mus musculus | 72 | 75 | 74 | 73.6. (0.9) | |
| Homo sapiens | 73 | 74 | 72 | 73.0. (0.6) | 74.1. (0.5) |
ver. BLASTP 2.2.1.4., settings: matrix = Blossum62, gap open = 11, gap extension = 1, x-drop-off = 50, expect = 10.0.0, wordsize = 3, and filter off,
denotes significantly different % similarity than rat, mouse, and human mean values (p < 0.0.5, one-way ANOVA followed by post-hoc Newman-Kuels test).
denotes group means (N=9) different for identity and similarity at p < 0.0.1, Student’s t-test.
The evolution of opioid receptors has a vector of increased type-selectivity as reflected in the greatest divergence of MOR, DOR, and KOR proteins in mammalian species. In other words, opioid receptors in earlier-evolved vertebrates are less type-selective. Given the balancing constraints of maintaining the common sequence for type membership and that of adaptive evolution for greater type-selectivity, it is likely that even minor differences in MOR, DOR, and KOR sequences between vertebrate species could be detected in the binding and efficacy of selective opioid ligands. This is supported by data from studies of the first non-mammalian opioid receptor, ccMOR, cloned from the white suckerfish, Catostomus commersoni (150). In this study, ccMOR expressed in HEK cells bound the non-selective opioid antagonist naloxone with high-affinity; however the mu-selective opioid agonist, DAMGO, displaced naloxone with surprisingly low affinity. Additionally, non-selective opioid ligands bound well to zebrafish, Danio rerio delta opioid receptor (ZFOR1, labeled drDOR in Figure 2) expressed in HEK cells but low affinity was observed for selective mu, delta, or kappa opioid ligands (171). Studies of opioid receptors cloned from a second amphibian species, the rough-skinned newt Taricha granulosa, led to the conclusion that the opioid type-selectivity of newt opioid receptors was less stringent than that in mammals (155). In data presented elsewhere, parallel studies of amphibian (rpMOR) and human (hMOR) opioid receptors expressed in CHO cells showed significant differences in the affinity and efficacy of selective opioid ligands (Brasel et al., submitted).
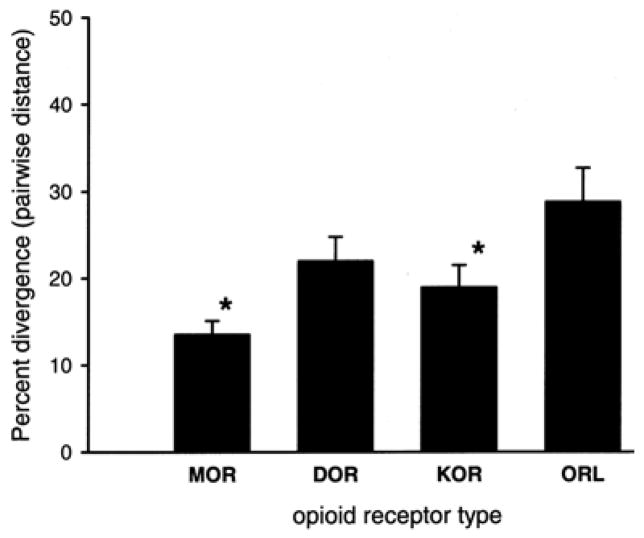
Besides comparing the MOR, DOR, KOR sequences to each other within species, which led to the discovery that the opioid receptors are less like each other in mammals than non-mammals (divergence), analysis of receptor types as a group shows that vertebrate MOR proteins are more similar to each other than ORL proteins are similar to each other (convergence). Similarly, the KOR proteins are also more similar to each other than are the ORL proteins. As shown in Figure 2 (boxed values), the mean distance of the MOR group is 13.5.% (which means that after aligning all six MOR primary sequences, 13.5.% of the sites were not identical), 18.9.% for the KOR group, 22.0.% for the DOR group, and for the group of vertebrate ORL sequences, 28.8.% of sites were not identical. Figure 3 graphs the values for each receptor type, showing that both MOR and KOR are significantly less divergent, thus more convergent, than the group of ORL proteins.
Figure 3.
The mean sequence divergence of vertebrate opioid receptor types. Pairwise distance was calculated for each group of six amino acid sequences following alignment by ClustalW (Poisson correction, uniform rate, gaps deleted; MEGA v. 4.0.). Mean plus SEM are plotted for each group. Asterisks indicate that the MOR and the KOR group significantly less divergent than the ORL group (one-way ANOVA followed by a post-hoc Newman-Keuls test, p=0.0.04; SigmaStat v. 3.1.)
The conclusion at this point from simple bioinformatic analyses is that vertebrate opioid receptors are less divergent in earlier-evolved vertebrates; however the groups of MOR and KOR proteins show signs of convergence throughout the vertebrate spectrum. MOR proteins are more similar to DOR proteins, with KOR more similar to ORL. Vertebrate ORL is most similar to rhodopsin. Next, data from the human genome and other results from the world of evolutionary biology are applied to the story of vertebrate opioid receptors.
6. THE HUMAN GENOME AND VERTEBRATE GENE DUPLICATION
This section reviews the recent data available since the completion of the human genome project, including opioid receptor gene location, the results on studies of receptor variation at single nucleotide positions (SNPs) and background information on the identification and characteristics of duplicate genes.
6.1. Opioid receptor genes in the human genome
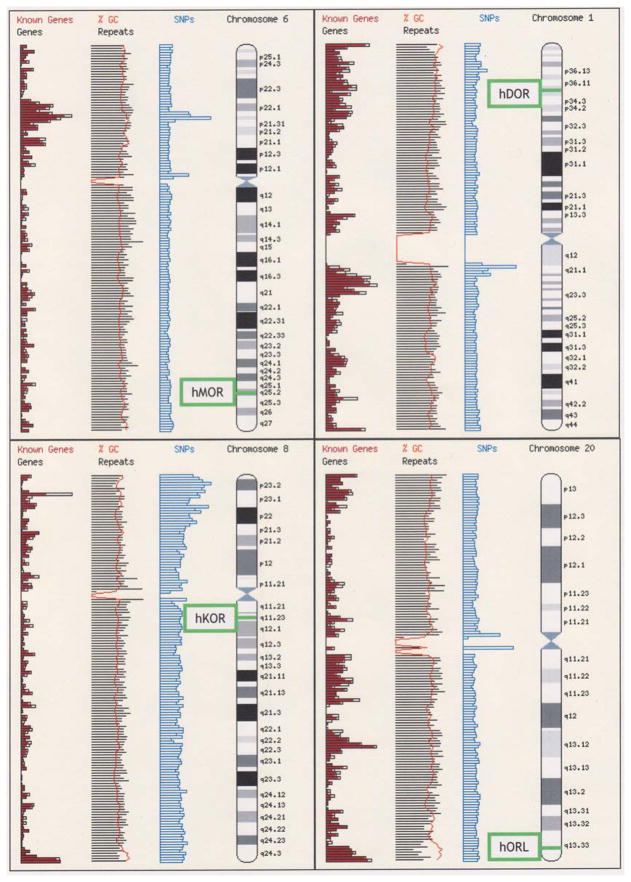
The three classical opioid receptor genes and the fourth member of the opioid receptor gene family, the ORL or nociceptin receptor gene, are mapped to four different chromosomes in the human genome (5), as shown in Fig 4. The classical opioid and ORL receptor gene loci are part of a broader set of paralogs, or common suites of genes across chromosomes that may indicate gene duplication.
Figure 4.
Chromosomal mapping of human opioid receptor genes. Image adapted from screen captures of the Ensembl Human MapView (http://www.ensembl.org/Homo_sapiens/index.html). Note that hMOR, hDOR, hKOR, and hORL map to chromosomes 6, 1, 8, and 20, respectively. Also shown in vertical columns are the density of known genes, the % GC content, and SNPs cataloged along the chromosomes. The genes are precisely located as follows: hMOR-Chr6.
The comparative or phylogenetic approach taken above uses sequence data from six key vertebrate species from three different classes of vertebrates. Simple bioinformatic analysis of this comparative dataset supported a vector of evolution such that the types of opioid receptors expressed in earlier-evolved vertebrates are more similar to each other than those found in mammals and humans. This suggests that one feature of vertebrate opioid receptor evolution is increased type-selectivity (divergence) which finds experimental support from behavioral and binding studies. Additionally, comparing the receptor sequences by type of opioid receptor among all vertebrates finds that MOR proteins are more similar to each other, followed by KOR proteins, then DOR, with ORL proteins the least similar among the different vertebrate species. Thus, these interspecific data suggest differential rates of adaptive evolution (natural or Darwinian selection) for each type of vertebrate opioid receptor.
6.2. Variation in human opioid receptor genes
The vast information available from human genomic data is beginning to allow studies of intraspecific variation due to differences in the nucleotide code from one human to the next. Generically called polymorphisms, variation data is obtaining by sampling the genome of numerous individuals from different population groups and cataloging sites where single nucleotide bases differ; i.e. single nucleotide polymorphisms or SNPs. Analysis of SNPs, or sets of associated SNPs called haplotypes, provides an insight into the evolution of particular genes and is especially useful in determining genetic defects leading to disease states or medical conditions (172–174). Although the majority of SNPs are found among intronic, repeatable sequences, or other lengths of non-coding DNA, SNPs found in coding or exonic DNA will either be classified as synonymous (mutating the nucleotide base does not change the amino acid, due to degeneracy in the codon table) or non-synonymous (changing the amino acid at that place on the protein).
Studies of polymorphism in human opioid receptor genes have focused on the association of particular SNPs with opioid-dependent and alcohol-dependent populations (175–178). Most studies have focused on the A118G allele of hMOR which changes the amino acid Asn to Asp (N40D) in the N-terminus of the hMOR protein. In general, and not without confounding data, these studies conclude that the population in question is likely to be associated with the A118G allele; the non-synSNP bringing about a ‘gain of function’ mutation. Along with some of the above studies, functional research from the Kreek laboratory team (most notably, the late Dr. LaForge) demonstrated that A118G-encoded hMOR binds beta-endorphin with a three-fold higher affinity than the canonical hMOR protein when tested in cell lines (177). Later studies with the A118G-hMOR showed little difference in beta-endorphin, but MOR-selective agonists showed reduced affinity and efficacy in stable cell lines (179). An earlier loss-of-function SNP in hMOR was described by Kieffer’s group (180). This SNP produced a Ser to Pro (S268P) switch in IL3 of hMOR. Due to the mutation in the region of the IL3 which is critical to G-protein interaction, they found that the S268P-hMOR lost signaling ability, with reduced potency and efficacy to all MOR-selective agonists tested. The location of numerous hMOR SNPs to the 5′ untranslated region (5′UTR) and upstream promoter regions as well as 3′ UTR regions of hMOR do not affect the receptor structure but do alter the expression of hMOR and may explain individual differences in pain sensitivity, drug-dependency, and the clinical efficacy of morphine (181–184). Altering the expression of genes, by adaptive evolution in non-coding DNA, is a viable outlet for Darwinian selection (185, 186). There are fewer studies of the hKOR polymorphism (187–188), hDOR (189) and only one on a single SNP in the promoter region of hORL (190). The above studies were done with small population samples (relative to the growing dbSNP at HapMap) but show that single amino acid differences in opioid receptor proteins have measurable effects on the structure-activity of the receptor and may be correlated to substance-abuse disorders.
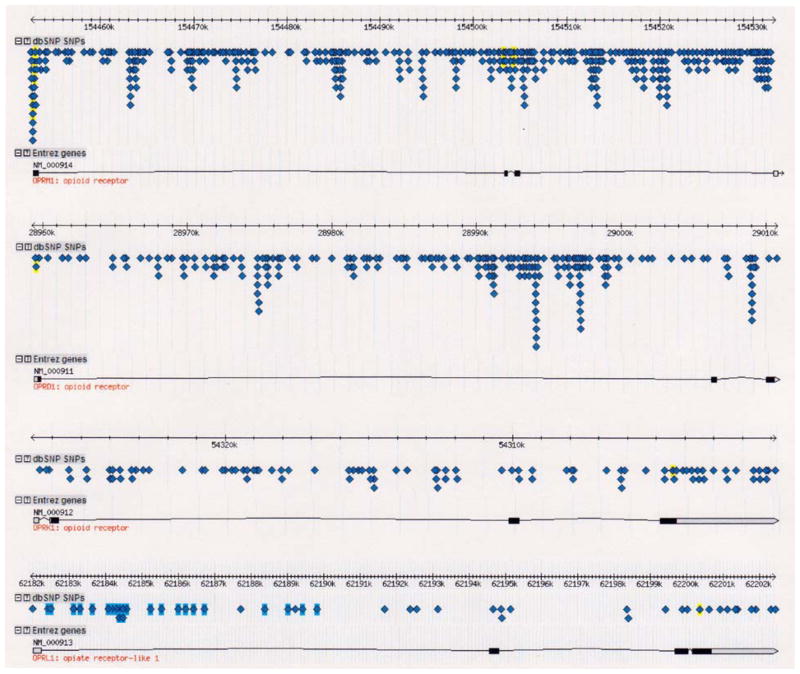
As shown in Table 3, hMOR has the greatest number of SNPs (Total SNPs column) found along its gene seuence, with 356 SNPs in the SNP reference database accessible from the HapMap project. This contrasts with the lowest number of SNPs observed along the hORL gene, with only 13 total SNPs. This table was constructed from data available from the HapMap website, using Release #21a, from January 2007 (NCBI build 35) and includes at least 3.8. million genotyped SNPs. While the population-based approach taken by the contributors to HapMap does not allow for complete genome sequencing of all individuals, there does not appear to be a sampling or ascertainment bias that could account for the results shown in Table 3 to explain the fact that hMOR has more identified SNPs (191). The rank order of total SNPs in the opioid receptor genes is hMOR >hDOR ≫hKOR>hORL. Of particular interest for the molecular evolution of opioid receptors with regard to primary structure are SNPs found within exonic regions of the receptor genes. There were 21 exonic SNPs for hMOR, three times higher than any other type of opioid receptor gene. Of these SNPs in exonic regions (coding SNP), 15 were found to be non-synonymous (non-synSNP) and six synonymous (synSNP). These values yielded a very high ratio of non-synSNP/synSNP for hMOR of 2.5.; i.e. 71 % of exonic SNPs in hMOR result in an amino acid substitution. The ratio of non-synSNP/synSNP for the other three opioid receptor genes ranged from 0.3.3 to 0.5.0. Figure 6 shows the total number of non-synononymous SNPs per human opioid receptor gene type.
Table 3.
Number and types of single-nucleotide polymorphisms (SNPs) found in human opioid receptor genes
| Gene | Total SNPs | Intron SNPs | Exon SNPs | Nonsyn SNPs | Syn SNPs | nonsynSNP/synSNP | % nonsynSNP |
|---|---|---|---|---|---|---|---|
| hMOR | 356 | 335 | 21 | 15 | 6 | 2.5.0 | 71.4.3 |
| hDOR | 351 | 248 | 4 | 2 | 2 | 0.5.0 | 50.0.0 |
| hKOR | 81 | 74 | 7 | 2 | 4 | 0.5.0 | 28.5.7 |
| hORL | 13 | 9 | 4 | 1 | 3 | 0.3.3 | 25.0.0 |
Figure 6.
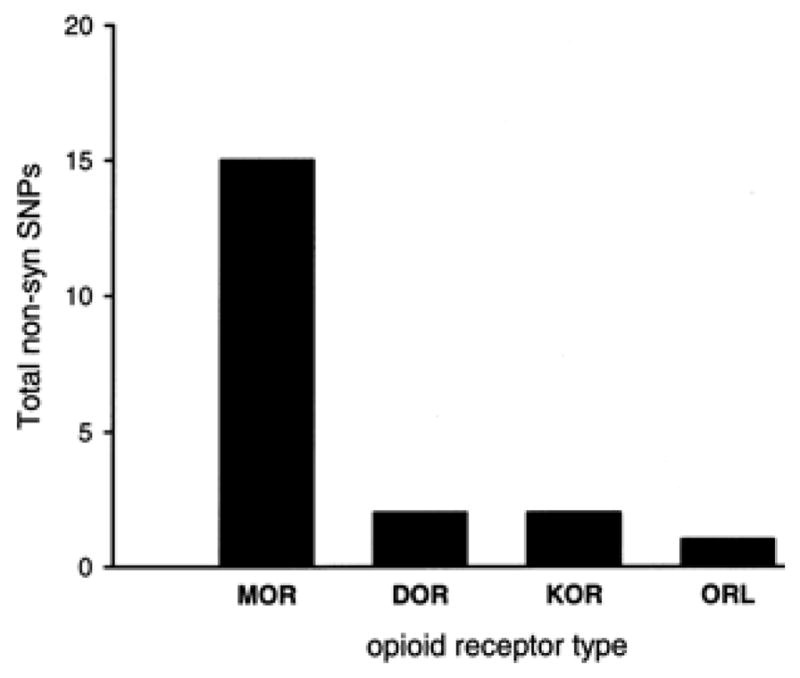
Plot of total non-synonymous single nucleotide polymorphisms (non-synSNPs) by type of human opioid receptor gene. See Table 3 for additional SNP data for human opioid receptor genes. There are a total of 15 non-syn SNPs in the exonic regions of hMOR, with 1 or 2 for the other types of opioid receptor genes.
The high degree of polymorphism in the hMOR gene resulting in amino acid changes compared to the other types of opioid receptor genes suggest that hMOR is under greater selection pressure and undergoing adaptive evolution. Under the nearly-neutral theory of evolution, all four opioid receptor genes should show rates of non-synonymous mutations much less than synonymous substitution yielding a non-synSNP/synSNP ration of ≪ 1.0. (192–195). Relatively speaking, the alleles in hMOR have not been fixed by purifying selection to the same degree as they are in hDOR, hKOR, and hORL. Comparing the interspecific differences (divergence) in vertebrate MOR, DOR, KOR, and ORL with the intraspecific variation (polymorphism) of Homo sapiens hMOR, hDOR, hKOR, and hORL is hampered by the lack of comprehensive SNP data from the five other vertebrate species. It is interesting however, that the most studied non-synonymous SNP in hMOR (A118G) is also found in the same alignment position as a non-synonymous SNP in the MOR gene of the rhesus monkey (196). This suggests that SNP data from the population studies of non-human genomes will be a fruitful approach for comparative analysis. It can be stated that in the most recently evolved species examined (humans), hMOR is the most polymorphic opioid receptor gene, followed by the group of hDOR, hKOR, and hORL. This finding suggests greater forces of adaptive evolution operating on MOR genes since the time of its birth by duplication, as outlined in the next section.
6.3. Duplication of vertebrate genes
Although not widely known among other scientists, it is accepted by most evolutionary biologists that two rounds of genome-wide duplication (paleoploidization) occurred early in vertebrate evolution, originally stated by Ohno and called the 2R hypothesis (197–199). Besides the initial explosion of quadrupled genes at the heart of vertebrate evolution, it is thought that duplicate genes (paralogs) have a gene-birth rate of 1 new pair of duplicates every million years (200). The duplication of whole genomes, parts or whole choromosomes, or individual genes are hypothesized to provide a rich source of genetic material for the evolution of new gene functions and diverse expression patterns.
The human genome contains at least 35% of its protein genes as paralogs (duplicated genes), and paralogous regions on chromosomes contain a string of similar genes correlating to one or more strings of similar genes on another chormosome (201, 202). Most paralogs do not survive, becoming pseudogenes or otherwise nonfunctional DNA sequence (203, 204). The fate of the duplicate genes that do survive is characterized by asymmetrical divergence such that one member of the pair continues as before and the other undergoes adaptive evolution as shown by a faster rate of molecular changes (205–208).
The phylogenetic pattern of (AB) (CD) in the ‘tree’ of vertebrate opioid receptor proteins (shown in Figure 2) is expected from the application of the 2R hypothesis to a single ancestral gene (209). Applied to the family of opioid receptor genes, the 2R hypothesis supports the idea that the genes encoding MOR, DOR, KOR, and ORL within a species are paralogs, i.e., genes related by duplication. The evidence for gene duplication for opioid receptors is also supported by the location of MOR, DOR, KOR and ORL genes on paralogous regions of human chromosomes (5). Human MOR and DOR genes are mapped to chromosome 6 and 1, respectively, while KOR and ORL genes map to chromosomes 8 and 20, as shown in Figure 4. The specific pairing of MOR with DOR, and KOR with ORL on separate branches of the sequence tree is supported by a genomic study whereby the greatest number of gene duplicates were found between paralogous regions on chromosomes 1 and 6, and between paralogous regions on chromosomes 8 and 20 (201).
Interestingly, there is evidence for another round of genome-wide duplication (3R) in the class of Pisces that led to the lineage of Teleost (bony fish) (210–213). In this case, it is hypothesized that there would be other types of opioid receptors (up to a total of 8) in bony fish species. In the first study cloning a full-length opioid receptor from a non-mammalian species, the white suckerfish, Catostomus commersoni, there is evidence of additional opioid receptor types in the suckerfish paper, although not discussed by the authors (150). Additional opioid receptor sequences are also predicted in the zebrafish, also a member of the class of Pisces, subclass Teleostei. Indeed, a recent paper from Rodriguez’s group in Spain reports on a second DOR-like protein cloned in the zebrafish (214).
7. THE EVOLUTION OF VERTEBRATE OPIOID RECEPTORS
A synthesis of the results from the above analysis comparing the sequences of vertebrate opioid receptors, by species and by type of opioid receptor, along with behavioral and binding data of opioids in non-mammalian species and recent data gleaned from human genome studies, suggests the following scenario for the evolution of vertebrate opioid receptors (Figure 9). Early in animal evolution, there existed a single opioid unireceptor gene5. The first round of genome duplication early in chordate evolution produced the ancestral DOR/MOR and ORL/KOR genes. A second round of genome duplication, led to the four opioid receptors present in all extant vertebrates today.
That the initial duplication led to an ancestral DOR/MOR gene is supported by the finding that the sequences of MOR and DOR are most identical to each other, as shown by above by pairwise comparison and graphically by the bifurcating pattern of the phylogenetic analysis shown in Figure 2. Likewise, the idea that KOR and ORL shared a common ancestral gene (KOR/ORL in the Figure) is supported by the above analysis as well as other data from ligand binding that suggest KOR and ORL maintain close ties. As mentioned above, it is known that duplicate genes undergo asymmetrical divergence such that one gene is under relaxed constraint, showing an increased rate of adaptive evolution (positive selection), while the other gene maintains ancestral structure and function (204–205). This asymmetry for the opioid receptor gene duplicates is noted by an ‘F’ for fast rate of adaptive evolution inside the chromosome, and conversely by an ‘S’ for the gene duplicate with a slower rate of evolution.
The gene encoding hMOR, and not any of the other opioid receptor types, was one of only nine genes controlling brain size or behavior that showed a significantly increased rate of protein evolution in the Homo sapiens genome compared to primate and rodent genomes (216). Thus, MOR is assigned an ‘F’ and conversely DOR is the ‘S’ member of the pair (see Figure 9). Likewise, because ORL protein is most closely related to the rhodopsin (RHO) sequences in the dendrogram, it is assigned an ‘S’ (maintaining more ancestral characteristics) and KOR is the faster evolving member of the pair. This is also supported by the finding that the vertebrate MOR, as a group, are more closely related to each other than vertebrate ORL (boxed values in Figure 2). Likewise, vertebrate KOR proteins share more common sequence as a group than do the vertebrate ORL proteins. Stated another way, the least divergence among all vertebrates is seen in the MOR proteins, followed by the KOR group, suggesting that adaptive evolution is greatest in the MOR genes (confirmed with the human genome studies above), less so for the KOR gene, and the least for DOR and ORL. Going back to the first duplication event, the finding that vertebrate ORL maintained most ancestral characteristics (most closely related to RHO) supports the assignment of the ORL/KOR duplicate gene as the ‘slow’ ancestor gene and DOR/MOR as the ‘fast’ duplicate.
Although many GPCR and other gene families in the human genome appear deficient in an even number of duplicate genes due to gene deletion or mutation into pseudogenes (217), it is possible that the opioid receptor gene family avoided this fate due to a gene dosage effect (increased signaling by doubling opioid receptor expression) that provided a selective advantage, as noted for other gene families (205). It is of greater interest to note that the target of most clinically used opioids is the mu opioid receptor (MOR) and selective mu opioid agonists are the most efficacious type of opioid analgesics in the clinic and in animal models. The rank order of opioid receptor efficacy in a number of (MOR ≫ KOR ≥ DOR > > ORL) approximates the rank order of the protein sequence identity of opioid receptor types among vertebrates, suggesting adaptive evolution of MOR (and to a lesser extent, KOR) in the six vertebrates examined. Thus, the molecular evolution of vertebrate opioid receptors may provide a striking example of Darwinian positive selection at the receptor level with convergent evolution in all vertebrate species leading to the most efficacious opioid receptor in the shape of the MOR protein.
8. SUMMARY AND PERSPECTIVE
The results presented here are a first approximation as the opioid receptor sequence dataset is limited. Complete cDNA sequences and the conceptual translation of the protein amino acids for MOR, DOR, KOR, and ORL are only available from six species representing three vertebrate classes. Specifically, opioid receptor sequences cloned from brain tissue cDNA of key species representing the classes of Reptilia or Aves are not available, not to mention the class of Agnatha (hagfish and lamprey). While new animal genomes are appearing online with increasing rapidity, these results are not of high-fidelity nor confirmed by expression in brain tissue of that species.
Understanding the evolution of vertebrate opioid receptor proteins contributes to the fundamental model of molecular attraction between opioid drugs and their receptor proteins. The primary structure of the protein sequence must be deterministic6 in each aspect of measured opioid function including ligand binding, conformational change, and signal transduction. The conceit of this evolutionary approach is not only to explain differences observed in the present opioid receptor proteins expressed in various vertebrate species but to seek the pattern of opioid receptor evolution to come. This will allow the design of engineered opioid receptors, most likely a variant of the MOR protein (super-MOR?) that could be usurped to provide unsurpassed analgesia, perhaps solely from endogenous opioid activation, once the inevitable arrival of gene therapy is secured.
Experiments can be performed to support the hypothesis of opioid receptor by gene duplication by using ancestral sequence analysis, as was done for steroid receptors (218). Using bioinformatics and available sequences, the most likely DOR/MOR protein sequence can be determined, a synthetic DOR/MOR gene made, transfected in CHO cells, and the ancestral receptor protein characterized. Predictions would include the binding of both MOR and DOR opioid agonists (but not KOR or ORL agonists) but perhaps less robust binding to the DOR/MOR receptor than to either MOR or DOR protein alone.
Many questions and analyses remain. Is there a relationship between number of splice variants and SNPs? The MOR protein has more of both compared to the other types. What specific amino acids determine the divergence in type-selectivity? However, it is still not known how mu, kappa, and delta opioid selectivity correlates to the primary amino acid structure of individual opioid receptor proteins. Specifically, it is not known what exact residues or domains of the primary amino acid sequence differ in each of the three proteins (MOR, DOR, and KOR) to produce mu, delta, or kappa opioid agonist type-selectivity. Continued analysis of the vertebrate dataset coupled with experimental data should provide some answers soon. What type of adaptive evolution is occurring? Certainly not purifying selection, but does the presence of a large number of SNPs in MOR signal positive or Darwinian selection, and if so, by balancing selection or diversifying selection? What are the selection pressures driving the convergent evolution of opioid receptors in different species, especially MOR and to a lesser extent KOR, and how are they balanced against the evolutionary vector of divergence of opioid receptor types? These and other questions must wait for now.
In summary, the degree of opioid receptor sequence divergence within species was correlated with vertebrate evolution. As primary amino acid sequence (structure) is a determinant of the selective binding (function) of opioid receptors, the correlation of sequence divergence with vertebrate evolution shows that opioid receptor proteins exhibit an evolutionary vector of increased type-selectivity. Additionally, the rapid rate of adaptive evolution of vertebrate MOR suggests an evolutionary vector of increased opioid receptor function. The selective pressure underlying the rapid adaptation of MOR is similar in all vertebrate species, an example of convergent evolution. It is of greater interest to note that the target of most clinically used opioids is the mu opioid receptor and selective mu opioids are the most potent type of opioid drug in the clinic and in animal models. This provides a striking example of Darwinian fitness at the molecular level should the selection pressure turn out to be a better opioid receptor. If confirmed, these findings provide a unique understanding of the pharmacology of vertebrate opioid receptors and the many other families of GPCR proteins encoded by duplicate genes.
Figure 5.
Chart of single-nucleotide polymorphisms for each human opioid receptor gene. Adapted from screen captures from the over 3 million database of SNPs (dbSNP) available at the HapMap website (http://www.hapmap.org ). Each triangular symbol represents a single SNP; the opioid receptor genes are noted by their database acronyms (OPRM1, OPRD1, OPRK1, and OPRL1 for hMOR, hDOR, hKOR, and hORL, respectively). Exons are designated by boxed regions along the line representing the genes; grayed boxes represent untranslated regions of the gene (5′ to the left, 3′ on the right) found on exon regions.
Figure 7.
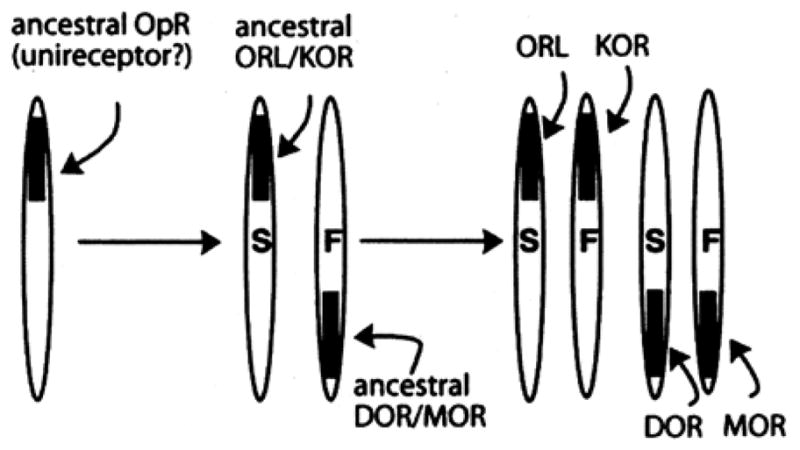
The molecular evolution of vertebrate opioid receptors. For simplicity, the genes are referred to by the same acronym as the opioid receptor proteins they encode. ‘S’ denotes slow and ‘F’ fast rate of adaptive evolution. See text for further details.
Acknowledgments
The author gratefully acknowledges the past and continued support of the National Institutes of Health,
NIDA, through research grants DA R15-12448 and DA R29-07326. Also much gratitude to my colleague, collaborator, and co-conspirator, Dr. Greg Sawyer; and to my wife and family for giving me the space mentally and physically to prepare this manuscript. Many thanks to my colleagues at the INRC for support and inspiration. This paper is dedicated to my father, Hugh E. Stevens, on the occasion of his 80th year birthday celebration.
Abbreviations
- MOR
mu opioid receptor
- DOR
delta opioid receptor
- KOR
kappa opioid receptor
- ORL
nociceptin receptor
- GPCR
G-protein coupled receptors
- beta-FNA
beta-funaltrexamine
- NTI
naltrindole
- nor-BNI
nor-binaltorphimine
- UTR
untranslated region
- SNP
single nucleotide positions
Footnotes
The convention for naming opioid receptor proteins here follows a hybrid system. For example, the mu opioid receptor in humans, mice and rats is hMOR, mMOR, and rMOR, but for all other species a representation of the Linnaean binomial taxonomy is used such that in the zebrafish, Danio rerio, the receptor is drMOR, in the leopard frog, Rana pipiens, rpMOR, and in the rough-skinned newt, Taricha granulosa, tgMOR. Likewise for the DOR, KOR, and ORL proteins. For simplification, the genes for these proteins are referred to using italicized acronyms; i.e. the gene that codes for hMOR is hMOR.
Dr. Goldstein was a founding member and Executive Secretary of the International Narcotics Research Conference (INRC) from 1972 to 1976. More about this scientific society for the study of opioids is available at www.inrcworld.org.
Ockham’s razor (also ‘Ockham’s law’) is a principle attributed to the 14th-century English intellectual and Franciscan monk, William of Ockham. The principle states that the explanation of a phenomenon should make as few assumptions as possible, disregarding any that do not change the observable predictions of the explanatory hypothesis.
The complete quote is: “The great tragedy of Science - the slaying of a beautiful hypothesis by an ugly fact.” --Thomas H. Huxley (1825–1895), English biologist and ardent supporter of Charles Darwin.
The unireceptor is likely a duplicate of a proto-unireceptor gene, perhaps from the time of the arthropod and chordate split, long before the posited 2R whole genome duplication at the root of the vertebrate evolution. There is even evidence that a 7 transmembrane (TM) proto-GPCR was formed by duplication of proto-gene encoding a helical-loop protein corresponding to TM regions 1–3, as there is surprising similarity in amino acids when this region is aligned with TM 5–7 regions (215).
There are, of course, other factors such as the surrounding milieu of the receptor protein in the membrane; lipid membrane constituents, levels of signal molecules, etc., that also modulate the function of the receptor. However, considerations limited to the differences in the primary amino acid sequences of vertebrate opioid receptors are a good place to start.
References
- 1.Attwood TK. A compendium of specific motifs for diagnosing GPCR subtypes. Trends Pharmacol Sci. 2001;22:162–165. doi: 10.1016/s0165-6147(00)01658-8. [DOI] [PubMed] [Google Scholar]
- 2.Perez DM. The evolutionarily triumphant g-protein-coupled receptor. Mol Pharmacol. 2003;63:1202–1205. doi: 10.1124/mol.63.6.1202. [DOI] [PubMed] [Google Scholar]
- 3.Perez DM. From plants to man: the GPCR “tree of life”. Mol Pharmacol. 2005;67:1383–1384. doi: 10.1124/mol.105.011890. [DOI] [PubMed] [Google Scholar]
- 4.Bockaert J, Pin JP. Molecular tinkering of G protein-coupled receptors: an evolutionary success. The EMBO Journal. 1999;18:1723–1729. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families: Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 6.Fredriksson R, Schiotch HB. The repertoire of g-protein-coupled receptors in fully sequenced genomes. Mol Pharmacol. 2005;67:1414–1425. doi: 10.1124/mol.104.009001. [DOI] [PubMed] [Google Scholar]
- 7.Bodnar RJ. Endogenous opiates and behavior: 2006. Peptides. 2007;28:2435–2513. doi: 10.1016/j.peptides.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin WR, Eades CG, Thompson JA, Huppler RE, Gilbert PE. The effects of morphine-and nalorphine-like drugs in the nondependent and morphine-dependent chronic spinal dog. J Pharmacol Exp Ther. 1976;197:517–532. [PubMed] [Google Scholar]
- 9.Lord JAH, Waterfield AA, Hughes J, Kosterlitz HW. Endogenous opioid peptides: multiple agonists and receptors. Nature. 1977;267:495–499. doi: 10.1038/267495a0. [DOI] [PubMed] [Google Scholar]
- 10.Portoghese PS, Sultana M, Takemori AE. Naltrindole, a highly selective and potent non-peptide delta opioid receptor antagonist. Eur J Pharm. 1988;146:185–186. doi: 10.1016/0014-2999(88)90502-x. [DOI] [PubMed] [Google Scholar]
- 11.Takemori AE, Ho BY, Naeseth JS, Portoghese PS. Nor-binaltorphimine, a highly selective kappa-opioid antagonist in analgesic and receptor binding assays. J Pharmacol Exp Ther. 1988;246:255–258. [PubMed] [Google Scholar]
- 12.Ward SJ, Portoghese PS, Takemori AE. Pharmacological profiles of beta-funaltrexamine (B-FNA) and Beta-Chlornaltrexamine (B-CNA) on the mouse vas deferens preparation. Eur J Pharmacol. 1982;80:377–384. doi: 10.1016/0014-2999(82)90083-8. [DOI] [PubMed] [Google Scholar]
- 13.Beckett AH, Casy AF. Synthetic analgesics: stereochemical considerations. J Pharm Pharmacol. 1954;6:986–1001. doi: 10.1111/j.2042-7158.1954.tb11033.x. [DOI] [PubMed] [Google Scholar]
- 14.Pert CB, Snyder SH. Opiate receptor: demonstration in nervous tissue. Science. 1973;179:1011–1014. doi: 10.1126/science.179.4077.1011. [DOI] [PubMed] [Google Scholar]
- 15.Simon EJ, Hiller JM, Edelman I. Stereospecific binding of the potent narcotic analgesic (3H) etorphine to rat-brain homogenate. Proc Natl Acad Sci USA. 1973;70:1947–1949. doi: 10.1073/pnas.70.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terenius L. Stereospecific interaction between narcotic analgesics and a synaptic plasma membrane fraction of rat cerebral cortex. Acta Pharmacol Toxicol. 1973;32:317–320. doi: 10.1111/j.1600-0773.1973.tb01477.x. [DOI] [PubMed] [Google Scholar]
- 17.Loh H, Smith A. Molecular characterization of opioid receptors. Annu Rev Pharmacol Toxicol. 1990;30:123–147. doi: 10.1146/annurev.pa.30.040190.001011. [DOI] [PubMed] [Google Scholar]
- 18.Simon EJ. Recent studies on opioid receptors: heterogeneity and purification. Ann New York Acad Sci. 1986;463:31–45. doi: 10.1111/j.1749-6632.1986.tb21501.x. [DOI] [PubMed] [Google Scholar]
- 19.Zaki PA, Bilsky EJ, Vanderah TW, Lai J, Evans CJ, Porreca F. Opioid receptor types and subtypes: The delta receptor as a model. Ann Rev Pharmacol Toxicol. 1996;36:379–401. doi: 10.1146/annurev.pa.36.040196.002115. [DOI] [PubMed] [Google Scholar]
- 20.Wollemann M, Benyhe S, Simon J. The kappa opioid receptor: evidence for the different subtypes. Life Sci. 1993;52:599–611. doi: 10.1016/0024-3205(93)90451-8. [DOI] [PubMed] [Google Scholar]
- 21.Kieffer BL, Befort K, Gaveriaux-Ruff C, Hirth CG. The delta-opioid receptor: Isolation of a cDNA by expression cloning and pharmacological characterization. Proc Natl Acad Sci USA. 1992;89:12048–12052. doi: 10.1073/pnas.89.24.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans CJ, Keith DE, Jr, Morrison H, Magendzo K, Edwards RH. Cloning of a delta opioid receptor by functional expression. Science. 1992;258:1952–1955. doi: 10.1126/science.1335167. [DOI] [PubMed] [Google Scholar]
- 23.Kano-Tanaka K, Higashida H, Fukami H, Tanaka T. Double minutes in mouse neuroblastoma cells and their hybrids. Cancer Genet Cytogenet. 1982;5:51–62. doi: 10.1016/0165-4608(82)90040-1. [DOI] [PubMed] [Google Scholar]
- 24.Wang JB, Johnson PS, Persico AM, Hawkins AL, Griffin CA, Uhl GR. Human mu opiate receptor. cDNA and genomic clones, pharmacologic characterization and chromosomal assignment. FEBS Lett. 1994;338:217–222. doi: 10.1016/0014-5793(94)80368-4. [DOI] [PubMed] [Google Scholar]
- 25.Knapp RJ, Malatynska E, Fang L, Li X, Babin E, Nguyen M, Santoro G, Varga EV, Hruby VJ, Roeske WR. Identification of a human delta opioid receptor: cloning and expression. Life Sci. 1994;54:L463–L469. doi: 10.1016/0024-3205(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 26.Simonin F, Befort K, Gaveriaux-Ruff C, Matthes H, Nappey V, Lannes B, Micheletti G, Kieffer B. The human delta-opioid receptor: genomic organization, cDNA cloning, functional expression, and distribution in human brain. Mol Pharmacol. 1994;46:1015–1021. [PubMed] [Google Scholar]
- 27.Mansson E, Bare L, Yang D. Isolation of a human kappa opioid receptor cDNA from placenta. Biochem Biophys Res Commun. 1994;202:1431–1437. doi: 10.1006/bbrc.1994.2091. [DOI] [PubMed] [Google Scholar]
- 28.Zhu J, Chen C, Xue JC, Kunapuli S, DeRiel JK, Liu-Chen LY. Cloning of a human kappa opioid receptor from the brain. Life Sci. 1995;56:L201–L207. doi: 10.1016/0024-3205(94)00507-o. [DOI] [PubMed] [Google Scholar]
- 29.Simonin F, Gaveriaux-Ruff C, Befort K, Matthes H, Lannes B, Micheletti G, Mattei MG, Charron G, Bloch B, Kieffer B. Kappa-opioid receptor in humans: cDNA and genomic cloning, chromosomal assignment, functional expression, pharmacology, and expression pattern in the central nervous system. Proc Natl Acad Sci USA. 1995;92:7006–7010. doi: 10.1073/pnas.92.15.7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mollereau C, Parmentier M, Mailleux P, Butour J-L, Moisand C, Chalon P, Caput D, Vassart G, Meunier J-C. ORL1, a novel member of the opioid receptor family: cloning, functional expression and localization. FEBS Lett. 1994;341:33–38. doi: 10.1016/0014-5793(94)80235-1. [DOI] [PubMed] [Google Scholar]
- 31.Fukuda K, Kato S, Mori K, Nishi M, Takeshima H, Iwabe N, Miyata T, Houtani T, Sugimoto T. cDNA cloning and regional distribution of a novel member of the opioid receptor family. FEBS Lett. 1994;343:42–46. doi: 10.1016/0014-5793(94)80603-9. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Fan Y, Liu J, Mestek A, Tian M, Kozak CA, Yu L. Molecular cloning, tissue distribution and chromosomal localization of a novel member of the opioid receptor gene family. FEBS Lett. 1994;347:279–283. doi: 10.1016/0014-5793(94)00560-5. [DOI] [PubMed] [Google Scholar]
- 33.Bunzow JR, Saez C, Mortrud M, Bouvier C, Williams JT, Low M, Grandy DK. Molecular cloning and tissue distribution of a putative member of the rat opioid receptor gene family that is not a mu, delta, or kappa opiod receptor type. FEBS Lett. 1994;347:284–288. doi: 10.1016/0014-5793(94)00561-3. [DOI] [PubMed] [Google Scholar]
- 34.Wang JB, Johnson PS, Imai Y, Persico AM, Ozenberger BA, Eppler CM, Uhl GR. cDNA cloning of an orphan opiate receptor gene family member and its splice variant. FEBS Lett. 1994;348:75–79. doi: 10.1016/0014-5793(94)00557-5. [DOI] [PubMed] [Google Scholar]
- 35.Lachowicz JE, Shen Y, Monsma FJ, Jr, Sibley DR. Molecular cloning of a novel G protein-coupled receptor related to the opiate receptor family. J Neurochem. 1995;64:34–40. doi: 10.1046/j.1471-4159.1995.64010034.x. [DOI] [PubMed] [Google Scholar]
- 36.Wick MJ, Minnerath SR, Lin X, Elde R, Law PY, Loh HH. Isolation of a novel cDNA encoding a putative membrane receptor with high homology to the cloned mu, delta, and kappa opioid receptors. Brain Res. 1994;27:37–44. doi: 10.1016/0169-328x(94)90181-3. [DOI] [PubMed] [Google Scholar]
- 37.Meunier J-C, Mollereau C, Toll L, Suaudeau C, Moisand C, Alvinerie P, Butour J-L, Guillemot JC, Ferrara P, Monsarrat B. Isolation and structure of the endogenous agonist of opioid receptor-like ORL1 receptor. Nature. 1995;377:532–535. doi: 10.1038/377532a0. [DOI] [PubMed] [Google Scholar]
- 38.Reinscheid RK, Nothacker HP, Bourson A, Ardati A, Henningsen RA, Bunzow JR, Grandy DK, Langen H, Monsma FJ, Civelli O. Orphanin FQ: a neuropeptide that activates an opioidlike G protein-coupled receptor. Science. 1995;270:792–794. doi: 10.1126/science.270.5237.792. [DOI] [PubMed] [Google Scholar]
- 39.Meunier J-C. Nociceptin/orphanin FQ and the opioid receptor-like ORL1 receptor. Eur J Pharmacol. 1997;340:1–15. doi: 10.1016/s0014-2999(97)01411-8. [DOI] [PubMed] [Google Scholar]
- 40.Mogil JS, Pasternak G. The molecular and behavioral pharmacology of the orphanin FQ/nociceptin peptide and receptor family. Pharmacol Rev. 2001;53:381–415. [PubMed] [Google Scholar]
- 41.Chen X, Liu-Chen LY, Tallarida RJ, Geller EB, de Riel JK, Adler MW. Use of a mu-antisense oligodeoxynucleotide as a mu opioid receptor noncompetitive antagonist in vivo. Neurochem Res. 1996;21:1363–1368. doi: 10.1007/BF02532377. [DOI] [PubMed] [Google Scholar]
- 42.Wang HQ, Kampine JP, Tseng L-F. Antisense oligodeoxynucleotide to a delta-opioid receptor messenger RNA selectively blocks the antinociception induced by intracerebroventricularly administered delta-, but not mu-, epsilon-, or kappa-opioid receptor agonists in the mouse. Neurosci. 1996;75:445–452. doi: 10.1016/0306-4522(96)00309-0. [DOI] [PubMed] [Google Scholar]
- 43.Pasternak GW, Standifer KM. Mapping of opioid receptors using antisense oligodeoxynucleotides: correlating their molecular biology and pharmacology. Trends Pharmacol Sci. 1995;16:344–350. doi: 10.1016/s0165-6147(00)89068-9. [DOI] [PubMed] [Google Scholar]
- 44.Uhl GR, Childers S, Pasternak G. An opiate-receptor gene family reunion. Trends Neurosci. 1994;17:89–93. doi: 10.1016/0166-2236(94)90110-4. [DOI] [PubMed] [Google Scholar]
- 45.Pan Y-X, Xu J, Bolan E, Abbadie C, Chang A, Zuckerman A, Rossi G, Pasternak GW. Identification and characterization of three new alternatively spliced mu-opioid receptor isoforms. Mol Pharmacol. 1999;56:396–403. doi: 10.1124/mol.56.2.396. [DOI] [PubMed] [Google Scholar]
- 46.Pan Y-X, Cheng J, Xu J, Rossi G, Jacobson E, Ryan-Moro J, Brooks AI, Dean GE, Standifer KM, Pasternak GW. Cloning and functional characterization through antisense mapping of a kappa3-related opioid receptor. Mol Pharmacol. 1995;47:1180–1188. [PubMed] [Google Scholar]
- 47.Pan YX, Xu J, Bolan E, Moskowitz HS, Xu M, Pasternak GW. Identification of four novel exon 5 splice variants of the mouse mu-opioid receptor gene: functional consequences of C-terminal splicing. Mol Pharmacol. 2005;68:866–875. doi: 10.1124/mol.105.011858. [DOI] [PubMed] [Google Scholar]
- 48.Pan YX, Xu J, Mahurter L, Xu M, Gilbert AK, Pasternak GW. Identification and characterization of two new human mu opioid receptor splice variants, hMOR-1O and hMOR-1X. Biochem Biophys Res Commun. 2003;301:1057–1061. doi: 10.1016/s0006-291x(03)00089-5. [DOI] [PubMed] [Google Scholar]
- 49.Pasternak GW. Molecular biology of opioid analgesia. J Pain Symptom Manag. 2005;29:S2–S9. doi: 10.1016/j.jpainsymman.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 50.Gaveriaux-Ruff C, Peluso J, Befort K, Simonin F, Zilliox C, Kieffer BL. Detection of opioid receptor mRNA by RT-PCR reveals alternative splicing for the delta- and kappa-opioid receptors. Mol Brain Res. 1997;48:298–304. doi: 10.1016/s0169-328x(97)00109-5. [DOI] [PubMed] [Google Scholar]
- 51.Wei LN, Law PY, Loh HH. Post-transcriptional regulation of opioid receptors in the nervous system. Front Biosci. 2004;9:1665–1679. doi: 10.2741/1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan YX, Xu J, Wan BL, Zuckerman A, Pasternak GW. Identification and differential regional expression of KOR-3/ORL-1 gene splice variants in mouse brain. FEBS Lett. 1998;435:65–68. doi: 10.1016/s0014-5793(98)01039-4. [DOI] [PubMed] [Google Scholar]
- 53.Nistri A, Pepeu G, Cammelli E, Spina L, De Bellis AM. Effects of morphine on brain and spinal acetylcholine levels and nociceptive threshold in the frog. Brain Res. 1974;80:199–209. doi: 10.1016/0006-8993(74)90684-2. [DOI] [PubMed] [Google Scholar]
- 54.Nistri A, Pepeu G. Increase in brain and spinal acetylcholine levels without antinociceptive actions following morphine administration in the frog. Br J Pharmacol. 1973;47:650P–651P. [PMC free article] [PubMed] [Google Scholar]
- 55.Pezalla PD. Morphine-induced analgesia and explosive motor behavior in an amphibian. Brain Res. 1983;273:297–305. doi: 10.1016/0006-8993(83)90854-5. [DOI] [PubMed] [Google Scholar]
- 56.Sei CA, Richard R, Dores RM. Steady-state levels of pro-dynorphin related end-products from the brain of the amphibian, Xenopus laevis. Brain Res. 1989;479:162–166. doi: 10.1016/0006-8993(89)91347-4. [DOI] [PubMed] [Google Scholar]
- 57.Dores RM, McDonald LK, Steveson TC, Sei CA. The molecular evolution of neuropeptides: prospects for the 90’s. Brain Behav Evol. 1990;36:80–99. doi: 10.1159/000115300. [DOI] [PubMed] [Google Scholar]
- 58.Dores RM, McDonald LK, Purdom LC, Sei CA. Reversed phase HPLC analysis of proenkephalin-related and prodynorphin-related end-products in the brain of a Urodele amphibian, Ambystoma tigrinum. Brain Behav Evol. 1993;42:69–76. doi: 10.1159/000114141. [DOI] [PubMed] [Google Scholar]
- 59.Lee J, Danielson P, Sollars C, Alrubaian J, Balm P, Dores RM. Cloning of a neoteleost (Oreochromis mossambicus) proopiomelanocortin (POMC) cDNA reveals a deletion of the gamma-melanotropin region and most of the joining peptide region: implications for POMC processing. Peptides. 1999;20:1391–1399. doi: 10.1016/s0196-9781(99)00148-5. [DOI] [PubMed] [Google Scholar]
- 60.Lecaude S, Alrubaian J, Sollars C, Propper C, Danielson P, Dores RM. Organization of proenkephalin in amphibians: cloning of a proenkephalin cDNA from the brain of the anuran amphibian, Spea multiplicatus. Peptides. 2000;21:339–344. doi: 10.1016/s0196-9781(00)00154-6. [DOI] [PubMed] [Google Scholar]
- 61.Danielson PB, Hoversten MT, Fitzpatrick M, Schreck C, Akil H, Dores RM. Sturgeon orphanin, a molecular “fossil” that bridges the gap between the opioids and orphanin FQ/nociceptin. J Biol Chem. 2001;276:22114–22119. doi: 10.1074/jbc.M011741200. [DOI] [PubMed] [Google Scholar]
- 62.Dores RM, Lecaude S, Bauer D, Danielson PB. Analyzing the evolution of the opioid/orphanin gene family. Mass Spectrum Rev. 2002;21:220–243. doi: 10.1002/mas.10029. [DOI] [PubMed] [Google Scholar]
- 63.Dores RM, Lecaude S. Trends in the evolution of the proopiomelanocortin gene. Gen Comp Endocrinol. 2005;142:81–93. doi: 10.1016/j.ygcen.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 64.Alrubaian J, Lecaude S, Barba J, Szynskie L, Jacobs N, Bauer D, Brown C, Kaminer I, Bagrosky B, Dores RM. Trends in the evolution of the preprodynorphin gene in teleosts: Cloning of eel and tilapia prodynorphin cDNAs. Peptides. 2006;27:797–804. doi: 10.1016/j.peptides.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 65.Stevens CW, Pezalla PD. A spinal site mediates opiate analgesia in frogs. Life Sci. 1983;33:2097–2103. doi: 10.1016/0024-3205(83)90333-8. [DOI] [PubMed] [Google Scholar]
- 66.Stevens CW, Pezalla PD. Naloxone blocks the analgesic action of levorphanol but not of dextrorphan in the leopard frog. Brain Res. 1984;301:171–174. doi: 10.1016/0006-8993(84)90418-9. [DOI] [PubMed] [Google Scholar]
- 67.Pezalla PD, Stevens CW. Behavioral effects of morphine, levorphanol, dextrorphan and naloxone in the frog, Rana pipiens. Pharmacol Biochem Behav. 1984;21:213–217. doi: 10.1016/0091-3057(84)90217-x. [DOI] [PubMed] [Google Scholar]
- 68.Stevens CW, Pezalla PD, Yaksh TL. Spinal antinociceptive action of three representative opioid peptides in frogs. Brain Res. 1987;402:201–203. doi: 10.1016/0006-8993(87)91069-9. [DOI] [PubMed] [Google Scholar]
- 69.Willenbring S, Stevens CW. Thermal, mechanical and chemical peripheral sensation in amphibians: opioid and adrenergic effects. Life Sci. 1996;58:125–133. doi: 10.1016/0024-3205(95)02265-1. [DOI] [PubMed] [Google Scholar]
- 70.Hamamoto DT, Forkey MW, Davis WL, Kajander KC. The role of pH and osmolarity in evoking the acetic acid-induced wiping response in a model of nociception in frogs. Brain Res. 2000;862:217–229. doi: 10.1016/s0006-8993(00)02138-7. [DOI] [PubMed] [Google Scholar]
- 71.Hamamoto DT, Simone DA. Characterization of cutaneous primary afferent fibers excited by acetic acid in a model of nociception in frogs. J Neurophysiol. 2003;90:566–577. doi: 10.1152/jn.00324.2003. [DOI] [PubMed] [Google Scholar]
- 72.Stevens CW, Kirkendall K. Time course and magnitude of tolerance to the analgesic effects of systemic morphine in amphibians. Life Sci. 1993;52:PL111–116. doi: 10.1016/0024-3205(93)90097-m. [DOI] [PubMed] [Google Scholar]
- 73.Pezalla PD, Dicig M. Stress-induced analgesia in frogs: evidence for the involvement of an opioid system. Brain Res. 1984;296:356–360. doi: 10.1016/0006-8993(84)90073-8. [DOI] [PubMed] [Google Scholar]
- 74.Stevens CW, Sangha S, Ogg BG. Analgesia produced by immobilization stress and an enkephalinase-inhibitor in amphibians. Pharmacol Biochem Behav. 1995;51:675–680. doi: 10.1016/0091-3057(94)00436-m. [DOI] [PubMed] [Google Scholar]
- 75.Willenbring S, Stevens CW. Spinal mu, delta, and kappa opioids alter chemical, mechanical and thermal sensitivities in amphibians. Life Sci. 1997;61:2167–2176. doi: 10.1016/s0024-3205(97)00919-3. [DOI] [PubMed] [Google Scholar]
- 76.Brenner GM, Klopp AJ, Deason LL, Stevens CW. Analgesic potency of alpha adrenergic agents after systemic administration in amphibians. J Pharmacol Exp Ther. 1994;270:540–545. [PubMed] [Google Scholar]
- 77.Stevens CW, Brenner GM. Spinal administration of adrenergic agents produces analgesia in amphibians. Eur J Pharmacol. 1996;316:205–210. doi: 10.1016/s0014-2999(96)00681-4. [DOI] [PubMed] [Google Scholar]
- 78.Mohan S, Stevens CW. Systemic and spinal administration of the mu opioid, remifentanil, produces antinociception in amphibians. Eur J Pharmacol. 2006;534:89–94. doi: 10.1016/j.ejphar.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stevens CW, Toth G, Borsodi A, Benyhe S. Xendorphin B1, a novel opioid-like peptide determined from a Xenopus laevis brain cDNA library, produces opioid antinociception after spinal administration in amphibians. Brain Res Bull. 2007;71:628–632. doi: 10.1016/j.brainresbull.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stevens CW, Klopp AJ, Facello JA. Analgesic potency of mu and kappa opioids after systemic administration in amphibians. J Pharmacol Exp Ther. 1994;269:1086–1093. [PubMed] [Google Scholar]
- 81.Stevens CW. Relative analgesic potency of mu, delta and kappa opioids after spinal administration in amphibians. J Pharmacol Exp Ther. 1996;276:440–448. [PubMed] [Google Scholar]
- 82.Stevens CW, Rothe KS. Supraspinal administration of opioids with selectivity for mu, delta, and kappa opioid receptors produces analgesia in amphibians. Eur J Pharmacol. 1997;331:15–21. doi: 10.1016/s0014-2999(97)01026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stevens CW. Opioid antinociception in amphibians. Brain Res Bull. 1988;21:959–962. doi: 10.1016/0361-9230(88)90034-2. [DOI] [PubMed] [Google Scholar]
- 84.Stevens CW. Opioid research in amphibians: an alternative pain model yielding insights on the evolution of opioid receptors. Brain Res Rev. 2004;46:204–215. doi: 10.1016/j.brainresrev.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stevens CW. Alternatives to the use of mammals for pain research. Life Sci. 1992;50:901–912. doi: 10.1016/0024-3205(92)90167-n. [DOI] [PubMed] [Google Scholar]
- 86.Stevens CW. Non-mammalian models for the study of pain. In: Conn PM, editor. Sourcebook of models for biomedical research. Humana Press; Totowa, New Jersey, USA: 2008. [Google Scholar]
- 87.Suckow M, Terril L, Grigdesby C, March P. Evaluation of hypothermia-induced analgesia and influence of opioid antagonists in leopard frogs (Rana pipiens) Pharmacol Biochem Behav. 1999;63:39–43. doi: 10.1016/s0091-3057(98)00237-8. [DOI] [PubMed] [Google Scholar]
- 88.Kanetoh T, Sugikawa T, Sasaki I, Muneoka Y, Minakata H, Takabatake I, Fujimoto M. Identification of a novel frog RFamide and its effect on the latency of the tail-flick response of the newt. Comp Biochem Physiol C Toxicol Pharmacol. 2003;134:259–266. doi: 10.1016/s1532-0456(02)00277-6. [DOI] [PubMed] [Google Scholar]
- 89.Jansen GA, Greene NM. Morphine metabolism and morphine tolerance in goldfish. Anesthesiol. 1970;32:231–235. doi: 10.1097/00000542-197003000-00011. [DOI] [PubMed] [Google Scholar]
- 90.Ehrensing RH, Michell GF, Kastin AJ. Similar antagonism of morphine analgesia by MIF-1 and naloxone in Carassius auratus. Pharmacol Biochem Behav. 1982;17:757–761. doi: 10.1016/0091-3057(82)90358-6. [DOI] [PubMed] [Google Scholar]
- 91.Stevens CW, Yaksh TL. Spinal action of dermorphin, an extremely potent opioid peptide from frog skin. Brain Res. 1986;385:300–304. doi: 10.1016/0006-8993(86)91076-0. [DOI] [PubMed] [Google Scholar]
- 92.Chervova LS, Kamenskii AA, Malyukhina GA, Baturina EY, Deigin VI, Yaroav EP. Study of dermorphin’s intranasal action mechanism in fish and mammal species. Zhurnal Evolyutsionnoi Biokhimii i Fiziologii. 1992;28:45–48. [PubMed] [Google Scholar]
- 93.Chervova LS, Lapshin DN, Kamenskii AA. Pain sensitivity of trout and analgesia induced by intranasal administration of dermorphine. Dokl Biol Sci. 1994;338:424–425. [PubMed] [Google Scholar]
- 94.Chervova LS. Pain sensitivity and behavior of fishes. J Ichthyol. 1997;37:98–102. [Google Scholar]
- 95.Chervova LS, Lapshin DN. Opioid modulation of pain threshold in fish. Dokl Biol Sci. 2000;375:703–704. doi: 10.1023/a:1026681519613. [DOI] [PubMed] [Google Scholar]
- 96.Sneddon LU, Braithwaite VA, Gentle MJ. Do fishes have nociceptors? Evidence for the evolution of a vertebrate sensory system. Proc Biol Sci. 2003;270:1115–1121. doi: 10.1098/rspb.2003.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sneddon LU. Evolution of nociception in vertebrates. comparative analysis of lower vertebrates. Brain Res Rev. 2004;46:123–130. doi: 10.1016/j.brainresrev.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 98.Sneddon LU, Braithwaite VA, Gentle MJ. Novel object test: examining nociception and fear in the rainbow trout. J Pain. 2003;4:431–440. doi: 10.1067/s1526-5900(03)00717-x. [DOI] [PubMed] [Google Scholar]
- 99.Sneddon LU. The evidence for pain in fish: the use of morphine as an analgesic. Appl Anim Behav Sci. 2003;83:153–162. [Google Scholar]
- 100.Mauk MD, Olson RD, LaHoste GJ, Olson GA. Tonic immobility produces hyperanalgesia and antagonizes morphine analgesia. Science. 1981;213:353–354. doi: 10.1126/science.7244620. [DOI] [PubMed] [Google Scholar]
- 101.Kanui TI, Hole K, Miaron JO. Nociception in crocodiles: capsaicin instillation, formalin and hot plate tests. Zool Sci. 1990;7:537–540. [Google Scholar]
- 102.Kanui TI, Hole K. Morphine and pethidine antinociception in the crocodile. J Vet Pharmacol Ther. 1992;15:101–103. doi: 10.1111/j.1365-2885.1992.tb00993.x. [DOI] [PubMed] [Google Scholar]
- 103.Hawkins MG. The use of analgesics in birds, reptiles, and small exotic mammals. J Exot Pet Med. 2006;15:177–192. [Google Scholar]
- 104.Schneider C. Effects of morphine-like drugs in chicks. Nature. 1961;191:607–608. doi: 10.1038/191607b0. [DOI] [PubMed] [Google Scholar]
- 105.Bardo MT, Hughes RA. Brief communication. Shock-elicited flight response in chickens as an index of morphine analgesia. Pharmacol Biochem Behav. 1978;9:147–149. doi: 10.1016/0091-3057(78)90027-8. [DOI] [PubMed] [Google Scholar]
- 106.Hughes RA, Sufka KJ. The ontogeny of thermal nociception in domestic fowl: thermal stimulus intensity and isolation effects. Dev Psychobiol. 1990;23:129–140. doi: 10.1002/dev.420230204. [DOI] [PubMed] [Google Scholar]
- 107.Hughes RA, Bowes M, Sufka KJ. Morphine hyperalgesic effects on developmental changes in thermal nociception and respiration in domestic fowl (Gallus gallus) Pharmacol Biochem Behav. 1992;42:535–539. doi: 10.1016/0091-3057(92)90151-5. [DOI] [PubMed] [Google Scholar]
- 108.Sufka KJ, Hughes RA. Dose and temporal parameters of morphine-induced hyperalgesia in domestic fowl. Physiol Behav. 1990;47:385–387. doi: 10.1016/0031-9384(90)90159-2. [DOI] [PubMed] [Google Scholar]
- 109.Sufka KJ, Hughes RA, Giordano J. Effects of selective opiate antagonists on morphine-induced hyperalgesia in domestic fowl. Pharmacol Biochem Behav. 1991;38:49–54. doi: 10.1016/0091-3057(91)90588-s. [DOI] [PubMed] [Google Scholar]
- 110.Sufka KJ, Hughes RA. Time-dependent codeine hypoalgesia and hyperalgesia in domestic fowl. Pharmacol Biochem Behav. 1992;41:349–353. doi: 10.1016/0091-3057(92)90109-s. [DOI] [PubMed] [Google Scholar]
- 111.Hughes RA, Sufka KJ. Morphine hyperalgesic effects on the formalin test in domestic fowl (Gallus gallus) Pharmacol Biochem Behav. 1991;38:247–251. doi: 10.1016/0091-3057(91)90273-5. [DOI] [PubMed] [Google Scholar]
- 112.Sufka KJ, Hoganson DA, Hughes RA. Central monoaminergic changes induced by morphine in hypoalgesic and hyperalgesic strains of domestic fowl. Pharmacol Biochem Behav. 1992;42:781–785. doi: 10.1016/0091-3057(92)90029-f. [DOI] [PubMed] [Google Scholar]
- 113.Roach JT, Sufka KJ. Characterization of the chick carrageenan response. Brain Res. 2003;994:216–225. doi: 10.1016/j.brainres.2003.09.038. [DOI] [PubMed] [Google Scholar]
- 114.Gentle MJ, Corr SA. Endogenous analgesia in the chicken. Neurosci Lett. 1995;201:211–214. doi: 10.1016/0304-3940(95)12181-1. [DOI] [PubMed] [Google Scholar]
- 115.Gentle MJ. Pain-related behaviour following sodium urate arthritis is expressed in decerebrate chickens. Physiol Behav. 1997;62:581–584. doi: 10.1016/s0031-9384(97)00164-9. [DOI] [PubMed] [Google Scholar]
- 116.Gentle MJ, Hocking PM, Bernard R, Dunn LN. Evaluation of intraarticular opioid analgesia for the relief of articular pain in the domestic fowl. Pharmacol Biochem Behav. 1999;63:339–343. doi: 10.1016/s0091-3057(99)00029-5. [DOI] [PubMed] [Google Scholar]
- 117.Stevens CW, Newman LC. Spinal administration of selective opioid antagonists in amphibians: evidence for an opioid unireceptor. Life Sci. 1999;64:PL125–PL130. doi: 10.1016/s0024-3205(99)00013-2. [DOI] [PubMed] [Google Scholar]
- 118.Stevens CW. Opioid research in amphibians: a unique perspective on mechanisms of opioid analgesia and the evolution of opioid receptors. Rev Analgesia. 2003;7:122–136. [Google Scholar]
- 119.Gonzalez-Nunez V, Barrallo A, Traynor JR, Rodriguez RE. Characterization of opioid binding sites in zebrafish brain. J Pharmacol Exp Ther. 2006;316:900–904. doi: 10.1124/jpet.105.093492. [DOI] [PubMed] [Google Scholar]
- 120.Brooks AI, Standifer KM, Cheng J, Ciszewska GR, Pasternak GW. Opioid binding in giant toad and goldfish brain. Receptor. 1994;4:55–62. [PubMed] [Google Scholar]
- 121.Bird DJ, Jackson M, Baker BI, Buckingham JC. Opioid binding sites in the fish brain: An autoradiographic study. Gen Comp Endocrinol. 1988;70:49–62. doi: 10.1016/0016-6480(88)90093-7. [DOI] [PubMed] [Google Scholar]
- 122.Buatti MC, Pasternak GW. Multiple opiate receptors: phylogenetic differences. Brain Res. 1981;218:400–405. doi: 10.1016/0006-8993(81)91319-6. [DOI] [PubMed] [Google Scholar]
- 123.Pert CB, Aposhian D, Snyder SH. Phylogenetic distribution of opiate binding. Brain Res. 1974;75:356–361. doi: 10.1016/0006-8993(74)90761-6. [DOI] [PubMed] [Google Scholar]
- 124.Xia Y, Haddad GG. Major difference in the expression of delta- and mu-opioid receptors between turtle and rat brain. J Comp Neurol. 2001;436:202–210. [PubMed] [Google Scholar]
- 125.Bakalkin GY, Pivovarov AS, Kobylyansky AG, Nesterenko PN, Yarygin KN. Lateralization of opioid receptors in turtle visual cortex. Brain Res. 1989;480:268–276. doi: 10.1016/0006-8993(89)90192-3. [DOI] [PubMed] [Google Scholar]
- 126.Kawashima M, Imai S, Takahashi T, Kamiyoshi M, Tanaka K. An opiate receptor in the neurohypophysis of laying hens. Poult Sci. 1995;74:716–722. doi: 10.3382/ps.0740716. [DOI] [PubMed] [Google Scholar]
- 127.Csillag A, Stewart MG, Szekely AD, Magloczky Z, Bourne RC, Steele RJ. Quantitative autoradiographic demonstration of changes in binding to delta opioid, but not mu or kappa receptors, in chick forebrain 30 minutes after passive avoidance training. Brain Res. 1993;613:96–105. doi: 10.1016/0006-8993(93)90459-z. [DOI] [PubMed] [Google Scholar]
- 128.Martin R, McGregor GP, Halbinger G, Falke N, Voigt KH. Methionine5-enkephalin and opiate binding sites in the neurohypophysis of the bird, Gallus domesticus. Regul Pept. 1992;38:33–44. doi: 10.1016/0167-0115(92)90070-b. [DOI] [PubMed] [Google Scholar]
- 129.Csillag A, Bourne RC, Stewart MG. Distribution of mu, delta, and kappa opioid receptor binding sites in the brain of the one-day-old domestic chick (Gallus domesticus): an in vitro quantitative autoradiographic study. J Comp Neurol. 1990;302:543–551. doi: 10.1002/cne.903020310. [DOI] [PubMed] [Google Scholar]
- 130.Simon EJ, Hiller JM, Groth J, Itzhak Y, Holland MJ, Beck SG. The nature of opiate receptors in toad brain. Life Sci. 1982;31:1367–1370. doi: 10.1016/0024-3205(82)90383-6. [DOI] [PubMed] [Google Scholar]
- 131.Zawilska J, Lajtha A, Borsodi A. Selective protection of benzomorphan binding sites against inactivation by N-ethylmaleimide. Evidence for kappa opioid receptors in frog brain. J Neurochem. 1988;51:736–739. doi: 10.1111/j.1471-4159.1988.tb01806.x. [DOI] [PubMed] [Google Scholar]
- 132.Benyhe S, Varga E, Hepp J, Magyar A, Borsodi A, Wollemann M. Characterization of kappa1 and kappa2 opioid binding sites in frog (Rana esculenta) brain membrane. Neurochem Res. 1990;15:899–904. doi: 10.1007/BF00965909. [DOI] [PubMed] [Google Scholar]
- 133.Simon J, Benyhe S, Hepp J, Khan A, Borsodi A, Szucs M, Medzihradszky K, Wollemann M. Purification of kappa-opioid receptor subtype from frog brain. Neuropept. 1987;10:19–28. doi: 10.1016/0143-4179(87)90085-0. [DOI] [PubMed] [Google Scholar]
- 134.Simon J, Benyhe S, Borsodi A, Szucs M, Wollemann M. Separation of kappa-opioid receptor subtype from frog brain. FEBS Lett. 1985;183:395–397. doi: 10.1016/0014-5793(85)80818-8. [DOI] [PubMed] [Google Scholar]
- 135.Benyhe S, Monory K, Farkas J, Toth G, Guerrini R, Salvadori S, Orosz G, Wollemann M, Borsodi A. Nociceptin binding sites in frog (Rana esculenta) brain membranes. Biochem Biophys Res Commun. 1999;260:592–596. doi: 10.1006/bbrc.1999.0907. [DOI] [PubMed] [Google Scholar]
- 136.Wollemann M, Farkas J, Toth G, Benyhe S. Comparison of the endogenous heptapeptide met-enkephalin-arg6-phe7 binding in amphibian and mammalian brain. Acta Biol Hung. 1999;50:297–307. [PubMed] [Google Scholar]
- 137.Benyhe S, Ketevan A, Simon J, Hepp J, Medzihradszky K, Borsodi A. Affinity labelling of frog brain opioid receptors by dynorphin (1–10) chloromethyl ketone. Neuropept. 1997;31:52–59. doi: 10.1016/s0143-4179(97)90020-2. [DOI] [PubMed] [Google Scholar]
- 138.Moitra J, Öktem HA, Borsodi A. Thermodynamic parameters of frog brain kappa-opioid receptors. J Neurochem. 1995;65:798–801. doi: 10.1046/j.1471-4159.1995.65020798.x. [DOI] [PubMed] [Google Scholar]
- 139.Benyhe S, Simon J, Borsodi A, Wollemann M, Barnard EA. (3H)Dynorphin 1–8 binding sites in frog (Rana esculenta) brain membranes. Neuropept. 1994;26:359–364. doi: 10.1016/0143-4179(94)90121-x. [DOI] [PubMed] [Google Scholar]
- 140.Wollemann M, Farkas J, Toth G, Benyhe S. Characterization of (3H) met-enkephalin-arg6-phe7 binding to opioid receptors in frog brain membrane preparations. J Neurochem. 1994;63:1460–1465. doi: 10.1046/j.1471-4159.1994.63041460.x. [DOI] [PubMed] [Google Scholar]
- 141.Benyhe S, Szucs M, Borsodi A, Wollemann M. Species differences in the stereoselectivity of kappa opioid binding sites for (3H)U-69593 and (3H) ethylketocyclazocine. Life Sci. 1992;51:1647–1655. doi: 10.1016/0024-3205(92)90309-d. [DOI] [PubMed] [Google Scholar]
- 142.Simon J, Benyhe S, Hepp J, Varga E, Medzihradszky K, Borsodi A, Wollemann M. Method for isolation of kappa-opioid binding sites by dynorphin affinity chromatography. J Neurosci Res. 1990;25:549–555. doi: 10.1002/jnr.490250412. [DOI] [PubMed] [Google Scholar]
- 143.Mollereau C, Pascaud A, Baillat G, Mazarguil H, Puget A, Meunier JC. Evidence for a new type of opioid binding site in the brain of the frog Rana ridibunda. Eur J Pharmacol. 1988;150:75–84. doi: 10.1016/0014-2999(88)90752-2. [DOI] [PubMed] [Google Scholar]
- 144.Newman LC, Wallace DR, Stevens CW. Characterization of 3H-diprenorphine binding in Rana pipiens: Observations of filter binding enhanced by naltrexone. J Pharmacol Toxicol Meth. 1999;41:43–48. doi: 10.1016/s1056-8719(99)00020-9. [DOI] [PubMed] [Google Scholar]
- 145.Newman LC, Wallace DR, Stevens CW. Selective opioid agonist and antagonist displacement of (3H)-naloxone binding in amphibian brain. Eur J Pharmacol. 2000;397:255–262. doi: 10.1016/s0014-2999(00)00265-x. [DOI] [PubMed] [Google Scholar]
- 146.Newman LC, Wallace DR, Stevens CW. Selective opioid receptor agonist and antagonist displacement of (3H)-naloxone binding in amphibian spinal cord. Brain Res. 2000;884:184–191. doi: 10.1016/s0006-8993(00)02967-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Newman LC, Sands SS, Wallace DR, Stevens CW. Characterization of mu, kappa, and delta opioid binding in amphibian whole brain tissue homogenates. J Pharmacol Exp Ther. 2002;301:364–370. doi: 10.1124/jpet.301.1.364. [DOI] [PubMed] [Google Scholar]
- 148.Li X, Keith DE, Jr, Evans CJ. Mu opioid receptor-like sequences are present throughout vertebrate evolution. J Mol Evol. 1996;43:179–184. doi: 10.1007/BF02338825. [DOI] [PubMed] [Google Scholar]
- 149.Li X, Keith DE, Jr, Evans CJ. Multiple opioid receptor-like genes are identified in diverse vertebrate phyla. FEBS Lett. 1996;397:25–29. doi: 10.1016/s0014-5793(96)01126-x. [DOI] [PubMed] [Google Scholar]
- 150.Darlison MG, Greten FR, Harvey RJ, Kreienkamp H, Stuhmer T, Zwiers H, Lederis K, Richter D. Opioid receptors from a lower vertebrate (Catostomus commersoni): Sequence, pharmacology, coupling to a G-protein-gated inward-rectifying potassium channel (GIRK1), and evolution. Proc Natl Acad Sci USA. 1997;94:8214–8219. doi: 10.1073/pnas.94.15.8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Barrallo A, González-Sarmiento R, Porteros A, Gracia-Isídoro M, Rodríguez RE. Cloning. molecular characterization, and distribution of a gene homologous to delta opoid receptor from zebrafish (Danio rerio) Biochem Biophys Res Commun. 1998;245:544–548. doi: 10.1006/bbrc.1998.8496. [DOI] [PubMed] [Google Scholar]
- 152.Barrallo A, González-Sarmiento R, Alvar F, Rodriguez RE. ZFOR2, a new opioid receptor-like gene from the teleost zebrafish (Danio rerio) Mol Brain Res. 2000;84:1–6. doi: 10.1016/s0169-328x(00)00152-2. [DOI] [PubMed] [Google Scholar]
- 153.Alvarez FA, Rodriguez-Martin I, Gonzalez-Nunez V, de Velasco EM, Gonzalez SR, Rodriguez RE. New kappa opioid receptor from zebrafish Danio rerio. Neurosci Lett. 2006;405:94–99. doi: 10.1016/j.neulet.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 154.Porteros A, Garcia-Isodoro M, Barrallo A, González-Sarmiento R, Rodriguez RE. Expression of ZFOR1, a delta-opioid receptor, in the central nervous system of the zebrafish (Danio rerio) J Comp Neurol. 1999;412:429–4358. doi: 10.1002/(sici)1096-9861(19990927)412:3<429::aid-cne4>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 155.Bradford CS, Walthers EA, Stanley DJ, Baugh MM, Moore FL. Delta and mu opioid receptors from the brain of a urodele amphibian, the rough-skinned newt, Taricha granulosa: Cloning, heterologous expression, and pharmacological characterization. Gen Comp Endocrinol. 2006;146:275–290. doi: 10.1016/j.ygcen.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 156.Bradford CS, Walthers EA, Searcy BT, Moore FL. Cloning, heterologous expression and pharmacological characterization of a kappa opioid receptor from the brain of the rough-skinned newt, Taricha granulosa. J Mol Endocrinol. 2005;34:809–823. doi: 10.1677/jme.1.01711. [DOI] [PubMed] [Google Scholar]
- 157.Walthers EA, Bradford CS, Moore FL. Cloning, pharmacological characterization and tissue distribution of an ORL1 opioid receptor from an amphibian, the rough-skinned newt, Taricha granulosa. J Mol Endocrinol. 2005;34:247–256. doi: 10.1677/jme.1.01687. [DOI] [PubMed] [Google Scholar]
- 158.Stevens CW, Brasel CM, Mohan S. Cloning and bioinformatics of amphibian mu, delta, kappa, and nociceptin opioid receptors expressed in brain tissue: Evidence for opioid receptor divergence in mammals. Neurosci Lett. 2007;419:189–194. doi: 10.1016/j.neulet.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Minami M, Satoh M. Molecular biology of the opioid receptors: structures, functions and distributions. Neurosci Res. 1995;23:121–145. doi: 10.1016/0168-0102(95)00933-k. [DOI] [PubMed] [Google Scholar]
- 160.Gether U. Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endo Rev. 2002;21:90–113. doi: 10.1210/edrv.21.1.0390. [DOI] [PubMed] [Google Scholar]
- 161.Shahrestanifar MS, Howells RD. Sensitivity of opioid receptor binding to N-substituted maleimides and methanethiosulfonate derivatives. Neurochem Res. 1996;21:1295–1299. doi: 10.1007/BF02532370. [DOI] [PubMed] [Google Scholar]
- 162.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Trong IL, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–746. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 163.Lomize AL, Pogozheva ID, Mosberg HI. Structural organization of G-protein-coupled receptors. J Comput Aided Mol Des. 1999;13:352–353. doi: 10.1023/a:1008050821744. [DOI] [PubMed] [Google Scholar]
- 164.Christopoulos A, Kenakin T. G protein-coupled receptor allosterism and complexing. Pharmacol Rev. 2002;54:323–374. doi: 10.1124/pr.54.2.323. [DOI] [PubMed] [Google Scholar]
- 165.Wang HL, Chang WT, Hsu CY, Huang PC, Chow YW, Li AH. Identification of two C-terminal amino acids, Ser (355) and Thr (357), required for short-term homologous desensitization of mu-opioid receptors. Biochem Pharmacol. 2002;15:257–266. doi: 10.1016/s0006-2952(02)01114-0. [DOI] [PubMed] [Google Scholar]
- 166.Chen C, Yin J, de Riel J, DesJarlais RL, Raveglia LF, Zhui J, Liu-Chen LY. Determination of the amino acid residue involved in (3H)beta-funaltrexamine covalent binding in the cloned rat mu opioid receptor. J Biol Chem. 1996;271:21422–21429. doi: 10.1074/jbc.271.35.21422. [DOI] [PubMed] [Google Scholar]
- 167.Ferguson DM, Kramer S, Metzger TG, Law PY, Portoghese PS. Isosteric replacement of acidic with neutral residues in extracellular loop-2 of the kappa-opioid receptor does not affect dynorphin A (1–13) affinity and function. J Biol Chem. 2000;43:1251–1252. doi: 10.1021/jm990583r. [DOI] [PubMed] [Google Scholar]
- 168.Chaturvedi K, Christoffers KH, Singh K, Howells RD. Structure and regulation of opioid receptors. Biopoly. 2000;55:334–346. doi: 10.1002/1097-0282(2000)55:4<334::AID-BIP1006>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 169.Stevens CW. Molecular evolution of vertebrate opioid receptor proteins. A preview. Recent Developments in Pain Research. In: Capasso A, editor. Research Signpost. Trivandrum, Kerala, India: 2005. [Google Scholar]
- 170.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 171.Rodríguez RE, Barrallo A, Garcia-Malvar F, McFadyen IJ, González-Sarmiento R, Traynor JR. Characterization of ZFOR1, a putative delta-opioid receptor from the teleost zebrafish. Neurosci Lett. 2000;288:207–210. doi: 10.1016/s0304-3940(00)01239-8. [DOI] [PubMed] [Google Scholar]
- 172.Flower DR, Attwood TK. Integrative bioinformatics for functional genome annotation: trawling for G protein-coupled receptors. Sem Cell Develop Biol. 2004;15:693–701. doi: 10.1016/j.semcdb.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 173.Hoehe MR. Haplotypes and the systematic analysis of genetic variation in genes and genomes. Pharmacogenom. 2003;4:547–570. doi: 10.2217/14622416.4.5.547. [DOI] [PubMed] [Google Scholar]
- 174.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nature Rev. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 175.Rommelspacher H, Smolka M, Schmidt LG, Samochowiec J, Hoehe MR. Genetic analysis of the mu-opioid receptor in alcohol-dependent individuals. Alcohol. 2001;24:129–135. doi: 10.1016/s0741-8329(01)00139-2. [DOI] [PubMed] [Google Scholar]
- 176.LaForge KS, Yuferov V, Kreek MJ. Opioid receptor and peptide gene polymorphisms: potential implications for addictions. Eur J Pharmacol. 2000;410:249–268. doi: 10.1016/s0014-2999(00)00819-0. [DOI] [PubMed] [Google Scholar]
- 177.Bond C, LaForge KS, Tian M, Melia D, Zhang S, Borg L, Gong J, Schluger J, Strong JA, Leal SM, Tischfield JA, Kreek MJ, Yu L. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci USA. 1998;95:9608–9613. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hoehe MR, Kopke K, Wendel B, Rohde K, Flachmeier C, Kidd KK, Berrettini WH, Church GM. Sequence variability and candidate gene analysis in complex disease: association of mu opioid receptor gene variation with substance dependence. Hum Mol Genet. 2000;9:2895–2908. doi: 10.1093/hmg/9.19.2895. [DOI] [PubMed] [Google Scholar]
- 179.Kroslak T, LaForge KS, Gianotti RJ, Ho A, Nielsen DA, Kreek MJ. The single nucleotide polymorphism A118G alters functional properties of the human mu opioid receptor. J Neurochem. 2007;103:77–87. doi: 10.1111/j.1471-4159.2007.04738.x. [DOI] [PubMed] [Google Scholar]
- 180.Befort K, Filliol D, Decaillot FM, Gaveriaux-Ruff C, Hoehe MR, Kieffer BL. A single nucleotide polymorphic mutation in the human mu-opioid receptor severely impairs receptor signaling. J Biol Chem. 2001;276:3130–3137. doi: 10.1074/jbc.M006352200. [DOI] [PubMed] [Google Scholar]
- 181.Mayer P, Hollt V. Pharmacogenetics of opioid receptors and addiction. Pharmacogenet Genomics. 2006;16:1–7. doi: 10.1097/01.fpc.0000182781.87932.0d. [DOI] [PubMed] [Google Scholar]
- 182.Klepstad P, Dale O, Skorpen F, Borchgrevink PC, Kaasa S. Genetic variability and clinical efficacy of morphine. Acta Anaesthesiol Scand. 2005;49:902–908. doi: 10.1111/j.1399-6576.2005.00772.x. [DOI] [PubMed] [Google Scholar]
- 183.Bayerer B, Stamer U, Hoeft A, Stuber F. Genomic variations and transcriptional regulation of the human mu-opioid receptor gene. Eur J Pain. 2007;11:421–427. doi: 10.1016/j.ejpain.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 184.Uhl GR, Sora I, Wang Z. The mu opiate receptor as a candidate gene for pain: polymorphisms, variations in expression, nociception, and opiate responses. Proc Natl Acad Sci USA. 1999;96:7752–7755. doi: 10.1073/pnas.96.14.7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gu J, Gu X. Induced gene expression in human brain after the split from chimpanzee. Trends Genet. 2003;19:63–65. doi: 10.1016/s0168-9525(02)00040-9. [DOI] [PubMed] [Google Scholar]
- 186.Jordan IK, Marino-Ramirez L, Koonin EV. Evolutionary significance of gene expression divergence. Gene. 2005;345:119–126. doi: 10.1016/j.gene.2004.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yuferov V, Fussell D, LaForge KS, Nielsen DA, Gordon D, Ho A, Leal SM, Ott J, Kreek MJ. Redefinition of the human kappa opioid receptor gene (OPRK1) structure and association of haplotypes with opiate addiction. Pharmacogenet. 2004;14:793–804. doi: 10.1097/00008571-200412000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Saito M, Ehringer MA, Toth R, Oros M, Szakall I, Sikela JM, Vadasz C. Variants of kappa-opioid receptor gene and mRNA in alcohol-preferring and alcohol-avoiding mice. Alcohol. 2003;29:39–49. doi: 10.1016/s0741-8329(02)00322-1. [DOI] [PubMed] [Google Scholar]
- 189.Kobayashi H, Hata H, Ujike H, Harano M, Inada T, Komiyama T, Yamada M, Sekine Y, Iwata N, Iyo M, Ozaki N, Itokawa M, Naka M, Ide S, Ikeda K, Numachi Y, Sora I. Association analysis of delta-opioid receptor gene polymorphisms in methamphetamine dependence/psychosis. Am J Med Genet B Neuropsychiatr Genet. 2006;141:482–486. doi: 10.1002/ajmg.b.30337. [DOI] [PubMed] [Google Scholar]
- 190.Ito E, Xie GX, Maruyama K, Palmer PP. A core-promoter region functions bi-directionally for human opioid-receptor-like gene ORL1 and its 5′-adjacent gene GAIP. J Mol Biol. 2000;304:259–270. doi: 10.1006/jmbi.2000.4212. [DOI] [PubMed] [Google Scholar]
- 191.Clark AG, Hubisz MJ, Bustamante CD, Williamson SH, Nielsen R. Ascertainment bias in studies of human genome-wide polymorphism. Gen Res. 2005;15:1496–1502. doi: 10.1101/gr.4107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Rana BK, Hewett-Emmett D, Jin L, Chang BH, Sambuughin N, Lin M, Watkins S, Bamshad M, Jorde LB, Ramsay M, Jenkins T, Li WH. High polymorphism at the human melanocortin 1 receptor locus. Genetics. 1999;151:1547–1557. doi: 10.1093/genetics/151.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hallböök F. Evolution of the vertebrate neurotrophin and Trk receptor gene families. Curr Opin Neurobiol. 1999;9:616–621. doi: 10.1016/S0959-4388(99)00011-2. [DOI] [PubMed] [Google Scholar]
- 194.Iwama H, Gojobori T. Identification of neurotransmitter receptor genes under significantly relaxed selective constraint by orthologous gene comparisons between humans and rodents. Mol Biol Evol. 2002;19:1891–1901. doi: 10.1093/oxfordjournals.molbev.a004013. [DOI] [PubMed] [Google Scholar]
- 195.McPartland JM, Norris RW, Kilpatrick CW. Tempo and mode in the endocannaboinoid system. J Mol Evol. 2007;65:267–276. doi: 10.1007/s00239-007-9004-1. [DOI] [PubMed] [Google Scholar]
- 196.Ikeda K, Ide S, Han W, Hayashida M, Uhl GR, Sora I. How individual sensitivity to opiates can be predicted by gene analyses. Trends Pharmacol Sci. 2005;26:311–317. doi: 10.1016/j.tips.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 197.Ohno S. George Allen and Unwin. London, England: 1970. Evolution by Gene Duplication. [Google Scholar]
- 198.Lundin LG, Larhammar D, Hallbook F. Numerous groups of chromosomal regional paralogies strongly indicate two genome doublings at the root of the vertebrates. J Struct Funct Genom. 2003;3:53–63. [PubMed] [Google Scholar]
- 199.Escriva H, Manzon L, Youson J, Laudet V. Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. Mol Biol Evol. 2002;19:1440–1450. doi: 10.1093/oxfordjournals.molbev.a004207. [DOI] [PubMed] [Google Scholar]
- 200.Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18:292–298. [Google Scholar]
- 201.McLysaght A, Hokamp K, Wolfe KH. Extensive genomic duplication during early chordate evolution. Nat Genet. 2002;31:200–204. doi: 10.1038/ng884. [DOI] [PubMed] [Google Scholar]
- 202.Hokamp K, McLysaght A, Wolfe KH. The 2R hypothesis and the human genome sequence. J Struct Funct Genom. 2003;3:95–110. [PubMed] [Google Scholar]
- 203.Krylov DM, Wolf YI, Rogozin IB, Koonin EV. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 2003;13:2229–2235. doi: 10.1101/gr.1589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- 205.Kondrashhov FA, Rogozin IB, Wolf YI, Koonin EV. Selection in the evolution of gene duplications. Genome Biol. 2002;3:0008.1–0008.9. doi: 10.1186/gb-2002-3-2-research0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Merritt TJ, Quattro JM. Evidence for a period of directional selection following gene duplication in a neurally expressed locus of triosephosphate isomerase. Genetics. 2001;159:689–697. doi: 10.1093/genetics/159.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zhang P, Gu Z, Li WH. Different evolutionary patterns between young duplicate genes in the human genome. Genome Biol. 2003;4:R56. doi: 10.1186/gb-2003-4-9-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Jordan IK, Wolf YI, Koonin EV. Duplicated genes evolve slower than singletons despite the initial rate increase. BMC Evol Biol. 2004;4:22. doi: 10.1186/1471-2148-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Makalowski W. Are we polyploids? A brief history of one hypothesis. Genome Res. 2001;11:667–670. doi: 10.1101/gr.188801. [DOI] [PubMed] [Google Scholar]
- 210.Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD) BioEssays. 2005;27:937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- 211.Christoffels A, Koh EG, Chia JM, Brenner S, Aparicio S, Venkatesh B. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol Biol Evol. 2004;21:1146–1151. doi: 10.1093/molbev/msh114. [DOI] [PubMed] [Google Scholar]
- 212.Woods IG, Wilson C, Friedlander B, Chang P, Reyes DK, Nix R, Kelly PD, Chu F, Postlethwait JH, Talbot WS. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res. 2005;15:1307–1314. doi: 10.1101/gr.4134305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Vandepoele K, De Vos W, Taylor JS, Meyer A, Van de Peer Y. Major events in the genome evolution of vertebrates: Paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc Natl Acad Sci USA. 2004;101 doi: 10.1073/pnas.0307968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Pinal-Seoane N, Martin IR, Gonzalez-Nunez V, de Velasco EM, Alvarez FA, Sarmiento RG, Rodriguez RE. Characterization of a new duplicate delta-opioid receptor from zebrafish. J Mol Endocrinol. 2006;37:391–403. doi: 10.1677/jme.1.02136. [DOI] [PubMed] [Google Scholar]
- 215.Taylor EW, Agarwal A. Sequence homology between bacteriorhodopsin and G-protein coupled receptors: exon shuffling or evolution by duplication? FEBS Lett. 1993;325:161–166. doi: 10.1016/0014-5793(93)81065-8. [DOI] [PubMed] [Google Scholar]
- 216.Dorus S, Vallender EJ, Evans PD, Anderson JR, Gilbert SL, Mahowald M, Wyckoff GJ, Malcom CM, Lahn BT. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 217.Wolfe KH, Li WH. Molecular evolution meets the genomics revolution. Nat Genet. 2003;33:255–265. doi: 10.1038/ng1088. [DOI] [PubMed] [Google Scholar]
- 218.Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]