Abstract
Tumor secreted substances (secretome), including extracellular matrix (ECM) components, act as mediators of tumor–host communication in the breast tumor microenvironment. Proteomic analysis has emphasized the value of the secretome as a source of prospective markers and drug targets for the treatment of breast cancers. Utilizing bioinformatics, our recent studies revealed global changes in protein expression after the activation of ECM-mediated signaling in breast cancer cells. A newly designed technique integrating a capillary ultrafiltration (CUF) probe with mass spectrometry was demonstrated to dynamically sample and identify in vivo and pure secretome from the tumor microenvironment. Such in vivo profiling of breast cancer secretomes may facilitate the development of novel drugs specifically targeting secretome.
Keywords: tumor microenvironment, breast cancer, secretome, extracellular matrix
1. Introduction
Breast cancer is the most common cancer and one of the leading causes of cancer-related death in women around the world. Because mortality from breast cancer is often due to distant metastasis, there is an urgent need to identify prospective biomarkers for early detection. Breast cancer cell proliferation and invasion involve the breaching of tissue barriers and the subsequent infiltration of cells into the surrounding tissue.1 The growth and progression of breast tumor cells depend not only on their malignant potential, but also on the multidirectional interactions of secreted substances (secretome), including extracelluar matrix (ECM), produced by all the cell types including tumor, stroma, endothelial cells, and immune system cells within the local microenvironment. Previous evidence demonstrated that a permissive tumor microenvironment is required for successful progression and metastasis of tumor cells.2 Secretomes are a rich source of new therapeutics and drug targets, and are becoming a major focus of drug discovery programs throughout the industry.3 Although many anticancer drugs targeting the secretome have been developed,4,5 a comprehensive secretome for breast cancer is still needed for further target identification and development of more potent and specific anticancer drugs. The identification of proteins or peptides released into the medium of tumor cells cultured in vitro is the most common method for determining a tumor secretome. However, the secretory pattern of cells in vitro is not always the same as the in vivo secretome. A novel technology using capillary ultrafiltration (CUF) probes is capable of sampling the secretome in vivo from various tissues at different time points.6–9 More importantly, this CUF probe sampling technique provides a unique modality for localized secretome sampling in the tumor microenvironment, thus, providing insight into the localized concentrations of secretome proteins that cannot be achieved via sampling from circulating blood plasma. Tumor secretomes are normally a mixture of multiple proteins and peptides released either from tumor or host cells.6,10 Mass spectrometry has been employed to identify several proteins in the CUF probe-collected secretome.6–9,11 Secretome ECM is a crucial component for the communication between the tumor cells and their microenvironment. In our recent research, a proteomics analysis of a peptide-derived drug mediating the integrin signaling cascade in breast cancer cells has emphasized the significance of treatments targeting the tumor microenvironment.12,13
2. Breast Tumor Microenvironment and Secretome
Studies of tumor microenvironment have emphasized evaluation of the tumor as an organ-like structure with complex, dynamic cross-talk.14 It is now known that tumor cells and their stroma co-evolve during tumorigenesis and progression. Stroma consists of cells, extracellular substances (secretome) including secreted proteins/petides, other molecules, and ECM. Tumor–stroma interactions in breast cancer are dynamic networks between epithelial cells and a microenvironment consisting of stromal cells that include fibroblasts, inflammatory cells, innate and adaptive immune cells, adipocytes, vasculature, glial cells, and specialized mesenchymal cells.14,15 Secretome in tumor microenvironment contains the ECM, constituted by proteins, receptors, proteoglycans, and adhesive molecules as well as a milieu of secreted proteins including cytokines, chemokines, growth factors, angiogenesis factors, and proteases at its surroundings.15,16 The ECM, abundantly secreted by fibroblasts, provides supportive and structural architecture to cells and tissues. Fibroblasts are the most abundant cellular component in tumor microenvironment, and the population is even greater than tumor cells in some specific cancers.17,18 The tumor-associated fibroblasts (TAFs) are phenotypically and functionally mimic to fibroblasts in wound healing but different from normal epithelial fibroblasts in the same tissue but not in the microenvironment.17,19 TAFs can produce high α-smooth muscle actin, which allows cells to recruit into inflammation region and contract for tissue reparation.20 On the other hand, fibroblast activation protein (FAP) is a type II membrane protein most prominent expressed on tumor stroma fibroblasts.17 FAP may represent an early detection biomarker, and a potential therapeutic target for breast cancer treatment.18 From large-scale gene expression profiles of normal breast tissue and in situ and invasive breast carcinomas, the unique CXCL12 overexpressed in tumor activated myofibroblasts and augmented their proliferation, invasion, and migration. It implied that chemokines play a key role in breast tumor progression by acting as paracrine factors.21
The inflammatory cell consists of monocytes/macrophages, neutrophils, eosinophils, mast cells, and lymphocytes which are recruited to breast tumors preferentially in necrosis and hypoxia areas.22 Tumor-infiltrating immune cells are originally regarded as cytotoxic to the tumor cells; however, current findings support that such tumor-associated leukocytes can contribute to cancer initiation, proliferation, metastasis due to the immune tolerance, or suppression associated with malignant disease.23,24 Of these immune cells, tumor-associated macrophages (TAMs) represent the largest population and the most multifunctional bioactivities.22 Circulating monocytes are attracted to tumor microenvironment by chemokines or chemoattractants, and once they have arrived at the tumor site, they differentiate into macrophages. TAMs belong to polarized M2 (F4/80+/CD206+) macrophage population and possess little cytotoxicity to tumor cells due to their restricted NO and proinflammatory cytokine productions. TAMs produce high interleukin (IL)-4, IL-13, and glucorticoids which are capable of tuning inflammatory stimuli and Th2 type immunity.23 TAMs can also promote tumor cell proliferation, matrix remodeling, intravasation, and spread by releasing epidermal growth factor (EGF), vascular endothelial growth factor (VEGF)-C, VEGF-D, VEGF receptor 3 (VEGFR-3), IL-8, matrix metalloproteinase (MMP)2, hepatocyte growth factor (HGF), platelet-derived growth factor (PDGF), vascular cell adhesion molecule (VCAM)-1, and transforming growth factor (TGF)-β.22,24 The chemokine, CCL5, was found to play a key role in recruitment of TAMs, and CCL5 blockage inhibited breast tumor growth.25
A vast amount of secretome including cytokines and growth factors is released both by the cancer and stromal cells. The secretome may reflect a variety of pathological conditions, thus, representing a rich source of biomarkers, which play critical roles in cell adhesion, intercellular interactions, and invasion at the level of cell and the whole organism. Thus, great interest is currently focused on the characterization of secretome from isolated cells and neoplastic tissues with the intention of identifying novel biomarkers. In 2007, Mbeunkui et al. have identified the secretome from a series of isogenic MCF10 breast cancer cell lines representing different aggressiveness by liquid chromatography–mass spectrometry (MS) MS (LC-MS/MS).26 The most significant changes were observed for alpha-1-antichymotrypsin and galectin-3-binding protein that were highly secreted from more malignant cell lines, yet undetected in the premalignant cell lines. Other proteins showing increasing abundance in the more aggressive cell lines included alpha-1-antitrypsin, cathepsin D, and lysyl oxidase. The results suggest the potential usefulness of the secretome for identifying prospective markers for the early detection and aggressiveness/progression of cancer.26 Celis et al. have demonstrated that the tumor interstitial fluid (TIF) perfusing the tumor microenvironment in invasive ductal carcinomas of the breast is a novel and potentially highly promising source of biomarkers.27 In addition to providing the global view of the TIF proteome, their findings offer the systematic search of diagnostic biomarkers and targets for therapeutic intervention as well.27 Shed proteins that perfuse the tumor microenvironment play a critical role in cell–cell and cell–matrix communication and may serve as a source of low molecular mass biomarkers for breast tumor. Proteins shed by vesicles contain matrix-degrading proteinases, beta 1 integrins, cathepsins B and D, IL-1 beta, BRCA1, and fibroblast growth factor (FGF)-2. Hanash et al. have indicated that heat shock proteins occur in the plasma via a nonclassical pathway.28 The member of S100 superfamily,S100A4, was shown to be involved in the metastasis process in human breast cancer. The protein is externalized by the stroma cells to the fluid that perfuses the tumor microenvironment. High expression of S100A4 seems strongly associated with poor prognosis.29 For the above studies, it should be stressed that the local microenvironment may be different in various stages of the tumor reflecting intratumor and intrastroma heterogeneity, and that new and more sensitive detection technologies in combination with tissue microdissection may be necessary to gain a better understanding of the biological events taking place in the local surroundings.
The complex network of cell types associated with tumor microenvironment produces either secreted or externalized proteins. The fluids generated in tumor lysates representing the cellular events and dynamic secretory processes affected in breast cancer may allow advanced cancer therapy, as well as further our understanding of the molecular mechanisms underlying breast cancer development and progression.30,31 Even though many secreted proteins still remain to be determined, the biological activities of the proteins identified so far have provided us with a glance of the biological processes taking place in the tumor microenvironment.
3. In Vivo Sampling of Secretome via Ultrafiltration
Many challenges make it difficult to obtain a comprehensive profile of tumor secretome. Identification of tumor secretome according to traditional methods is to obtain the released proteins/peptides from the medium of in vitro tumor cell culture and then analyze their properties in vivo. However, the data from the in vivo animal systems rarely matches well with those that are in vitro. One alternative is to homogenize and spin down the tumor masses, and then harvest the tumor secretome from supernatants. Unfortunately, tumor secretomes collected from supernatants generally are contaminated by other proteins/peptides, which leak from the homogenization-damaged tumor or host cells. Additionally, it generally requires sacrificing many animals to obtain a dynamic secretion pattern of secretome. Microdialysis and ultrafiltration are two major methods that have been applied for dynamically sampling of in vivo secretome.32
Microdialysis has been used for sampling growth factors33 and measuring interstitial tumor 5-fluorouracil (5-FU) pharmacokinetics in breast cancer.34 Microdialysis sampling depends on the passive diffusion of substances across a semipermeable hollow membrane driven by a concentration gradient.35 It is a commonly used means to obtain macromolecule-free samples from the extracellular fluids, although there is a challenge for microdialysis sampling to detect peptides and proteins. Peptide/proteins have relatively larger size and smaller aqueous diffusion coefficients, resulting in a lower recovery due to mass transport limitations through the semipermeable hollow membranes.36 Recently, it has been shown that the addition of affinity-based trapping agents into the microdialysis perfusion fluid considerably enhanced the relative recovery via the binding reaction of larger substances such as peptides/proteins.35 Furthermore, microdialysis method has been currently used for in vivo sampling of cytokines and growth factors. Because of the high biological activity of most cytokines, their concentration in extracellular fluids is usually in low-picomolar concentrations. Under normal circumstances, cytokines are typically undetectable in extracellular fluids or tissues. Cytokines are highly localized secreted proteins (approximately 8–80 kDa) produced by various cell types as part of an immunological response. Elevated expression of cytokines is a sign of activation of cytokine pathways associated with inflammation or disease progression. Enzyme linked immunoabsorbent assays (ELISAs) using a ‘two-site’ sandwich detection assay, that is, one antibody is used to capture cytokine antigen(s) and another detection antibody, are the most broadly used methods to quantify cytokines in biological matrices because of their acceptable specificity, sensitivity, rapid turnaround time, convenience, the ease of performance, and a relatively low cost. However, a typical ELISA can only measure one cytokine at a time and requires at least 100 µL sample volumes. It is important to note that microdialysis sampling endows with localized-sampling and thus insight into localized concentrations of cytokines that cannot be accomplished via sampling from blood plasma.37 This has been recently illustrated with the IL-6 where its interstitial fluid concentration was 100-fold higher than that found in the plasma.38 Previous studies demonstrated that intraperitoneal implantation of a microdialysis probe into a lipopolysaccharide-induced inflammation mouse is capable of sampling cytokines in vivo.36 The microdialysis probe was perfused with mouse inflammation cytokine antibody coated microspheres. The use of cytokine antibody coated microspheres as a means for microdialysis sampling significantly improves the detection of cytokines including tumor necrosis factor (TNF)-α, and monocyte chemoattractant protein (MCP)-1 and IL-6.35 Online coupling of in vivo microdialysis with mass spectrometry has been also demonstrated. 36 Jakubowski and co-workers developed a miniature hollow-fiber microdialysis device optimized for desalting small-volume neuronal samples online, with the device directly linked to a dynamic nanoelectrospray ionization assembly interfaced with an ion trap mass spectrometer.39 The device using online microdialysis-dynamic nanoelectrospray mass spectrometry was able to detect acidic peptide, acidic peptide 1–24, and delta-bag cell peptide that were secreted from the peptidergic bag cell neurons of the marine mollusk, Aplysia californica. Although the microdialysis method has been extensively applied for in vivo sampling of proteins, there are still several concerns when it was employed for sampling. Primarily, fluids collected by microdialysis did not directly reflect the tissue concentration. Since perfusion fluid was injected when microdialysis was performed, the tissue concentration of the samples taken by microdialysis has to be calculated by taking into account the diffusive characteristics of the membranes, the composition of the perfusion fluid, the flow rate of fluid through the membranes, and the recovery of the samples. Furthermore, since microdialysis sampling is mainly based on diffusion of the analyte into the dialysate, sampling of proteins with larger molecular weights is always a challenge.
Like microdialysis, ultrafiltration techniques also employ semipermeable membranes to separate substances.40 Substances pass through the ultrafiltration membrane by convection together with the fluids in which they are dissolved. Thus, sample concentration in the ultrafiltration-collected fluids directly reflects the tissue concentration. The ultrafiltration sampling technique applies a vacuum to semipermeable membranes to extract fluids containing secretome from the extracellular space.41–44 There are several advantages of ultrafiltration sampling in comparison with in vivo sampling with microdialysis. First, the ultrafiltration collects a small volume sample and thus allows samples to be taken more frequently. Second, it is potentially better for in vivo monitoring because no dilution factor has to be considered.32 In the past, ultrafiltration sampling has been utilized to collect various ions32 such as calcium, potassium, and sodium to monitor glucose from subcutaneous tissue, blood, and saliva as well as to detect cytokines and drug metabolites in various biological fluids (Table 1). Recently, our laboratory linked this technology to mass spectrometry for in vivo sampling of peptides and proteins.6–9
Table 1.
Ultrafiltration Sampling
A CUF probe has been newly developed in our laboratory based on the ultrafiltration technology. A semipermeable hollow membrane fiber, a key component of CUF probe, is positioned at the front of CUF probe (Figure 1) and connected to a polytetrafluroethylene (PTFE) tube. The semipermeable hollow membrane fibers with various molecular weight cutoffs (MWCOs) can be made with different surface-charged materials. One end of the semipermeable hollow membrane fiber was attached to the PTFE tubing through a small section of fused silica capillary, while the other end was sealed with epoxy. A sharpened needle was attached to the end of the PTFE ultramicrobore tube. The probe was connected to a vacutainer so that negative pressure would drive the ultrafiltration process to collect extracellular fluids. Distinct membrane fibers with a broad range of MWCO(s) and varying electro-negativities can be accommodated to CUF probes. We have implanted the CUF probes into mice and collected secretomes in vivo from ear skins,8 skin wounds,7 sodium lauryl sulfate-treated skins9 and solid tumors.6 In our laboratory, a regressive tumor model was used to examine the efficiency of CUF probe in vivo sampling. CUF probes were implanted into tumor masses 1 (progressive stage) and 3 weeks (regressive stage) after C3H/HeN mice were injected with ultraviolet (UV)-induced fibrosarcoma UV-2240 cell lines. The UV-2240 tumor cell line is highly antigenic and is routinely rejected when transplanted into normal syngeneic mice, but grows progressively in immune-deficient mice.6,45 The CUF probe was implanted into the central part of tumor masses to collect in vivo tumor secretome for 6 h. During sample collection, the semipermeable membrane fiber in the front end of CUF probe was entirely enclosed by a tumor mass. Implantation of the CUF probe involved inserting a 22-gauge needle into the tumor mass as a guide and then feeding the probe through the needle. The needle was then removed. The CUF probe-collected secretome which contains a multiple protein/peptide mixture was digested with trypsin. Subsequently, the complex mixture of tryptic digests was directly subjected to matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) and quadrupole time-of-flight (Q-TOF) MS/MS for protein identification. Five secreted proteins (cyclophilin-A, S100A4, profilin-1, thymosin beta 4 and 10), previously associated with tumor progression, were identified from tumor masses at the progressive stage. Five secreted proteins including apolipoprotein A-I, apolipoprotein C-I, and three protease inhibitors (fetuin-A, alpha-1 antitrypsin 1–6, and contrapsin) were identified from tumor masses at the regressive stage. Although ultrafiltration separation has been applied for the in vitro preparation of breast tumor samples for proteomics studies,46 this technology has never been used for sampling in vivo secretome in breast tumor microenvironments. Thus, the CUF probe may be an excellent alternative for sampling ductal lavage from breast tumors.47
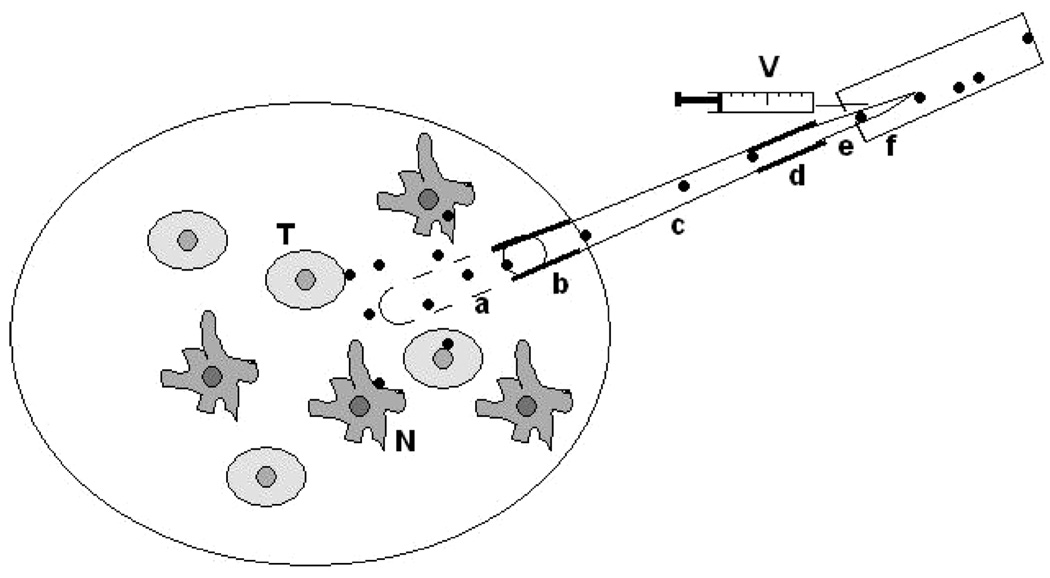
Figure 1.
Sampling tumor secretome in vivo via a CUF probe. A CUF probe can be implanted into a growing tumor mass. The implanted probe collects the in vivo tumor secretome released either from tumor cells (T) or nontumor cells (N). The CUF probe extracts tumor secretome by using a vacuum applied to semipermeable hollow membrane fiber (a). The vacuum can be created by withdrawing a syringe from the vacutainer (V). The extraction efficiency of the CUF probe relies on the various pore sizes and surface charges of membrane fibers as well as the nature of proteins/peptides. As a general rule, large sizes of proteins/peptides are retained, while small proteins/peptides pass through the membrane. The CUF probes can be fabricated to fit the various sizes of tumor masses. The semipermeable hollow membrane fiber at the front end of the probe is entirely covered after implantation of CUF probe into a tumor mass. One end of the semipermeable hollow membrane fiber is glued with a small section of fused silica capillary (b) and attached to a PTFE tubing (c), while the other end is completely sealed with epoxy. A sharpened needle (e) is attached to the end of the PTFE ultramicrobore tube (d) and inserted into the vacutainer. Solid dots: tumor secretome.
The probe can be positioned in the different locations of tumor microenvironments in order to obtain a distinctive expression pattern of secretome. A tumor microenvironment is generally consisted of various types of cells and, as a whole, heterogeneous. Previous studies have revealed that the expression of many proteins was varied in the different distributions of tumor tissues.48,49 Gathering CUF probe-collected secretomes from different portions of breast tumor microenvironment may be able to reflect the overall features of tumor secretions. Although various lengths of semipermeable membranes in CUF probes can be fabricated to fit in different sizes of tumor masses, the probes also can be implanted nearby tumor initiation sites where host cells interact with the early stage tumors. CUF probes have been connected to a freely moving arm system for long-term and dynamic sampling collection.9 The connection will allow continuously sampling of the biomarkers at the early stages of tumor formation. The semipermeable membrane in the front end of CUF probe is a key component for probe sampling. It has been demonstrated that cellular debris and hemoglobin were predominantly collected by a CUF probe without a semipermeable membrane, leading to the failure in the protein identification by mass spectrometry.8 Although implantation of CUF probes is a minimally invasive sampling method, control experiments to subtract the effect of implantation-induced minor tissue damages are required. Proteins collected by CUF probes are a complex mixture derived from secreted proteins, cell matrix proteins, as well as proteins from interstitial fluid or plasma. Incorporation of affinity-based trapping agents35 into CUF probes may enhance the protein selectivity. Furthermore, the establishment of comprehensive proteomes of tumor and nontumor cells as well as plasma in the future will assist to distinguish the cell sources of secretomes collected by CUF probes.
4. Tumor ECM in Microenvironment
The paradoxical role of immune system developing during tumorigenesis is increasingly clear but still needs more experimental confirmation that the neoplastic immune response consists of humoral B cells, chronic inflammatory cells, and that Th2 cells lead to tumor initiation and progression.28,50 On the other hand, when cytotoxic T cells, Th1 cells, and natural killer (NK) cells are profoundly expressed in environment, it would protect against tumor development.51–53 In addition, malignant areas can infiltrate regulatory T cells (Tregs) leading to immunosuppression effect on cytotoxic T cells.52,54 IL-2 depleted Tregs can cause markedly immune-mediated rejection of breast cancers without concomitant lymphopenia.55,56 The neoplastic cells have to avoid cytotoxic T lymphocyte rejection for survival.57 This can be partially achieved by IL-10, known for blunting antitumor activity by cytotoxic T lymphocytes, secreted both by TAMs and tumor cells.
Hyaluronan, also called hyaluronic acid (HA), an abundant glycosaminoglycan in ECM, is highly overexpressed in malignant tumors and involved in enhancing these cells’ motility and invasion.58 HA interacts with tumor cells through its surface receptor, CD44, and hyaluronan-binding protein (RHAMM) by which it promotes tumor cell growth, survival, and migration. HA promotes tumor-associated angiogenesis likely by a mask that protects cancer cell from cytotoxic T cells and chemotherapeutic drugs.58 Increased HA and hyaluronan synthase 2 expression levels can contribute to highly metastatic breast carcinomas.59 In contrast, breast cancer metastasis is blocked through antisense-mediated suppression of hyaluronan synthase 2 and CD44.60,61 More recently, the masking of the erythroblastic leukemia viral oncogene homologue 2 (ErbB2) in trastuzumab-resistant breast cancer are positively associated with CD44 and HA expressions. The hyaluronan synthase suppressor, 4-methylumbelliferone, greatly inhibited HA synthesis levels resulting in promotion of trastuzumab binding to ErbB2, and significant ErbB2 reduction.62 Heparanase is a β-endoglucuronidase, which degrades heparin sulfate in the cancer cell ECM. Heparanase breaks down heparin sulfate leading to FGF2 and hepatocyte growth factor secretion, and increase breast tumor growth, invasion, and angiogenesis.63,64 Osteopontin (OPN) is a phosphorylated, integrin-binding glycoprotein and commonly overexpressed in ECM of many types of cancers.65 OPN, which can mediate cell–cell and cell–matrix communication promotes cell adhesion, angiogenesis, invasion, and metastasis. OPN mainly interacts with integrins through a specific RGD motif, and as well as nonintegrin receptor, CD44.66 Overexpressed OPN can highly upregulate hyaluronan synthase 2 and stimulate HA production leading to metastastic breast cancer.67 OPN also plays a role in recruiting macrophages, leukocytes to a tumor environment, and ECM remodeling. Overexpression of OPN is not only found in cancer cells relevant to breast, lung, colorectal, stomach, and ovary cancers and melanoma but also serum and plasma from such patients.68 These lend OPN great potential to be a prognostic and therapeutic target for breast cancer and others. Tenascin-C is an ECM glycoprotein, highly expressed in most tumors and early stages of development, but much restricted in developed tissues.69,70 Overexpression of tenascin-C has been correlated with tumorigenesis and angiogenesis through regulation of oncogenes, tumor suppressor genes, genome integrity genes, and evading of immune surveillance.71 Tenascin-C may inactivate cell cycle checkpoint genes in tumor cell to facilitate neoplastic phenotype expansion.72 Breast cancers are currently found to produce syncytin, an endogenous retroviral envelope glycoprotein, which is involved in fusions between placental trophoblasts.73,74 It is generally accepted that breast cancer can spontaneously fuse with normal endothelial cells, and may abrogate the tumor or transit into more aggressive phenotype according to the net expression level of tumor suppressor genes.75 Syncytin stimulates IL-1 beta and nitric oxide synthase expression, and may modulate host immune system to regulate cell apoptosis and metastasis. From current clinical data, syncytin presents a positive prognostic factor in breast cancer because it inhibits tumor cell penetration of endothelial cell hindrance.76
Because of the imbalance between cytotoxic, humoral immune system, and tumor microenvironment, the neoplastic cells could possibly evade immune-mediated elimination and may favor a more aggressive type. The ECM constituted of tumor cell is very different from the surrounding normal cell. The ECM-derivative glycoprotein may have immuno-modulating ability to make normal cell into tumor-prone type.77 These may imply that a crucial and urgent topic is how we globally understand the scientific impact of glycan or post-translational modification information of ECM protein on breast cancer tumorigensis and metastasis.
5. Integrins as a Therapeutic Target in Breast Cancer Metastasis
Cell adhesive interactions play important roles during many normal physiological processes such as embryonic development and wound repair, and also during the progression of diseases such as cancer. Cell adhesion is mediated by the specific interactions of cell surface receptors with extracellular glycoproteins. The best characterized cell adhesion receptors are the integrins.78 Integrins are heterodimeric cell surface receptors whose extracellular domains bind matrix proteins,79,80 including laminin-1, laminin-5 (epiligrin), fibronectin, collagen, and entactin.81 Functions of integrins have been shown to be involved in several important cellular processes, including cell differentiation, angiogenesis, apoptosis, cell migration, and tumor growth.82 The best characterized integrin ligand is fibronectin which is a multifunctional glycoprotein comprised of three different types of homologous repeating units, type I, type II, and type III. Fibronectin has one cell adhesive region which is located near the center of the polypeptide chain in the ninth and tenth type III modules and binds to the alpha 5 beta 1 integrin. The biological function of the central cell adhesive region requires two critical amino acid sequences, an Arg-Gly-Asp (RGD) sequence and a Pro-His-Ser-Arg-Asn (PH-SRN) sequence.78 Their function in synergy is for optimal binding to the alpha 5 beta 1 integrin.78 Since integrin mediated proliferation,12,13,83 adhesion,84 migration,85 and invasion86 of breast cancer cells in response to ECM, targeting these integrins to modulate integrin–ECM interactions in tumor microenvironment may be a prospective approach to reduce the dissemination of breast carcinoma in vivo.
The tripeptide sequence RGD (Arg-Gly-Asp) is a common cell-recognition motif, which is part of integrin binding ligands, like fibronectin, fibrinogen, and vitronectin. This sequence has been used as a lead compound for developing different integrin antagonists.87 RGD-containing peptides have been found to be the efficient inhibitors of integrin–ligand interactions in studies of cell adhesion, migration, growth, and differentiation. Recently, the RGD-containing peptides have been further discovered to be able to induce cell apoptosis mediated by activation of caspase-3 (an important molecule standing in the cell death pathway).88 Therefore, it is highly worth to develop more potent RGD-containing peptides as drugs for cancer therapy.
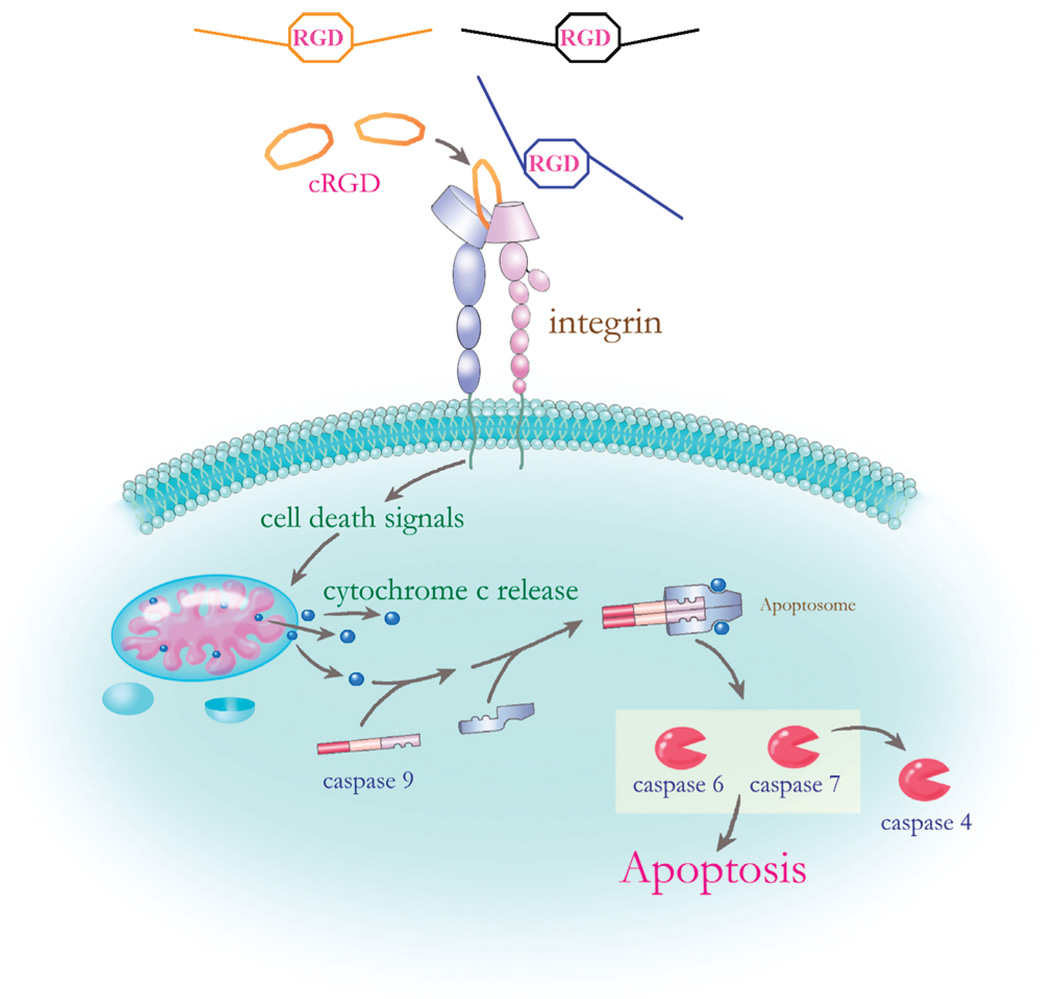
In our recent research, we synthesized a cyclic RGD, (Tpa-RGDWPC, cRGD), which can mimic the RGD motif of integrin-binding ligands to bind to integrin. This binding could not induce the signal pathway involved in cell growth and proliferation as indicated by the observed growth factor receptor-bound protein 2 (GRB2) behavior. GRB2 is a component in the integrin signal cascade and was found to be down-regulated after cRGD treatment. As a result, the breast cancer cells, MCF-7, proceeded to apoptosis pathways. These findings showed that cRGD acts as an anticancer compound to decrease the proliferation of breast cancer MCF-7 cells.12 On the other hand, the gene network results indicate that the cell death caused by cRGD may be triggered through activation of caspase pathway.13 In Figure 2, we show the cRGD-induced apoptosis hypothetical pathway, proposed signaling pathway by which interaction of cRGD with integrin α5β3 in the plasma membrane leads to cancer cell apoptosis via caspase-9 activation. Caspase-9, located at the upstream, activates downstream effector caspase-7. The activation of caspase-9 is believed to be a well-defined outcome of the release of mitochondrial cytochrome c into the cytoplasm and its subsequent association with the Apaf-1 protein. The assembly of cytochrome c, Apaf-1, and procaspase-9, called apoptosome, triggers the activation initiated by the Apaf-1–caspase-9 apoptosome.89 These results show that RGD-containing peptides can be potential drugs for breast cancer therapy.
Figure 2.
The cRGD-induced apoptosis hypothetical pathway. The tripeptide RGD is a common domain in ECM ligands (such as fibronectin, fibrinogen, and vitronectin) for integrin binding. cRGD is a synthetic tripeptide with a similar structure with RGD in the integrin binding ligands. This is a proposed signaling pathway by which interaction of cRGD with integrin α5β3 in the plasma membrane leads to cancer cell apoptosis via caspase-9 activation.
6. Drugs Targeting Tumor Microenvironment
Many studies proved the crucial role that ECM played in tumor development. Among the activation of PDGF,90 TGF-β,91 or other growth factors, precursor fibroblasts will differentiate into myofibroblasts which contribute to dense and rigid ECM network around tumors.18 The remodeling microenvironment leads to the elevated interstitial fluid pressure (IFP) that reduces drug uptake of cancer cells and thus confers them chemoresistance. To enhance the efficiency of cancer therapy, some researchers designed drugs that can “normalize” IFP and increase the drug delivery to cancer cells (Table 2).92 One of the strategies is blocking the activation of TAF. PDGF receptor tyrosine kinase is associated with the TAF activation and thus becomes the target of anticancer drug design.93 Imatinib (ST1157),94,95 the first generation drug with the PDGFR tyrosine kinase inhibitory effects, was found to decrease IFP around tumors. Imatinib-driven IFP decrease benefitted the elevated sensitivity to anticancer agents of subcutaneous growth thryroid carcinomas. Beside imatinib, other PDGF receptor inhibitors, like SU1124896 and BAY43–9006,97 have been developed and tested in clinic study. These drugs mentioned above have multitargets, and can also block other target receptors, like c-kit. CDP86098 is a polyethylene glycol conjugated Fab molecule, which binds to the PDGFR-β more specifically and inhibits its activity. The other target of drug design is FAP.99 FAP is overexpressed in tumor-associated fibroblasts including breast cancer but rarely occurs in health tissue. This property makes it an attractive candidate in cancer-directed prodrug design. Sibrotuzumab,100 a monoclonal antibody directed against FAP, has been demonstrated to specifically accumulate in tumors but not in normal tissues. The conjugation of toxic agent with sibrotuzumab may enhance the efficiency of chemotherapy. Besides, FAP also functions as a serine protease which can cleave gelatin and collagen and contributes to ECM remodeling.99 A DNA vaccine has been developed based on this concept. This vaccine successfully inhibited the growth of primary cancer cells and metastasis of murine colon and breast cancers.101 On the other hand, the FAP protease inhibitor, (PT-100),102 elevated the anticancer activity of immune system by enhancing the production of granulocyte-colony stimulating factor (G-CSF) in stromal cells. TAM also plays an important role in tumor development.25 Cancer cells can make VEGF and enhance the infiltration of macrophage into tumor areas.103 In cancer microenvironment, TGF-β, secreted by cancer or stromal cells, will activate TAM to enhance the expression of VEGF, MMP-9,104 and other cytokines (like IL-1 beta) and result in tumor progression. Prohibition of the migration or activation of TAM seems to be a novel approach to interfere with tumor development. Bevacizumab,103 an anti-VEGF monoclonal antibody, could neutralize the tumor-derived VEGF and reduce macrophage density in xenograft human anaplastic thyroid carcinoma. Furthermore, tumor extracellular fluid volume, tumor-vessel leakage of plasma protein, and IFP were decreased concomitantly. Another agent targeted to TAM is soluble TGF-β receptor type II-murine Fc:IgG2A chimeric protein (Fc:TβRII).91 Fc:TβRII could counteract TGF-β and reverse IFP elevated by TAM. The growth inhibition effect of doxorubicin was increased significantly in vivo. The other strategy aims at TAM destruction. A DNA vaccine against legumain, an endopeptidase overexpressed by TAM, has been proven to emerge CD8+ T cell responsive to TAM, and increase the survival rate in murine model.24
Table 2.
Anticancer Agents Targeting Tumor Microenvironmenta
| targeted cell | targeted molecule | agent |
|---|---|---|
| TAF | PDGFR | Imatinib (ST1157) |
| SU11248 | ||
| BAY43-9006 | ||
| CDP860 | ||
| FAP | Sibrotuzumab | |
| DNA vaccine | ||
| PT-100 | ||
| TAM | VEGF | Anti-VEGF antibody |
| TGF-β | Fc:TβRII | |
| Legumain | DNA vaccine |
TAF, tumor-associated fibroblasts; TAM, tumor-associated macrophages; PDGFR, plated-derived growth factor receptors; FAP, fibroblast activation protein; VEGF, vascular endothelial growth factor; TGF- β, transforming growth factor β.
7. Conclusions and Perspectives
There is a dynamic interaction within the breast tumor microenvironment, where host and tumor cells compete with each other to maximize their own survival.105 Although many theories have been proposed, the mechanisms underlying the dynamic interaction between tumor and host cells has not been fully explored. The loss of surface antigens, such as major histocompatibility (MHC) class I, is an independent indicator of good prognosis in breast cancer.106 It has also been well-documented that cancer patients’ sera contain an impressive variety of immunosuppressive proteins, indicating that many immunosuppressive substances may be secreted from either tumor or host cells.107 These substances, secreted in the tumor microenvironment, can be defined as part of the tumor secretome. Results from our current proteomics analysis provide a molecular explanation for the properties of ECM in breast cancer cells. ECM is one of components in the tumor secretome and can modulate the host immune system, thus, regulating tumor cell progression. Unraveling the tumor secretome will facilitate the development of antibreast cancer drugs specifically targeting secreted proteins/peptides. CUF probe sampling provides a promising method to dynamically obtain pure tumor secretome samples in vivo. More importantly, CUF probes locally sample the secretome directly in the tumor microenvironment. Mass spectrometry integrated with CUF probe sampling facilitates the secretome identification. Profiling the in vivo secretome within tumor microenvironments of breast cancers at different stages by CUF probes linked to mass spectrometry may be a powerful approach to identifying biomarkers for early detection.
Acknowledgment
This work was supported by National Institutes of Health Grants (R01-AI067395-01, R21-R022754-01, and R21-I58002-01). We thank R. A. Dorschner for critical reading of the manuscript.
Abbreviations
- CUF
capillary ultrafiltration
- ECM
extracellular matrix
- EGF
epidermal growth factor
- ELISAs
enzyme linked immunoabsorbent assays
- ErbB2
erythroblastic leukemia viral oncogene homologue 2
- FAP
fibroblast activation protein
- FGF
fibroblast growth factor
- HA
hyaluronic acid
- 5-FU
5-fluorouracil
- G-CSF
granulocyte-colony stimulating factor
- GRB2
growth factor receptor-bound protein 2
- HGF
hepatocyte growth factor
- IFP
interstitial fluid pressure
- IL
interleukin
- LC
liquid chromatography
- MALDI-TOF MS
matrix-assisted laser desorption/ionization time of flight MS
- MCP
monocyte chemoattractant protein
- MHC
major histocompatibility
- MMP
matrix metalloproteinases
- MS
mass spectrometry
- MWCOs
molecular weight cutoffs
- NK
natural killer
- OPN
osteopontin
- PDGF
platelet-derived growth factor
- PTFE
polytetrafluroethylene
- Q-TOF MS/MS
quadrupole time of flight MS/MS
- RHAMM
hyaluronan-binding protein
- TAFs
tumor-associated fibroblasts
- TAMs
tumor-associated macrophages
- TGF
transforming growth factor
- TIF
tumor interstitial fluid
- Tregs
regulatory T cells
- TNF
tumor necrosis factor
- UV
ultraviolet
- VCAM
vascular cell adhesion molecule
- VEGF
vascular endothelial growth factor
References
- 1.Denardo DG, Coussens LM. Inflammation and breast cancer Balancing immune response: crosstalk between adaptive and innate immune cells during breast cancer progression. Breast Cancer Res. 2007;9:212. doi: 10.1186/bcr1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vernon AE, Bakewell SJ, Chodosh LA. Deciphering the molecular basis of breast cancer metastasis with mouse models. Rev. Endocr. Metab. Disord. 2007;8(3):199–213. doi: 10.1007/s11154-007-9041-5. [DOI] [PubMed] [Google Scholar]
- 3.Bonin-Debs AL, Boche I, Gille H, Brinkmann U. Development of secreted proteins as biotherapeutic agents. Expert Opin. Biol. Ther. 2004;4:551–558. doi: 10.1517/14712598.4.4.551. [DOI] [PubMed] [Google Scholar]
- 4.Shinkaruk S, Bayle M, Lain G, Deleris G. Vascular endothelial cell growth factor (VEGF), an emerging target for cancer chemotherapy. Curr. Med. Chem. Anticancer Agents. 2003;3:95–117. doi: 10.2174/1568011033353452. [DOI] [PubMed] [Google Scholar]
- 5.Ohe Y, Kasai T, Heike Y, Saijo N. Clinical trial of IL-12 for cancer patients. Gan to Kagaku Ryoho. 1998;25:177–184. [PubMed] [Google Scholar]
- 6.Huang CM, Ananthaswamy HN, Barnes S, Ma Y, Kawai M, Elmets CA. Mass spectrometric proteomics profiles of in vivo tumor secretomes: capillary ultrafiltration sampling of regressive tumor masses. Proteomics. 2006;6:6107–6116. doi: 10.1002/pmic.200600287. [DOI] [PubMed] [Google Scholar]
- 7.Huang CM, Wang CC, Barnes S, Elmets CA. In vivo detection of secreted proteins from wounded skin using capillary ultrafiltration probes and mass spectrometric proteomics. Proteomics. 2006;6:5805–5814. doi: 10.1002/pmic.200600163. [DOI] [PubMed] [Google Scholar]
- 8.Huang CM, Wang CC, Kawai M, Barnes S, Elmets CA. In vivo protein sampling using capillary ultrafiltration semipermeable hollow fiber and protein identification via mass spectrometry-based proteomics. J. Chromatogr., A. 2006;1109:144–151. doi: 10.1016/j.chroma.2005.11.104. [DOI] [PubMed] [Google Scholar]
- 9.Huang CM, Wang CC, Kawai M, Barnes S, Elmets CA. Surfactant sodium lauryl sulfate enhances skin vaccination: molecular characterization via a novel technique using ultrafiltration capillaries and mass spectrometric proteomics. Mol. Cell. Proteomics. 2006;5:523–532. doi: 10.1074/mcp.M500259-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Hathout Y. Approaches to the study of the cell secretome. Expert Rev. Proteomics. 2007;4:239–248. doi: 10.1586/14789450.4.2.239. [DOI] [PubMed] [Google Scholar]
- 11.Huinink KD, Venema K, Roelofsen H, Korf J. In vitro sampling and storage of proteins with an ultrafiltration collection device (UCD) and analysis with absorbance spectrometry and SELDI-TOF-MS. Analyst. 2005;130:1168–1174. doi: 10.1039/b503136b. [DOI] [PubMed] [Google Scholar]
- 12.Juan HF, Wang IH, Huang TC, Li JJ, Chen ST, Huang HC. Proteomics analysis of a novel compound: cyclic RGD in breast carcinoma cell line MCF-7. Proteomics. 2006;6:2991–3000. doi: 10.1002/pmic.200500435. [DOI] [PubMed] [Google Scholar]
- 13.Huang TC, Huang HC, Chang CC, Chang HY, Ou CH, Hsu CH, Chen ST, Juan HF. An apoptosis-related gene network induced by novel compound-cRGD in human breast cancer cells. FEBS Lett. 2007;581:3517–3522. doi: 10.1016/j.febslet.2007.06.067. [DOI] [PubMed] [Google Scholar]
- 14.Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 15.Wiseman BS, Werb Z. Stromal effects on mammary gland development and breast cancer. Science. 2002;296:1046–1049. doi: 10.1126/science.1067431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pupa SM, Menard S, Forti S, Tagliabue E. New insights into the role of extracellular matrix during tumor onset and progression. J. Cell Physiol. 2002;192:259–267. doi: 10.1002/jcp.10142. [DOI] [PubMed] [Google Scholar]
- 17.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desmouliere A, Guyot C, Gabbiani G. The stroma reaction myofibroblast: a key player in the control of tumor cell behavior. Int. J. Dev. Biol. 2004;48:509–517. doi: 10.1387/ijdb.041802ad. [DOI] [PubMed] [Google Scholar]
- 19.Silzle T, Randolph GJ, Kreutz M, Kunz-Schughart LA. The fibroblast: sentinel cell and local immune modulator in tumor tissue. Int. J. Cancer. 2004;108:173–180. doi: 10.1002/ijc.11542. [DOI] [PubMed] [Google Scholar]
- 20.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat. Rev. Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 21.Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, Porter D, Hu M, Chin L, Richardson A, Schnitt S, Sellers WR, Polyak K. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 22.Leek RD, Harris AL. Tumor-associated macrophages in breast cancer. J. Mammary Gland Biol. Neoplasia. 2002;7:177–189. doi: 10.1023/a:1020304003704. [DOI] [PubMed] [Google Scholar]
- 23.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 24.Luo Y, Zhou H, Krueger J, Kaplan C, Lee SH, Dolman C, Markowitz D, Wu W, Liu C, Reisfeld RA, Xiang R. Targeting tumor-associated macrophages as a novel strategy against breast cancer. J. Clin. Invest. 2006;116:2132–2141. doi: 10.1172/JCI27648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin EY, Pollard JW. Tumor-associated macrophages press the angiogenic switch in breast cancer. Cancer Res. 2007;67:5064–5066. doi: 10.1158/0008-5472.CAN-07-0912. [DOI] [PubMed] [Google Scholar]
- 26.Mbeunkui F, Metge BJ, Shevde LA, Pannell LK. Identification of differentially secreted biomarkers using LC-MS/MS in isogenic cell lines representing a progression of breast cancer. J. Proteome Res. 2007;6:2993–3002. doi: 10.1021/pr060629m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Celis JE, Gromov P, Cabezón T, Moreira JM, Ambartsumian N, Sandelin K, Rank F, Gromova I. Proteomic characterization of the interstitial fluid perfusing the breast tumor microenvironment: a novel resource for biomarker and therapeutic target discovery. Mol. Cell. Proteomics. 2004;3:327–344. doi: 10.1074/mcp.M400009-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Hagemann T, Balkwill F, Lawrence T. Inflammation and cancer: a double-edged sword. Cancer Cell. 2007;12:300–301. doi: 10.1016/j.ccr.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cabezón T, Celis JE, Skibshøj I, Klingelhöfer J, Grigorian M, Gromov P, Rank F, Myklebust JH, Maelandsmo GM, Lukanidin E, Ambartsumian N. Expression of S100A4 by a variety of cell types present in the tumor microenvironment of human breast cancer. Int. J. Cancer. 2007;121:1433–1444. doi: 10.1002/ijc.22850. [DOI] [PubMed] [Google Scholar]
- 30.Dombkowski AA, Cukovic D, Novak RF. Secretome analysis of microarray data reveals extracellular events associated with proliferative potential in a cell line model of breast disease. Cancer Lett. 2006;241:49–58. doi: 10.1016/j.canlet.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 31.Haslam SZ, Woodward TL. Host microenvironment in breast cancer development: epithelial-cell-stromal-cell interactions and steroid hormone action in normal and cancerous mammary gland. Breast Cancer Res. 2003;5:208–215. doi: 10.1186/bcr615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leegsma-Vogt G, Janle E, Ash SR, Venema K, Korf J. Utilization of in vivo ultrafiltration in biomedical research and clinical applications. Life Sci. 2003;73:2005–2018. doi: 10.1016/s0024-3205(03)00569-1. [DOI] [PubMed] [Google Scholar]
- 33.Dabrosin C. Microdialysis—an in vivo technique for studies of growth factors in breast cancer. Front. Biosci. 2005;10:1329–1335. doi: 10.2741/1622. [DOI] [PubMed] [Google Scholar]
- 34.Müller M, Mader RM, Steiner B, Steger GG, Jansen B, Gnant M, Helbich T, Jakesz R, Eichler HG, Blochl-Daum B. 5-fluorouracil kinetics in the interstitial tumor space: clinical response in breast cancer patients. Cancer Res. 1997;57:2598–2601. [PubMed] [Google Scholar]
- 35.Duo J, Fletcher H, Stenken JA. Natural and synthetic affinity agents as microdialysis sampling mass transport enhancers: current progress and future perspectives. Biosens. Bioelectron. 2006;22:449–457. doi: 10.1016/j.bios.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Ao X, Stenken JA. Microdialysis sampling of cytokines. Methods. 2006;38:331–341. doi: 10.1016/j.ymeth.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 37.Price KE, Vandaveer SS, Lunte CE, Larive CK. Tissue targeted metabonomics: metabolic profiling by microdialysis sampling and microcoil NMR. J. Pharm. Biomed. Anal. 2005;38:904–909. doi: 10.1016/j.jpba.2005.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sopasakis VR, Sandqvist M, Gustafson B, Hammarstedt A, Schmelz M, Yang X, Jansson PA, Smith U. High local concentrations and effects on differentiation implicate interleukin-6 as a paracrine regulator. Obes. Res. 2004;12:454–460. doi: 10.1038/oby.2004.51. [DOI] [PubMed] [Google Scholar]
- 39.Jakubowski JA, Hatcher NG, Sweedler JV. Online microdialysis-dynamic nanoelectrospray ionization-mass spectrometry for monitoring neuropeptide secretion. J. Mass Spectrom. 2005;40:924–931. doi: 10.1002/jms.869. [DOI] [PubMed] [Google Scholar]
- 40.Janle EM, Kissinger PT. Microdialysis and ultrafiltration. Adv. Food Nutr. Res. 1996;40:183–196. doi: 10.1016/s1043-4526(08)60028-5. [DOI] [PubMed] [Google Scholar]
- 41.Odland RM, Kizziar R, Rheuark D, Simental A. The effect of capillary ultrafiltration probes on skin flap edema. Otolaryngol. Head Neck Surg. 2003;128:210–214. doi: 10.1067/mhn.2003.69. [DOI] [PubMed] [Google Scholar]
- 42.Janle EM, Sojka JE. Use of ultrafiltration probes in sheep to collect interstitial fluid for measurement of calcium and magnesium. Contemp. Top. Lab. Anim. Sci. 2000;39:47–50. [PubMed] [Google Scholar]
- 43.Imsilp K, Whittem T, Koritz GD, Zachary JF, Schaeffer D. Inflammatory response to intramuscular implantation of polyacrylonitrile ultrafiltration probes in sheep. J. Vet. Res. 2000;31:623–634. doi: 10.1051/vetres:2000145. [DOI] [PubMed] [Google Scholar]
- 44.Linhares MC, Kissinger PT. Capillary ultrafiltration: in vivo sampling probes for small molecules. Anal. Chem. 1992;64:2831–2835. doi: 10.1021/ac00046a029. [DOI] [PubMed] [Google Scholar]
- 45.Fernandez A, Chen PW, Aggarwal BB, Ananthaswamy HN. Resistance of Ha-ras oncogene-induced progressor tumor variants to tumor necrosis factor and interferon-gamma. Lymphokine Cytokine Res. 1992;11:79–85. [PubMed] [Google Scholar]
- 46.Chernokalskaya E, Gutierrez S, Pitt AM, Leonard JT. Ultrafiltration for proteomic sample preparation. Electrophoresis. 2004;25:2461–2468. doi: 10.1002/elps.200405998. [DOI] [PubMed] [Google Scholar]
- 47.Rivers A, Newman LA. Ductal lavage for breast cancer risk assessment. Surg. Oncol. Clin. N. Am. 2005;14:45–68. doi: 10.1016/j.soc.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 48.Hlubek F, Brabletz T, Budczies J, Pfeiffer S, Jung A, Kirchner T. Heterogeneous expression of Wnt/beta-catenin target genes within colorectal cancer. Int. J. Cancer. 2007;121:1941–1948. doi: 10.1002/ijc.22916. [DOI] [PubMed] [Google Scholar]
- 49.Larsson LI, Holck S. Occurrence of thymosin beta4 in human breast cancer cells and in other cell types of the tumor microenvironment. Hum. Pathol. 2007;38:114–119. doi: 10.1016/j.humpath.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 50.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balkwill F, Charles KA, Mantovani A. Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cell. 2005;7:211–217. doi: 10.1016/j.ccr.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 52.Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat. Rev. Cancer. 2005;5:263–274. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]
- 53.Cronin SJ, Penninger JM. From T-cell activation signals to signaling control of anti-cancer immunity. Immunol. Rev. 2007;220:151–168. doi: 10.1111/j.1600-065X.2007.00570.x. [DOI] [PubMed] [Google Scholar]
- 54.de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat. Rev. Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 55.Knutson KL, Dang Y, Lu H, Lukas J, Almand B, Gad E, Azeke E, Disis ML. IL-2 immunotoxin therapy modulates tumor-associated regulatory T cells and leads to lasting immune-mediated rejection of breast cancers in neu-transgenic mice. J. Immunol. 2006;177:84–91. doi: 10.4049/jimmunol.177.1.84. [DOI] [PubMed] [Google Scholar]
- 56.Colombo MP, Piconese S. Regulatory T-cell inhibition versus depletion: the right choice in cancer immunotherapy. Nat. Rev. Cancer. 2007;7:880–887. doi: 10.1038/nrc2250. [DOI] [PubMed] [Google Scholar]
- 57.Lang K, Entschladen F, Weidt C, Zaenker KS. Tumor immune escape mechanisms: impact of the neuroendocrine system. Cancer Immunol. Immunother. 2006;55:749–760. doi: 10.1007/s00262-006-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gotte M, Yip GW. Heparanase, hyaluronan, and CD44 in cancers: a breast carcinoma perspective. Cancer Res. 2006;66:10233–10237. doi: 10.1158/0008-5472.CAN-06-1464. [DOI] [PubMed] [Google Scholar]
- 59.Udabage L, Brownlee GR, Nilsson SK, Brown TJ. The overexpression of HAS2, Hyal-2 and CD44 is implicated in the invasiveness of breast cancer. Exp. Cell Res. 2005;310:205–217. doi: 10.1016/j.yexcr.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 60.Udabage L, Brownlee GR, Waltham M, Blick T, Walker EC, Heldin P, Nilsson SK, Thompson EW, Brown TJ. Antisense-mediated suppression of hyaluronan synthase 2 inhibits the tumorigenesis and progression of breast cancer. Cancer Res. 2005;65:6139–6150. doi: 10.1158/0008-5472.CAN-04-1622. [DOI] [PubMed] [Google Scholar]
- 61.Li Y, Li L, Brown TJ, Heldin P. Silencing of hyaluronan synthase 2 suppresses the malignant phenotype of invasive breast cancer cells. Int. J. Cancer. 2007;120:2557–2567. doi: 10.1002/ijc.22550. [DOI] [PubMed] [Google Scholar]
- 62.Palyi-Krekk Z, Barok M, Isola J, Tammi M, Szollo si J, Nagy P. Hyaluronan-induced masking of ErbB2 and CD44-enhanced trastuzumab internalisation in trastuzumab resistant breast cancer. Eur. J. Cancer. 2007;43:2423–2433. doi: 10.1016/j.ejca.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 63.Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 64.Delehedde M, Lyon M, Sergeant N, Rahmoune H, Fernig DG. Proteoglycans: pericellular and cell surface multireceptors that integrate external stimuli in the mammary gland. J. Mammary Gland Biol. Neoplasia. 2001;6:253–273. doi: 10.1023/a:1011367423085. [DOI] [PubMed] [Google Scholar]
- 65.Allan AL, George R, Vantyghem SA, Lee MW, Hodgson NC, Engel CJ, Holliday RL, Girvan DP, Scott LA, Postenka CO, Al-Katib W, Stitt LW, Uede T, Chambers AF, Tuck AB. Role of the integrin-binding protein osteopontin in lymphatic metastasis of breast cancer. Am. J. Pathol. 2006;169:233–246. doi: 10.2353/ajpath.2006.051152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tuck AB, Chambers AF, Allan AL. Osteopontin overexpression in breast cancer: Knowledge gained and possible implications for clinical management. J. Cell Biochem. 2007;102:859–868. doi: 10.1002/jcb.21520. [DOI] [PubMed] [Google Scholar]
- 67.Cook AC, Chambers AF, Turley EA, Tuck AB. Osteopontin induction of hyaluronan synthase 2 expression promotes breast cancer malignancy. J. Biol. Chem. 2006;281:24381–24389. doi: 10.1074/jbc.M602694200. [DOI] [PubMed] [Google Scholar]
- 68.Rodrigues LR, Teixeira JA, Schmitt FL, Paulsson M, Lindmark-Mansson H. The role of osteopontin in tumor progression and metastasis in breast cancer. Cancer Epidemiol. Biomarkers Prev. 2007;16:1087–1097. doi: 10.1158/1055-9965.EPI-06-1008. [DOI] [PubMed] [Google Scholar]
- 69.Jahkola T, Toivonen T, von Smitten K, Blomqvist C, Virtanen I. Expression of tenascin in invasion border of early breast cancer correlates with higher risk of distant metastasis. Int. J. Cancer. 1996;69:445–447. doi: 10.1002/(SICI)1097-0215(19961220)69:6<445::AID-IJC4>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 70.Chiquet-Ehrismann R, Chiquet M. Tenascins: regulation and putative functions during pathological stress. J. Pathol. 2003;200(4):488–499. doi: 10.1002/path.1415. [DOI] [PubMed] [Google Scholar]
- 71.Orend G. Potential oncogenic action of tenascin-C in tumorigenesis. Int. J. Biochem. Cell Biol. 2005;37:1066–1083. doi: 10.1016/j.biocel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 72.Ilunga K, Nishiura R, Inada H, El-Karef A, Imanaka-Yoshida K, Sakakura T, Yoshida T. Co-stimulation of human breast cancer cells with transforming growth factor-beta and tenascin-C enhances matrix metalloproteinase-9 expression and cancer cell invasion. Int. J. Exp. Pathol. 2004;85:373–379. doi: 10.1111/j.0959-9673.2004.00406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miller FR, McInerney D, Rogers C, Miller BE. Spontaneous fusion between metastatic mammary tumor subpopulations. J. Cell Biochem. 1988;36:129–136. doi: 10.1002/jcb.240360204. [DOI] [PubMed] [Google Scholar]
- 74.Larsson LI, Bjerregaard B, Wulf-Andersen L, Talts JF. Syncytin and cancer cell fusions. Sci. World J. 2007;7:1193–1197. doi: 10.1100/tsw.2007.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bjerregaard B, Holck S, Christensen IJ, Larsson LI. Syncytin is involved in breast cancer-endothelial cell fusions. Cell. Mol. Life Sci. 2006;63:1906–1911. doi: 10.1007/s00018-006-6201-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Larsson LI, Holck S, Christensen IJ. Prognostic role of syncytin expression in breast cancer. Hum. Pathol. 2007;38:726–731. doi: 10.1016/j.humpath.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 77.Kong HJ, Mooney DJ. Microenvironmental regulation of biomacromolecular therapies. Nat. Rev, Drug Discovery. 2007;6:455–463. doi: 10.1038/nrd2309. [DOI] [PubMed] [Google Scholar]
- 78.Akiyama SK. Integrins in cell adhesion and signaling. Hum. Cell. 1996;9:181–186. [PubMed] [Google Scholar]
- 79.Lin HY, Lansing L, Merillon JM, Davis FB, Tang HY, Shih A, Vitrac X, Krisa S, Keating T, Cao HJ, Bergh J, Quackenbush S, Davis PJ. Integrin alphaVbeta3 contains a receptor site for resveratrol. FASEB J. 2006;20:1742–1744. doi: 10.1096/fj.06-5743fje. [DOI] [PubMed] [Google Scholar]
- 80.Takagi J. Structural basis for ligand recognition by integrins. Curr. Opin. Cell Biol. 2007;19:557–564. doi: 10.1016/j.ceb.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 81.Morini M, Mottolese M, Ferrari N, Ghiorzo F, Buglioni S, Mortarini R, Noonan DM, Natali PG, Albini A. The alpha 3 beta 1 integrin is associated with mammary carcinoma cell metastasis, invasion, and gelatinase B (MMP-9) activity. Int. J. Cancer. 2000;87:336–342. [PubMed] [Google Scholar]
- 82.Pignatelli M, Cardillo MR, Hanby A, Stamp GW. Integrins and their accessory adhesion molecules in mammary carcinomas: loss of polarization in poorly differentiated tumors. Hum. Pathol. 1992;23:1159–1166. doi: 10.1016/0046-8177(92)90034-z. [DOI] [PubMed] [Google Scholar]
- 83.Yao ES, Zhang H, Chen YY, Lee B, Chew K, Moore D, Park C. Increased beta1 integrin is associated with decreased survival in invasive breast cancer. Cancer Res. 2007;67:659–664. doi: 10.1158/0008-5472.CAN-06-2768. [DOI] [PubMed] [Google Scholar]
- 84.Guo W, Pylayeva Y, Pepe A, Yoshioka T, Muller WJ, Inghirami G, Giancotti FG. Beta 4 integrin amplifies ErbB2 signaling to promote mammary tumorigenesis. Cell. 2006;126:489–502. doi: 10.1016/j.cell.2006.05.047. [DOI] [PubMed] [Google Scholar]
- 85.Katz M, Amit I, Citri A, Shay T, Carvalho S, Lavi S, Milanezi F, Lyass L, Amariglio N, Jacob-Hirsch J, Ben-Chetrit N, Tarcic G, Lindzen M, Avraham R, Liao YC, Trusk P, Lyass A, Rechavi G, Spector NL, Lo SH, Schmitt F, Bacus SS, Yarden Y. A reciprocal tensin-3-cten switch mediates EGF-driven mammary cell migration. Nat. Cell Biol. 2007;9(8):961–969. doi: 10.1038/ncb1622. [DOI] [PubMed] [Google Scholar]
- 86.Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat. Cell Biol. 2007;9:893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 87.Sulyok GA, Gibson C, Goodman SL, Holzemann G, Wiesner M, Kessler H. Solid-phase synthesis of a nonpeptide RGD mimetic library: new selective alphavbeta3 integrin antagonists. J. Med. Chem. 2001;44:1938–1950. doi: 10.1021/jm0004953. [DOI] [PubMed] [Google Scholar]
- 88.Buckley CD, Pilling D, Henriquez NV, Parsonage G, Threlfall K, Scheel-Toellner D, Simmons DL, Akbar AN, Lord JM, Salmon M. RGD peptides induce apoptosis by direct caspase-3 activation. Nature. 1999;397:534–539. doi: 10.1038/17409. [DOI] [PubMed] [Google Scholar]
- 89.Twiddy D, Cohen GM, Macfarlane M, Cain K. Caspase-7 is directly activated by the approximately 700-kDa apoptosome complex and is released as a stable XIAP-caspase-7 approximately 200-kDa complex. J. Biol. Chem. 2006;281:3876–3888. doi: 10.1074/jbc.M507393200. [DOI] [PubMed] [Google Scholar]
- 90.Board R, Jayson GC. Platelet-derived growth factor receptor (PDGFR): a target for anticancer therapeutics. Drug Resist. Updates. 2005;8:75–83. doi: 10.1016/j.drup.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 91.Salnikov AV, Roswall P, Sundberg C, Gardner H, Heldin NE, Rubin K. Inhibition of TGF-beta modulates macrophages and vessel maturation in parallel to a lowering of interstitial fluid pressure in experimental carcinoma. Lab. Invest. 2005;85:512–521. doi: 10.1038/labinvest.3700252. [DOI] [PubMed] [Google Scholar]
- 92.Bouzin C, Feron O. Targeting tumor stroma and exploiting mature tumor vasculature to improve anti-cancer drug delivery. Drug Resist. Updates. 2007;10:109–120. doi: 10.1016/j.drup.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 93.Millette E, Rauch BH, Kenagy RD, Daum G, Clowes AW. Platelet-derived growth factor-BB transactivates the fibroblast growth factor receptor to induce proliferation in human smooth muscle cells. Trends Cardiovasc. Med. 2006;16:25–28. doi: 10.1016/j.tcm.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 94.Pietras K, Ostman A, Sjoquist M, Buchdunger E, Reed RK, Heldin CH, Rubin K. Inhibition of platelet-derived growth factor receptors reduces interstitial hypertension and increases transcapillary transport in tumors. Cancer Res. 2001;61:2929–2934. [PubMed] [Google Scholar]
- 95.Pietras K, Rubin K, Sjoblom T, Buchdunger E, Sjoquist M, Heldin CH, Ostman A. Inhibition of PDGF receptor signaling in tumor stroma enhances antitumor effect of chemotherapy. Cancer Res. 2002;62:5476–5484. [PubMed] [Google Scholar]
- 96.Mendel DB, Laird AD, Xin X, Louie SG, Christensen JG, Li G, Schreck RE, Abrams TJ, Ngai TJ, Lee LB, Murray LJ, Carver J, Chan E, Moss KG, Haznedar JO, Sukbuntherng J, Blake RA, Sun L, Tang C, Miller T, Shirazian S, McMahon G, Cherrington JM. In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet-derived growth factor receptors: determination of a pharmacokinetic/pharmacodynamic relationship. Clin. Cancer Res. 2003;9:327–337. [PubMed] [Google Scholar]
- 97.Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, Chen C, Zhang X, Vincent P, McHugh M, Cao Y, Shujath J, Gawlak S, Eveleigh D, Rowley B, Liu L, Adnane L, Lynch M, Auclair D, Taylor I, Gedrich R, Voznesensky A, Riedl B, Post LE, Bollag G, Trail PA. BAY 43–9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004;64:7099–7109. doi: 10.1158/0008-5472.CAN-04-1443. [DOI] [PubMed] [Google Scholar]
- 98.Jayson GC, Parker GJ, Mullamitha S, Valle JW, Saunders M, Broughton L, Lawrance J, Carrington B, Roberts C, Issa B, Buckley DL, Cheung S, Davies K, Watson Y, Zinkewich-Peotti K, Rolfe L, Jackson A. Blockade of platelet-derived growth factor receptor-beta by CDP860, a humanized, PEGylated di-Fab’, leads to fluid accumulation and is associated with increased tumor vascularized volume. J. Clin. Oncol. 2005;23:973–981. doi: 10.1200/JCO.2005.01.032. [DOI] [PubMed] [Google Scholar]
- 99.Kelly T. Fibroblast activation protein-alpha and dipeptidyl peptidase IV (CD26): cell-surface proteases that activate cell signaling and are potential targets for cancer therapy. Drug Resist. Updates. 2005;8:51–58. doi: 10.1016/j.drup.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 100.Kloft C, Graefe EU, Tanswell P, Scott AM, Hofheinz R, Amelsberg A, Karlsson MO. Population pharmacokinetics of sibrotuzumab, a novel therapeutic monoclonal antibody, in cancer patients. Invest. New Drugs. 2004;22:39–52. doi: 10.1023/b:drug.0000006173.72210.1c. [DOI] [PubMed] [Google Scholar]
- 101.Loeffler M, Kruger JA, Niethammer AG, Reisfeld RA. Targeting tumor-associated fibroblasts improves cancer chemotherapy by increasing intratumoral drug uptake. J. Clin. Invest. 2006;116:1955–1962. doi: 10.1172/JCI26532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Adams S, Miller GT, Jesson MI, Watanabe T, Jones B, Wallner BP. PT-100, a small molecule dipeptidyl peptidase inhibitor, has potent antitumor effects and augments antibody-mediated cytotoxicity via a novel immune mechanism. Cancer Res. 2004;64:5471–5480. doi: 10.1158/0008-5472.CAN-04-0447. [DOI] [PubMed] [Google Scholar]
- 103.Salnikov AV, Heldin NE, Stuhr LB, Wiig H, Gerber H, Reed RK, Rubin K. Inhibition of carcinoma cell-derived VEGF reduces inflammatory characteristics in xenograft carcinoma. Int. J. Cancer. 2006;119:2795–2802. doi: 10.1002/ijc.22217. [DOI] [PubMed] [Google Scholar]
- 104.Fridman R, Toth M, Chvyrkova I, Meroueh SO, Mobashery S. Cell surface association of matrix metalloproteinase-9 (gelatinase B) Cancer Metastasis Rev. 2003;22:153–166. doi: 10.1023/a:1023091214123. [DOI] [PubMed] [Google Scholar]
- 105.Poggi A, Zocchi MR. Mechanisms of tumor escape: role of tumor microenvironment in inducing apoptosis of cytolytic effector cells. Arch. Immunol. Ther. Exp. 2006;54:323–333. doi: 10.1007/s00005-006-0038-7. [DOI] [PubMed] [Google Scholar]
- 106.Madjd Z, Spendlove I, Pinder SE, Ellis IO, Durrant LG. Total loss of MHC class I is an independent indicator of good prognosis in breast cancer. Int. J. Cancer. 2005;117:248–255. doi: 10.1002/ijc.21163. [DOI] [PubMed] [Google Scholar]
- 107.Ochsenbein AF. Principles of tumor immunosurveillance and implications for immunotherapy. Cancer Gene Ther. 2002;9:1043–1055. doi: 10.1038/sj.cgt.7700540. [DOI] [PubMed] [Google Scholar]
- 108.Rosenbloom AJ, Ferris RL, Sipe DM, Riddler SA, Connolly NC, Abe K, Whiteside TL. In vitro and in vivo protein sampling by combined microdialysis and ultrafiltration. J. Immunol. Methods. 2006;309:55–68. doi: 10.1016/j.jim.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 109.Uchino S, Bellomo R, Morimatsu H, Goldsmith D, Davenport P, Cole L, Baldwin I, Panagiotopoulos S, Tipping P, Morgera S, Neumayer HH, Goehl H. Cytokine dialysis: an ex vivo study. ASAIO J. 2002;48:650–653. doi: 10.1097/00002480-200211000-00013. [DOI] [PubMed] [Google Scholar]
- 110.Linhares MC, Kissinger PT. Pharmacokinetic monitoring in subcutaneous tissue using in vivo capillary ultrafiltration probes. Pharm. Res. 1993;10:598–602. doi: 10.1023/a:1018914522749. [DOI] [PubMed] [Google Scholar]
- 111.Lam H, Davies M, Lunte CE. Vacuum ultrafiltration sampling for determination of plasma protein binding of drugs. J. Pharm. Biomed. Anal. 1996;14:1753–1757. doi: 10.1016/0731-7085(96)01811-0. [DOI] [PubMed] [Google Scholar]