SUMMARY
Two early-acting components of the cellular ESCRT pathway, ESCRT-I and ALIX, participate directly in HIV-1 budding. The membrane fission activities of ESCRT-III subunits are also presumably required, but humans express 11 different CHMP/ESCRT-III proteins whose functional contributions are not yet clear. We therefore depleted cells of each of the different CHMP proteins and protein families, and examined the effects on HIV-1 budding. Virus release was profoundly inhibited by co-depletion of either CHMP2 or CHMP4 family members, resulting in ≥100-fold titer reductions. CHMP2A and CHMP4B proteins bound one another and this interaction was required for budding. By contrast, virus release was reduced only modestly by depletion of CHMP3 and CHMP1 proteins (2-8-fold titer reductions) and was unaffected by depletion of other human ESCRT-III proteins. HIV-1 budding therefore requires only a subset of the known human ESCRT-III proteins, with the CHMP2 and CHMP4 families playing key functional roles.
INTRODUCTION
The ESCRT pathway mediates membrane fission in several important biological processes including intralumenal vesicle formation at the multivesicular body (MVB), abscission during cytokinesis, and budding of enveloped viruses from the plasma membrane (for recent reviews, see (Bieniasz, 2009; Hurley, 2010; Hurley and Hanson, 2010; Raiborg and Stenmark, 2009; Saksena and Emr, 2009)), In each case, the ESCRT machinery functions from the cytoplasmic face of the bilayer and draws the closing membrane neck toward itself. This ability to catalyze “reverse topology” membrane fission events appears to explain why many enveloped viruses, including HIV-1, have evolved to usurp the cellular ESCRT machinery to bud from cells.
Although the proteins and complexes of the ESCRT pathway must work together, they are recruited sequentially and perform distinct functions. Early acting factors such as ESCRT-I and ALIX recognize and help remodel the appropriate membranes, concentrate vesicular cargoes, and recruit the later acting factors (Hurley, 2010). Proteins of the ESCRT-III complex, in turn, help mediate membrane fission and recruit the terminal VPS4 ATPase complex. VPS4 then completes the cycle by using the energy of ATP hydrolysis to remove the assembled ESCRT machinery from the membrane (Babst et al., 1998; Hurley and Hanson, 2010; Wollert et al., 2009).
The mechanism of membrane fission is not yet understood in detail, but current models hold that the soluble ESCRT-III proteins are deposited within the necks of budding particles, where they polymerize into filaments that spiral closed to form a “dome” that draws the opposing membranes together (Fabrikant et al., 2009; Hurley and Hanson, 2010). ESCRT-III proteins share a common helical core domain comprising an extended helical hairpin and two shorter helices that pack at the open end of the hairpin. ESCRT-III proteins can adopt “closed” and “open” configurations in which their extended C-termini either fold back on the core to autoinhibit polymerization (closed conformation), or extend to allow core polymerization and expose binding sites for VPS4, ALIX and other proteins (open conformation) (Bajorek et al., 2009b; Lata et al., 2008; Muziol et al., 2006; Shim et al., 2007).
ESCRT-III proteins can be divided into different families based upon primary sequence similarities (see Figure S1). Sacharaomyces cerevisiae expresses four essential ESCRT-III proteins and three “regulatory” ESCRT-III-like proteins that function primarily in Vps4p recruitment and regulation (Babst et al., 2002; Hurley, 2010; Saksena and Emr, 2009). The four essential ESCRT-III proteins are recruited sequentially, in the following order (with human protein names in parentheses). First, two Vps20p (CHMP6) subunits appear to nucleate the assembly of two spiraling Vps32p/Snf7p CHMP4A-C) filaments within the bud necks (Teis et al., 2010). These filaments are then capped by Vps24p (CHMP3) and Vps2p (CHMP2A-B). Vps24p is required to complete membrane fission, and Vps2p recruits Vps4p (Babst et al., 2002; Hurley and Hanson, 2010; Saksena and Emr, 2009; Wollert et al., 2009). Hence, all four essential yeast ESCRT-III proteins are required for ESCRT-mediated fission.
Humans express at least 12 ESCRT-III-like proteins (termed CHMP1-7 and IST1, see Figure S1) that can be grouped into seven families corresponding to each of the seven different yeast ESCRT-III proteins, plus an additional protein (CHMP7) not found in yeast. Although their functional roles in virus budding are not yet well defined, many ESCRT-III proteins can potently inhibit virus budding when overexpressed as fusion proteins (Martin-Serrano et al., 2003; Strack et al., 2003; von Schwedler et al., 2003) or as truncations that remove the C-terminal autoinhibitory region (Zamborlini et al., 2006). However, CHMP6 fusions do not inhibit virus budding (Martin-Serrano et al., 2003; von Schwedler et al., 2003), and HIV-1 can reportedly bud efficiently from cells that lack IST1 (Agromayor et al., 2009; Bajorek et al., 2009a), CHMP5 (Ward et al., 2005), or CHMP6 (Langelier et al., 2006). Here, we have systematically examined the requirements for each of the different human CHMP proteins and families in HIV-1 budding, and thereby determined their redundancy and functional importance.
RESULTS
Requirements for Individual ESCRT-III Proteins in HIV-1 Budding
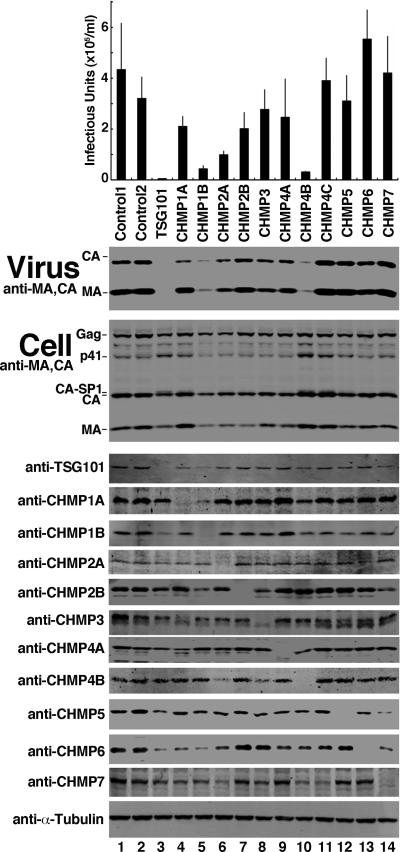
We depleted each of the 11 different human CHMP proteins and observed HIV-1 release and infectivity. A twelfth human ESCRT-III protein family member, IST1, was omitted because it is dispensable for HIV-1 budding (Agromayor et al., 2009; Bajorek et al., 2009a). Effects on HIV-1 release and infectivity were tested using at least two different siRNAs that targeted each CHMP protein (Morita et al., 2010). The data were consistent in all cases, and data for a single siRNA is therefore typically reported. Each siRNA depleted the endogenous CHMP protein efficiently (Figure 1, lanes 4-14, panels 5-14 and see (Morita et al., 2010)), with the exception of siRNA targeting CHMP4C, which were validated by demonstrating efficient depletion of an exogenously expressed, epitope-tagged protein because suitable antibodies were not available (Morita et al., 2010). Two irrelevant siRNA constructs were used as negative controls (lanes 1 and 2), and an siRNA against the essential budding factor, TSG101, was used as a positive control (lane 3).
Figure 1. HIV-1 Release and Infectivity Can Be Modestly Affected by Depletion of Individual ESCRT-III Proteins.
Viral titers produced by cells expressing HIV-1 and treated with control siRNAs (panel 1, lanes 1 and 2), with siRNAs that targeted TSG101 (positive control, lane 3) or the designated ESCRT-III proteins (lanes 4-14) (n = 6 titer measurements from two independent experiments ± S.D.). Lower western blots show virion-associated viral Gag proteins released into the supernatant (panel 2, “Virus”, anti-CA and anti-MA), intracellular viral Gag protein levels (panel 3, “Cell”, anti-CA and anti-MA), endogenous cellular ESCRT proteins (panels 4-14), or α-Tubulin (panel 15, loading controls). (see also Figure S1).
To analyze effects on HIV-1 release, infectious proviral expression constructs were co-transfected into siRNA-treated HEK 293T cells. Viral proteins were allowed to accumulate for 48 h, and then viral titers (panel 1) and levels of virion-associated viral CA and MA proteins released into the media (panel 2) were measured. As expected, TSG101 depletion strongly inhibited viral infectivity (Figure 1, panel 1, 76-fold reduction in infectivity vs. control siRNA treatments) and virion release (panel 2, see virion-associated CA and MA protein levels in lane 3 vs. lanes 1 and 2). By contrast, HIV-1 release was not dramatically affected by depletion of most individual ESCRT-III proteins, although moderate reductions were observed in several cases. The strongest reduction occurred upon depletion of CHMP4B, which reduced virion release and infectivity 12-fold, without altering intracellular Gag, CA, or MA protein levels (panel 3, compare lane 10 to lanes 1 and 2). The next greatest effect was seen upon depletion of CHMP1B, which reduced viral titers 8-fold. However, CHMP1B depletion also diminished cellular Gag, CA, and MA protein levels slightly, which could have contributed to the reduction in virus release and infectivity (panel 3, compare lane 5 to lanes 1 and 2). Depletion of CHMP2A, CHMP2B, and CHMP1A caused modest reductions in virus release and infectivity (4-, 2- and 2-fold reductions, respectively), and virus release was not appreciably diminished by depletion of any of the other ESCRT-III proteins. Thus, depletion of individual ESCRT-III proteins, particularly CHMP4B, can inhibit virus release, but the effects are much weaker than for depletion of the essential virus budding factor TSG101.
The CHMP2 and CHMP4 Families Are Required for HIV-1 Budding
We next tested the possibility that the modest phenotypes seen in our initial survey might reflect functional redundancy within the different CHMP protein families. Here, we analyzed the effects of simultaneously depleting multiple proteins within each family: CHMP1 (CHMP1A and 1B), CHMP2 (CHMP2A and 2B), and CHMP4 (CHMP4A, 4B and 4C). Co-depletion of CHMP1A and CHMP1B reduced virus release and infectivity only modestly (>2-fold, Figure S2, compare lane 6 to lanes 1 and 2). Thus, CHMP1 family members may participate in virus budding but are not absolutely essential.
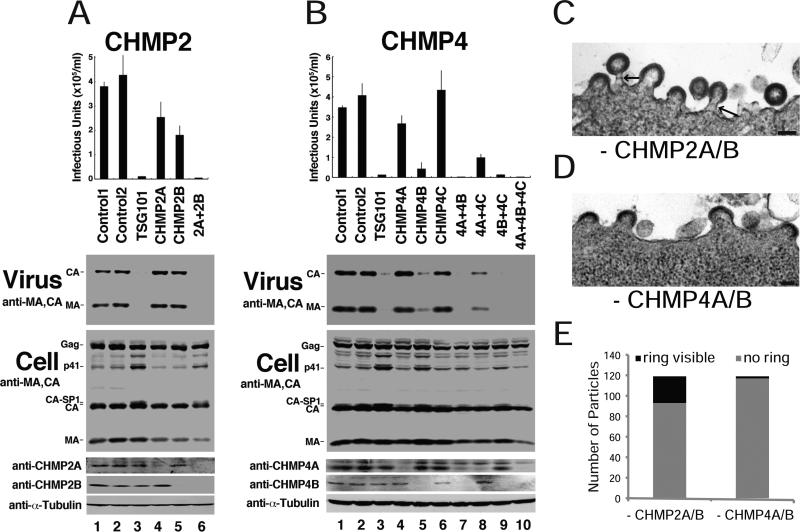
Co-depletion of either CHMP2A and CHMP2B or CHMP4A, CHMP4B and CHMP4C resulted in dramatic reductions in HIV-1 release and infectivity (95- and 166-fold, respectively; Figure 2A, compare lane 6 to lanes 1 and 2, and Figure 2B, compare lane 10 to lanes 1 and 2). These data imply that individual CHMP2 and CHMP4 family members can function redundantly, and that HIV-1 budding requires the presence of at least one CHMP2 family member and at least one CHMP4 family member. For individual CHMP4 family members, the relative functional importance (4B>4A>4C, compare lanes 5, 4, and 6, respectively) parallels their reported cellular abundance (4B>4A>4C)(Katoh et al., 2004), with CHMP4C making a negligible contribution to the phenotype.
Figure 2. CHMP2 and CHMP4 Protein Families Are Required for HIV-1 Release and Infectivity.
(A,B) Viral titers, virion-associated Gag proteins, and intracellular protein levels in 293T cells expressing HIV-1 and depleted or co-depleted of the designated (A) CHMP2 or (B) CHMP4 family members. Figure panels are equivalent to those shown in Figure 1 (n = 3 titer measurements ± S.D.). (C,D) Thin-sectioned, transmission electron micrographs of arrested HIV-1 budding particles in cells lacking CHMP2A/B (C) or CHMP4A/B (D). Scale bars are 100 nm. (E) Quantification of arrested budding particles without (grey) or with (black) visible striations within their necks (see arrows in Figure 2C for examples). (see also Figure S2).
CHMP2 and CHMP4 Are Required for Late Stages of Virus Assembly
Thin section transmission electron microscopic analyses confirmed that partially budded virions accumulated at the plasma membranes of cells that lacked either CHMP2A/B (Figure 2C) or CHMP4A/B (Figure 2D). In both cases, the virions had immature morphologies, and ranged from half shells to nearly complete spheres. The two cases differed, however, in that ring-like striations were frequently visible in the necks of virions that budded from cells that lacked CHMP2A/B (arrows in Figure 2C, 22% of late budding virions, see Figure 2E for quantification), but rarely in cells that lacked CHMP4A/B (2% of budding virions).
CHMP3 Is Not Essential for HIV-1 Budding
During MVB vesicle formation in S. cerevisiae, the homolog of CHMP3 (Vps24p) forms an essential bridge between Vps2p (CHMP2) and Vps32p/Snf7p (CHMP4) (Babst et al., 2002; Wollert et al., 2009). It was therefore surprising that depletion of the only known human CHMP3 protein reduced virus release and infectivity less than 2-fold (Figure 1, compare lane 8 to lanes 1 and 2). To confirm this result, we tested the effects of ten additional CHMP3 siRNA constructs. As shown in Figure S3, five different siRNA constructs depleted endogenous CHMP3 very efficiently (Panel 4, compare lane 1 to lanes 2-6), but none had major effects on virion release (Panel 2) or viral titers (Panel 1). In two cases, viral titers were reduced 2-4-fold (siRNA constructs 474 and 461) and in the other three cases the titers changed less than 2-fold, whereas co-depletion of either CHMP2A/B or CHMP4A/B reduced viral titers more than 75-fold (positive controls). Hence, CHMP3 depletion could impair virus infectivity, release, and Gag processing slightly (Panel 3, compare the CA-SP1 bands in lanes 2-6 to lane 1), but the effects were much weaker than depletion of the CHMP2 or CHMP4 families.
Yeast Two-hybrid Analyses of CHMP2:CHMP4 Interactions
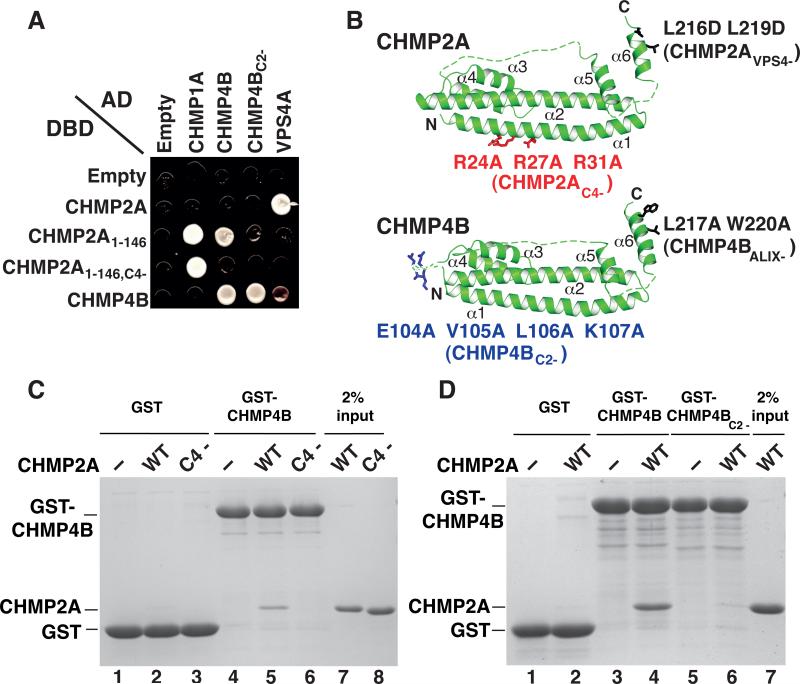
The relative unimportance of CHMP3 in our virus release and infectivity assays suggested that CHMP2:CHMP4 interactions might bypass the functional requirement for CHMP3. We used the yeast two-hybrid assay as a convenient method to screen for possible CHMP2:CHMP4 interactions. As reported previously, we did not detect an interaction between the full length CHMP2A and CHMP4B proteins in either possible orientation ((Martin-Serrano et al., 2003; von Schwedler et al., 2003), Figure 3A and data not shown), even though our CHMP4B and CHMP2A constructs showed positive interactions with known binding partners in control experiments (e.g., CHMP4B:CHMP4B and VPS4A:CHMP2A). However, a construct that lacked the C-terminal autoinhibitory region of CHMP2A (CHMP2A1-146) did interact with CHMP4B in these assays. The truncated CHMP2A1-146 construct also gained a CHMP1A interaction and lost the VPS4A interaction (because it lacked the C-terminal VPS4A MIT domain binding site (Stuchell-Brereton et al., 2007)). Thus, removing the CHMP2A C-terminal autoinhibitory region promoted interactions with other ESCRT-III proteins, consistent with previous reports (e.g., (Lata et al., 2008; Lin et al., 2005; Shim et al., 2007)).
Figure 3. Identification and Characterization of CHMP2A:CHMP4B Interactions.
(A) Panel showing yeast two-hybrid interactions between the designated ESCRT proteins fused to activation (AD) or DNA binding (DBD) domains (or with empty vector controls). The following interactions were judged to be positive (AD fusions listed first): CHMP1A:CHMP2A1-146, CHMP1A:CHMP2A1-146,C4-, CHMP4B:CHMP2A1-146, CHMP4B:CHMP4B, CHMP4BC2-:CHMP4B, VPS4A:CHMP2A. VPS4A also interacted very weakly with CHMP4B in this assay.
(B) Upper: homology model of CHMP2A showing mutations that blocked binding of CHMP4B (designated CHMP2AC4-) or VPS4A and B (CHMP2AVPS4-). Lower: homology model of CHMP4B showing mutations that blocked binding of CHMP2A (CHMP4BC2-) or ALIX (CHMP4BALIX-). Homology models were generated by threading the CHMP2A and CHMP4B sequences onto the known CHMP3 structure (pdb 3FRT) (Bajorek et al., 2009b; Muziol et al., 2006) using SWISS-MODEL (Arnold et al., 2006) and COOT (Emsley and Cowtan, 2004), and displayed using PYMOL (DeLano, 2008). (C,D) GST pull-down analyses demonstrating that recombinant CHMP2A and CHMP4B proteins interact and that the mutations in the CHMP2AC4- (C) and CHMP4BC2- (D) constructs inhibit these interactions. (C) SDS-PAGE/Coomassie detection of matrix-bound proteins following incubation of immobilized GST (lanes 1-3, negative controls) or GST-CHMP4B proteins (lanes 4-6) with no added proteins (lanes 1 and 4, negative controls), or with purified wild type CHMP2A (lanes 2 and 5) or CHMP2AC4- (lanes 3 and 6). Input CHMP2A proteins are shown in lanes 7 and 8 (2% of total). (D) SDS-PAGE/Coomassie detection of matrix-bound proteins following incubation of immobilized GST (lanes 1 and 2, negative controls), wild type GST-CHMP4B (lanes 3 and 4) or GST-CHMP4BC2- (lanes 5 and 6) with no added proteins (lanes 1, 3 and 5) or with purified wild type CHMP2A (lanes 2, 4, and 6). Input CHMP2A is shown in lane 7 (2% of total). (see also Figure S3).
Homology models were used to guide the design of surface alanine scanning mutations for both CHMP2A1-146 (52 mutants) and CHMP4B (118 mutants) that might inhibit the CHMP2A1-146:CHMP4B interaction while retaining other control interactions. These analyses identified: 1) a triple point mutation in three successive Arg residues on the exposed face of CHMP2A1-146 helix 1 (R24R27R31 to AAA, termed CHMP2AC4-) that inhibited the interaction with CHMP4B but retained the CHMP1A interaction (Figures 3A and 3B), and 2) a quadruple point mutation on the exposed loop between helices 2 and 3 of CHMP4B (EVLK104-107 to AAAA, termed CHMP4BC2-) that inhibited the CHMP2A interaction, but retained the CHMP4B interaction, Figures 3A and 3B). Thus, we identified a truncated CHMP2A construct that could interact with both CHMP4B and CHMP1A, and point mutations within either CHMP2A1-146 or CHMP4B that eliminated the CHMP2A1-146:CHMP4B interaction.
Recombinant CHMP2A and CHMP4B Protein Interactions
Overexpressed CHMP2A and CHMP4 proteins can be co-immunoprecipitated, indicating that these two proteins can interact in cells (Martin-Serrano et al., 2003). However, such CHMP2A:CHMP4 interactions may depend upon membrane binding and/or other cellular proteins. We performed GST pulldown experiments to test whether full length, recombinant CHMP2A and CHMP4B proteins could interact specifically. As shown in Figure 3C, pure recombinant CHMP2A protein bound weakly, but reproducibly, to immobilized GST-CHMP4B and not to a control matrix (compare lanes 2 and 5). Furthermore, the same CHMP2A mutation that inhibited CHMP4B binding in the yeast two-hybrid experiments also inhibited CHMP2AC4- binding to GST-CHMP4B, demonstrating the specificity of this interaction (compare lanes 5 and 6). Similarly, the GST-CHMP4BC2- mutation blocked the binding of pure recombinant CHMP2A protein (Figure 3D, compare lanes 4 and 6). Thus, recombinant full-length CHMP2A and CHMP4B proteins can interact in vitro, and the CHMP2AC4- and CHMP4BC2- mutations inhibit this interaction.
The CHMP2A:CHMP4B Interaction Is Required for HIV-1 Budding
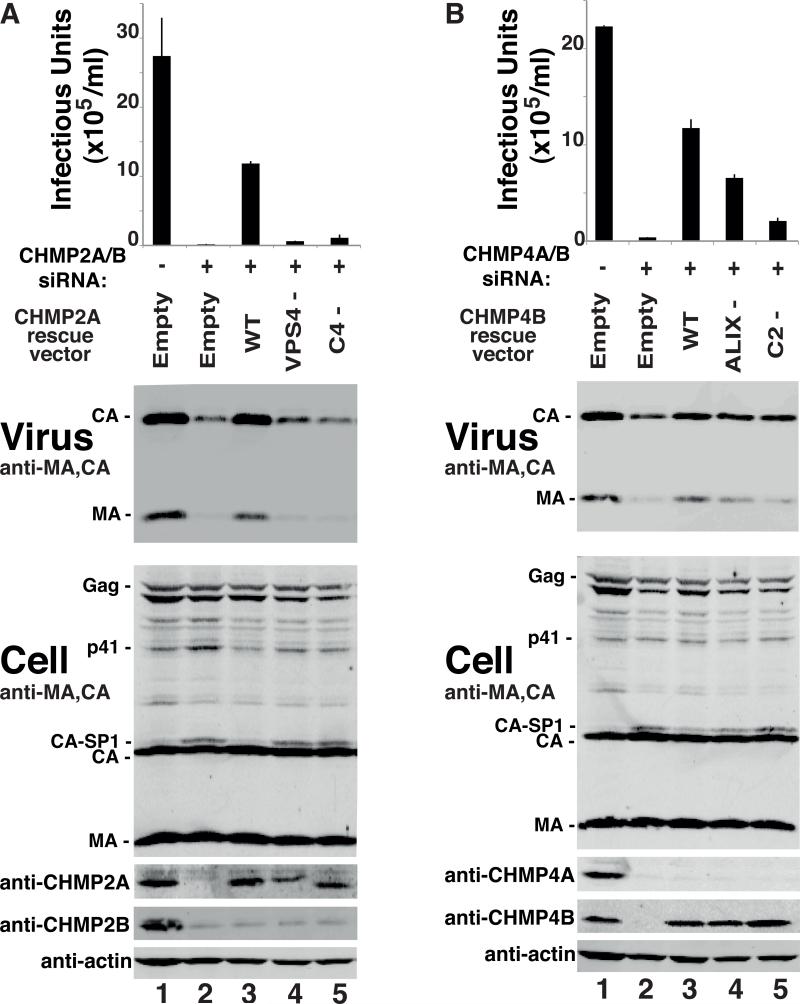
To examine the functional importance of the CHMP2A-CHMP4B interaction, we tested whether point mutations that inhibited this interaction also inhibited HIV-1 release and infectivity using siRNA depletion and rescue assays. As expected, HIV-1 release and infectivity were strongly inhibited by co-depletion of CHMP2A and CHMP2B (Figure 4A, panels 1 and 2, 167-fold infectivity reduction, compare lanes 1 and 2). This inhibition was substantially, though not completely rescued by over-expression of either a wild type, siRNA-resistant CHMP2A construct (72-fold rescue, compare lanes 2 and 3) or a wild type, siRNA-resistant CHMP2B (data not shown). In contrast, mutant CHMP2A constructs that could not bind either the VPS4 ATPases (CHMP2AVPS4-) or CHMP4B (CHMP2AC4-), were unable to rescue the defects in virus infectivity, release, or Gag processing (compare lanes 4 and 5 to lane 3), even though both mutant proteins expressed normally (Figure 4A, panel 4). Thus, CHMP2A mutations that inhibited VPS4 or CHMP4 binding also impaired HIV-1 budding.
Figure 4. Point Mutations that Inhibit CHMP2A-CHMP4B Interactions also Block HIV-1 Budding.
(A) Viral titers, virion-associated Gag proteins, and intracellular protein levels in 293T cells expressing HIV-1 (lanes 1-5) and treated with a control siRNA (lane 1), or with siRNAs that co-depleted CHMP2A/B (lanes 2-5), together with an empty control vector (lane 2) or with vectors expressing siRNA-resistant wild type CHMP2A (lane 3) CHMP2AVPS4- (lane 4) or CHMP2AC4- (lane 5). Figure panels are equivalent to those in Figures 1 and 2 (n = 9 titer measurements from 3 independent experiments ± S.D.). (B) This experiment is equivalent to Figure 4A except that cells were co-depleted of CHMP4A/B (lanes 2-5) and the siRNA-resistant rescue constructs expressed wild type CHMP4B (lane 3), CHMP4BALIX- (lane 4) or CHMP4BC2- (lane 5) (n = 6-12 titer measurements from 2-4 independent experiments ± S.D.). (see also Figure S4).
Analogous experiments were used to test the functional importance of the CHMP2A binding site on CHMP4B. Co-depletion of CHMP4A and CHMP4B reduced viral infectivity 76-fold (Figure 4B, compare lanes 1 and 2), and this defect was substantially rescued by overexpression of an siRNA-resistant CHMP4B protein (compare lanes 2 and 3, 40-fold infectivity rescue). In contrast, CHMP4B mutants that could not bind ALIX (CHMP4BALIX-) or CHMP2A (CHMP4BC2-) rescued virus budding less well (22-fold and 7-fold, respectively, compare lanes 4 and 5 to lane 3) and also failed to rescue the accumulation of intracellular CA-SP1 processing intermediates (panel 3). Thus, inactivating point mutations on either side of the CHMP2A:CHMP4B interface inhibited HIV-1 budding.
DISCUSSION
We have shown that efficient HIV-1 budding requires at least one member of both the CHMP2 and CHMP4 ESCRT-III protein families, and different members of both families can function redundantly. Viral infectivity and Gag processing were also modestly inhibited by depletion of CHMP1 and CHMP3 proteins (Figures 1, S2 and S3), suggesting that these two ESCRT-III families may also participate in HIV budding, but are not essential. By contrast, virus budding proceeded efficiently in the absence of CHMP5, CHMP6, CHMP7 and IST1 (Figure 1 and (Agromayor et al., 2009; Bajorek et al., 2009a; Langelier et al., 2006; Ward et al., 2005)). These results contrast with the requirements for cytokinesis, where each human ESCRT-III protein, with the possible exception of CHMP6, plays a significant functional role (Morita et al., 2010). Thus, efficient HIV-1 budding requires only a subset of the known human ESCRT-III proteins and functions.
siRNA Depletion Experiments
As in all siRNA depletion experiments, negative or weak budding phenotypes could have reflected insufficient protein depletion and/or functional redundancy. However, our siRNA protocols depleted CHMP1A, CHMP1B, CHMP3, and CHMP5-7 sufficiently to induce cell division defects (Morita et al., 2010), and multiple different siRNAs that depleted CHMP3 efficiently all failed to inhibit HIV-1 budding substantially (Figure S3). Furthermore, we tested all possible pairwise knockdowns of CHMP1A, CHMP1B, CHMP3, and CHMP5-7, and did not observe synergistic inhibition of HIV-1 budding in any case (Figure S2 and data not shown). We conclude that none of these proteins function redundantly or provide critical functions in HIV-1 budding.
The strong budding defects induced by depletion of endogenous CHMP2A/B or CHMP4A/B were substantially rescued by expression of exogenous, siRNA-resistant CHMP2A, CHMP2B, and CHMP4B proteins, ruling out off-target effects (Figure 4 and data not shown). Furthermore, point mutations that abrogated the CHMP2A:VPS4, CHMP2A:CHMP4B, and CHMP4:ALIX interactions also inhibited virus budding, indicating that each of these interactions is functionally significant. Although it remains possible that sequestration of ESCRT factors on endosomal membranes could have contributed indirectly to the observed budding defects, our siRNA depletion experiments are the most direct functional tests of ESCRT-III protein requirements in HIV-1 budding reported to date. Indeed, it appears likely that the dominant inhibition of HIV-1 budding reported previously for overexpression of CHMP3 and several other ESCRT-III constructs may have resulted from indirect sequestration or inactivation of CHMP2, CHMP4, and/or other ESCRT factors, rather than from the direct loss of critical budding activities (Martin-Serrano et al., 2003; Strack et al., 2003; von Schwedler et al., 2003; Zamborlini et al., 2006).
Comparisons to Yeast MVB Vesicle Formation
Yeast MVB vesicle formation serves as an important paradigm for understanding ESCRT-III assembly and function (Hurley and Hanson, 2010; Saksena and Emr, 2009). However, we have demonstrated that the ESCRT-III requirements for yeast MVB vesicle formation and HIV-1 budding differ in several important ways. Firstly, human CHMP3 is dispensable for HIV budding, whereas yeast Vps24p (CHMP3) provides a critical link between Vps2p (CHMP2) and Snf7p (CHMP4)(Babst et al., 2002; Wollert et al., 2009), both in vivo and in a reconstituted yeast MVB vesicle system (Wollert et al., 2009). We found instead that human CHMP2 and CHMP4 can interact, which apparently obviates the strict requirement for a bridging CHMP3 protein. Similarly, although CHMP2A/CHMP3 complexes can co-assemble into helical domes in vitro (Bajorek et al., 2009b; Lata et al., 2008), CHMP3 does not appear to be an obligate CHMP2 binding partner during HIV budding.
Secondly, HIV budding efficiency was unaltered by CHMP6 depletion (Figures 1 and S4). We have previously reported that ESCRT-II, a protein complex that can recruit CHMP6, is also dispensable for HIV-1 budding (Langelier et al., 2006). This situation again differs from yeast MVB formation, where the ESCRT-II/Vps20 (CHMP6) interaction is required to nucleate Snf7p(CHMP4) assembly (Teis et al., 2010). Thus, HIV-1 virions and yeast MVB vesicles must initiate ESCRT-III recruitment and assembly using fundamentally different sets of protein-protein interactions.
ESCRT-III Protein Recruitment
We cannot yet fully explain the mechanisms by which HIV-1 recruits ESCRT-III proteins to sites of virus budding. The intermediate activity of the CHMP4BALIX- protein implies that the CHMP4B:ALIX interaction contributes to the efficiency of HIV-1 budding, but is not absolutely required (Figure 4B). Similarly, depleting ALIX from 293T cells (or mutating the ALIX binding site within p6Gag) typically reduces viral infectivity ~2-fold (e.g., (Fujii et al., 2009) and see Figure S4, compare lanes 1 and 2). These observations all indicate that ALIX contributes to, but is not absolutely essential for, HIV-1 budding under conditions where p6Gag can also bind TSG101/ESCRT-I.
The critical importance of the CHMP2 and CHMP4 protein families implies that ESCRT-I can recruit these two ESCRT-III protein families independently of ALIX. We hypothesized that sequential interactions between ESCRT-I, ESCRT-II and CHMP6 might serve to recruit CHMP4 proteins, by analogy to the well-established sequential recruitment steps during yeast MVB formation. This model was attractive because human ESCRT-I:II, ESCRT-II:CHMP6, and CHMP6:CHMP4 interactions have all been documented (e.g., (Langelier et al., 2006; Martin-Serrano et al., 2003; von Schwedler et al., 2003)), and because redundant pathways for CHMP4 recruitment (i.e., via p6Gag:ALIX or via p6Gag:ESCRT-I:ESCRT-II:CHMP6) could, in principle, explain why HIV-1 can still bud from cells that lack either ESCRT-II, CHMP6, or ALIX. However, co-depletion of CHMP6 did not further exacerbate the 2-fold reduction in virus release and infectivity seen for depletion of ALIX alone (Figure S4). Thus, HIV-1 Gag and/or ESCRT-I appear to recruit CHMP4 using additional protein-protein or protein-membrane interactions that remain to be identified.
Minimal ESCRT-III Requirements for Efficient HIV -1 Budding
Although ESCRT-III protein activities and functions remain to be characterized more fully, our data are consistent with a model in which CHMP4 proteins form rings within the necks of budding particles and recruit CHMP2 proteins, which in turn recruit the VPS4 ATPases and promote membrane fission by helping to create dome-like structures that bring the opposing membranes together (Fabrikant et al., 2009). Consistent with this model, we frequently observed ring-like structures within the necks of arrested particles that lacked CHMP2 (Figure 2), others have shown that overexpressed CHMP4 proteins can form helical plasma membrane protrusions (Hanson et al., 2008), and the CHMP2 proteins contain binding sites for the MIT domains of VPS4 proteins (Stuchell-Brereton et al., 2007). Thus, the ability to assemble a constricting dome within the bud neck and to recruit VPS4 ATPase(s) may represent the minimal activities required for efficient HIV-1 budding.
EXPERIMENTAL PROCEDURES
Cell Culture
293T and HeLa-TZM reporter cells were obtained from Drs. J. C. Kappes and X. Wu through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH and maintained in DMEM supplemented with 10% FCS.
siRNA and Expression Vectors
siRNAs were designed using the Dharmacon siDESIGN Center (Thermo Fisher Scientific Inc.), selected as described in (Morita et al., 2010), and listed in the Supplemental Material along with expression vectors.
HIV-1 Production from Cells Lacking ESCRT Proteins
siRNA depletion/reconstitution, virion production, and quantification procedures have been described (Kieffer et al., 2008), and detailed protocols are provided in the Supplemental Material.
Yeast Two-Hybrid Experiments
Directed two-hybrid assays with human ESCRT genes were performed using the Matchmaker GAL4 Yeast Two-Hybrid 3 system (Clontech). pGADT7 and pGBKT7-based constructs (500 ng each, see Table S2) were co-transformed into 10 μl of competent yeast strain PJ69-4A in the presence of 38 μl of 40% PEG3350, 0.1 M lithium acetate, 10 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.0. Transformation efficiencies were tested on selective -Leu, -Trp plates and two-hybrid interactions were tested on selective -Leu, -Trp, -Ade, -His plates.
GST Pulldown Experiments
Protocols for CHMP2A protein expression and purification and for GST pulldown experiments are provided in the Supplemental Experimental Methods.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Nancy Chandler in the University of Utah Health Sciences Center Electron Microscopy Core Facility for technical assistance, Heidi Schubert for assistance with homology modeling and mutagenesis, and Chris Hill and Mary Anne Karren for critical reading of this manuscript. This work was supported by NIH grant AI051174 (to WIS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
- Loss of the CHMP2 or CHMP4 families of ESCRT-III proteins blocked HIV-1 budding.
- Individual CHMP2 or CHMP4 family members functioned redundantly.
- Point mutations that inhibited CHMP2:CHMP4 interactions blocked virus budding.
REFERENCES
- Agromayor M, Carlton JG, Phelan JP, Matthews DR, Carlin LM, Ameer-Beg S, Bowers K, Martin-Serrano J. Essential role of hIST1 in cytokinesis. Mol Biol Cell. 2009;20:1374–1387. doi: 10.1091/mbc.E08-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Babst M, Katzmann D, Estepa-Sabal E, Meerloo T, Emr S. Escrt-III. An endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. Embo J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M, Morita E, Skalicky JJ, Morham SG, Babst M, Sundquist WI. Biochemical analyses of human IST1 and its function in cytokinesis. Mol Biol Cell. 2009a;20:1360–1373. doi: 10.1091/mbc.E08-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, Stubblefield WM, Uter NT, Myszka DG, Hill CP, Sundquist WI. Structural basis for ESCRT-III protein autoinhibition. Nat Struct Mol Biol. 2009b;16:754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz PD. The cell biology of HIV-1 virion genesis. Cell Host Microbe. 2009;5:550–558. doi: 10.1016/j.chom.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific LLC; Palo Alto, CA: 2008. [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fabrikant G, Lata S, Riches JD, Briggs JA, Weissenhorn W, Kozlov MM. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput Biol. 2009;5:e1000575. doi: 10.1371/journal.pcbi.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii K, Munshi UM, Ablan SD, Demirov DG, Soheilian F, Nagashima K, Stephen AG, Fisher RJ, Freed EO. Functional role of Alix in HIV-1 replication. Virology. 2009;391:284–292. doi: 10.1016/j.virol.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH. The ESCRT complexes. Crit Rev Biochem Mol Biol. 2010;45:463–487. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Shibata H, Hatta K, Maki M. CHMP4b is a major binding partner of the ALG-2-interacting protein Alix among the three CHMP4 isoforms. Arch Biochem Biophys. 2004;421:159–165. doi: 10.1016/j.abb.2003.09.038. [DOI] [PubMed] [Google Scholar]
- Kieffer C, Skalicky JJ, Morita E, De Domenico I, Ward DM, Kaplan J, Sundquist WI. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev Cell. 2008;15:62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Langelier C, von Schwedler U, Fisher RD, De Dominco I, White PL, Hill CP, Kaplan J, Ward D, Sundquist WI. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. Journal of Virology. 2006 doi: 10.1128/JVI.01049-06. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Kimpler LA, Naismith TV, Lauer JM, Hanson PI. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J Biol Chem. 2005;280:12799–12809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J, Yaravoy A, Perez-Caballero D, Bieniasz PD. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci U S A. 2003;100:12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Colf LA, Karren MA, Sandrin V, Rodesch CK, Sundquist WI. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc Natl Acad Sci U S A. 2010;107:12889–12894. doi: 10.1073/pnas.1005938107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Gottlinger H, Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Saksena S, Emr SD. ESCRTs and human disease. Biochem Soc Trans. 2009;37:167–172. doi: 10.1042/BST0370167. [DOI] [PubMed] [Google Scholar]
- Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S, Kimpler LA, Hanson PI. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX Is a Binding Partner for HIV-1 p6 and EIAV p9 Functioning in Virus Budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- Teis D, Saksena S, Judson BL, Emr SD. ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. EMBO J. 2010;29:871–883. doi: 10.1038/emboj.2009.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Ward DM, Vaughn MB, Shiflett SL, White PL, Pollock AL, Hill J, Schnegelberger R, Sundquist WI, Kaplan J. The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. J Biol Chem. 2005;280:10548–10555. doi: 10.1074/jbc.M413734200. [DOI] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamborlini A, Usami Y, Radoshitzky SR, Popova E, Palu G, Gottlinger H. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc Natl Acad Sci U S A. 2006;103:19140–19145. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.