Abstract
Indole-3-acetic acid (IAA), the main auxin in higher plants, has profound effects on plant growth and development. Both plants and some plant pathogens can produce IAA to modulate plant growth. While the genes and biochemical reactions for auxin biosynthesis in some plant pathogens are well understood, elucidation of the mechanisms by which plants produce auxin has proven to be difficult. So far, no complete pathway of de novo auxin biosynthesis in plants has been firmly established. However, recent studies have led to the discoveries of several genes in tryptophan dependent auxin biosynthesis pathways. Recent findings have also revealed that local auxin biosynthesis plays essential roles in many developmental processes including gametogenesis, embryogenesis, seedling growth, vascular patterning, and flower development. In this review, I summarize the recent advances in dissecting auxin biosynthetic pathways and how the understanding of auxin biosynthesis provides a different angle for analyzing the mechanisms of plant development.
Keywords: Arabidopsis, tryptophan, YUCCA, TAA1, flavin monooxygenase
Introduction
Auxin was identified as a plant growth hormone because of its ability to stimulate differential growth in response to gravity or light stimuli. The in vitro bioassay in which auxin-containing agar blocks stimulated the growth of oat coleoptile segments led to the identification of indole-3-acetic acid (IAA) as the main naturally occurring auxin in plants. Applications of IAA or synthetic auxins to plants cause profound changes in plant growth and development (6). Much of the knowledge of the physiological roles of auxin in plants is derived from studies on how plants respond to excess exogenous auxin. However, an equally important aspect of auxin biology is to characterize the developmental defects caused by auxin deficiency, which cannot be achieved without a clear grasp of auxin biosynthetic pathways.
In contrast to the great progress made in understanding auxin signaling and transport (40, 64), much less is known about how auxin is produced in plants. Elucidation of the molecular mechanisms of auxin biosynthesis will have a great impact on defining the roles of auxin in plant development, understanding auxin transport, and studying the mechanisms of auxin in regulating plant development. Despite its apparent importance in auxin biology, auxin biosynthesis has remained an elusive target for plant biologists. Only recently have several key genes in de novo auxin biosynthesis been identified with molecular genetic approaches.
Auxin biosynthesis in plants is extremely complex. It is likely that multiple pathways contribute to de novo auxin production. IAA can also be released from IAA conjugates by hydrolytic cleavage of IAA-amino acids, IAA-sugar, and IAA-methyl ester (3, 36, 67, 72). Furthermore, although plants share evolutionary conserved core mechanisms for auxin biosynthesis, different plant species may also have unique strategies and modifications to optimize their IAA biosynthesis. In this review, I will devote my discussions solely to the mechanisms of tryptophan (trp) dependent auxin biosynthesis. Discussions on trp independent auxin biosynthesis pathway and auxin conjugation / modification can be found elsewhere (3, 67). I will focus on the auxin biosynthesis genes identified in Arabidopsis, but also briefly discuss relevant studies in other species. I will also include two examples to demonstrate that knowledge of auxin biosynthesis provides a genetic basis for solving key plant developmental questions.
Lessons from auxin biosynthesis in plant pathogens
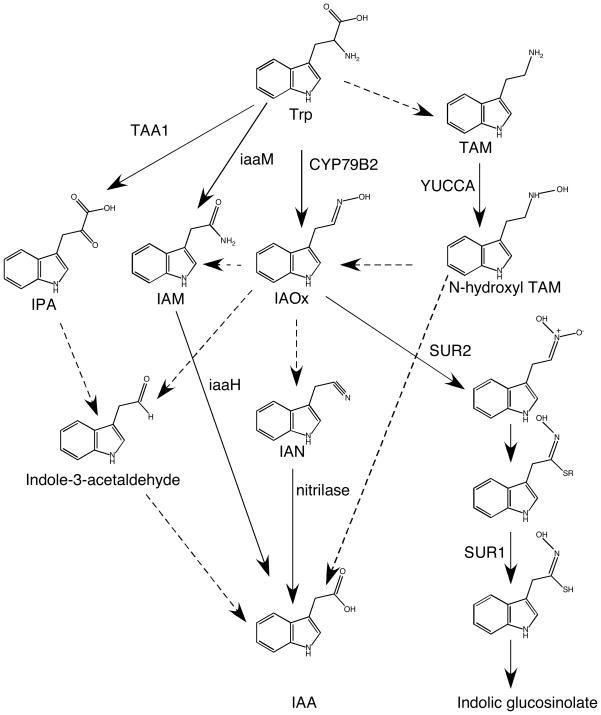
Plant pathogens such as Agrobacterium produce auxin to hijack plant cells for nutrient production. Pseudomonas and Agrobacterium use a tryptophan-2-monooxygeanse called iaaM to convert tryptophan to indole-3-acetamide (IAM), which is subsequently hydrolyzed into IAA by a hydrolase iaaH (7, 13) (Figure 1). So far, the iaaM / iaaH pathway is the only known complete trp dependent IAA biosynthesis pathway. It is generally believed that plants do not use the iaaM / iaaH pathway to make IAA. However, IAM exists in plant extracts and has been suggested as a key intermediate in converting Indole-3-acetaldoxime (IAOx) to IAA (58) (Figure 1). Furthermore, a family of amidases that can hydrolyze IAM into IAA has been identified in Arabidopsis (47), suggesting that IAM can be an intermediate for IAA biosynthesis in plants. However, the biochemical reactions for IAM production in plants have not been solved.
Figure 1. Trp dependent auxin biosynthesis pathways.
Solid arrow: genes responsible for the steps have been identified in plants or microbes. Dashed arrow: proposed steps, but genes for the steps have not been conclusively determined. IAOx: indole-3-acetaldoxime ; IPA: indole-3-pyruvate; IAM: indole-3-acetamide; IAN: indole-3-acetonitrile ; TAM: tryptamine.
The understanding of the iaaM / iaaH pathway has been instrumental to recent progress in dissecting auxin biosynthesis in plants. The bacterial iaaM gene also provides a useful way to manipulate auxin levels in transgenic plants. Overexpression of the iaaM gene alone in petunia (32), tobacco (53), and Arabidopsis (49) leads to auxin overproduction phenotypes, suggesting that plants have enzymes for the hydrolysis of IAM. Transgenic Arabidopsis plants that overexpress the iaaM gene under the control of the CaMV 35S promoter are much taller than wild type plants when grown in light (49). The iaaM overexpression lines define the characteristics of auxin overproduction in Arabidopsis, thus providing important traits for identifying plant auxin biosynthetic genes (see below). Tissue specific expression of the iaaM gene enables the supply of auxin locally, which provides a key piece of evidence that demonstrates the roles of a family of flavin monooxygenases in auxin biosynthesis (see below).
IAA can also be produced from trp through the indole-3-pyruvate pathway (IPA) (Figure 1) found in some microbes. Unlike the iaaM / iaaH pathway, the IPA pathway has not been completely solved in microbes. The IPA decarboxylase, which catalyzes the conversion of IPA to indole-3-acetaldehyde, has been cloned from Enterobacter cloacae (33) and Azospirillum brasilense (14), but the genes responsible for converting tryptophan to IPA and enzymes for catalyzing indole-3-acetaldehyde to IAA have not been conclusively identified in microbes. Whether the IPA decarboxylase from microbes is functional in plants has not been investigated. Recently, enzymes involved in converting trp to IPA have been isolated in Arabidopsis (see below). The microbe system may be useful for identifying the other genes involved in the IPA pathway in plants. For example, screening of plant cDNA libraries for genes that are able to complement the IPA decarboxylase function in microbes may help define the next step in the IPA pathway in Arabidopsis.
Early molecular genetics studies on auxin biosynthesis
Early physiological studies on auxin biosynthesis were reviewed comprehensively (3). Physiological and stable isotope labeling studies established that trp is a precursor for de novo IAA biosynthesis in plants. In addition, all the defined auxin biosynthesis pathways in microbes so far have been trp dependant. Therefore, it was logical that early molecular genetics studies on auxin biosynthesis were centered on analyzing trp deficient mutants. Surprisingly, there were no differences in free IAA levels between wild type and trp mutants (42, 69). In fact, the trp auxotroph mutants actually produced more IAA-conjugates (42, 69). Consistent with the IAA measurement results, the Arabidopsis trp mutants used for the IAA analysis experiments did not show developmental defects as dramatic as those observed in some known auxin mutants including pin1 (24) and monopteros (48). Further feeding experiments with [15N]anthranilate and [2H5]tryptophan led to the hypothesis that IAA is mainly produced through a trp independent pathway in Arabidopsis and in Maize (42, 69). Although the early studies on trp mutants were informative, they did not identify the genes responsible for auxin biosynthesis in plants. Furthermore, interpretation of the experiments with trp mutants is not straightforward. It is difficult to determine whether the growth defects in the trp mutants are caused by problems in the synthesis of proteins, auxin, other trp metabolites, or a combination of several processes. The trp mutants used in early auxin research were not true trp deficient mutants because they still made some trp. The residual trp synthesis activity might complicate the interpretations of analytic biochemistry experiments. The flux of trp to different metabolic pathways may also be changed in a trp mutant.
Auxin biosynthesis pathways defined by Arabidopsis genetic studies
There have been no reported forward genetic screens conducted systematically for the purpose of isolating auxin deficient mutants in any systems. Part of the reason is the lack of knowledge of the developmental consequences associated with auxin deficiency. Therefore, no robust auxin deficient trait was known for genetic screens. Another difficulty is caused by genetic redundancy in auxin biosynthesis. Auxin can be synthesized from a trp independent pathway as well as from several trp dependent pathways. The recently identified auxin biosynthesis genes all belong to gene families, which explains why no auxin deficient mutants came up from many screens for developmental defects in Arabidopsis. Some trp biosynthesis mutants including anthranilate synthase were isolated from screens for weak ethylene resistance mutants and from the methyl jasmonate insensitive mutant screens (55, 59). The hormone-resistant phenotypes of anthranilate synthase mutants (asa1/wei2, and asb1/wei7) were attributed to defects in auxin production. Recently, a rice tryptophan deficient dwarf mutant, tdd1 that encodes the β subunit of anthranilate synthase, was found to have a reduced level of IAA and strong defects in floral and embryonic development (51). The identified anthranilate synthase mutants demonstrated that auxin plays a critical role in plant development, but did not solve the mechanisms of converting trp to IAA.
Identification of the first key trp dependent auxin biosynthesis genes was originated from the characterization of several auxin overproduction mutants in Arabidopsis. Analysis of the auxin overproduction mutants has led to the identification of two trp-dependent IAA biosynthesis routes (Figure 1). Recent characterization of mutants that are defective in shade avoidance and ethylene responses, has identified an aminotransferase important for the production of indole-3-pyruvate (IPA) (Figure 1).
The IAOx and glucosinolate pathway
This pathway was defined by three auxin overproduction mutants, superroot1 (sur1) (5), superroot2 (sur2) (2, 16), and CYP79B2 overexpression lines (29, 74). CYP79B2 and its close homolog CYP79B3 convert trp to IAOx (Figure 1) while SUR1 and SUR2 are involved in converting IAOx to indolic glucosinolates.
The mutant sur1 is the first identified auxin overproduction mutant in Arabidopsis that displayed dramatic developmental defects (5). Light-grown sur1 seedlings have long hypocotyls and epinastic cotyledons while dark-grown sur1 seedlings have short hypocotyls and lack an apical hook. In addition, sur1 produces massive adventitious roots from hypocotyls. SUR1 encodes a C-S lyase that catalyzes the conversion of S-alkylthiohydroximate to thiohydroximic acid, a key reaction in indolic glucosinolates biosynthesis (39) (Figure 1). Inactivation of SUR1 disrupts glucosinolate biosynthesis and likely leads to the accumulation of upstream intermediates including IAOx (Figure 1). Given the fact that sur1 is recessive and a loss-of-function allele, the auxin overproduction phenotypes of sur1 are likely caused by funneling excess IAOx into IAA biosynthesis.
Like sur1, sur2 also produces many adventitious roots from hypocotyls (16). Overall, sur2 is phenotypically very similar to sur1. SUR2 encodes the cytochrome P450 CYP83B1, an enzyme that synthesizes 1-aci-nitro-2-indolyl-ethane from IAOx (2) (Figure 1). SUR2 defines the first step in making indolic glucosinolates from IAOx (Figure 1). Loss-of-function sur2/cyp83B1 mutants block the production of glucosinolates from IAOx, leading to an increased IAOx flux for IAA biosynthesis.
Unlike sur1 and sur2, which are recessive and loss-of-function, inactivation of CYP79B2 does not cause auxin overproduction. Instead, it is the overexpression of CYP79B2 in Arabidopsis that leads to auxin overproduction (74). CYP79B2 was isolated as an enzyme that metabolizes trp from a screen for Arabidopsis cDNAs that can confer resistance to 5-methyl trp in yeast (29). CYP79B2 catalyzes the conversion of trp to IAOx in vitro (29) (Figure 1). Overexpression of CYP79B2 in Arabidopsis probably leads to an overproduction of IAOx, thus increasing the flux of IAOx to IAA biosynthesis.
Further evidence for the IAOx pathway comes from the observations that the cyp79b2 cyp79b3 double loss-of-function mutants show measurably lower levels of free IAA than wild type and display phenotypes consistent with lower levels of auxin at high temperatures (74). Another piece of evidence supporting the IAOx pathway is that asa1 wei2 and asb1/wei7 mutants, which are defective in trp biosynthesis, suppress the phenotypes of sur1 and sur2 (55), presumably by decreasing the flux from trp to IAOx, and thereby restoring IAOx to normal levels in sur1 and sur2.
Production of IAOx from CYP79B2/B3 is probably not the main IAA biosynthesis pathway in plants for several reasons. First, IAOx was not detected in monocots such as rice and maize (58). Second, there are no apparent orthologs of CYP79B2 and CYP79B3 in rice and maize while the Arabidopsis cyp79b2 cyp79b3 have no detectable levels of IAOx (58). Third, the phenotypes of cyp79b2 cyp79b3 double mutants were very subtle compared to known auxin signaling or transport mutants.
It is clear that IAOx can be converted to IAA in Arabidopsis on the basis of the observed phenotypes of sur1, sur2, and CYP79B2 overexpression lines, but the exact biochemical mechanisms that convert IAOx to IAA have not been worked out. In theory, IAOx can be used to make indole-3-acetonitrile (IAN) and indole-3-acetaldehye, which can be further converted to IAA by nitrilases (43) and aldehyde oxidases (15, 52), respectively. Recent biochemical analysis of cyp79b2 cyp79b2 double mutants suggests that IAM is probably also an important intermediate in converting IAOx to IAA, but the genes and enzymes for making IAM from IAOx are not known (58).
The YUC pathway
The Arabidopsis mutant yucca (renamed as yuc1D) was isolated from an activation-tagging screen for long hypocotyl mutants defective in light signaling (73). But it was soon realized that it was more likely that yuc1-D altered hormone homeostasis than light signaling because it had long hypocotyls in all light wavelengths. The phenotypes of yuc1D are very similar to those of known auxin overproduction mutants. Additional evidence further demonstrates that yuc1D is indeed an auxin overproduction mutant. Weak alleles of yuc1D contains 50% more free IAA than wild type and explants of yuc1D mutants produces massive roots in the absence of any exogenous plant hormones (73). Known auxin-inducible genes are up regulated in yuc1D. Overexpression of iaaL, which conjugates free IAA to the amino acid lysine, partially suppresses yuc1D. Furthermore yuc1D is resistant to toxic trp analogs such as 5-methyl trp, suggesting that yuc1D overproduces auxin through a trp-dependent pathway (73).
The auxin overproduction phenotypes of yuc1D are caused by overexpression of a flavin monooxygenase-like (FMO) enzyme (73). Because yuc1D is dominant and gain-of-function, it is likely that YUC1 catalyzes a rate-limiting step in auxin biosynthesis. In vitro assay indicates that YUC1 is capable of catalyzing the conversion of tryptamine into N-hydroxyl tryptamine, which can proceed to IAA through IAOx or other intermediates (Figure 1).
YUC1 belongs to a family with 11 members in Arabidopsis and a subset of the members in the family may have overlapping functions (8). It is known that overexpression of other members of the YUC family in Arabidopsis also leads to similar auxin overproduction phenotypes (8, 31, 38, 68), indicating that the YUC-like genes can participate in similar reactions. As expected, single loss-of-function mutants of any YUC genes do not display any obvious developmental defects. However, some combinations of yuc mutants in which two or more YUC genes are simultaneously inactivated display dramatic developmental defects (8, 9). For example, yuc1 yuc4 double mutants produce abnormal flowers that do not have any functional male or female floral organs. The yuc1 yuc4 double mutants also make fewer vascular strains in leaves and flowers (8). Overall, the YUC genes appear to have both overlapping and unique functions during plant development. Analysis of different yuc mutant combinations clearly demonstrates that YUC genes are essential for embryogenesis, seedling growth, leaf and flower initiation, and vascular formation.
The yuc mutants are the first identified Arabidopsis mutants that are partially auxin deficient with dramatic developmental defects. In many ways, the developmental defects of yuc mutant combinations are very similar to those of known auxin signaling or transport mutants. For example, yuc1 yuc4 yuc10 yuc11 quadruple mutants fail to make the basal part of the embryo during embryogenesis (9), a phenotype also observed in monopteros / arf5 (27), tir1 afb1 afb2 afb3 (17), and pin quadruple mutants (21). Inactivation of YUC genes leads to down regulation of the auxin reporter DR5-GUS. Loss-of-function yuc mutants display synergistic interactions with known auxin mutants such as pin1 and pid (9). More importantly, the developmental defects of yuc1 yuc4 are rescued by expression of the bacterial auxin biosynthetic gene iaaM under the control of the YUC1 promoter, which presumably produces auxin in situ (8).
Unlike the CYP79B2/B3 genes that have so far only been detected in Brassica, the YUC genes appear to have a much broader existence. Genes homologous to YUC1 have been identified in all of the plant genomes with available sequence data including moss and rice. Some YUC genes in petunia (61), rice (66, 71), corn (23), and tomato (19) have been functionally characterized and they play similar roles in auxin biosynthesis and plant development. Therefore, the YUC pathway appears to be highly conserved throughout the plant kingdom.
Genetically, it is unambiguous that YUC genes are key auxin biosynthesis genes. Overexpression of YUC genes leads to auxin overproduction while loss-of-function yuc mutants display developmental defects that are rescued by in situ auxin production. However, the detailed biochemical mechanisms of YUC flavin monooxygenases are still not solved. It is clear that YUC flavin monooxygenases catalyze a rate-limiting step in a trp dependent auxin biosynthesis pathway. YUC was tentatively placed to the step from tryptamine to N-hydroxyl tryptamine on basis of the in vitro biochemical studies (19, 31, 73). Because flavin monooxygenases are known to have broad substrate specificities in vitro (75), further investigations are needed to determine whether tryptamine is the in vivo substrate for YUCs.
The IPA pathway
Indole-3-pyruvate has long been suggested as an intermediate for IAA biosynthesis (Figure 1). However, only recently has the role of IPA in plant auxin biosynthesis and development been revealed (56, 60). Three independent genetic studies have identified an Arabidopsis aminotransferase that can convert Trp to IPA in vitro. Tao et al. initiated a genetic screen for Arabidopsis mutants defective in shade avoidance response, a process in which plants adapt to light quality changes by elongating stems and petioles (60). The shade avoidance (sav) mutant sav3 fails to elongate after being transferred to simulated shade conditions. SAV3 encodes a protein homologous to aminotransferases and later is shown to catalyze the production of IPA from trp; thus SAV3 is renamed as TAA1 (TRYPTOPHAN AMINOTRANSFERASE of ARABIDOPSIS). Several pieces of evidence indicate that TAA1 is involved in auxin biosynthesis. The taa1 mutants contain 60% less free IAA and show a decreased IAA synthesis rate when transferred to shade conditions (60). Some known auxin inducible genes are down regulated in taa1 plants grown in shade conditions (60). Furthermore taa1 is partially rescued by a synthetic auxin picloram and taa1 is hypersensitive to the toxic trp analog 5-methyl tryptophan (60).
TAA1 was also isolated from a genetic screen for weak ethylene insensitive mutants (wei mutants) in Arabidopsis (56). The taa1 / wei8 mutants have elongated roots in the presences of ACC, an ethylene biosynthesis precursor, while wild type root elongation is inhibited under the same conditions. The wei8 mutants show altered expression of the auxin reporter DR5-GUS, decreased level of free IAA, and partial rescue of the ethylene defects with exogenous IAA. More importantly, simultaneously inactivating TAA1 and two of its close homologs (TAR1 and TAR2) leads to developmental defects similar to those in monopteros and pinoid, two well-known auxin mutants.
TAA1 was also identified as tir2 (70), a mutant resistant to auxin transport inhibitor NPA that inhibits the elongation of Arabidopsis roots. The tir2 mutant has short hypocotyl, a phenotype that can be rescued by IPA and IAA, indicating that TIR2 may be involved in the IPA auxin biosynthetic pathway. Furthermore, TIR2 is required for temperature dependent hypocotyl elongation in Arabidopsis. Taken together, the TAA1 and its close homologs play important roles in auxin biosynthesis and plant development.
TAA1 is a PLP-dependent enzyme and appears to have a wide distribution throughout the plant kingdom, suggesting that the IPA pathway is also highly conserved. Functional characterization of TAA1-like genes in other species has not been reported. The step catalyzed by TAA1 may not be a rate-limiting step in auxin biosynthesis because overexpression of TAA1 under the control of the 35S promoter does not cause auxin overproduction phenotypes (56, 60). Alternatively, TAA1 may not be regulated at transcriptional level.
The relationships among the proposed auxin biosynthesis pathways
The intermediate IAOx was proposed as a shared intermediate among the CYP79B2 /B3, YUC, and glucosinolate biosynthesis pathways (74) (Figure 1). Recent biochemical analysis demonstrated that the bulk of IAOx is produced from the CYP79B2/B3 pathway in Arabidopsis (58). A caveat of this study is that the production of IAOx and conversion of IAOx to IAA may be coupled while this may not be the case for IAOx to glucosinolate biosynthesis. In addition, YUC genes are mainly involved in local auxin biosynthesis (see below) and the analysis of bulk IAOx may not be indicative.
The relationship between the YUC pathway and the IPA pathway is even more intriguing. Inactivation of either YUC genes or multiple TAA1 genes leads to similar phenotypes. For example, yuc1 yuc4 yuc10 yuc11 quadruple mutants do not make hypocotyls and roots, a phenotype that is also displayed in taa1 tar1 tar2 triple mutants (9, 56). The phenotypic similarities between yuc and taa mutants suggest that the two gene families may participate in the same pathway. One possibility is that YUC genes act downstream of IPA, but it is not obvious how a flavin-containing monooxygenase, which catalyzes the hydroxylation of hetero-atoms in organic compounds (75), may be involved in reactions downstream of IPA that do not contain any hetero-atoms such as nitrogen and sulfur. Alternatively, TAA1 could catalyze the reaction of converting IPA to trp, a reaction that may be important for intracellular trp transport and thus feed the YUC pathway. It is known that a bacterial trp aminotransferase, which reversibly catalyzes the transamination of trp to yield IPA, has a 138-fold lower Km value for IPA than for trp (34). Silico-docking experiments with Arabidopsis TAA1 crystal structure have also shown that IPA scored better than trp (60), demonstrating that IPA could be a better substrate than trp for TAA1.
The YUC pathway and the IPA pathway could simply be two independent pathways as currently suggested. Some developmental processes such as vascular development may just need a threshold level of auxin (9). Disruption of either pathway could potentially lower the IAA levels below the threshold to cause similar phenotypes. Apparently, the two pathways are not redundant in other developmental processes such as shade avoidance (60). Further experiments are needed to address the relationship between the two pathways. Analysis of the various potential intermediates including IPA, IAN, IAM, and IAOx in various mutant backgrounds and feeding conditions will be essential. Conducting genetic interaction studies among various auxin mutants will be helpful as well.
Auxin biosynthesis is local and non-cell autonomous
It is surprising to find that de novo auxin production is highly localized and that local auxin biosynthesis plays a key role in shaping local auxin gradients (8, 9, 56, 60). The predominant view in the auxin field has been that polar auxin transport is responsible for generating auxin gradients and auxin maxima, which are known to be essential for proper plant development. The location of auxin biosynthesis has been regarded as unimportant, and in some cases irrelevant (25). Mathematical modeling of auxin-regulated developmental processes including root development (25), vascular formation (18, 20), and phyllotaxis (4) are based exclusively on the analyses of auxin transport. However, recent findings demonstrate that auxin biosynthesis is regulated both temporally and spatially. The expression of both YUC genes and TAA1 genes is restricted to a small group of discrete cells. For example, during embryogenesis both YUC1 and TAA1 are initially expressed in the apical region at the globular stage, then gradually are concentrated at the apical meristem at the heart stage, and finally are restricted only to the apical meristem in the mature embryo (9, 56, 60).
The location of auxin biosynthesis appears to be an important aspect of overall regulation of auxin functions. Different combinations of yuc mutants have different phenotypes, which often correlate with the YUC gene expression patterns. The yuc1 yuc4 double mutants have a decreased expression of DR5-GUS in the cells where YUC1/4 are expressed, but DR5-GUS activities in non-YUC1/4 expressing cells are not affected, suggesting that auxin peaks are mainly generated locally (9). TAA1 and its homolog TAR2 are expressed in the root tips in Arabidopsis and the DR5-GUS expression is dramatically reduced in the root tip of taa1 tar2 double mutants (56). Local auxin biosynthesis was also recently shown to modulate gradient-directed planar polarity in root hair development in Arabidopsis (30).
For a long time, the shoot was believed to be the only source of auxin biosynthesis. It was also thought that the other parts of a plant were dependent on polar auxin transport to supply auxin. Now it is clear that both shoot and root can produce auxin (9, 46, 56). TAA1 is expressed in both shoot and root (56). Inactivation of TAA1 and its homologs affect both root and shoot development. YUC genes are also expressed in all organs including flowers, leaves, and roots in Arabidopsis. Each organ appears to be self-sufficient in terms of controlling auxin gradients for development. For example, auxin produced in other floral organs cannot compensate for the effects of inactivation of YUC2 and YUC6 in stamens (8). Within an organ, YUC genes appear to be non-cell autonomous (61), which means auxin synthesized by YUCs in one cell is certainly used by other cells.
The finding that auxin biosynthesis is local and the YUC genes are non-cell autonomous raises interesting questions regarding the relationship between biosynthesis and polar transport and the relative contributions of the two processes to local auxin gradient generation and maintenance. These questions are difficult to address because of the existence of various regulatory loops. For example, auxin is known to regulate PIN genes at transcriptional level and to affect intracellular vesicle traffic that is important for proper targeting of PIN proteins (44, 65). However, genetic studies of interactions between yuc mutants and auxin transport mutants have shed some light on this important question. The transport mutant pin1 showed synergistic interactions with yuc1 yuc4 double mutants (9). The triple mutants completely abolished the formation of true leaves, a phenotype not displayed in either pin1 alone or in the yuc1 yuc4 double mutants, demonstrating a complex relationship between auxin biosynthesis and transport.
Regulation of auxin biosynthesis
Auxin biosynthesis is regulated by both environmental and developmental signals. For example, when plants are transferred from normal growth conditions to shade conditions, auxin levels and biosynthesis rates are up regulated (60). With the identification of several key auxin biosynthetic genes, it is also feasible to monitor auxin biosynthesis by analyzing gene expression changes in response to various signals or in different mutant backgrounds. There are several reports that document the regulation of YUC gene expression during plant development. A recent report identified a family of transcription factors (SHI, SHORT INTENOTES) that regulate the expression of YUC4 (54). STY1 (STYLISH1), one of the SHI genes, was initially isolated from genetic screens for Arabidopsis mutants defective in style development (54). Inactivation of STY1 leads to abnormal style morphology and vascular patterning. When the expression of STY1 is induced, YUC4 mRNA levels increase and auxin levels also increase accordingly (54). It is still not clear whether STY1 binds directly to the YUC4 promoter.
Another family of transcription factors called NGATHAs (NGA) was recently found to participate in regulating style development as well (1, 63). NGA genes act redundantly to control style development in a dosage-dependent manner and the quadruple nga mutant completely abolishes the formation of style and stigma tissues. The nga mutant phenotypes were partially attributed to the failure of activating two YUC genes (YUC2 and YUC4) in the apical domain of Arabidopsis gynoecium (63). LEAFY COTYLEDON2 (LEC2), a central regulator of embryogenesis, is also found to activate the auxin biosynthetic genes YUC2 and YUC4 (57). Moreover, LEC2 binds to the YUC4 promoter as revealed by ChIP experiments, suggesting that YUC4 may be a direct target of LEC2 (57). Unlike LEC2, NGA, and SHI that are positive regulators of YUC expression, SPOROCYTELESS (SPL) appears to be a negative regulator (37). Overexpression of SPL led to a significant repression of the expression of YUC2 and YUC6. The TAA genes display unique expression patterns, but the transcription factors responsible for TAA expression have not been identified.
From auxin biosynthesis to developmental mechanisms
Recent progress in auxin biosynthesis opens a new line of research in mechanisms of how auxin controls plant development. Understanding auxin biosynthesis provides tools to manipulate auxin levels in plants with temporal and spatial precisions. The available auxin biosynthetic mutants also provide sensitized backgrounds for genetically isolating key components that are responsible for auxin-mediated development.
Auxin biosynthesis regulates female gametophyte development in Arabidopsis
The female gametophyte in Arabidopsis is a seven-cell structure with four different cell types including an egg cell, a central cell, two synergid cells, and three antipodal cells (45). Fertilization of the egg cell is the first step in embryogenesis while fertilization of the central cell directs the development of the endosperm. Development of the female gametophyte in Arabidopsis consists of the specification of cell types and the formation of a particular pattern that dictates the relative positions of the cells within the embryo sac. The underlying mechanisms of female gametophyte development have remained elusive until recently when auxin gradients were discovered in the developing embryo sac (45). Normal auxin distribution and signaling are required for the proper pattern formation in the embryo sac and for the specification of gametic and non-gametic cell identities (45). The auxin reporter DR5-GFP is asymmetrically distributed with the maxima located at the micropylar pole of the developing embryo sac (45). The observed auxin reporter gradient is surprisingly not generated nor maintained by the efflux-dependent auxin transport because the PIN genes are not expressed during the stages of gametophyte development (45). The identification of YUC genes as key auxin biosynthesis enzymes made it feasible to investigate whether the auxin gradient in the embryo sac is correlated with local auxin production. Indeed, the expression of both YUC1 and YUC2 genes localizes to the micropylar pole of the gametophyte (45), where the DR5-GFP maxima were observed. Furthermore, mis-expression of YUC1 in the embryo sac induces cell identity switch between gametic and non-gametic cells, providing strong evidence that auxin is a morphogenic signal (45).
Understanding of auxin biosynthesis provides new opportunities to genetically dissect developmental mechanisms
Analyses of Arabidopsis mutants defective in auxin signaling or transport are instrumental in elucidating the molecular mechanisms of auxin action in plant growth and development (50, 64). Previous genetic screens for auxin signaling mutants in Arabidopsis have taken advantage of the observation that primary root elongation is greatly inhibited in the presence of exogenous auxin (35). Mutants defective in auxin uptake and signaling are less sensitive to exogenous auxin and thus have longer primary roots than wild type on auxin-containing media. Because the previous screens relied on the development of roots, some of the key auxin genes may have been missed if the genes were not expressed in the root, or were essential for root development. For example, mp and bdl, two key components of auxin signaling in specifying root meristem during embryogenesis, would not be isolated from a root-based auxin-resistant screen (26, 27). Some other known auxin mutants including pin1 (24) and pid (12) mainly affect shoot development and do not display auxin-resistance in a root elongation assay.
Recent progress in auxin biosynthesis makes it possible to isolate mutants that can overcome or become oversensitive to partial auxin deficiency. Such screens offer an exciting opportunity to identify new components responsible for auxin action in plant development. Perhaps due to complications from polar auxin transport, exogenous IAA treatment and auxin overproduction differ phenotypically. All known auxin overproduction mutants have long hypocotyls while the main phenotypic readout of IAA treatment is the inhibition of primary root elongation. Furthermore, the developmental defects of yuc mutants cannot be rescued by exogenous auxin treatments, but are instead rescued by producing IAA from the iaaM gene under the control of a corresponding YUC promoter (8). Therefore, the auxin biosynthetic mutants provide a different angle for genetically dissecting auxin action in plant development.
The successful identification of the yuc1 yuc4 enhancer, npy1 (naked pins in yuc mutants 1), clearly demonstrates the power of using auxin biosynthesis mutants as starting materials for isolating new auxin components (10). The yuc1 yuc4 double mutants produce abnormal flowers. When the NPY1 gene is inactivated in the yuc1 yuc4 background, the resulting yuc1 yuc4 npy1 triple mutants develop pin-like inflorescences, a phenotype that is also observed in known auxin mutants pin1, pid, and mp (10). Formation of pin-like inflorescences is a hallmark of defective auxin related processes. The npy1 mutant is allelic to enp1 / mab4 that was isolated as an enhancer of pid (22, 62). The npy1/enp1and pid double mutants do not make cotyledons, indicating that NPY1 / ENP1 / MAB4 plays a general role in organogenesis. Furthermore, the yuc1 yuc4 pid triple mutants phenocopy npy1 pid double mutants. Genetic analyses have put YUC, PID, and NPY1 in a linear developmental pathway.
NPY1 is a novel protein, but shares significant homology with NON-PHOTOTROPIC HYPOCOTYL 3 (NPH3) (41). Both NPY1 and NPH3 belong to a plant-specific superfamily with 32 members in the Arabidopsis genome (11). The mechanisms of auxin-mediated organogenesis, particularly flower formation, appear to be analogous to those of phototropic responses. NPY1 is homologous to NPH3 and inactivation of NPY1 and its close homologs NPY3 and NPY5 leads to pin-like inflorescences (10, 11). Inactivation of PID, which is a ser / thr kinase and is homologous to the photoreceptor PHOT1, causes phenotypes similar to npy1 npy3 npy5 trip mutants. Another similarity between these two pathways is that both require an auxin response factor ARF5/MONOPTEROS. Disruption of ARF5 causes the formation of pin-like inflorescences while inactivation of ARF7 abolishes phototropic responses (28). Identification of NPY1 in a genetic screen for yuc1 yuc4 enhancers demonstrates that genetic screens for modifiers of auxin biosynthesis mutants will probably lead to the discoveries of additional components in the pathway.
Conclusions
Tremendous progress has been made in understanding auxin biosynthesis in plants in the past few years. Several key auxin biosynthetic genes and their roles in plant development have been discovered. It is clear that auxin biosynthesis takes place locally in response to both environmental and developmental signals. De novo auxin biosynthesis plays an essential role in many developmental processes. However, there are still many outstanding questions regarding auxin biosynthesis and its role in plant growth and development. The current understanding of auxin biosynthesis is still fragmented and no complete de novo biosynthesis pathway has been defined. It will likely take a combination of genetic studies and biochemical analyses of auxin biosynthetic intermediates with isotope labeling experiments to fill out the gaps in auxin production in plants. It is also not clear how the expression patterns of auxin biosynthesis genes are generated and how they are regulated.
To understand the molecular mechanisms by which auxin regulates plant development, integration of findings from auxin biosynthesis, conjugation, transport and signaling will be necessary. Results from auxin biosynthesis probably should be incorporated in various mathematical models of plant development. Recent progress in auxin biosynthesis makes it practical to alter auxin levels with temporal and spatial precision, providing exciting tools to tackle complex questions regarding the mechanisms of how auxin controls plant development.
Summary Points.
Auxin biosynthesis in plants is complex and several pathways contribute to de novo IAA production.
De novo auxin biosynthesis plays essential roles in virtually every aspect of plant development
Auxin biosynthesis is temporally and spatially regulated. Local auxin biosynthesis contributes to the generation and maintenance of local auxin gradients.
Understanding of auxin biosynthesis provides new tools for solving difficult plant development questions.
Future Issues.
There is still no complete de novo auxin biosynthesis pathway in plants. Enzymes that function in the immediate upstream and downstream steps of the YUCs and TAAs need to be identified.
The relationship among the proposed tryptophan dependent auxin biosynthesis pathways and the relative contribution of each pathway to overall IAA production need to be determined
Identification of the transcription factors that bind to the promoters of the key auxin biosynthesis genes will help to understand how auxin biosynthesis is regulated by developmental and environmental signals.
Identification of the transcription factors that bind to the promoters of the key auxin biosynthesis genes will help to understand how auxin biosynthesis is regulated by developmental and environmental signals.
Acknowledgments
I would like to thank Jose Alonso, Allison Zhao, and members of the Zhao lab for critical reading of this manuscript. Work in my lab is supported by the NIH (R01GM068631) and the NSF (Plant Genome DBI-0820729).
References
- 1.Alvarez JP, Goldshmidt A, Efroni I, Bowman JL, Eshed Y. The NGATHA Distal Organ Development Genes Are Essential for Style Specification in Arabidopsis. Plant Cell. 2009 doi: 10.1105/tpc.109.065482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barlier I, Kowalczyk M, Marchant A, Ljung K, Bhalerao R, et al. The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc Natl Acad Sci U S A. 2000;97:14819–24. doi: 10.1073/pnas.260502697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel B. Auxin Biosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:51–66. doi: 10.1146/annurev.arplant.48.1.51. [DOI] [PubMed] [Google Scholar]
- 4.Bayer EM, Smith RS, Mandel T, Nakayama N, Sauer M, et al. Integration of transport-based models for phyllotaxis and midvein formation. Genes Dev. 2009;23:373–84. doi: 10.1101/gad.497009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, et al. Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell. 1995;7:1405–19. doi: 10.1105/tpc.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonner J, Bandurski RS. Studies of the Physiology, Pharmacology, and Biochemistry of the Auxins. Annual Review of Plant Physiology. 1952;3:59–86. [Google Scholar]
- 7.Camilleri C, Jouanin L. The TR-DNA region carrying the auxin synthesis genes of the Agrobacterium rhizogenes agropine-type plasmid pRiA4: nucleotide sequence analysis and introduction into tobacco plants. Mol Plant Microbe Interact. 1991;4:155–62. doi: 10.1094/mpmi-4-155. [DOI] [PubMed] [Google Scholar]
- 8.Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20:1790–9. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Y, Dai X, Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell. 2007;19:2430–9. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng Y, Qin G, Dai X, Zhao Y. NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc Natl Acad Sci U S A. 2007;104:18825–9. doi: 10.1073/pnas.0708506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng Y, Qin G, Dai X, Zhao Y. NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc Natl Acad Sci U S A. 2008;105:21017–22. doi: 10.1073/pnas.0809761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen SK, Dagenais N, Chory J, Weigel D. Regulation of auxin response by the protein kinase PINOID. Cell. 2000;100:469–78. doi: 10.1016/s0092-8674(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 13.Comai L, Kosuge T. Cloning characterization of iaaM, a virulence determinant of Pseudomonas savastanoi. J Bacteriol. 1982;149:40–6. doi: 10.1128/jb.149.1.40-46.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costacurta A, Keijers V, Vanderleyden J. Molecular cloning and sequence analysis of an Azospirillum brasilense indole-3-pyruvate decarboxylase gene. Mol Gen Genet. 1994;243:463–72. doi: 10.1007/BF00280477. [DOI] [PubMed] [Google Scholar]
- 15.Dai X, Hayashi K, Nozaki H, Cheng Y, Zhao Y. Genetic and chemical analyses of the action mechanisms of sirtinol in Arabidopsis. Proc Natl Acad Sci U S A. 2005;102:3129–34. doi: 10.1073/pnas.0500185102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delarue M, Prinsen E, Onckelen HV, Caboche M, Bellini C. Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 1998;14:603–11. doi: 10.1046/j.1365-313x.1998.00163.x. [DOI] [PubMed] [Google Scholar]
- 17.Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9:109–19. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 18.Dimitrov P, Zucker SW. A constant production hypothesis guides leaf venation patterning. Proc Natl Acad Sci U S A. 2006;103:9363–8. doi: 10.1073/pnas.0603559103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Expósito-Rodríguez M, Borges AA, Borges-Pérez AB, Hernández M, Pérez JA. Cloning and Biochemical Characterization of ToFZY, a Tomato Gene Encoding a Flavin Monooxygenase Involved in a Tryptophan-dependent Auxin Biosynthesis Pathway. Journal of Plant Growth Regulation. 2007;26:329–40. [Google Scholar]
- 20.Feugier FG, Iwasa Y. How canalization can make loops: a new model of reticulated leaf vascular pattern formation. J Theor Biol. 2006;243:235–44. doi: 10.1016/j.jtbi.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 21.Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, et al. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426:147–53. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 22.Furutani M, Kajiwara T, Kato T, Treml BS, Stockum C, et al. The gene MACCHI-BOU 4/ENHANCER OF PINOID encodes a NPH3-like protein and reveals similarities between organogenesis and phototropism at the molecular level. Development. 2007;134:3849–59. doi: 10.1242/dev.009654. [DOI] [PubMed] [Google Scholar]
- 23.Gallavotti A, Barazesh S, Malcomber S, Hall D, Jackson D, et al. sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc Natl Acad Sci U S A. 2008;105:15196–201. doi: 10.1073/pnas.0805596105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galweiler L, Guan C, Muller A, Wisman E, Mendgen K, et al. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282:2226–30. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- 25.Grieneisen VA, Xu J, Maree AF, Hogeweg P, Scheres B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature. 2007;449:1008–13. doi: 10.1038/nature06215. [DOI] [PubMed] [Google Scholar]
- 26.Hamann T, Benkova E, Baurle I, Kientz M, Jurgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002;16:1610–5. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–11. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, et al. The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell. 2000;12:757–70. doi: 10.1105/tpc.12.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hull AK, Vij R, Celenza JL. Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci U S A. 2000;97:2379–84. doi: 10.1073/pnas.040569997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikeda Y, Men S, Fischer U, Stepanova AN, Alonso JM, et al. Local auxin biosynthesis modulates gradient-directed planar polarity in Arabidopsis. Nat Cell Biol. 2009;11:731–8. doi: 10.1038/ncb1879. [DOI] [PubMed] [Google Scholar]
- 31.Kim JI, Sharkhuu A, Jin JB, Li P, Jeong JC, et al. yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol. 2007;145:722–35. doi: 10.1104/pp.107.104935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klee H, Horsch RB, Hinchee MB, Hein MB, Hoffmann NL. The effects of overproduction of two Agrobacterium tumefaciens T-DNA auxin biosynthetic gene products in transgenic petunia plants. Genes Dev. 1987;1:86–96. [Google Scholar]
- 33.Koga J, Adachi T, Hidaka H. Purification and characterization of indolepyruvate decarboxylase. A novel enzyme for indole-3-acetic acid biosynthesis in Enterobacter cloacae. J Biol Chem. 1992;267:15823–8. [PubMed] [Google Scholar]
- 34.Koga J, Syono K, Ichikawa T, Adachi T. Involvement of L-tryptophan aminotransferase in indole-3-acetic acid biosynthesis in Enterobacter cloacae. Biochim Biophys Acta. 1994;1209:241–7. doi: 10.1016/0167-4838(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 35.Leyser O. Molecular genetics of auxin signaling. Annu Rev Plant Biol. 2002;53:377–98. doi: 10.1146/annurev.arplant.53.100301.135227. [DOI] [PubMed] [Google Scholar]
- 36.Li L, Hou X, Tsuge T, Ding M, Aoyama T, et al. The possible action mechanisms of indole-3-acetic acid methyl ester in Arabidopsis. Plant Cell Rep. 2008;27:575–84. doi: 10.1007/s00299-007-0458-9. [DOI] [PubMed] [Google Scholar]
- 37.Li LC, Qin GJ, Tsuge T, Hou XH, Ding MY, et al. SPOROCYTELESS modulates YUCCA expression to regulate the development of lateral organs in Arabidopsis. New Phytol. 2008;179:751–64. doi: 10.1111/j.1469-8137.2008.02514.x. [DOI] [PubMed] [Google Scholar]
- 38.Marsch-Martinez N, Greco R, Van Arkel G, Herrera-Estrella L, Pereira A. Activation tagging using the En-I maize transposon system in Arabidopsis. Plant Physiol. 2002;129:1544–56. doi: 10.1104/pp.003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mikkelsen MD, Naur P, Halkier BA. Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 2004;37:770–7. doi: 10.1111/j.1365-313x.2004.02002.x. [DOI] [PubMed] [Google Scholar]
- 40.Mockaitis K, Estelle M. Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol. 2008;24:55–80. doi: 10.1146/annurev.cellbio.23.090506.123214. [DOI] [PubMed] [Google Scholar]
- 41.Motchoulski A, Liscum E. Arabidopsis NPH3: A NPH1 photoreceptor-interacting protein essential for phototropism. Science. 1999;286:961–4. doi: 10.1126/science.286.5441.961. [DOI] [PubMed] [Google Scholar]
- 42.Normanly J, Cohen JD, Fink GR. Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc Natl Acad Sci U S A. 1993;90:10355–9. doi: 10.1073/pnas.90.21.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Normanly J, Grisafi P, Fink GR, Bartel B. Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell. 1997;9:1781–90. doi: 10.1105/tpc.9.10.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paciorek T, Zazimalova E, Ruthardt N, Petrasek J, Stierhof YD, et al. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature. 2005;435:1251–6. doi: 10.1038/nature03633. [DOI] [PubMed] [Google Scholar]
- 45.Pagnussat GC, Alandete-Saez M, Bowman JL, Sundaresan V. Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science. 2009;324:1684–9. doi: 10.1126/science.1167324. [DOI] [PubMed] [Google Scholar]
- 46.Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, et al. An Auxin Gradient and Maximum in the Arabidopsis Root Apex Shown by High-Resolution Cell-Specific Analysis of IAA Distribution and Synthesis. Plant Cell. 2009 doi: 10.1105/tpc.109.066480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pollmann S, Duchting P, Weiler EW. Tryptophan-dependent indole-3- acetic acid biosynthesis by 'IAA-synthase' proceeds via indole-3-acetamide. Phytochemistry. 2009;70:523–31. doi: 10.1016/j.phytochem.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 48.Przemeck GK, Mattsson J, Hardtke CS, Sung ZR, Berleth T. Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta. 1996;200:229–37. doi: 10.1007/BF00208313. [DOI] [PubMed] [Google Scholar]
- 49.Romano CP, Robson PR, Smith H, Estelle M, Klee H. Transgene-mediated auxin overproduction in Arabidopsis: hypocotyl elongation phenotype and interactions with the hy6–1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Mol Biol. 1995;27:1071–83. doi: 10.1007/BF00020881. [DOI] [PubMed] [Google Scholar]
- 50.Santner A, Estelle M. Recent advances and emerging trends in plant hormone signalling. Nature. 2009;459:1071–8. doi: 10.1038/nature08122. [DOI] [PubMed] [Google Scholar]
- 51.Sazuka T, Kamiya N, Nishimura N, Ohmae K, Sato Y, et al. A rice tryptophan deficient dwarf mutant, tdd1, contains a reduced level of indole acetic acid and develops abnormal flowers and organless embryos. The Plant J. 2009 doi: 10.1111/j.1365-313X.2009.03952.x. Published Online: 15 Jun 2009. [DOI] [PubMed] [Google Scholar]
- 52.Seo M, Akaba S, Oritani T, Delarue M, Bellini C, et al. Higher activity of an aldehyde oxidase in the auxin-overproducing superroot1 mutant of Arabidopsis thaliana. Plant Physiol. 1998;116:687–93. doi: 10.1104/pp.116.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sitbon F, Hennion S, Sundberg B, Little CH, Olsson O, Sandberg G. Transgenic Tobacco Plants Coexpressing the Agrobacterium tumefaciens iaaM and iaaH Genes Display Altered Growth and Indoleacetic Acid Metabolism. Plant Physiol. 1992;99:1062–9. doi: 10.1104/pp.99.3.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sohlberg JJ, Myrenas M, Kuusk S, Lagercrantz U, Kowalczyk M, et al. STY1 regulates auxin homeostasis and affects apical-basal patterning of the Arabidopsis gynoecium. Plant J. 2006;47:112–23. doi: 10.1111/j.1365-313X.2006.02775.x. [DOI] [PubMed] [Google Scholar]
- 55.Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM. A Link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell. 2005;17:2230–42. doi: 10.1105/tpc.105.033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–91. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 57.Stone SL, Braybrook SA, Paula SL, Kwong LW, Meuser J, et al. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: Implications for somatic embryogenesis. Proc Natl Acad Sci U S A. 2008;105:3151–6. doi: 10.1073/pnas.0712364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sugawara S, Hishiyama S, Jikumaru Y, Hanada A, Nishimura T, et al. Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:5430–5. doi: 10.1073/pnas.0811226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun J, Xu Y, Ye S, Jiang H, Chen Q, et al. Arabidopsis ASA1 Is Important for Jasmonate-Mediated Regulation of Auxin Biosynthesis and Transport during Lateral Root Formation. Plant Cell. 2009 doi: 10.1105/tpc.108.064303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–76. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tobena-Santamaria R, Bliek M, Ljung K, Sandberg G, Mol JN, et al. FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev. 2002;16:753–63. doi: 10.1101/gad.219502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Treml BS, Winderl S, Radykewicz R, Herz M, Schweizer G, et al. The gene ENHANCER OF PINOID controls cotyledon development in the Arabidopsis embryo. Development. 2005;132:4063–74. doi: 10.1242/dev.01969. [DOI] [PubMed] [Google Scholar]
- 63.Trigueros M, Navarrete-Gomez M, Sato S, Christensen SK, Pelaz S, et al. The NGATHA Genes Direct Style Development in the Arabidopsis Gynoecium. Plant Cell. 2009 doi: 10.1105/tpc.109.065508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136:1005–16. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 65.Vieten A, Vanneste S, Wisniewska J, Benkova E, Benjamins R, et al. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development. 2005;132:4521–31. doi: 10.1242/dev.02027. [DOI] [PubMed] [Google Scholar]
- 66.Woo YM, Park HJ, Su'udi M, Yang JI, Park JJ, et al. Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol Biol. 2007;65:125–36. doi: 10.1007/s11103-007-9203-6. [DOI] [PubMed] [Google Scholar]
- 67.Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Ann Bot (Lond) 2005;95:707–35. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woodward C, Bemis SM, Hill EJ, Sawa S, Koshiba T, Torii KU. Interaction of auxin and ERECTA in elaborating Arabidopsis inflorescence architecture revealed by the activation tagging of a new member of the YUCCA family putative flavin monooxygenases. Plant Physiol. 2005;139:192–203. doi: 10.1104/pp.105.063495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wright AD, Sampson MB, Neuffer MG, Michalczuk L, Slovin JP, Cohen JD. Indole-3-Acetic Acid Biosynthesis in the Mutant Maize orange pericarp, a Tryptophan Auxotroph. Science. 1991;254:998–1000. doi: 10.1126/science.254.5034.998. [DOI] [PubMed] [Google Scholar]
- 70.Yamada M, Greenham K, Prigge MJ, Jensen PJ, Estelle M. The TRANSPORT INHIBITOR RESPONSE2 (TIR2) gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol. 2009 doi: 10.1104/pp.109.138859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T. Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol. 2007;143:1362–71. doi: 10.1104/pp.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang Y, Xu R, Ma CJ, Vlot AC, Klessig DF, Pichersky E. Inactive methyl indole-3-acetic acid ester can be hydrolyzed and activated by several esterases belonging to the AtMES esterase family of Arabidopsis. Plant Physiol. 2008;147:1034–45. doi: 10.1104/pp.108.118224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–9. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- 74.Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J, et al. Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 2002;16:3100–12. doi: 10.1101/gad.1035402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ziegler DM. Flavin-containing monooxygenases: catalytic mechanism and substrate specificities. Drug Metab Rev. 1988;19:1–32. doi: 10.3109/03602538809049617. [DOI] [PubMed] [Google Scholar]