Abstract
Nuclear factor erythroid-2 related factor-2 (Nrf2) is a redox-sensitive transcription factor that regulates the expression of electrophile and xenobiotic detoxification enzymes and efflux proteins, which confer cytoprotection against oxidative stress and apoptosis in normal cells. Loss of function mutations in the Nrf2 inhibitor, Kelch-like ECH-associated protein (Keap1), results in constitutive activation of Nrf2 function in non-small-cell lung cancer (NSCLC). In this study, we demonstrate that constitutive activation of Nrf2 in lung cancer cells promotes tumorigenicity and contributes to chemoresistance by upregulation of glutathione, thioredoxin and the drug efflux pathways involved in detoxification of electrophiles and broad spectrum of drugs. RNAi-mediated reduction of Nrf2 expression in lung cancer cells induces generation of reactive oxygen species, suppresses tumor growth and results in increased sensitivity to chemotherapeutic drug induced cell death in vitro and in vivo. Inhibiting Nrf2 expression using naked siRNA duplexes in combination with carboplatin significantly inhibits tumor growth in a subcutaneous model of lung cancer. Thus, targeting Nrf2 activity in lung cancers, particularly those with Keap1 mutations, could be a promising strategy to inhibit tumor growth and circumvent chemoresistance.
Keywords: Nrf2, Keap1, lung cancer, drug resistance, ROS, RNAi
Introduction
Lung cancer is the leading cause of cancer deaths in the United States and worldwide for men and women. Despite considerable progress over the last 25 years in the systemic therapy of lung cancer, intrinsic and acquired resistance to chemotherapeutic agents remains a challenge (1). Most patients with small-cell-lung cancer (SCLC) have a favorable initial response to chemotherapy but the majority relapse and their tumors tend to be largely refractory to further treatment. Non-small-cell-lung cancers (NSCLC) are intrinsically resistant and are generally non-responsive to initial chemotherapy. Frequently, resistance is intrinsic to the cancer, but as the therapy becomes increasingly effective, acquired resistance has also become common (1).
Formation of reactive oxygen species (ROS) is important for induction of apoptosis for commonly used chemotherapy agents such as cisplatin, bleomycin, paclitaxel, adriamycin and etoposide (2, 3). Many chemotherapeutic agents depend on oxidative insult to cancer cells for their mode of action. Xenobiotic metabolism enzymes in conjunction with drug efflux proteins act to detoxify cancer drugs, whereas antioxidants confer cytoprotection by attenuating drug-induced oxidative stress and apoptosis. Several studies have shown that the expression of xenobiotic metabolism genes [glutathione-S-transferases (GSTs)], antioxidants [glutathione (GSH)], and drug efflux proteins [multidrug resistance protein (MRP) family] are increased in NSCLC (4–6). Cancer cells exhibit a superior defense system against electrophiles as compared with normal cells due to the upregulation of genes involved in electrophile detoxification. In addition, lung cancer cells have greater expression of multidrug resistance proteins which confer chemoresistance (7).
In normal cells, Nrf2, a cap ‘n’ collar basic leucine zipper transcription factor regulates a transcriptional program that maintains cellular redox homeostasis and protects against toxic xenobiotics. Keap1 is a cytoplasmic anchor of Nrf2 and maintains steady-state levels of Nrf2 and Nrf2-dependent transcription by targeting it for proteasomal degradation (8–10). Keap1 constitutively suppresses Nrf2 activity in the absence of stress. Oxidants, xenobiotics and electrophiles hamper the Keap1-mediated proteasomal degradation of Nrf2, which results in increased nuclear accumulation and transcriptional induction of target genes. The Nrf2-regulated transcriptional program includes a broad spectrum of genes, including genes encoding antioxidants (e.g., the glutathione system: γ-glutamyl cysteine synthetase modifier subunit [GCLm], γ-glutamyl cysteine synthetase catalytic subunit [GCLc], glutathione synthetase [GSS], glutathione reductase [GSR], glutathione peroxidase [GPX] and the cysteine/glutamate transporter [SLC7A11]); the thioredoxin system: thioredoxin-1 [TXN], thioredoxin reductase [TXNRD1] and peroxiredoxins [PRDX], xenobiotic metabolism enzymes (e.g., NADP[H] quinone oxidoreductase 1 [NQO1], UDP-glucuronosyltransferase) and members of the glutathione-S-transferase family [GSTs]), and several ATP-dependent multidrug resistant drug efflux pumps (e.g., ABCC1 and ABCC2) (11–18). Nrf2 also protects against apoptosis induced by oxidants and FAS ligand (16, 19, 20). Downregulation of Nrf2 using anti-sense RNA resulted in cell sensitization to apoptosis (19). Thus, Nrf2 promotes survival against stress caused by exposure to electrophiles and xenobiotics.
Recently, we have reported point mutations in the Keap1 gene leading to non-conservative amino acid substitutions and nonsense mutations in a high percentage of the lung cancer cell lines and tumors (21). Somatic mutations in Keap1 coding region in Japanese patients with lung cancer as well as breast cancer have been reported by other groups (22–24). Loss of Keap1 activity due to mutations leads to constitutive activation of Nrf2. In the present study, we determined the functional consequences of increased Nrf2 activity in lung cancer using two lung cancer cell lines (A549 and H460) with complete loss of functional Keap1 activity (21) and developed a novel strategy for counteracting therapeutic resistance by targeting Nrf2-Keap1 pathway.
Materials and Methods
Cell Culture and Reagents
A549 and H460 cells were purchased from American Type Culture Collection (Manassas) and cultured under recommended conditions. All transfections were carried out using Lipofectamine 2000 (Invitrogen).
Generation of lung cancer cell lines stably expressing Nrf2 shRNA
To inhibit the expression of Nrf2, we designed a short hairpin RNA (shRNA) targeting the 3’ end of the Nrf2 transcript as described in our previous reports (21, 25). The Nrf2-shRNA duplex with the following sense and antisense sequences was used: 5'GATCCGTAAGAAGCCAGATGTTAATTCAAGAGACATTCTTCGGTCTACAATTTTTTTTGGAAA-3' (sense) and 5'-AGCTTTTCCAAAAAAAATTGTAGACCGAAGAATGTCTCTTGAATTAACATCTGGCTTCTTACG-3' (antisense) (21). Short hairpin RNA cassette was subcloned into pSilencer vector and transfected into A549 and H460 cells. A short hairpin RNA targeting luciferase gene was used as control. Stable cell clones with reduced Nrf2 expression were generated.
For the in vivo experiments, all siRNA compounds were chemically synthesized being stabilized by 2O’-Me modifications (Biospring). The sequence of siRNA targeting human Nrf2 used for in vivo experiments is 5’-UCCCGUUUGUAGAUGACAA-3’ (sense) and 5’-UUGUCAUCUACAAACGGGA-3’ (antisense). The sequence of control siRNA targeting GFP is 5’-GGCUACGUCCAGGAGCGCACC-3’ (sense) and 5’-GGUGCGCUCCUGGACGUAGC-3’ (antisense) (26).
Real Time RT-PCR
Total RNA was extracted from tumors and cells using the RNeasy kit (Qiagen). The reverse transcription reaction was performed using high capacity cDNA synthesis kit (Applied Biosystems). Quantitative real time RT-PCR analyses of Human Nrf2, GCLc, GCLm, GSR, xCT, G6PDH, PRDX1, GSTM4, MGST1, NQO1, HO-1, TXN1, TXNRD1, ABCC1, and ABCC2 were performed by using assay on demand primers and probe sets from Applied Biosystems. β-ACTIN was used for normalization.
Western Blot Analysis
Immunoblot analysis was performed by following the protocol described by Singh and colleagues (21). Following antibodies were used for immunoblotting: anti-Nrf2, anti-TXNRD1 and anti-actin (SantaCruz Biotechnology), anti-GAPDH (Imgenex), anti-Caspase-3 (BD Biosciences) and anti-TXN1 (American Diagnostica).
Measurement of ROS levels
Cells were incubated with 10 µM c-H2DCFDA (Molecular probes) for 30mins at 37°C to assess the ROS mediated oxidation to the fluorescent compound c-H2DCF. Fluorescence of oxidized c-H2DCF was measured at an excitation wavelength of 488nM and an emission wavelength of 525nM using a FAC Scan flow cytometer (BD Biosciences).
Enzyme Assay
Enzyme activities of GST, GSR and NQO1 and G6PDH were determined in the total protein lysates by following methods previously described (27).
Drug Accumulation Assay
A549-Nrf2shRNA and H460-Nrf2shRNA cells as well as their respective control cells expressing Luc-shRNA were seeded at a density of 0.3 × 106 cells/ ml in 6-well plates. After 12h, growth medium was aspirated and replaced with 1.5ml of RPMI-1640 containing 0.2µM of [3H] Etoposide (646 mCi/mmol; Moravek Biochemicals) and [14C] Carboplatin (53 mCi/mmol; Amersham Biosciences). Cells were incubated with radiolabeled drug for indicated period of time and then cooled on ice, washed with ice-cold PBS, and solubilized with 1.0 ml of 1% SDS. The radioactivity in each sample was determined by scintillation counting. Results are presented as means ± SD. Comparisons were made by paired t- test.
MTS Cell Viability Assay
The in vitro drug sensitivity to etoposide and carboplatin was assessed using Cell Titer 96-Aqueous assay kit (Promega). Cells were plated at a density of 5,000 cells/well in 96-well plates, allowed to recover for 12 h and then exposed to various concentrations of etoposide and carboplatin for 72–96 h. Drug cytotoxicity was evaluated by adding 40µl of 3-(4,5-dimethylthiazol-2-yl)-5-)3-Carboxymethoxyphenyl)-2-(sulfophenyl)-2H-tetrazolium solution. The plates were incubated at 37°C for 2h and absorbance at 490nM was measured. Each combination of cell line and drug concentration was set up in eight replicate wells, and the experiment was repeated three times. Each data point represents a mean ± SD and normalized to the value of the corresponding control cells.
Cell Proliferation assay
Cellular proliferation was analyzed using the colorimetric MTS assay kit (Promega). Briefly, H460 cells (1000 cells/well) and A549 cells (1500 cells/well) were plated in 96-well plates and the growth rate was measured.
Soft agar growth assay
A549 and H460 cells (2×104) stably expressing Nrf2-shRNA or the control Luc-shRNA were diluted in 4ml of DMEM medium containing 10% serum and 0.4% low melting point (LMP) agarose. This mixture was subsequently placed over 5ml of hardened DMEM medium containing 10% serum and 1% LMP and allowed to harden at room temperature. The cells were allowed to grow for 2–3 weeks, after which visible colonies containing greater than 50 cells were counted.
Tumor Xenografts and siRNA Treatment
We injected A549 cells (5 × 106) and H460 cells (2×106) subcutaneously into the flank of athymic nude mice and measured the tumor dimensions by caliper once every week. The tumor volumes were calculated using the following formula: [length (mm) × width (mm) × width (mm) × 0.52]. For in vivo delivery of siRNA into tumors, siRNA duplexes diluted in PBS were injected into the tumors using insulin syringes at a concentration of 10µg of siRNA/ 50mm3 of tumor volume. Intraperitoneal injections of carboplatin were given at a dose of 40mg/kg body weight. Both siRNA and carboplatin were administered weekly twice for 4 weeks. Upon termination, tumors were harvested and weighted. All experimental protocols conducted on the mice were performed in accordance with NIH guidelines and were approved by the Johns Hopkins University Animal Care and Use Committee.
Immunohistochemistry
Formalin-fixed, paraffin embedded tissue sections were treated with anti-Ki-67 antibody at a dilution of 1:100 for 1hr and stained using LSAB+System-HRP kit (DakoCytomation) according to the manufacturer’s instructions. Non-immune rabbit IgG (Jackson ImmunoResearch Laboratories) was used as a negative control. Ki-67 positive cells/ 200 cancer cells were counted in randomly selected visual fields at 20X magnification and the mean of 5 fields were calculated.
Statistical Analysis
Statistical comparisons were performed by Student’s t-tests, Wilcoxon rank-sum test or Chi-square test. A value of p<0.05 was considered statistically significant. Tumor weights and changes in tumor volume were summarized using descriptive statistics. Differences in tumor measures between treatment groups were examined using linear regression models with generalized estimating equations (GEE). The distributions of both tumor measurements were skewed, so log transformations were used.
Results
Generation of lung cancer cell lines stably expressing Nrf2shRNA
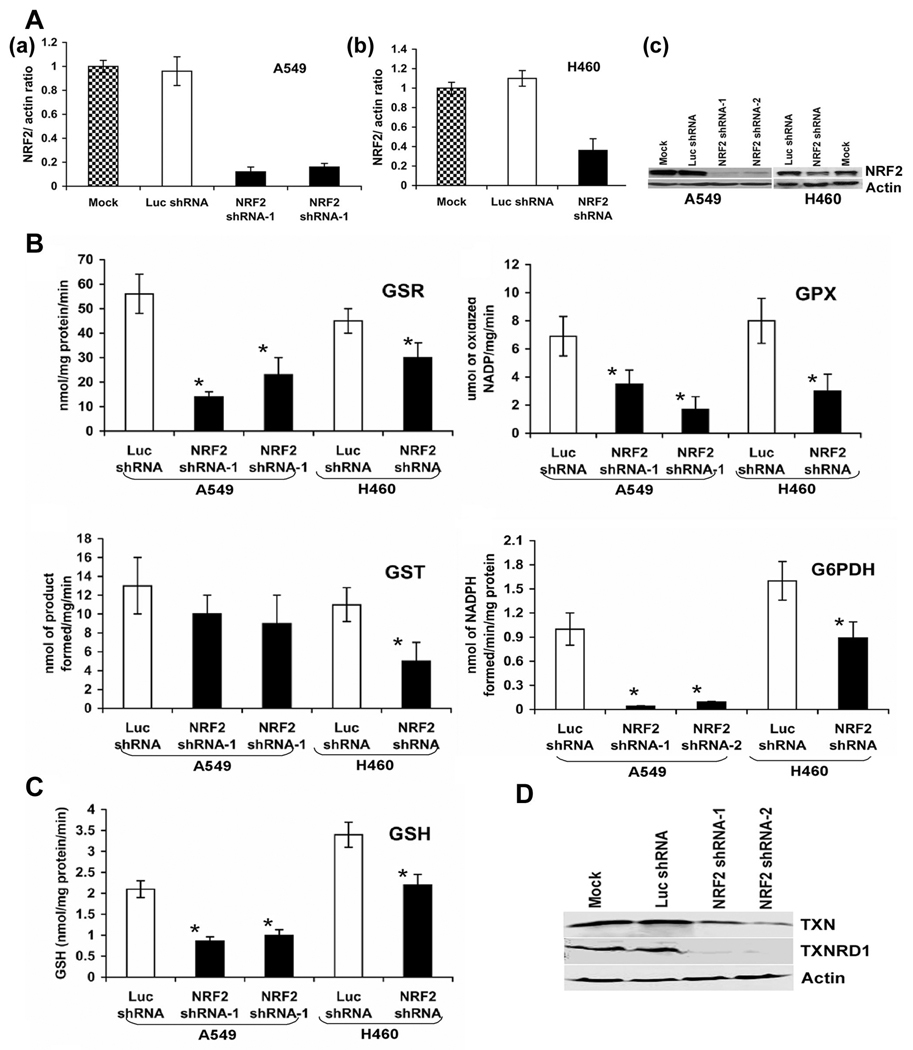
To inhibit the expression of Nrf2, we designed a short hairpin RNA targeting the 3’ end of the Nrf2 transcript as described in our previous reports (21, 25). Short hairpin RNA cassette was subcloned into pSilencer vector and transfected into A549 and H460 cells. A short hairpin RNA targeting luciferase gene was used as control. Stable cell clones with reduced Nrf2 expression were generated. We screened 15 clone’s transfected with Nrf2-shRNA and 10 clone’s transfected with luciferase shRNA for each cell line by real time quantitative PCR and immunoblotting. After initial screening, we selected two independent clones of A549 cells expressing Nrf2-shRNA, which demonstrated a stable 85% downregulation of Nrf2 mRNA (Fig. 1a). A single clone expressing Nrf2 shRNA derived from H460 cells demonstrated 70% inhibition of Nrf2 mRNA (Fig. 1b). Measurement of Nrf2 protein by western blotting showed similar decrease in protein levels. The expression of Nrf2 did not change between the control cells transfected with luciferase shRNA and the untransfected cancer cells (Fig. 1c).
Figure. 1.
(A) Generation of cell lines stably expressing Nrf2 shRNA. (a–c) Real time RT-PCR analysis of Nrf2 expression in A549 and H460 cells stably expressing Nrf2 shRNA. Total RNA from stable clones harboring Nrf2 shRNA or non-targeting luciferase shRNA were analyzed for expression of Nrf2. GAPDH was used as normalization control. (c) Immunoblot detection of Nrf2 in A549 and H460 cells stably transfected with shRNAs targeting Nrf2. (B–C) Comparison of GSR, GPX, GST, G6PDH enzyme activities and total GSH levels between cells expressing Nrf2 shRNA and control cells expressing luciferase shRNA. Data represent mean ± SE (n = 3). *, p < 0.05 relative to the cells expressing luciferase shRNA (by t-test). (D) Western blot analysis of TXN1 and TXNRD1 levels in A549 cells stably transfected with the Nrf2 shRNA and control cells expressing luciferase shRNA.
Lowering Nrf2 expression in A549 and H460 cells causes global decrease in expression of electrophile and drug detoxification system
Lowering of Nrf2 protein level leads to a decline in the expression of electrophile and drug detoxification genes in normal cells. The expression of selected electrophile and drug detoxification genes were determined in two clones of A549 and one clone of H460 cells stably expressing Nrf2 shRNA using real time RT-PCR (Table-1).
Table. 1.
Table showing the list of genes downregulated in A549-Nrf2shRNA and H460-Nrf2 shRNA cells in response to Nrf2 inhibition. The expressions of several Nrf2 dependent genes were quantified using real time RT-PCR. Cells stably expressing luciferase shRNA were used as baseline control to calculate the fold changes. All the represented fold change values of Nrf2 shRNA expressing cells are significant compared to the control cells transfected with luciferase shRNA.
| Gene Title/ Group | Accession No. |
A549 | H460 | |
|---|---|---|---|---|
| Real time data | ||||
| Nrf2shRNA-1 | Nrf2shRNA-2 | Nrf2shRNA | ||
| %inhibition (±SE) | ||||
| Glutathione pathway genes | ||||
| glutamate-cysteine ligase, modifier subunit | NM_002061.1 | −80% ±5 | −79% ±5 | −68% ±4 |
| glutamate-cysteine ligase, catalytic subunit | NM_001498.1 | −72% ±8 | −75% ±6 | −60% ±5 |
| glutathione reductase | NM_000637.1 | −70% ±9 | −69% ±6 | −50% ±7 |
| solute carrier family 7, (cationic amino acid transporter, y+ system) member 11 | NM_14331.1 | −68% ±7 | −72% ±6 | −58% ±4 |
| Microsomal glutathione S-transferase 1 | AV705233 | −75% ±6 | −68% ±5 | −58% ±5 |
| glutathione S-transferase M4 | NM_000850.1 | −66% ±4 | −60% ±3 | −50% ±6 |
| glutathione peroxidase 2 (gastrointestinal) | NM_002082.1 | −85% ±6 | −83% ±7 | −74% ±3 |
| glutathione peroxidase 3 (plasma) | NM_02084.2 | −68% ±4 | −65% ±4 | −48% ±4 |
| Thioredoxin pathway genes | ||||
| thioredoxin | NM_003329.1 | −65% ±4 | −60% ±6 | −50% ±3 |
| thioredoxin reductase 1 | NM_003330.1 | −60% ±6 | −62% ±4 | −51% ±7 |
| NADPH regenerating enzymes | ||||
| glucose-6-phosphate dehydrogenase | NM_000402.1 | −80% ±8 | −78% ±6 | −50% ±6 |
| malic enzyme 1 | NM_002395.1 | −70% ±5 | −71% ±6 | −58% ±3 |
| Other antioxidants | ||||
| NAD(P)H dehydrogenase, quinone 1 | NM_000903.1 | −70% ±9 | −59% ±8 | −46% ±8 |
| heme oxygenase (decycling) 1 | NM_002133.1 | −75% ±6 | −70% ±8 | −60% ±4 |
| Peroxiredoxin 1 | NM_002574.2 | −60% ±4 | −62% ±5 | −50% ±5 |
| Drug Transporters | ||||
| ATP-binding cassette, sub-family C (CFTR/MRP), member 1 | NM_004996.2 | −60% ±3 | −57% ±4 | −53% ±4 |
| ATP-binding cassette, sub-family C (CFTR/MRP), member 2 | NM_000392.1 | −65% ±4 | −60% ±7 | −60% ±5 |
Lowering Nrf2 expression by RNAi in the A549 and H460 cells decreased the mRNA expression of the genes that constitute the glutathione system [γ-glutamyl cysteine synthetase modifier and catalytic subunit (GCLM, GCLC), glutathione reductase (GSR), and the cysteine/glutamate transporter (SLC7A11) that transports cysteine for synthesis of glutathione] as well as the glutathione-dependent enzymes Glutathione peroxidase 2 (GPx2), Glutathione peroxidase 3 (GPx3) and Glutathione S-transferase’ s (MGST1 and GSTM4) (Table-1).
Enzyme activity measurements for selected gene products (GSR, GPX and GST) were carried out to determine the extent to which their transcriptional inhibition paralleled changes in their activities. There was significant decrease in activities of all of these enzymes in the A549-Nrf2shRNA and H460-Nrf2shRNA cells relative to the cells expressing luciferase shRNA (Fig.1B). Direct measurement of intracellular GSH concentration by Teitz assay demonstrated a decrease in GSH levels in Nrf2 depleted A549 cells and in H460 cells (Fig.1C). Pretreatment of A549-LucshRNA and A549-Nrf2shRNA cells with NAC increased GSH levels in both A549-LucshRNA and A549-Nrf2shRNA cells but the total GSH levels in NAC treated A549-Nrf2shRNA remained much less than the vehicle treated A549-LucshRNA cells (Supplementary Fig. S1). A549-LucshRNA cells responded rapidly and their GSH levels increased after 1hr of NAC treatment. A549-Nrf2RNA cells demonstrated an increase in GSH levels only after 24hrs of NAC treatment suggesting that GSH synthesis in response to NAC treatment is impaired in A549-Nrf2RNA cells.
Lowering of Nrf2 in A549 and H460 cell caused significant decrease in the mRNA for TXN1 and TXNRD1 which constitutes the thioredoxin system and has been associated with therapeutic resistance. Protein levels of TXN1 and TXNRD1 were decreased in A549-Nrf2shRNA cells but did not change between control A549 cells expressing Luc-shRNA and the untransfected cells (Fig.1D).
NADPH is required to provide reducing equivalents for the regeneration of reduced glutathione and thioredoxin by GSR and TXNRD1. Expression of genes encoding the NADPH biosynthesis enzymes, such as G6PDH and malic enzyme 1 (ME1) were downregulated in the A549-Nrf2shRNA and H460-Nrf2shRNA cells suggesting the dependence of these genes on Nrf2 for their expression (Table-1). Consistent with low transcript levels, G6PDH enzyme activity was significantly downregulated in A549-Nrf2shRNA and H460-Nrf2shRNA (Fig.1C).
We also found that other antioxidant genes such as NQO1, HO-1 and PRDX1 were downregulated as a result of lowering of Nrf2 by shRNA in cancer cells (Table 1). Furthermore, the transcript levels of multidrug resistance protein like ATP-binding cassette, sub family C, member 1 (ABCC1) and ATP-binding cassette, sub family C, member 2 (ABCC2), were significantly downregulated in cells expressing Nrf2 shRNA. Thus, downregulation of Nrf2 profoundly decreased the expression of antioxidant enzymes and electrophile and drug detoxification systems in cancer cells with constitutive activation of Nrf2 function.
Enhanced production of ROS in cells stably transfected with Nrf2 shRNA
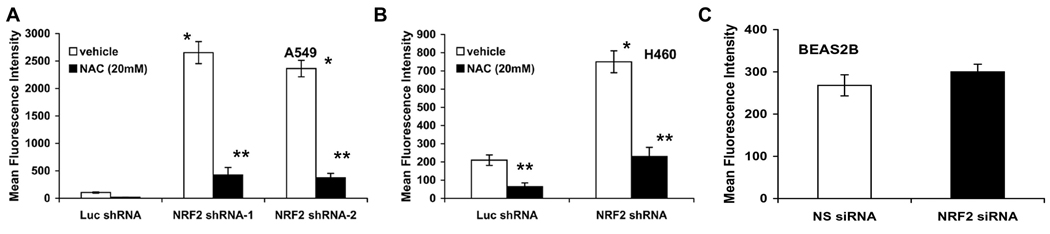
To determine the degree of overall increase in oxidative stress as a result of global decrease in the expression of electrophile detoxification system by downregulating Nrf2, intracellular ROS levels were monitored using 2’, 7’-dichlorodihydrofluorescein diacetate (c-H2DCFDA) and flow cytometry. The results demonstrated an increase in fluorescence in both A549-Nrf2shRNA and H460-Nrf2shRNA cells (Fig. 2A–B). A549-Nrf2 shRNA cells demonstrated a pronounced 25-fold increase in ROS level where as H460-Nrf2shRNA cells demonstrated a 3.5-fold increase in ROS levels. Treatment of these cells with nonspecific free radical scavenger NAC for 30mins reduced ROS production and attenuated the mean fluorescent intensity in A549-Nrf2shRNA and H460-Nrf2shRNA by 85% and 75% in A549 and H460 cells respectively. These results suggest that the generation of ROS at a steady state is relatively increased in Nrf2 shRNA transfectants than in control Luc-shRNA cells. Interestingly, inhibition of Nrf2 activity in non-tumorigenic BEAS2B cells did not show a significant increase in ROS (Fig. 2C). Thus, constitutive Nrf2 activity is indispensable for maintaining redox balance in cancer cells unlike normal cells in the absence of stress.
Figure. 2.
Inhibition of Nrf2 activity leads to ROS accumulation in A549-Nrf2shRNA and H460-Nrf2shRNA cells. (A–B) Comparison of ROS levels in A549 and H460 cells stably expressing Nrf2 shRNA. Cells expressing non-targeting Luc shRNA were used as control. Pretreatment with 20mM NAC decreased the ROS levels. ROS levels in cells expressing luciferase shRNA were same as the control untransfected cells. (C) ROS levels did not change significantly between the BEAS2B cells transfected with Nrf2 siRNA and the control non-targeting NS siRNA. *, p < 0.01 relative to the cells expressing luciferase shRNA; **, p < 0.01 relative to the cells pretreated with NAC.
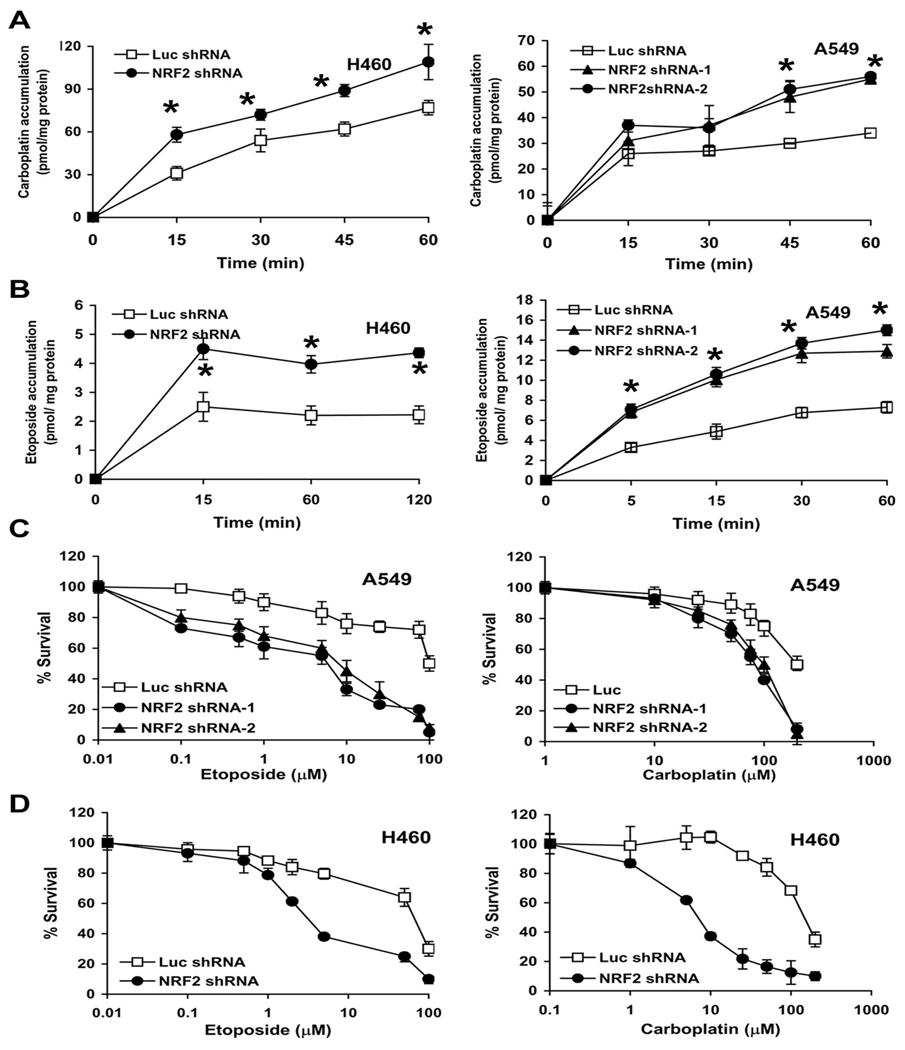
Decrease in Nrf2 expression by shRNA leads to increased drug accumulation and enhanced chemosensitivity in cancer cells
Since Nrf2 shRNA causes decrease in expression of drug detoxification enzymes as well as drug efflux pumps, we measured drug accumulation in cancer cells (H460 and A549) stably transfected with shRNA targeting Nrf2. A non-specific shRNA targeting luciferase (Luc shRNA) was used as control. To analyze drug accumulation, cells were incubated with radiolabeled drug and intracellular drug content was assayed at various time points. Drug retention was substantially higher in Nrf2shRNA cells (~1.5–2 fold) at 30–120 mins as compared with the control Luc-shRNA cells. The difference in intracellular drug content remained same or increased with time. Increased drug accumulation in cells with low levels of Nrf2 protein suggests that Nrf2 plays an important role in regulating the accumulation of drug in the cancer cells (Fig. 3A–B)
Figure. 3.
Overexpression of Nrf2 confers drug resistance. (A–B) Effect of Nrf2 inhibition on drug accumulation in lung cancer cells. Tritium (3H) labeled etoposide and 14C labeled carboplatin accumulation in A549-Nrf2shRNA and H460-Nrf2shRNA cells was measured at regular time intervals (15–120 mins) after incubation with the drug. A non-targeting luciferase shRNA was used as control. Data are mean of 3 independent replicates, combined to generate the mean ± SE for each concentration. Drug accumulation was significantly higher in cells expressing Nrf2 shRNA. ‘*’, P<0.05 relative to Luc shRNA. (C–D) Enhanced sensitivity of A549-Nrf2shRNA and H460–Nrf2shRNA cells to carboplatin and etoposide. Cells were exposed to drugs for 72h– 96h and viable cells were determined by MTS/ phenazine methosulfate assay. Data is represented as percentage of viable cells relative to the vehicle treated control. Data are mean of 8 independent replicates, combined to generate the mean ± SD for each concentration. Representative experiments are shown.
To study whether targeting Nrf2 expression enhances the sensitivity of lung cancer cells to chemotherapeutic drugs like carboplatin and etoposide, we used same A549 and H460 cells stably expressing Nrf2 shRNA. We treated these cell populations with escalating concentrations of carboplatin and etoposide. The concentrations were selected after pilot experiments to determine the maximum amount of drug that revealed survival differences between A549 and H460 cells expressing control shRNA and its derivatives expressing anti-Nrf2 shRNA. We found that lowering of Nrf2 expression in both A549 and H460 cell lines greatly enhanced the cytotoxicity (~30–70%) of these drugs resulting in increased cell death compared to the control shRNA group (Fig. 3C–D). The IC50 doses of carboplatin and etoposide was followed by a reduction in the number of viable cells to 50% as compared with vehicle treated control cells. The IC50 for carboplatin and etoposide decreased in both A549-Nrf2shRNA and H460-Nrf2shRNA cells when compared with their respective control cells expressing luciferase shRNA. In summary, decreasing Nrf2 activity in cancer cells enhanced the cytotoxicity of drugs. Pretreatment with 5mM NAC for 2hr significantly protected the A549 and H460 control cells as well as Nrf2-shRNA cells against the toxicity of carboplatin (Supplementary Fig. S2) but not etoposide (data not shown)..
To further demonstrate that increased Nrf2 activity confers resistance against carboplatin and etoposide induced apoptosis and inhibition of Nrf2 can sensitize cells to drug induced apoptosis, we carried out Caspase-3 cleavage assays. H460-LucshRNA cells were resistant to carboplatin (25µM and 50µM) and etoposide (1µM) mediated apoptosis and did not show any caspase-3 cleavage in response to drug treatment. In contrast, H460-Nrf2 shRNA cells displayed the presence of cleaved caspase-3 product, indicative of apoptosis after treatment with identical concentrations of carboplatin and etoposide (Supplementary Fig. S3).
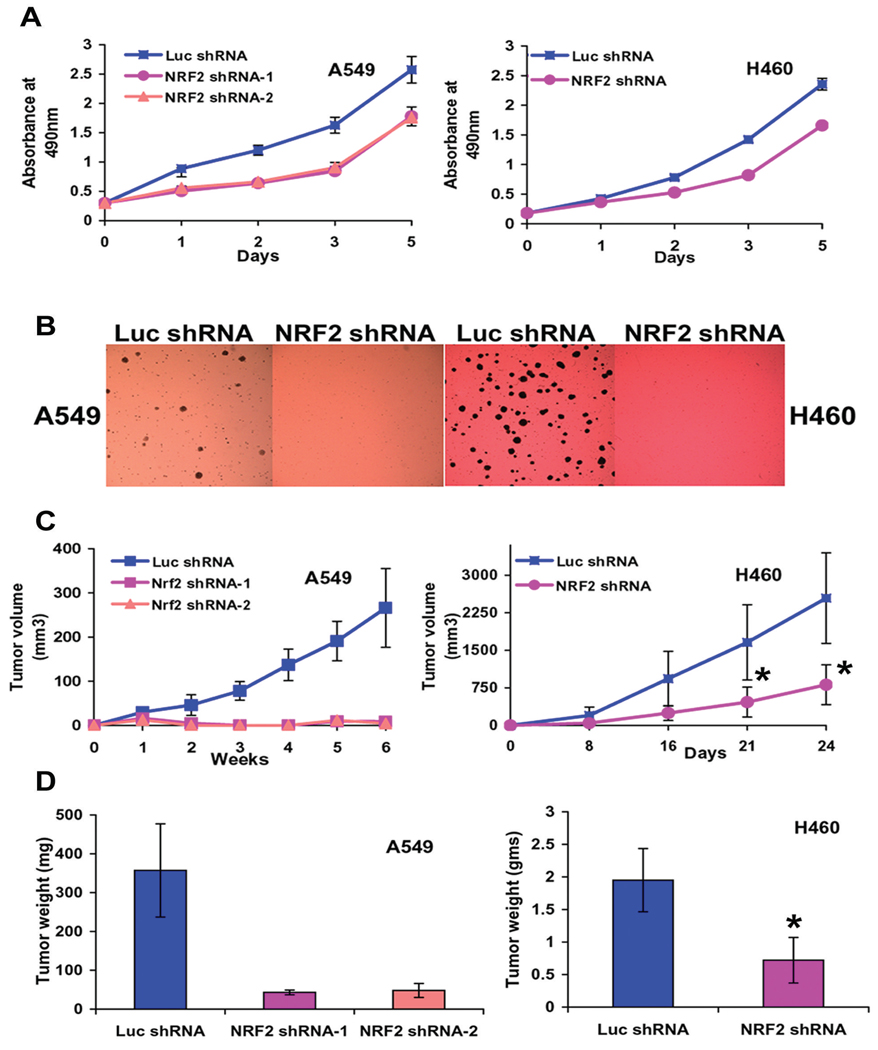
Nrf2 is required for anchorage independent growth and tumor formation in vivo
The misexpression of Nrf2 prompted us to examine its significance in the tumorigenic properties of the non-small-cell-lung-cancer cells. Depletion of Nrf2 in both the cancer cell lines resulted in a pronounced decrease in cellular proliferation as measured by MTS assay (Fig. 4A). We also determined the ability of A549-Nrf2shRNA and H460-Nrf2shRNA cells to form colonies in soft agar. Suppression of Nrf2 in H460 and A549 cell lines resulted in a substantial reduction on colony formation in soft agar compared to the control cells expressing luciferase shRNA (Fig. 4B). In order to further examine the affect of Nrf2 suppression on lung tumorigenesis, we injected A549-Nrf2shRNA and H460-Nrf2shRNA and their corresponding control cells expressing Luc-shRNA into the flank of athymic nude mice and monitored the increase in tumor volume over a 4–6 week period. Weight of the tumor was recorded at the termination of the experiment. Significantly, suppression of Nrf2 in the A549 cells resulted in complete inhibition of tumor formation whereas H460 cells showed a less dramatic yet significant and reproducible reduction in tumor volume (Fig. 4C). Mean difference in tumor weight between the Luc-shRNA and Nrf2 shRNA expressing H460 cells was 1.24 gms (95% CI=0.773 to 1.71; P=0.0001) (Fig. 4D). Data was analyzed using two-sample Wilcoxon rank-sum (Mann-Whitney) test. These data indicate that Nrf2 is required for maintenance of the transformed phenotype in vitro and in vivo.
Figure. 4.
Nr2 ablation leads to reduced tumorigenic properties in vitro and in vivo. (A) Nrf2 promotes lung cancer cell proliferation. A549-Nrf2shRNA (1500 cells) and H460-Nrf2shRNA (1000 cells) cells were plated in 96 well plates and cellular proliferation was analyzed using the colorimetric MTS assay over the indicated time course. Cancer cells expressing Luc-shRNA were used as control. (B) A549-Nrf2shRNA and H460-Nrf2shRNA expressing cells were also analyzed for anchorage-independent growth. (C–D) A549-Nrf2shRNA and H460-Nrf2shRNA cells were injected in the flank of male athymic nude mice (n = 7 for H460, n=6 for A549). A549 and H460 cells expressing Luc-shRNA were used as control. Weekly measurements were taken from the tumors, and the mean tumor volume was determined after 4–6 weeks. Weight of the tumor was recorded at the termination of the experiment. Mean difference in tumor weight between the Luc-shRNA and Nrf2 shRNA expressing H460 cells was 1.24 gms (95% CI=0.773 to 1.71; “*” P=0.0001). Data was analyzed using two-sample Wilcoxon rank-sum (Mann-Whitney) test. A549-Nrf2 shRNA cells did not form any tumor in nude mice.
Therapeutic efficacy of Nrf2 siRNA in combination with carboplatin in vivo
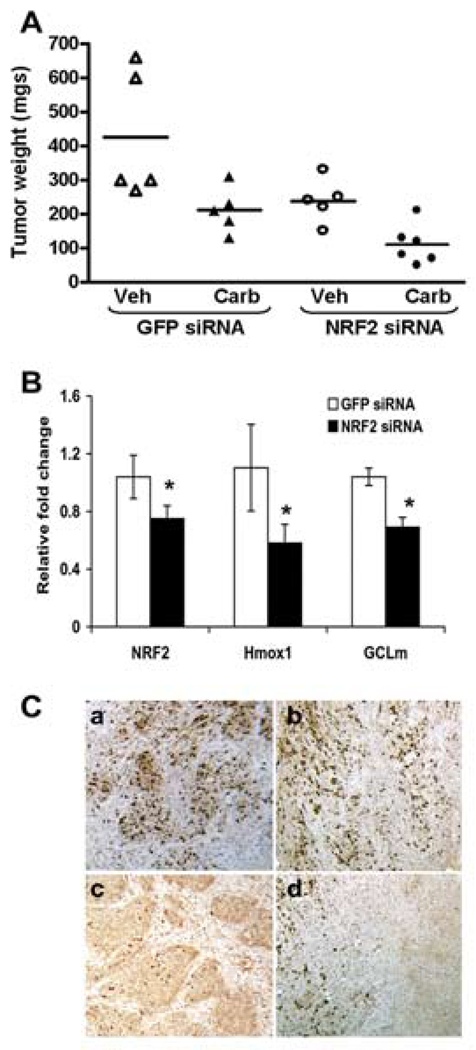
To elucidate whether the potential synergistic mode of action of Nrf2 shRNA and carboplatin observed in cell culture occurs in vivo, we performed a xenograft experiment with A549 cells. Mice bearing subcutaneous tumors were randomly allocated to one of the following groups with therapy beginning 15 days after tumor cell injection: GFP siRNA, GFP siRNA+ carboplatin, Nrf2 siRNA and Nrf2 siRNA+ carboplatin. Mice were treated with siRNA and carboplatin twice a week for 4 weeks and tumor volume was measured biweekly. Tumor weight was measured at the termination of the experiment (Fig. 5A) (Supplementary Table-1). Treatment with control non-targeting siRNA did not inhibit tumor growth as compared to control mice treated with PBS alone (data not shown). The change in tumor volume was significantly different between GFP siRNA and Nrf2 siRNA treated tumors (P<0.0001). Tumor weights were significantly higher in the GFP siRNA treated tumors compared to Nrf2 siRNA treated tumors (ratio of weights = 2.09, 95%CI: [1.41, 3.10], p=0.0002), and siRNA compared to siRNA + carboplatin treated tumors (2.13, 95%CI: [1.44, 3.16], p=0.001) (Fig. 5A). The change in tumor volume was significantly different between Nrf2 siRNA and GFP siRNA treated tumors (ratio of differences = 0.46, 95%CI: [0.31, 0.68], p=0.0001) and siRNA + carboplatin and siRNA tumors (0.45, 95%CI: [0.29, 0.71], p=0.0005). The difference in the change in tumor volume was larger between GFP siRNA + carboplatin and Nrf2 siRNA + carboplatin (differences of 352.34 and 58.78) than it was for GFP siRNA and Nrf2 siRNA (differences of 532.94 and 249.17), (ratio of differences = 2.38, 95%CI: [1.03, 5.48], p=0.042). Data from the second set of experiments validating the same findings is presented in the supplement. (Supplementary Fig. S4) (Supplementary Table-2). Gene expression analysis of randomly selected tumors from GFP siRNA and Nrf2 siRNA groups demonstrated significant decrease in Nrf2 and its downstream target gene expression (Fig. 5B).
Figure. 5.
Therapeutic efficacy of Nrf2 siRNA in combination with carboplatin. (A) Nude mice were injected subcutaneously with A549 cells and randomly allocated to one of the following groups with therapy beginning 15 days after tumor cell injection: GFP siRNA, GFP siRNA+ carboplatin, Nrf2 siRNA and Nrf2 siRNA+ carboplatin. Mice were treated for 4 weeks and then sacrificed. A dot plot shows the tumor weights upon termination by treatment group. Weights of the GFP siRNA treated tumors were significantly higher compared to Nrf2 siRNA treated tumors (ratio of weights = 2.09, 95% CI: [1.41, 3.10], p = 0.0002), and siRNA treated compared to siRNA+ carboplatin treated tumors (2.13, 95% CI: [1.44, 3.16], p = 0.001). (B) Delivery of naked Nrf2 siRNA duplex into tumor inhibited the expression of Nrf2 and its downstream target genes (HO-1 and GCLm). ‘*’,P<0.05 (Wilcoxon rank-sum test). (C) The proliferative index based on Ki-67 immunoreactivity in A549 tumors. Part “a”, shows large fraction of Ki-67 positive cells in GFP siRNA treated A549 tumors. Part ’b’ shows large number of Ki-67 stained cells in GFP siRNA+ carboplatin treated tumors. Part ‘c’ shows very few Ki-67 positive cells in Nrf2 siRNA treated tumors. Part ‘d’ shows ki-67 stained cells in Nrf2 siRNA+ carboplatin treated A549 tumors. Note that ‘d’ has area of extensive cell death (approximately the right half of the panel), and this massive cellular death was not seen in the other samples.
Immunohistochemical staining of GFP siRNA treated A549 tumors demonstrated 48% Ki-67 positive cells in tumors treated with GFP siRNA alone and 46% positive cells in tumors treated with GFP siRNA and carboplatin, suggesting that carboplatin treatment alone does not significantly affect the rate of cell replication (Fig. 5a–b). On the other hand, tumors treated with Nrf2 siRNA alone showed only 18% of cells labeling with Ki-67, and tumors treated with a combination of Nrf2 siRNA and carboplatin showed only 19% of cells labeling with Ki-67, suggesting that blocking Nrf2 expression does reduce the rate of cell replication (Figures 5c and 5d). In addition, the tumors treated with a combination of Nrf2 siRNA and carboplatin showed areas of massive cell death, corresponding to the greatly reduced size of tumors seen in this treatment group (Fig. 5d). Number of Ki-67 positive cells in Nrf2 siRNA treated tumors were significantly lower than the GFP siRNA treated tumors (P<0.001). There were no significant differences in Ki-67 labeled cells between the GFP siRNA and GFP+carboplatin or Nrf2 siRNA and Nrf2 siRNA+ carboplatin treated groups.
Discussion
Therapeutic resistance in cancer cells occurs as a result of genetic aberrations that confer tumorigenic potential and survival advantage against chemotherapy. Nrf2, a redox sensitive bZIP transcription factor, activates cytoprotective pathways against oxidative injury, inflammation and apoptosis through transcriptional induction of a broad spectrum of genes involved in electrophile /drug detoxification and antioxidant protection (10, 16, 27). Keap1 negatively regulates Nrf2 activity by targeting it for proteasomal degradation. We and others have shown that there is increased Nrf2 activity in NSCLC cells due to somatic mutations in Keap1 gene (21–23). Loss of Keap1 activity leading to constitutive activation of Nrf2 in lung cancer cells upregulates the expression of antioxidants, electrophile and drug detoxification enzymes and efflux proteins (21, 22). To understand the functional significance of the gain of Nrf2 function in lung cancer, we used an RNAi approach and selected two lung cancer cell lines (A549 and H460) that have been previously reported to have complete loss of Keap1 activity (21).
Inhibition of Nrf2 in A549 and H460 cells resulted in marked decrease in the expression of genes that constitute the glutathione system (GSH biosynthesizing enzymes, GPX, GSR, GST’s), the thioredoxin system (TXNRD1, TXN and PRDX), NADPH regenerating system (G6PDH), antioxidants, and drug efflux pumps (10, 27). In corroboration with gene expression, enzyme activities of GSR, GPX, GST and G6PDH as well total GSH levels were significantly reduced in A549-Nrf2shRNA and H460-Nrf2shRNA cells when compared with Luc-shRNA control cells. Thus, downregulation of Nrf2 expression profoundly decreased the expression of key antioxidant enzymes and electrophile/ drug detoxification systems in lung cancer cells with constitutive activation Nrf2 function.
Increased reactive oxygen species (ROS) is common in cancer cells and is believed to be attributable at least in part to high metabolism and hyperactive glycolytic metabolism driven by oncogenic proliferative signals (7). The intrinsic ROS associated with oncogenic transformation renders the cancer cell highly dependent on antioxidant systems to maintain redox balance, and thus, vulnerable to agents that impair antioxidant capacity. The downregulation of Nrf2 pathway resulted in dramatic accumulation of intracellular ROS in A549-Nrf2shRNA and H460-Nrf2shRNA cells. Treatment of these cells with non-specific free radical scavenger NAC reduced ROS production in both A549-Nrf2shRNA and H460-Nrf2shRNA cells. These results suggest that steady state generation of ROS is relatively increased in Nrf2-depleted cells as compared to control cancer cells and it provides a biochemical basis to develop new therapeutic strategies to preferentially increase ROS to a toxic level in cancer cells and selectively eradicate them. Interestingly, basal levels of ROS did not differ between wild type and nrf2−/− mouse embryonic fibroblasts (28).
Depletion of Nrf2 in both the cancer cell lines resulted in a pronounced decrease in cellular proliferation. Suppression of Nrf2 in the H460 and A549 cells resulted in a substantial reduction in colony formation on soft agar compared to the control cells. Significantly, decreased Nrf2 levels in the A549 cells resulted in complete inhibition of tumor formation in athymic mice whereas H460 cells showed significant reduction in tumor volume and weight. These data indicate that Nrf2 is essential for growth of cancer cells in vitro and in vivo. Recently, Reddy et al (29) reported that type-II epithelial cells isolated from nrf2−/− mice lungs display defects in cell proliferation and GSH supplementation rescues these phenotypic defects (29). We hypothesize that decreased antioxidant capacity leading to increased ROS levels in A549-Nrf2shRNA and H460-Nrf2shRNA cells inhibited the growth of these cells in vitro and in vivo compared to the control A549 and H460 cells expressing Luc-shRNA. Thus, unlike normal cells, constitutive activation of Nrf2 is indispensable for maintaining the redox balance and growth of lung cancer cells under homeostatic conditions.
Anticancer drugs like cisplatin, carboplatin, and oxaliplatin are commonly used intravenous platinating agents (30, 31). Treatment with these agents is characterized by resistance, both acquired and intrinsic. This resistance can be caused by a number of cellular adaptations including reduced uptake, inactivation by glutathione and other antioxidants and increased levels of DNA repair. Since Meister (32) claimed that the cellular metabolism of glutathione could affect the fate of chemotherapeutic agents, several reports have shown that glutathione content is increased in several drug resistant cancer cell lines (33, 34). Glutathione, a non-protein thiol, can interact with the reactive site of a drug, resulting in conjugation of the drug with glutathione. The conjugate is less active and more water soluble and it is excluded from the cell with the participation of transporter proteins named GS-X (including multidrug resistance proteins). Increased levels of glutathione were found in cell lines resistant to alkylating agents (e.g. nitrogen mustard, chlorambucil, melphalan, cyclophosphamide carmustine, vincristine and anthracyclines) (5, 35). Glutathione-S-transferases catalyze the interactions between glutathione and alkylating drugs thereby increasing the rate of drug detoxification. Thus, activation of these enzymes can cause cellular drug resistance (5, 36). Expression of thioredoxin, another important thiol, increases in many human cancers and is a validated target associated with resistance to standard chemotherapeutic agents like cisplatin and etoposide and decreased patient survival (37–39). Inhibition of Nrf2 activity by shRNA-mediated gene silencing debilitated the expression of antioxidants and drug detoxification genes thereby increasing the accumulation of etoposide and carboplatin in lung cancer cells and enhanced the cytotoxicity of these drugs. Increased accumulation of these drugs in Nrf2 shRNA expressing cells supports the idea that Nrf2 contributes to drug resistance by modulating the expression of several drug detoxification enzymes and efflux proteins.
Pretreatment with NAC, a potent thiol antioxidant, reversed the cytotoxicity of carboplatin in A549 and H460 parent cells as well as Nrf2 depleted cells and significantly increased the survival. However, NAC pretreatment did not restore GSH levels to normal in Nrf2 knockdown cells because the genes involved in GSH biosynthesis are positively regulated by Nrf2. NAC can protect cells in this context by covalent binding of the molecule to the platinum, producing an inactive complex (40–42). Schweitzer (1993) showed that sulfur-containing compounds may prevent cisplatin from interacting with target molecules, displacing platinum after it is bound (43). Therefore, the NAC-mediated protective effect against carboplatin was most likely due to its function as a direct thiol containing antioxidant rather than as a precursor for GSH synthesis (44, 45). However, pretreatment with NAC had no effect on etoposide induced apoptotic cell death. Similar observations have been reported by Garder et al., where antioxidants failed to prevent etoposide provoked apoptosis, although etoposide induced ROS production and glutathione depletion (46, 47). Etoposide, a topoisomerase-II inhibitor, can be metabolized to DNA inactivating catechol, ortho-quinone and semi-quinone free radical derivatives which may contribute to its cytotoxicity (48). GSH protects against semiquinones and orthoquinone free radicals of etoposide by GST-mediated conjugation with these species (48). The GS-X conjugates of etoposide are pumped out of the cell by GS-X pumps encoded by ABCC1, ABCC2, ABCC3 etc (49). Depletion of GSH levels by D,L-buthionine-S,R-sulfoximine treatment enhanced the cytotoxicity of etoposide (50). Thus it appears that total cellular GSH content plus phase-II and phase-III drug detoxification enzymes may be a determinant for etoposide toxicity.
To elucidate whether suppression of Nrf2 expression can potentiate the cytotoxicity of carboplatin in vivo, we performed a xenograft experiment with A549 cells. Mice bearing A549 subcutaneous tumors were treated with Nrf2 siRNA and carboplatin and tumor volume as well as weight were measured. Treatment with Nrf2 siRNA alone reduced mean tumor weight by 53% (±20% SD) compared to the control group. When Nrf2 siRNA was combined with carboplatin, there was an even greater reduction in mean tumor weight. Thus, combination of Nrf2 siRNA with carboplatin led to a significant reduction in tumor growth compared with either agent alone. Treatment with Nrf2 siRNA inhibited cell proliferation as demonstrated by reduced Ki-67 staining. Interestingly, tumors treated with combination of Nrf2siRNA and carboplatin displayed areas of massive cellular death that was not seen in the other samples. These findings suggest that Nrf2, but not carboplatin, affects cell replication, and that the combination of the two agents’ results in extensive cell death in addition to the effects on cell replication caused by Nrf2. Thus, Nrf2 siRNA inhibitors may present a novel therapeutic approach for lung cancer with chemoresistance.
In conclusion, we demonstrate that constitutive activation of Nrf2 plays an important transcriptional role in the activation of genes involved in protection against oxidative stress and thereby promotes tumorigenecity and chemoresistance. These studies provide a potentially novel therapeutic strategy to circumvent therapeutic resistance in lung cancer.
Supplementary Material
Acknowledgements
We thank Krishnamachari Balaji, Ellen Tully and Juliana Cuervo Rojas at Johns Hopkins University, for help with mice experiments, immunohistochemical staining and statistical analysis respectively. We also thank the Paul Fallon and Jay Bream at Becton Dickinson Immune Function Laboratory, Johns Hopkins School of Public health, for help with the flow cytometry work. We thank Igor Mett and Hagar Kalinski from Quark Pharmaceuticals, Inc. for Nrf2 siRNA design. This work was supported by NIH grants NCI SPORE P50 CA058184 (SB, EG), R01 HL081205 (SB), P30ES03819, R01 CA104253 (FB), a research grant from the Maryland Cigarette Restitution fund (SB), Quark Pharmaceuticals, Inc (SB), Maryland State Stem Cell Pilot grant (SB) and young clinical innovator award (SB) and young clinical scientist award to AS from Flight Attendant Medical Research Institute. SKR was supported by Raman research fellowship from Council for Scientific and Industrial Research, India.
Abbreviations
- ARE
antioxidant response element
- RNAi
RNA interference
- shRNA
short hairpin RNA
- siRNA
short interfering RNA
- GSH
glutathione
- GCLc
γ-glutamyl cysteine synthetase catalytic subunit
- GCLm
γ-glutamyl cysteine synthetase modifier subunit
- GSR
glutathione reductase
- GST
glutathione-S-transferase
- ROS
reactive oxygen species
- KEAP1
Kelch-like ECH-associated protein 1
- Nrf2
nuclear factor erythroid-2 related factor 2
- NSCLC
non-small-cell lung cancer
- GSH
glutathione
- GSR
glutathione reductase
- GST
glutathione-S-transferase
- G6PDH
Glucose-6-phosphate dehydrogenase
- TXN1
Thioredoxin-1
- TXNRD1
Thioredoxin reductase 1
- PRDX
peroxiredoxin
- ABCC1
ATP-binding cassette, sub family C, member 1
- ABCC2
ATP-binding cassette, sub family C, member 2
- ABCC3
ATP-binding cassette, sub family C, member 3
- NAC
N-acetyl-L-cysteine
References
- 1.Nadkar A, Pungaliya C, Drake K, Zajac E, Singhal SS, Awasthi S. Therapeutic resistance in lung cancer. Expert Opin Drug Metab Toxicol. 2006;2:753–777. doi: 10.1517/17425255.2.5.753. [DOI] [PubMed] [Google Scholar]
- 2.Kurosu T, Fukuda T, Miki T, Miura O. BCL6 overexpression prevents increase in reactive oxygen species and inhibits apoptosis induced by chemotherapeutic reagents in B-cell lymphoma cells. Oncogene. 2003;22:4459–4468. doi: 10.1038/sj.onc.1206755. [DOI] [PubMed] [Google Scholar]
- 3.Masuda H, Tanaka T, Takahama U. Cisplatin generates superoxide anion by interaction with DNA in a cell-free system. Biochem Biophys Res Commun. 1994;203:1175–1180. doi: 10.1006/bbrc.1994.2306. [DOI] [PubMed] [Google Scholar]
- 4.Soini Y, Napankangas U, Jarvinen K, Kaarteenaho-Wiik R, Paakko P, Kinnula VL. Expression of gamma-glutamyl cysteine synthetase in nonsmall cell lung carcinoma. Cancer. 2001;92:2911–2919. doi: 10.1002/1097-0142(20011201)92:11<2911::aid-cncr10105>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 5.Tew KD. Glutathione-associated enzymes in anticancer drug resistance. Cancer Res. 1994;54:4313–4320. [PubMed] [Google Scholar]
- 6.Yang P, Ebbert JO, Sun Z, Weinshilboum RM. Role of the glutathione metabolic pathway in lung cancer treatment and prognosis: a review. J Clin Oncol. 2006;24:1761–1769. doi: 10.1200/JCO.2005.02.7110. [DOI] [PubMed] [Google Scholar]
- 7.Trachootham D, Zhou Y, Zhang H, Demizu Y, Chen Z, Pelicano H, Chiao PJ, Achanta G, Arlinghaus RB, Liu J, Huang P. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell. 2006;10:241–252. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi A, Suzuki H, Itoh K, Yamamoto M, Sugiyama Y. Transcription factor Nrf2 is required for the constitutive and inducible expression of multidrug resistance-associated protein 1 in mouse embryo fibroblasts. Biochem Biophys Res Commun. 2003;310:824–829. doi: 10.1016/j.bbrc.2003.09.086. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 13.Vollrath V, Wielandt AM, Iruretagoyena M, Chianale J. Role of Nrf2 in the regulation of the Mrp2 (ABCC2) gene. Biochem J. 2006;395:599–609. doi: 10.1042/BJ20051518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim YJ, Ahn JY, Liang P, Ip C, Zhang Y, Park YM. Human prx1 gene is a target of Nrf2 and is up-regulated by hypoxia/reoxygenation: implication to tumor biology. Cancer Res. 2007;67:546–554. doi: 10.1158/0008-5472.CAN-06-2401. [DOI] [PubMed] [Google Scholar]
- 15.Lee TD, Yang H, Whang J, Lu SC. Cloning and characterization of the human glutathione synthetase 5'-flanking region. Biochem J. 2005;390:521–528. doi: 10.1042/BJ20050439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rangasamy T, Cho CY, Thimmulappa RK, Zhen L, Srisuma SS, Kensler TW, Yamamoto M, Petrache I, Tuder RM, Biswal S. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J Clin Invest. 2004;114:1248–1259. doi: 10.1172/JCI21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rangasamy T, Guo J, Mitzner WA, Roman J, Singh A, Fryer AD, Yamamoto M, Kensler TW, Tuder RM, Georas SN, Biswal S. Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. J Exp Med. 2005;202:47–59. doi: 10.1084/jem.20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thimmulappa RK, Scollick C, Traore K, Yates M, Trush MA, Liby KT, Sporn MB, Yamamoto M, Kensler TW, Biswal S. Nrf2-dependent protection from LPS induced inflammatory response and mortality by CDDO-Imidazolide. Biochem Biophys Res Commun. 2006;351:883–889. doi: 10.1016/j.bbrc.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotlo KU, Yehiely F, Efimova E, Harasty H, Hesabi B, Shchors K, Einat P, Rozen A, Berent E, Deiss LP. Nrf2 is an inhibitor of the Fas pathway as identified by Achilles' Heel Method, a new function-based approach to gene identification in human cells. Oncogene. 2003;22:797–806. doi: 10.1038/sj.onc.1206077. [DOI] [PubMed] [Google Scholar]
- 20.Morito N, Yoh K, Itoh K, Hirayama A, Koyama A, Yamamoto M, Takahashi S. Nrf2 regulates the sensitivity of death receptor signals by affecting intracellular glutathione levels. Oncogene. 2003;22:9275–9281. doi: 10.1038/sj.onc.1207024. [DOI] [PubMed] [Google Scholar]
- 21.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV, Biswal S. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 23.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 24.Ohta KI Tsutomu, Miyamoto Mamiko, Nakahara Izumi, Tanaka Hiroshi, Ohtsuji Makiko, Suzuki Takafumi, Kobayashi Akira, Yokota Jun, Sakiyama Tokuki, Shibata Tatsuhiro, Yamamoto Masayuki, Hirohashi Setsuo. Loss of Keap1 Function Activates Nrf2 and Provides Advantages for Lung Cancer Cell Growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 25.Singh A, Rangasamy T, Thimmulappa RK, Lee H, Osburn WO, Brigelius-Flohe R, Kensler TW, Yamamoto M, Biswal S. Glutathione peroxidase 2, the major cigarette smoke-inducible isoform of GPX in lungs, is regulated by Nrf2. Am J Respir Cell Mol Biol. 2006;35:639–650. doi: 10.1165/rcmb.2005-0325OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamar P, Song E, Kokeny G, Chen A, Ouyang N, Lieberman J. Small interfering RNA targeting Fas protects mice against renal ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2004;101:14883–14888. doi: 10.1073/pnas.0406421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–5203. [PubMed] [Google Scholar]
- 28.Osburn WO, Wakabayashi N, Misra V, Nilles T, Biswal S, Trush MA, Kensler TW. Nrf2 regulates an adaptive response protecting against oxidative damage following diquat-mediated formation of superoxide anion. Arch Biochem Biophys. 2006;454:7–15. doi: 10.1016/j.abb.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reddy NM, Kleeberger SR, Cho HY, Yamamoto M, Kensler TW, Biswal S, Reddy SP. Deficiency in Nrf2-GSH signaling impairs type II cell growth and enhances sensitivity to oxidants. Am J Respir Cell Mol Biol. 2007;37:3–8. doi: 10.1165/rcmb.2007-0004RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartmann JT, Lipp HP. Toxicity of platinum compounds. Expert Opin Pharmacother. 2003;4:889–901. doi: 10.1517/14656566.4.6.889. [DOI] [PubMed] [Google Scholar]
- 31.Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33:9–23. doi: 10.1016/j.ctrv.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meister A. Selective modification of glutathione metabolism. Science. 1983;220:472–477. doi: 10.1126/science.6836290. [DOI] [PubMed] [Google Scholar]
- 33.Byun SS, Kim SW, Choi H, Lee C, Lee E. Augmentation of cisplatin sensitivity in cisplatin-resistant human bladder cancer cells by modulating glutathione concentrations and glutathione-related enzyme activities. BJU Int. 2005;95:1086–1090. doi: 10.1111/j.1464-410X.2005.05472.x. [DOI] [PubMed] [Google Scholar]
- 34.Godwin AK, Meister A, O'Dwyer PJ, Huang CS, Hamilton TC, Anderson ME. High resistance to cisplatin in human ovarian cancer cell lines is associated with marked increase of glutathione synthesis. Proc Natl Acad Sci U S A. 1992;89:3070–3074. doi: 10.1073/pnas.89.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinha BK, Mimnaugh EG, Rajagopalan S, Myers CE. Adriamycin activation and oxygen free radical formation in human breast tumor cells: protective role of glutathione peroxidase in adriamycin resistance. Cancer Res. 1989;49:3844–3848. [PubMed] [Google Scholar]
- 36.Zhang K, Chew M, Yang EB, Wong KP, Mack P. Modulation of cisplatin cytotoxicity and cisplatin-induced DNA cross-links in HepG2 cells by regulation of glutathione-related mechanisms. Mol Pharmacol. 2001;59:837–843. doi: 10.1124/mol.59.4.837. [DOI] [PubMed] [Google Scholar]
- 37.Powis G, Kirkpatrick DL. Thioredoxin signaling as a target for cancer therapy. Curr Opin Pharmacol. 2007;7:392–397. doi: 10.1016/j.coph.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 38.Sasada T, Iwata S, Sato N, Kitaoka Y, Hirota K, Nakamura K, Nishiyama A, Taniguchi Y, Takabayashi A, Yodoi J. Redox control of resistance to cisdiamminedichloroplatinum (II) (CDDP): protective effect of human thioredoxin against CDDP-induced cytotoxicity. J Clin Invest. 1996;97:2268–2276. doi: 10.1172/JCI118668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yokomizo A, Ono M, Nanri H, Makino Y, Ohga T, Wada M, Okamoto T, Yodoi J, Kuwano M, Kohno K. Cellular levels of thioredoxin associated with drug sensitivity to cisplatin, mitomycin C, doxorubicin, and etoposide. Cancer Res. 1995;55:4293–4296. [PubMed] [Google Scholar]
- 40.Dickey DT, Wu YJ, Muldoon LL, Neuwelt EA. Protection against cisplatin-induced toxicities by N-acetylcysteine and sodium thiosulfate as assessed at the molecular, cellular, and in vivo levels. J Pharmacol Exp Ther. 2005;314:1052–1058. doi: 10.1124/jpet.105.087601. [DOI] [PubMed] [Google Scholar]
- 41.Fuertes MA, Castilla J, Alonso C, Perez JM. Novel concepts in the development of platinum antitumor drugs. Curr Med Chem Anticancer Agents. 2002;2:539–551. doi: 10.2174/1568011023353958. [DOI] [PubMed] [Google Scholar]
- 42.Neuwelt EA, Pagel MA, Hasler BP, Deloughery TG, Muldoon LL. Therapeutic efficacy of aortic administration of N-acetylcysteine as a chemoprotectant against bone marrow toxicity after intracarotid administration of alkylators, with or without glutathione depletion in a rat model. Cancer Res. 2001;61:7868–7874. [PubMed] [Google Scholar]
- 43.Schweitzer VG. Ototoxicity of chemotherapeutic agents. Otolaryngol Clin North Am. 1993;26:759–789. [PubMed] [Google Scholar]
- 44.Aruoma OI, Halliwell B, Hoey BM, Butler J. The antioxidant action of N-acetylcysteine: its reaction with hydrogen peroxide, hydroxyl radical, superoxide, and hypochlorous acid. Free Radic Biol Med. 1989;6:593–597. doi: 10.1016/0891-5849(89)90066-x. [DOI] [PubMed] [Google Scholar]
- 45.Moldeus P, Cotgreave IA, Berggren M. Lung protection by a thiol-containing antioxidant: N-acetylcysteine. Respiration. 1986;50 Suppl 1:31–42. doi: 10.1159/000195086. [DOI] [PubMed] [Google Scholar]
- 46.Garcia-Bermejo L, Perez C, Vilaboa NE, de Blas E, Aller P. cAMP increasing agents attenuate the generation of apoptosis by etoposide in promonocytic leukemia cells. J Cell Sci. 1998;111(Pt 5):637–644. doi: 10.1242/jcs.111.5.637. [DOI] [PubMed] [Google Scholar]
- 47.Gardner A, Xu FH, Fady C, Sarafian T, Tu Y, Lichtenstein A. Evidence against the hypothesis that BCL-2 inhibits apoptosis through an anti-oxidant effect. Cell Death Differ. 1997;4:487–496. doi: 10.1038/sj.cdd.4400264. [DOI] [PubMed] [Google Scholar]
- 48.Mans DR, Lafleur MV, Westmijze EJ, Horn IR, Bets D, Schuurhuis GJ, Lankelma J, Retel J. Reactions of glutathione with the catechol, the ortho-quinone and the semi-quinone free radical of etoposide Consequences for DNA inactivation. Biochem Pharmacol. 1992;43:1761–1768. doi: 10.1016/0006-2952(92)90708-q. [DOI] [PubMed] [Google Scholar]
- 49.Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 50.Mans DR, Schuurhuis GJ, Treskes M, Lafleur MV, Retel J, Pinedo HM, Lankelma J. Modulation by D,L-buthionine-S,R-sulphoximine of etoposide cytotoxicity on human non-small cell lung, ovarian and breast carcinoma cell lines. Eur J Cancer. 1992;28A:1447–1452. doi: 10.1016/0959-8049(92)90541-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.