Abstract
Genes influencing resting energy expenditure (REE) and respiratory quotient (RQ) represent candidate genes for obesity and the metabolic syndrome because of the involvement of these traits in energy balance and substrate oxidation. We aim to explore the molecular basis for individual variation in REE and fuel partitioning as reflected by RQ. We performed microarray studies in human vastus lateralis muscle biopsies from 40 healthy subjects with measured REE and RQ values. We identified 2,392 and 1,115 genes significantly correlated with REE and RQ, respectively. Genes correlated with REE and RQ encompass a broad array of functions, including carbohydrate and lipid metabolism, gene expression, mitochondrial processes, and membrane transport. Microarray pathway analysis revealed that REE was positively correlated with upregulation of G protein-coupled receptor signaling (meet criteria/total genes: 65 of 283) involved in autonomic nervous system functions, including those receptors mediating adrenergic, dopamine, γ-aminobutyric acid (GABA), neuropeptide Y (NPY), and serotonin action (meet criteria/total genes: 46 of 176). Reduced REE was associated with an increase in genes participating in ubiquitin-proteasome-dependent proteolytic pathways (58 of 232). Serine-type peptidase activity (9 of 76) was positively correlated with RQ, while genes involved in the protein phosphatase type 2A complex (4 of 9), mitochondrial function and cellular respiration (38 of 315), and unfolded protein binding (19 of 97) were associated with reduced RQ values and a preference for lipid fuel metabolism. Individual variations in whole body REE and RQ are regulated by differential expressions of specific genes and pathways intrinsic to skeletal muscle.
Keywords: microarray, human, skeletal muscle, G protein-coupled receptor, ubiquitin, protein phosphatase 2A, mitochondrial function, respiratory quotient
obesity increases the risk of type 2 diabetes and the metabolic syndrome, a trait cluster consisting of insulin resistance, obesity, glucose intolerance, upper body fat distribution, hypertension, dyslipidemia, and dysfibrinolysis (23, 41). The pathophysiology of obesity represents an imbalance between a high-energy intake and/or low-energy expenditure. Prospective studies have shown that a reduced rate of energy expenditure is a risk factor for body weight gain and obesity (10, 13, 56, 59). Total energy expenditure can be divided into three major components, namely, resting metabolic rate (RMR) or resting energy expenditure (REE), thermic effect of exercise (TEE), and thermic effect of food (TEF). Each component can be increased or decreased in response to environmental changes. REE is measured when the individual is at rest under basal conditions and comprises 60–75% of total energy expenditure. There is marked individual variability in REE, which is due in large part to differences in body weight (56, 75) and lean body mass (17, 24, 37). However, even when REE is normalized by lean body mass, there remains significant variability in REE among individuals. The molecular basis for interindividual variability in energy expenditure is largely unknown.
Skeletal muscle is a major insulin target tissue (21) and is responsible for ∼80% of insulin-responsive glucose uptake. It is also involved in glycogen storage, lipid oxidation, protein turnover, and nonshivering thermogenesis (19–21). Thus muscle metabolism not only accounts for a significant proportion of total REE but also is a major determinant of individual variability in REE normalized for lean mass (78). It is not surprising then that skeletal muscle metabolism plays a role in the pathogenesis of obesity since individuals with a “low” metabolic rate may be at higher risk of gaining weight (78). Several factors might influence the variation in muscle energy expenditure, including thyroid hormone, muscle tone, sympathetic innervation, catecholamine levels (78), and muscle fiber type composition (73). Most studies in skeletal muscle have focused on changes in metabolic flux and enzyme activities. A few studies have addressed the impact of specific gene expression on energy expenditure (15, 53, 55). For example, uncoupling proteins (UCPs) play important roles in adaptive thermogenesis and energy expenditure in rodents (22) and can be induced by cold exposure, food intake, β-adrenergic stimulation, and thyroid hormone (31, 57, 58, 63), while glucocorticoids inhibit UCP expression (71). These studies in rodents should only be extrapolated cautiously to humans. In humans, muscle UCP2 and UCP3 mRNA levels are not related to REE (7), and the role of these proteins in energy metabolism is not well understood (54). The full extent and importance of alterations in gene transcription in human skeletal muscle relevant to REE are still unknown, and no systemic analysis of the changes in muscle gene expression correlated to REE has been reported.
The respiratory quotient (RQ), i.e., the ratio of the volume of carbon dioxide released to the volume of oxygen consumed by a body tissue, is used as an index of the ratio of carbohydrate to fat oxidation. RQ can be used to estimate fuel partitioning between fat and carbohydrate, as preferred substrates for energy generation and fuel preferences to generate REE also exhibit individual variation. We have reported that an exon 6 splice-junction polymorphism in the gene of UCP3 increases RQ and reduces basal lipid metabolism in Gullah-speaking African Americans (4). However, differences in gene expression that could explain individual variation in RQ have not been extensively studied.
Genes influencing REE and RQ could represent candidate genes for obesity, metabolic syndrome, and type 2 diabetes due to the involvement of these traits in energy balance and substrate oxidation. To examine this possibility, we performed high-throughput microarray analyses of human skeletal muscle biopsied from 40 healthy nondiabetic individuals with REE and resting substrate oxidation rates measured by indirect calorimetry. We report here: 1) REE and RQ are correlated with differential expressions of specific subsets of genes and pathways in skeletal muscle, and molecular regulation intrinsic to skeletal muscle can predict differences in whole body REE and RQ; 2) REE is enhanced via the upregulation of receptors and pathways that accommodate a greater effect of the sympathetic nervous system (SNS), whereas decreased REE is associated with greater activity of catabolic processes; 3) major determinants of fuel partitioning include serine-type peptidase activity, protein phosphatase 2A (PP2A), mitochondrial function and cell respiration, and unfolded protein binding.
METHODS AND MATERIALS
Human Subjects
Microarray studies were performed in 40 healthy normoglycemic individuals, age 21–55 yr (37.15 ± 9.08 yr). Detailed clinical characteristics of subjects are shown in Table 1. The mean body mass index (BMI) was 27.04 ± 4.07 kg/m2. None of the subjects were taking medications that would affect metabolism or glucose homeostasis. Each volunteer had a normal physical examination, an unremarkable medical history, and was verified by blood chemistry examination to have normal hepatic, renal, and thyroid functions. Before metabolic studies, subjects were equilibrated for 3 days on an isocaloric diet (28–32 kcal·kg−1·day−1) consisting of 50% carbohydrates, 30% fat, and 20% protein. Subjects were instructed to maintain usual activity levels, and no subjects engaged in regular exercise. The patients were admitted to the General Clinical Research Center (GCRC) at the University of Alabama at Birmingham (UAB) Hospital and underwent inpatient evaluation as described below. Protocols were approved by the UAB Institutional Review Board, and written informed consent was obtained from every subject.
Table 1.
Descriptive characteristics of the study subjects
| Variable | Total | Men | Women |
|---|---|---|---|
| n | 40 | 25 | 15 |
| Age, yr | 37.2 ± 9.1 | 35.1 ± 8.9 | 40.6 ± 8.5 |
| Ethnic groups | 2H/25E/13A | 1H/15E/9A | 1H/10E/4A |
| GDR, mg · kg−1 · min−1 | 13.6 ± 3.2 | 13.0 ± 3.3 | 14.4 ± 3.1 |
| Weight, kg | 80.0 ± 14.7 | 85.3 ± 14.1 | 71.3 ± 11.4 |
| BMI, kg/m2 | 27.0 ± 4.1 | 27.5 ± 3.9 | 26.7 ± 4.3 |
| Percent body fat, % | 28.4 ± 9.0 | 23.7 ± 5.9 | 36.4 ± 8.3* |
| LBM, kg | 54.9 ± 11.5 | 61.5 ± 8.1 | 43.0 ± 2.9* |
| Fasting glucose, mg/dl | 85.4 ± 7.1 | 86.8 ± 7.5 | 84.4 ± 6.9* |
| Mean RQ, % | 0.86 ± 0.05 | 0.86 ± 0.05 | 0.86 ± 0.05 |
| CHO, g · kg−1 · day−1 | 3.21 ± 1.59 | 3.22 ± 1.75 | 3.19 ± 1.35 |
| FAT, g · kg−1 · day−1 | 1.18 ± 0.54 | 1.21 ± 0.53 | 1.13 ± 0.58 |
| REE, kcal/day | 1,615.4 ± 329.9 | 1,807.1 ± 260.9 | 1,308.8 ± 141.5† |
Values are means ± SD; n = no. of study subjects. GDR, glucose disposal rate; H, Hispanic American; E, European American; A, African American; CHO, carbohydrate oxidation; FAT, fat oxidation; LBM, lean body mass; REE, resting energy expenditure. Significantly different from men:
P < 0.001,
P < 0.05.
Body Composition and Indirect Calorimetry
Total body fat and lean body mass (LBM) were measured by dual-energy X-ray absorptiometry (Lunar Radiation, Madison, WI), as previously described (38). After an overnight fast, and while at the GCRC, REE was measured by indirect calorimetry using a Deltratrac metabolic monitor (Deltratrac II, SensorMedics, Yorba Linda, CA). Measurements were performed in supine position and began 30 min after each subject was awakened from overnight sleep. The instrument was calibrated by ethanol combustion tests on a monthly basis and against standard gases before each test. Expired air was collected using the adult-size ventilated canopy system for 20 min after a 10-min equilibration. Whole body oxygen consumption (V̇o2) and CO2 production (V̇co2) were calculated by measuring gradients across the face and the flow rates of air using Haldane transformation. In our laboratory, the coefficient of variation between REE measured in a walk-in room calorimeter and the Deltratrac metabolic monitor is 7.3%. Energy expenditure and substrate oxidation rates were determined from the RQ value and the tables of Lusk and Du Bois (43). Under aerobic conditions, respiration of fat gives an RQ of 0.7, respiration of protein gives an RQ of 0.9, and respiration of carbohydrate gives an RQ of 1.0. A mixed diet of fat and carbohydrate results in an average value between these numbers.
Insulin Sensitivity
In vivo insulin sensitivity was assessed using the euglycemic-hyperinsulinemic glucose-clamp technique at a maximally effective steady-state serum insulin concentration as previously described (30). After a 12-h fast, a catheter was inserted into the brachial vein to administer insulin, glucose, and K3PO4. A dorsal hand vein was cannulated in a retrograde manner and kept in a warming device (65°C) to provide arterialized venous blood for sampling. For maximally stimulating glucose uptake and suppressing hepatic glucose production, regular insulin (Humulin; Eli Lilly, Indianapolis, IN) was administered at a rate of 200 mU·m−2·min−1, producing a mean steady-state insulin concentration of 3,480 ± 138 pmol/l, which is maximally effective for stimulating glucose uptake into skeletal muscle. Plasma glucose was clamped at 5.0 mmol/l for at least 3 h, and maximal glucose uptake for each individual was calculated from the mean glucose infusion rate over the final three 20-min intervals. Whole body glucose uptake was calculated on the basis of the glucose infusion rate corrected for changes in the glucose pool size, assuming a distribution volume of 19% body weight and a pool fraction of 0.65. Glucose uptake was normalized per kilogram of lean body mass (excluding bone mass) determined by dual-energy X-ray absorptiometry to yield the glucose disposal rate (GDR) per kilogram of lean body mass.
Measurement of Plasma Glucose and Serum Insulin Levels
Plasma glucose was measured by the glucose oxidase method using a glucose analyzer (YSI 2300; Yellow Springs Instruments, Yellow Springs, OH). Serum insulin levels were measured using an electrochemiluminescence immunoassay (Roche Diagnostics, Mannheim, Germany).
Muscle Biopsies
Percutaneous needle biopsies of the vastus lateralis (∼400 mg) were performed as previously described (28, 29, 76). Muscle tissue was then blotted on a sterile cloth, placed immediately into liquid N2, and then transferred to a −80°C freezer for storage.
RNA Extraction and Microarray Hybridization
RNA was isolated by phenol/chloroform extraction using the Ultraspec RNA isolation Kit (Biotecx Laboratories, Houston, TX) followed by a DNase I treatment. RNA quality from each biopsy was examined by the A260/A280 absorbance ratio and by electrophoresis of RNA on agarose formaldehyde gels. Our RNA samples had an A260/A280 ratio of 1.8–2.1, and no evidence of RNA degradation was detected on agarose gels in the samples that underwent analysis. In vitro transcription was accomplished using double-stranded cDNA and the MEGAscript T7 System (Ambion, Houston, TX) in the presence of biotinylated UTP and CTP to generate labeled antisense RNA. cRNA was separated from unincorporated nucleotides using RNeasy spin columns (Qiagen, Valencia, CA). The cRNA was then resuspended in diethyl pyrocarbonated-treated (DEPC) water for microarray study. Microarray analyses of human skeletal muscle biopsies were conducted using the Affymetrix Hu95A chips as previously described (76).
Statistical Data Analysis
Clinical data.
To explore the optimal approach for normalizing REE measurements, we performed simple ordinary least-squares (OLS) regression analysis to determine the associations between REE and different anthropometric and metabolic variables, including BMI, total fat mass, percentage fat, LBM, and RQ. This was followed by multiple regression models using REE as the dependent variable and BMI, total fat mass, percentage fat, LBM, and RQ as independent variables.
Microarray data.
We first normalized the data using quantile normalization (11) and then obtained the probe set intensity using robust multi-array average (RMA) (35) as implemented in the affy package in Bioconductor (http://www.bioconductor.org/). We also generated present/absent calls for each probe set based on the Wilcoxon signed-rank test using the mass5call function in the affy package to filter out low-expression genes based on present calls (45).
For identifying differentially expressed genes, we used ordinary least-squares regression analysis to test for correlation between gene expression and REE or RQ values. The gene expression values were regressed on these two clinical phenotypes at each probe set separately with or without sex and age as covariates. The regression analysis indicated that P values from the unadjusted and adjusted analyses are nearly the same, which suggests strongly that sex and age do not significantly affect the expression level of genes that are involved in regulating resting energy expenditure and respiratory quotient (data not shown). Therefore, we chose to present the analysis data without adjustment for sex and age. The individual was dropped from that regression analysis if either phenotype or expression was missing. For pathway analyses, nominal P value <0.05 was deemed as statistically significant. We also computed the posterior true positive probabilities (PTP) and posterior false positive probabilities (PFP) based on a mixture model approach using the High Dimension Biology Statistical Analysis (HDBStat!) software (http://www.ssg.uab.edu/hdbstat/) (3). The association between gene expression data and the human clinical data analysis was conducted using SAS statistical software (version 9.1.3).
Genes that reached statistical significance for differential expression were subjected to PubMed search for their identified functions. Functional inferences are based on the authors' interpretation of literature referenced in Online Mendelian Inheritance in Man (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM&itool=toolbar) and Stanford Source (http://smd.stanford.edu) pertaining to each gene. For further standardized analyses of gene function, we used Gene Ontology (GO) for gene annotations (www.geneontology.org). GoArray and FuncAssociate analysis programs were used for statistical analyses of effects on gene categories in the context of the domain knowledge represented in the Gene Ontology Database (http://goarray.med.yale.edu/GOArray/) (5, 9, 52, 77). The schematic overview of the data analysis is shown in Fig. 1.
Fig. 1.
Schematic overview figure of data analysis. EDR, expected discovery rate; FDR, false discovery rate; REE, resting energy expenditure; RQ, respiratory quotient.
Quantitative Reverse Transcriptase-Polymerase Chain Reaction (qPCR)
Total RNA was extracted from tissues using RNeasy mini kit and treated with DNase I (Qiagen, CA). RNA quality from each biopsy was examined by the A260/A280 absorbance ratio and by electrophoresis of RNA on agarose formaldehyde gels. RNA quality was assessed using the RNA 6000 Nano LabChip kit (Agilent Bioanalyser 2100, Agilent Technologies). Our RNA samples have an A260/A280 ratio of 1.8–2.1. A total of 1 μg of RNA was reverse transcribed per reaction using first-strand complementary DNA synthesis with random primers (SuperScriptTM III reverse transcriptase kit, Invitrogen). qPCR was performed using the ABI Prism 7700 (Applied Biosystem, CA) and sequence-specific PCR primers and TaqMan probes for: O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) (OGT) [probe (5′-ACCCCCATGGTGACTATGCCAGGA-3′) and primers (5′-CACACCACAGGGATGGATGTC-3′ and 5′-TGCAACTCGAGAAGCAAGAGTCT-3′)]; thyroid hormone receptor, α (THRA) [probe (5′-AGCAGAGCCACTTCCGTGTCATCCA-3′) and primers (5′-CGAACTGGGCAAGTCACTCTCT-3′ and 5′-CCGAGCGGTCTGTTGACATT-3′)]; nuclear receptor subfamily 1, group D, member 1 (NR1D1) [probe (5′-CCCACACACTTTACACAGTAACACCATGCCAT-3′) and primers (5′-CACCAGCAACATCACCAAGCT-3′ and 5′-CAGGCGTGCACACCGTAGT-3′)]; and protein phosphatase 1, regulatory (inhibitor) subunit 3A (PPP1R3A) [probe (5′-CACAGCAGATGTGTCTCATTCACCAAGGA-3′) and primers (5′-CTGAGCGAACATACCGCAATC-3′ and 5′-CTAAAGCGCTGCCTTCACTAGTC-3′)]. To quantify transcripts for genes of interest, we used GAPDH transcript as an internal control, and each sample was normalized with respect to GAPDH transcript content (Applied Biosystem). All assays were performed in duplicate, and the average values were used for analysis.
RESULTS
In simple regression analyses using demographic, anthropometric, and metabolic characteristics of the study subjects, REE was highly correlated with LBM (r2 = 0.85, P < 0.001) and also with BMI (r2 = 0.48, P < 0.01), but the correlation between REE and percentage fat did not reach statistical significance. When both LBM and BMI were included in the same multiple regression model, BMI lost significance (P > 0.05) while LBM explained most of the variance in REE (r2 = 0.73, P < 0.001). Mean RQ was not correlated with either REE (r2 = −0.12), body adiposity (r2 = −0.07), or LBM (r2 = −0.14) (P = NS). For purposes of examining the relationship between energy expenditure and muscle gene expression, REE rates were normalized per kilogram of LBM.
Number of Gene Transcripts Associated with REE and RQ
We have deposited the raw microarray data into GEO public database following MIAME standards (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE22309). To identify genes that were differentially expressed as a function of REE or RQ, we used a regression model written as gene expression j = β0j + β1j × X (phenotype, either REE or RQ), where j represents probe set number. Using the mixture model approach of Allison et al. (3), the number of genes that were truly significantly correlated with REE or RQ was estimated by fitting the mixture model to the distribution of P values. Supplementary Tables 1A and 1B, available with the online version of this article, show significant positive and negative correlations, respectively, between gene expression values and REE/LBM, including P values from the regression model, posterior true positive probabilities (PTP), and posterior false positive probabilities (PFP). For a particular gene, PTP is an estimate of the proportion of genes that are truly differentially expressed among the genes with an observed P value smaller than the observed P value of this particular gene. PFP is an estimate of the proportion of genes that do not have a true differential expression among the genes with an observed P value smaller than the observed P value of this particular gene. Supplementary Tables 2A and 2B, available with the online version of this article, show significant positive and negative correlations between gene expression values and RQ, respectively.
Differential Expression by Functional Categories
The microarrays used in the present work allowed us to analyze mRNA levels encoding 12,626 genes and expressed sequence tags (ESTs) detected in human skeletal muscle. Of these genes, 1,280 were positively correlated to REE/LBM, while 1,112 were negatively correlated to REE/LBM. The number of genes significantly correlated with RQ was much smaller than those with REE/LBM: 442 genes were positively correlated to RQ, whereas 673 genes were negatively correlated to RQ.
Gene Ontology annotations, Stanford Source (http://smd.stanford.edu), and PubMed (www.ncbi.nlm.nih.gov), were used to assign these candidate genes into different functional categories. The full lists of the gene classifications are shown in Table 2. Genes correlated with REE/LBM encode proteins involved in transcriptional regulation (13%), receptors/carriers/transporters (11.9%), intracellular signaling (10.7%), cell cycle/apoptosis/oncogenes/tumor suppressor genes (9%), and translational regulation (8.8%). Other functional categories consisted of genes involved in intermediary and energy metabolism (2.9%), cytoskeleton (3.6%), vesicle trafficking (1.1%), mitochondrial function (1.8%), and immune response (3.4%). In addition, 810 genes (490 up, 320 down) did not belong to any of these major categories. The chief functional categories of genes correlating with RQ were gene transcription (11.8%), protein metabolism (13.6%), receptors, channels, carriers, and transporters (9.9%), and cell cycle/apoptosis/oncogenes/tumor suppressor genes (10.6%).
Table 2.
Functional classification of genes related to REE/LBM or to RQ
| Functional Classification | Total Genes/LBM | Negative Correlation | Positive Correlation |
|---|---|---|---|
| Genes related to REE/LBM (adjusted P < 0.05) | |||
| Carbohydrate and lipid metabolism | 69 (2.9%) | 31 | 38 |
| Protein metabolism | 210 (8.8%) | 129 | 81 |
| Gene expression | 311 (13.0%) | 199 | 112 |
| Vesicle trafficking | 27 (1.1%) | 16 | 11 |
| Muscle fiber, cytoskeleton and microtubule | 86 (3.6%) | 48 | 38 |
| Cell cycle/apoptosis/oncogenes/tumor suppressor genes | 216 (9.0%) | 119 | 97 |
| Mitochondria | 42 (1.8%) | 28 | 14 |
| Receptors, channels, and transporter | 284 (11.9%) | 97 | 187 |
| Signaling | 256 (10.7%) | 97 | 159 |
| Cytokines/secreted proteins/extracellular proteins | 81 (3.4%) | 28 | 53 |
| Miscellaneous/EST/hypothetical proteins | 810 | 320 | 490 |
| Total | 2,392 | 1,112 | 1,280 |
| Genes related to RQ (adjusted P < 0.05) | |||
| Carbohydrate and lipid metabolism | 37 (3.3%) | 32 | 5 |
| Protein metabolism | 152 (13.6%) | 109 | 43 |
| Gene expression | 132 (11.8%) | 95 | 37 |
| Vesicle trafficking | 11 (0.9%) | 9 | 2 |
| Muscle fiber, cytoskeleton and microtubule | 27 (2.4%) | 15 | 12 |
| Cell cycle/apoptosis/oncogenes/tumor suppressor genes | 118 (10.6%) | 70 | 48 |
| Mitochondria | 28 (2.5%) | 24 | 4 |
| Receptors, channels, and transporter | 110 (9.9%) | 50 | 60 |
| Signaling | 78 (7.0%) | 53 | 25 |
| Cytokines/secreted proteins/extracellular proteins | 47 (4.2%) | 21 | 26 |
| Miscellaneous/EST/hypothetical proteins | 375 | 195 | 180 |
| Total | 1,115 | 673 | 442 |
REE, resting energy expenditure; RQ, respiratory quotient; EST, expressed sequence tag; LBM, lean body mass.
To examine the determinants of REE and RQ in a more standardized and systematic manner, all genes were classified to different categories corresponding to molecular processes, biochemical functions/pathways, or subcellular components by using the Gene Ontology (GO) program. The GoArray and FuncAssociate Gene Ontology programs were then used to statistically identify gene categories that overrepresented genes that were positively or negatively correlated with REE (or RQ). Only those gene categories that were top-ranked by both GoArray and FuncAssociate programs were considered in our data interpretation to give higher confidence in the results. These gene categories were then analyzed in relation to REE and RQ. Table 3 summarizes gene categories that were significantly correlated with REE/LBM, as does Table 4 for RQ. In addition, the full lists of genes significantly associated with REE/LBM (positively and negatively) identified from regression analysis and mixed model are shown in Supplementary Tables 1A and 1B, respectively, and in Supplementary Tables 2A and 2B for positive or negative correlation with RQ (each available with the online version of this article).
Table 3.
Gene categories identified by both the GoArray program and the FuncAssociate program in which gene expression was positively or negatively correlated with REE/LBM
| DAG Accession | DAG Node Name | z-Score | P Value | (Meet Criteria)/Total Genes | DAG Node Depth | False Positive for Node |
|---|---|---|---|---|---|---|
| Gene expression positively correlated with REE/LBM | ||||||
| Molecular function | ||||||
| GO:0001584 | Rhodopsin-like receptor activity | 5.811 | 6.18e-09 | 38/135 | 7 | 6/1,000 |
| Biological process | ||||||
| GO:0007186 | G-protein coupled receptor protein signaling pathway | 5.726 | 1.02e-08 | 65/283 | 7 | 0/1,000 |
| GO:0007268 | Synaptic transmission | 5.812 | 6.15e-09 | 46/176 | 7 | 0/1,000 |
| Gene expression negatively correlated with REE/LBM | ||||||
| Molecular function | ||||||
| GO:0016790 | Thiolester hydrolase activity | 4.686 | 2.79e-06 | 13/36 | 6 | 6/1,000 |
| GO:0004221 | Ubiquitin thiolesterase activity | 5.258 | 1.45e-07 | 12/28 | 9 | 5/1,000 |
| Biological process | ||||||
| GO:0043285 | Biopolymer catabolic process | 3.757 | 1.72e-04 | 28/128 | 6 | 5/1,000 |
| GO:0006512 | Ubiquitin cycle | 6.559 | 5.41e-11 | 58/232 | 8 | 7/1,000 |
| GO:0008380 | RNA splicing | 5.774 | 7.73e-09 | 28/92 | 8 | 5/1,000 |
GO (www.geneontology.org) was used to annotate genes, and GO numbers are indicated for each function and pathway. P value was calculated as previously described (50). A z-score is assigned to each term based on the number of genes associated with that term or any of its children relative to the total numbers of gene of interest (GOI) and non-GOI (NGOI). DAG, directed acyclic graph; URLs: GoArray program, http://goarray.med.yale.edu/GOArray; FuncAssociate program, http://llama.med.harvard.edu/cgi/func/funcassociate).
Table 4.
Gene categories identified by both the GoArray program and the FuncAssociate program in which gene expression was positively or negatively correlated with RQ
| DAG Accession | DAG Node Name | z-Score | P Value | (Meet Criteria)/Total Genes | DAG Node Depth | False Positive for Node |
|---|---|---|---|---|---|---|
| Gene expression positively correlated with RQ | ||||||
| Molecular function | ||||||
| GO:0008236 | Serine-type peptidase activity | 3.407 | 6.55e-04 | 9/76 | 6 | 7/1000 |
| Gene expression negatively correlated with RQ | ||||||
| Molecular function | ||||||
| GO:0051082 | Unfolded protein binding | 4.697 | 2.64e-06 | 19/97 | 5 | 12/1000 |
| Cellular component | ||||||
| GO:0000159 | Protein phosphatase type 2A complex | 4.309 | 1.64e-05 | 4/9 | 7 | 26/1000 |
| GO:0005739 | Mitochondrion | 3.309 | 9.35e-04 | 38/315 | 9 | 7/1000 |
| Biological process | ||||||
| GO:0045333 | Cellular respiration | 4.008 | 6.11e-05 | 7/25 | 7 | 24/1000 |
See Table 3 legend for program URLs, abbreviations, and further details.
Pathways and effector systems linked positively to regulation of REE include G protein-coupled receptor protein signaling pathway (meet criteria genes/total genes, 65 of 283, P value = 1.02e-08), rhodopsin-like receptor activity (38 of 135, P value = 6.18e-09), and synaptic transmission (46 of 176, P value = 6.15e-09). The basis for the involvement of the G protein-coupled receptor protein signaling pathway involved differential expression of receptors binding norepinephrine, epinephrine, dopamine, gamma-aminobutyric acid (GABA)-A, 5-hydroxytryptamine (serotonin), opioids, and glutamate. These receptors mediate effects of the sympathetic nervous system and other neuroregulatory systems. Categories that were negatively correlated with REE include the ubiquitin cycle (58 of 232, P value = 5.41e-11), RNA splicing (28 of 92, P value = 7.73e-09), ubiquitin thiolesterase activity (12 of 28, P value = 1.45e-07), thiolester hydrolase activity (13 of 36, P value = 2.79e-06), and biopolymer catabolic process (28 of 128, P value = 1.72e-04). In particular, there appeared to be a broad-based upregulation of ubiquitin-related pathways, in which the genes in this category were negatively related to REE (Supplemental Table 1B) including ubiquitination factors (UBE1C, UBE4A, UBE4B), ubiquitin conjugating enzymes (ARIH1, UBE2B, UBE2D3, UBE2G1, UBE2J1, UBE2N, UBE2V2, WWP2), ubiquitin-specific proteases (USP1, USP7, USP8, USP9X, USP10, USP14, USP15, USP33, USP52), ubiquitin esterase (UCHL3), and ubiquitin like protein 1.
Skeletal muscle is able to switch between carbohydrates and fat as fuel substrates depending on energy requirement and substrate availability. Serine-type peptidase activity (9 of 76, P value = 6.55e-04) was the only Gene Ontology gene category that was shown by both GoArray and FuncAssociate programs to be positively correlated with RQ values, thus promoting carbohydrate as the preferred fuel substrate over fat. On the other hand, cell systems negatively correlated to RQ include protein phosphatase type 2A complex (4 of 9, P value = 1.64e-05), the mitochondrion (38 of 315, P value = 9.35e-04), cellular respiration (7 of 25, P value = 6.11e-05), and unfolded protein binding (19 of 97, P value = 2.64e-06).
mRNA Level of Gene Transcripts Measured by qPCR Related to REE/LBM and RQ
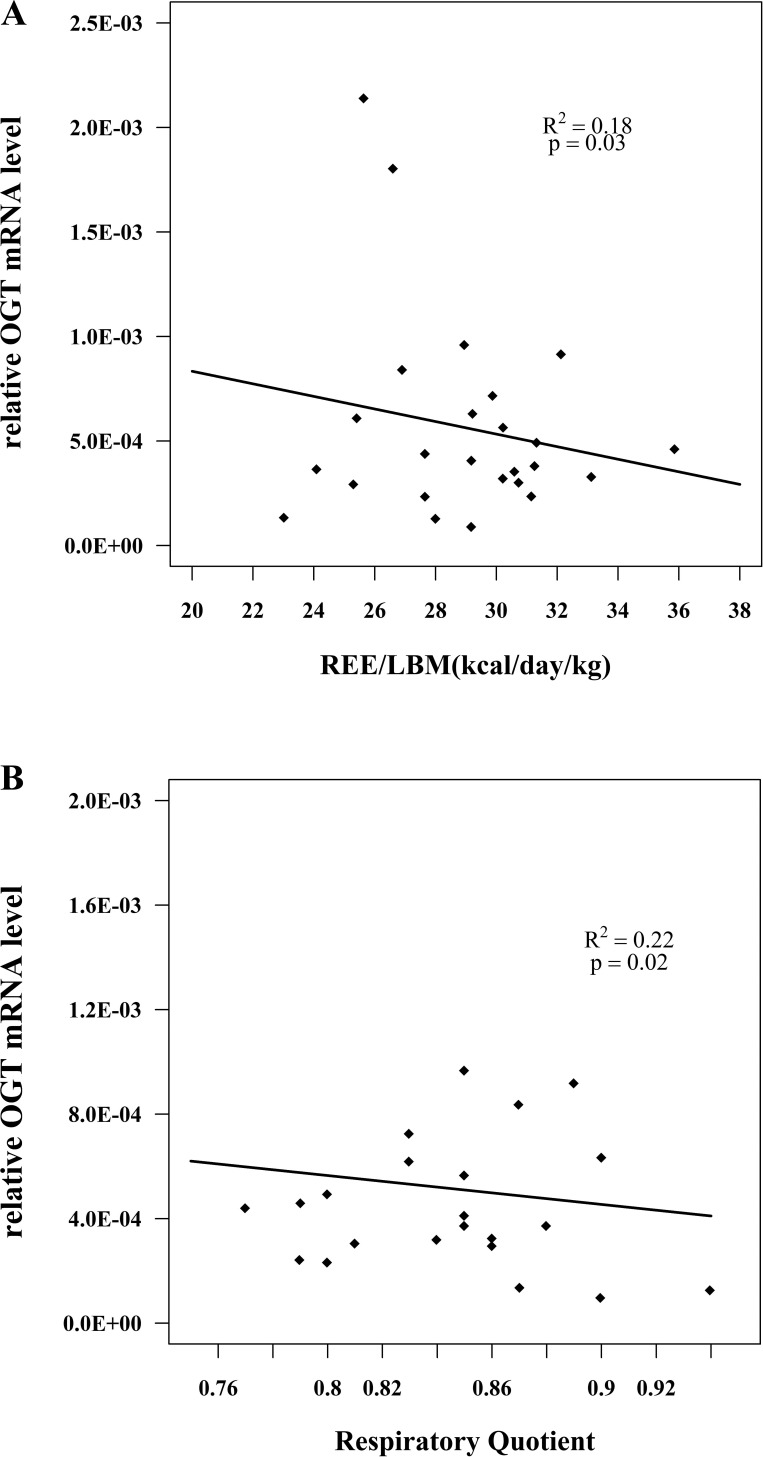
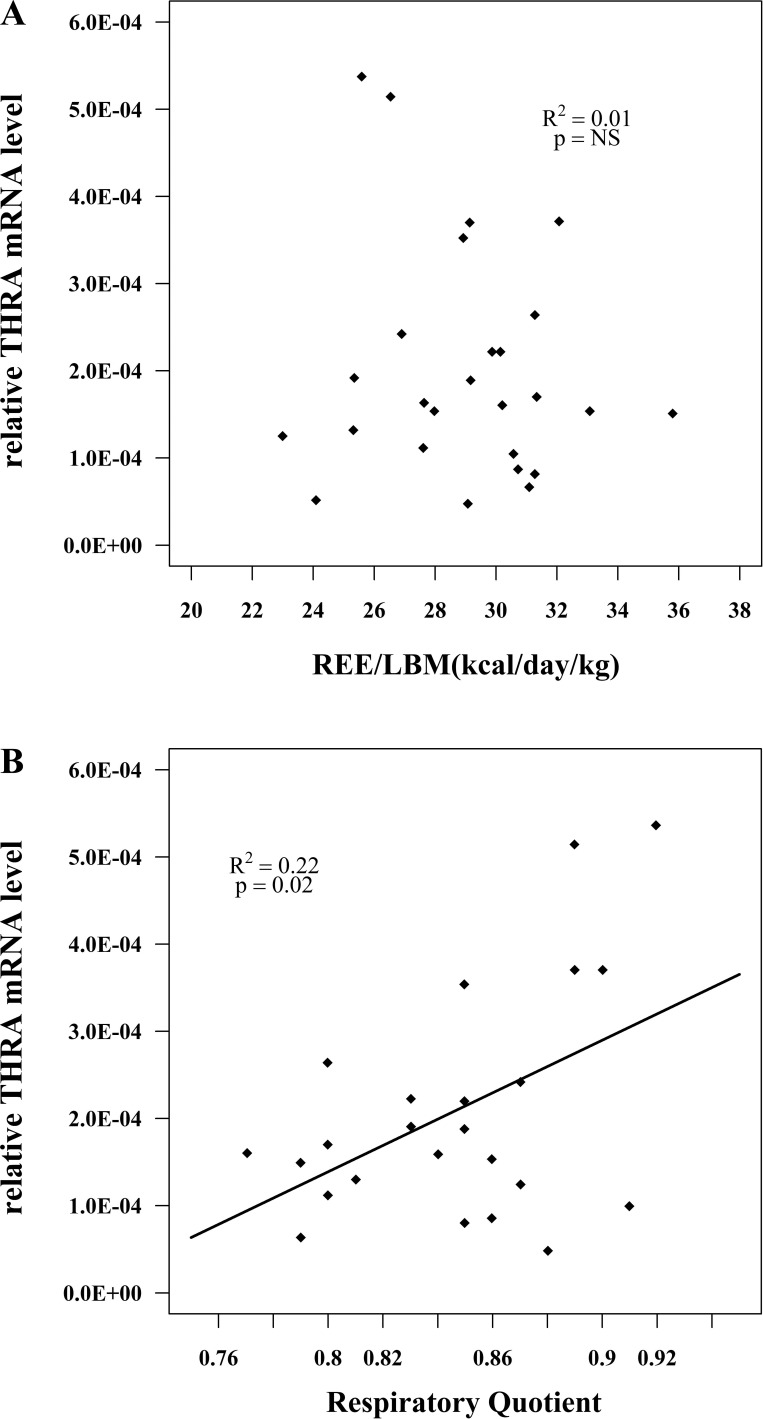
To confirm microarray results, we measured the mRNA level of four gene transcripts: OGT, THRA, NR1D1 and PPP1R3A. Since REE/LBM and RQ are continuous variables, to further assess the relationship between these four genes and REE/LBM as well as RQ, we correlated the values of mRNA level in human subjects with REE/LBM and RQ. We observed a statistically significant inverse relationship between OGT and REE/LBM (r2 = 0.18, P < 0.05) and between OGT and RQ (r2 = 0.22, P < 0.05) (Fig. 2, A and B). We also observed a statistically significant positive relationship between THRA and RQ (r2 = 0.22, P < 0.05), whereas there was no significant correlation between THRA and REE/LBM (r2 = 0.01, P > 0.05) (Fig. 3, A and B). These correlations observed using qPCR to assess mRNA levels confirmed the same significant correlations observed in the microarray data. Furthermore, we tested two genes, NR1D1 and PPP1R3A, whose expression level showed no significant correlation to either REE/LBM or RQ in the microarray study. The results from qPCR similarly showed lack of a correlation in confirmation of the microarray data (data not shown).
Fig. 2.
Relationships between muscle O-linked N-acetylglucosamine transferase (OGT) mRNA expression and resting energy expenditure normalized by lean body mass (REE/LBM) (A) and respiratory quotient (B). OGT mRNA level was determined by qPCR. Data were based on studies of healthy normoglycemic individuals. The correlation coefficient (r) and P values are shown for each graph. These relationships were similarly demonstrated to be statistically significant in the microarray experiments.
Fig. 3.
Relationships between muscle thyroid hormone receptor-α (THRA) mRNA expression and resting energy expenditure normalized by lean body mass (REE/LBM) (A) and respiratory quotient (B). THRA mRNA level was determined by qPCR. Data were based on studies of healthy normoglycemic individuals. The correlation coefficient (r) and P values are shown for each graph. These relationships were similar to those demonstrated by microarray.
DISCUSSION
This is the first high-throughput study to explore the genetic determinants of energy metabolism and fuel utilization using microarray technology to comprehensively assess the transcriptional profile in human skeletal muscle. Because individual variability in REE is heavily dependent on differences in LBM, REE values were normalized for LBM in each subject. More than 85% of genes on the microarray did not significantly correlate to REE. Approximately 15% of the genes were differentially expressed as a function of REE/LBM in a statistically significant manner, indicating possible bases for orchestrated regulation of cellular functions that determine variation in energy metabolism. The analysis of these data has elucidated patterns of transcriptional regulation and biochemical pathways in skeletal muscle that affect whole body REE and fuel partitioning.
Positive Regulation of REE
Our microarray data indicate that increased gene expression of receptors that are involved in rhodopsin-like receptor and G protein-coupled receptor signaling pathways is associated with increased REE. This includes genes encoding adrenergic receptors, dopamine receptors, G protein-coupled receptors, serotonin receptors, opioid receptors, GABA-A receptors, glutamate receptors, and nicotinic cholinergic receptors. Most of these receptors function in synaptic transmission processes. In particular, genetic expression of α-adrenergic receptors (ADRA1β) and β-adrenergic receptors (ADRB3) was found to be positively correlated to REE. These receptors are keys for promulgating effects of the SNS. Activation of SNS pathways is known to augment each of the major components of energy expenditure: REE (26, 65), the thermic effect of food (61), spontaneous physical activity (16), and 24-h RQ (64). In particular, β-adrenergic receptor signaling influences total daily energy expenditure via modulation of REE in humans (8, 42, 47, 74). Catecholamines can increase REE via β3-adrenergic receptor (51), and β3-agonists act on cognate receptors in skeletal muscle (6), brown adipose tissue (27), and white adipose tissue to promote weight loss and improve insulin sensitivity (33, 51, 60). Systemic administration of isoproterenol (a sympathomimetic β-adrenergic receptor agonist) also results in a dose-dependent increase in energy expenditure in humans, accompanied by an increase in RQ and carbohydrate oxidation rate (12). Intravenous propranolol (a β-adrenergic receptor antagonist) infusion caused an acute decrease in REE (47). It is also important to consider that obesity is generally characterized by reduced SNS activity (51, 69). For example, Pima Indians prone to obesity have lower rates of muscle sympathetic activity compared with weight-matched Caucasians (65) along with lower energy expenditure (67), and low SNS activity was associated with poor weight loss outcomes in obese individuals treated with a dietary restriction intervention (68). The current data indicate that it is not only the activity of effluent SNS pathways that can regulate energy metabolism; in addition, the end-organ response (i.e., skeletal muscle) can also impact REE via intrinsic changes in gene regulation, namely the upregulation of adrenergic receptors in muscle.
Regarding other receptors, dopamine receptors D2 and D4 were also positively correlated with REE. This is consistent with the observation that bromocriptine, a dopamine receptor D2 agonist, has been shown to increase oxygen consumption and REE in humans (40). In addition, a loss-of-function mutation of the human dopamine receptor D2 gene has been associated with reduced energy expenditure (66). Serotinin receptor expression (HTR1B, HTR1D, HTR1E, HTR4, HTR7) was also positively correlated with REE. While the mechanism linking serotonin receptor upregulation and REE is not clear, this would be predicted to augment serotonin (or 5-hydroxytryptamine) action, which has been shown to positively regulate glucose transport in rat skeletal muscle (32). Similarly, the association between expression of opioid receptors (opioid receptor kappa 1, opioid receptor mu 1, opioid receptor sigma 1, opioid growth factor receptor) and REE is deserving of more research, given that opioid receptors in the central nervous system regulate both appetite (44) and REE (36). We also noted that upregulation of genes encoding receptor ligands influences REE. For example, increased expression of both neuropeptide Y (NPY) and neuropeptide FF-amide peptide precursor (NPFF) were associated with increased energy expenditure. NPY is an important sympathetic neurotransmitter involved in local neurovascular regulation of skeletal muscle energy metabolism, and NPFF plays a role in the regulation of heart rate and blood pressure. Thus norepinephrine and NPY have been shown to act synergistically in causing a reduction in body weight associated with an increase in resting metabolic rate (1).
Negative Regulation of REE
Ubiquitin-proteasome-dependent proteolytic pathways were negatively correlated to REE, including genes encoding ubiquitin-conjugating enzymes, ubiquitin-specific proteases, and proteasomal proteins. In skeletal muscle, the ubiquitin-proteasome pathway is rapidly up- and downregulated during cycles of muscle atrophy and muscle accretion (72) and is the major mechanism leading to increased muscle protein breakdown during starvation (25). Starvation is also characterized by reduced energy expenditure and substrate metabolism by muscle (34), and prolonged starvation can lead to decrements in metabolic rates of up to 20–30% (14, 48). Upregulation of genes related to the ubiquitin-proteasome system in 24-h-fasted rats suggests that the ubiquitin-proteasome system in starvation is rapidly activated in skeletal muscle even before overt proteolysis (18, 49). Our results in isocaloric individuals suggest that the ubiquitin-proteasome system could negatively modulate REE and explain differences in REE among healthy individuals, as well as mediate both a further decrement in REE and protein catabolism during periods of starvation.
Positive Regulation of RQ
The skeletal muscle is able to switch between carbohydrates and fat as fuel substrates depending on energy requirements. However, even under basal conditions, there exists wide individual variation in fuel preferences. Apart from the impact of diet composition, RQ is also influenced by recent energy balance, sex, adiposity, and, importantly, familial aggregation of obesity (70, 79). Genes that are positively correlated with RQ will alter macronutrient fuel partitioning in favor of carbohydrates by promoting carbohydrate oxidation or impairing lipid oxidation. Increased expression of genes encoding serine-type peptidase activity is associated with high RQ values, including serine proteases (PRSS3, PRSS16, PRTN3), dipeptidylpeptidase (DPP6), chymotrypsin (CTRL), and elastase 2 (ELA2, ELA2A). Serine proteases cleave and inactivate musclin, a muscle-derived protein that impairs glucose metabolism (50), but any relevance to our current results is speculative. To our knowledge, we are the first to report a correlation between serine-type peptidases and muscle fuel partitioning, and this is an area deserving of future study.
Negative Regulation of RQ
Genes that favor lipid oxidation or reduce carbohydrate oxidation in basal muscle will lower RQ. We observed that the major functional categories responsible for preferential fat oxidation included the protein phosphatase 2A complex, mitochondrial function, and cellular respiration, due to differential expression of genes including protein phosphatase 2A (PP2A), ATP synthase, H+ transporters, mitochondrial F0 complex, NADH dehydrogenase (ubiquinone), succinate-CoA ligase (alpha subunit of mitochondrial succinyl CoA synthetase), translocase of outer mitochondrial membrane, ubiquinol-cytochrome c reductase, and heat shock protein. PP2A acts to dephosphorylate signaling molecules and can blunt insulin stimulation of glucose metabolism (i.e., glucose uptake, glycogen synthesis). The genes pertaining to mitochondrial function and cell respiration would clearly increase capacity for fatty acid oxidation, resulting in reduced RQ values.
There are two notable limitations of the study that deserve mention. First, it is important to keep in mind that the muscle biopsy contains cell types other than myocytes, including cells comprising the microvasculature, adipocytes, and connective tissue. Thus, while mRNA from muscle cells will predominate, differential gene expression could be due to changes in other cell types. Second, the method used for normalization of Affymetrix array data can have a major impact on the results (39, 46, 62). We used RMA in our manuscript, which is the most commonly chosen normalization method (2). However, a gold standard method for microarray data normalization has not been yet defined. And third, we only chose to present the gene categories that are identified by both GoArray and FuncAssociate pathway programs, which could have resulted in an overconservative analysis. In any event, the data identified biochemical pathways and processes in skeletal muscle that predict individual differences in resting energy expenditure and fuel partitioning. Importantly, neuroregulation of REE does not only depend on central (autonomic) nervous system activity, but also, individual variation in REE is determined by differential expressions of neurotransmitter receptors intrinsic to skeletal muscle, which can affect the end-organ metabolic response. The correlations we investigated looked for linear associations between the response and gene expression. More complicated associations might either be missed or exaggerated when fitting a linear model.
GRANTS
This work was supported by grants from the National Institutes of Health (DK-038764, DK-083562, and HL-055782 to W. T. Garvey), by an American Heart Association Beginning Grant-in-Aid (X. Wu), and by the Merit Review program of the Department of Veterans Affairs (W. T. Garvey). We gratefully acknowledge the support of the research core facilities of the University of Alabama at Birmingham (UAB) Center for Clinical and Translational Science (UL1-RR-025777), the UAB Nutrition and Obesity Research Center (P30-DK-56336), and the UAB Diabetes Research and Training Center (P60-DK-079626).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
We thank Jelai Wang and Tapan Shirish Mehta for excellent technical assistance. We are grateful for the participation of our research volunteers.
Glossary
- BMI
Body mass index
- CHO
Carbohydrate oxidation rate
- DEPC water
Diethyl pyrocarbonated-treated water
- EDR
Expected discovery rate
- EST
Expressed sequence tag
- GABA
γ-Aminobutyric acid
- GDR
Glucose disposal rate
- LBM
Lean body mass
- NPY
Neuropeptide Y
- NR1D1
Nuclear receptor subfamily 1, group D, member 1
- OGT
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
- PFN
Posterior false negative probabilities
- PFP
Posterior false positive probabilities
- PPP1R3A
Protein phosphatase 1, regulatory (inhibitor) subunit 3A
- PTP
Posterior true positive probabilities
- REE
Resting energy expenditure
- RQ
Respiratory quotient
- TEE
Thermic effect of exercise
- TEF
Thermic effect of food
- THRA
Thyroid hormone receptor, α
REFERENCES
- 1. al-Arabi A, Andrews JF. Synergistic action by neuropeptide Y (NPY) and norepinephrine (NE) on food intake, metabolic rate, and brown adipose tissue (BAT) causes remarkable weight loss in the obese (fa/fa) Zucker rat. Biomed Sci Instrum 33: 216–225, 1997. [PubMed] [Google Scholar]
- 2. Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet 7: 55–65, 2006. [DOI] [PubMed] [Google Scholar]
- 3. Allison DB, Gadbury GL, Heo M, Fernandez JR, Lee CK, Prolla TA, Weindruch R. A mixture model approach for the analysis of microarray gene expression data. Comput Statistics Data Analysis 39: 1–20, 2002. [Google Scholar]
- 4. Argyropoulos G, Brown AM, Peterson R, Likes CE, Watson DK, Garvey WT. Structure and organization of the human uncoupling protein 2 gene and identification of a common biallelic variant in Caucasian and African-American subjects. Diabetes 47: 685–687, 1998. [DOI] [PubMed] [Google Scholar]
- 5. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Astrup A, Bulow J, Madsen J, Christensen NJ. Contribution of BAT and skeletal muscle to thermogenesis induced by ephedrine in man. Am J Physiol Endocrinol Metab 248: E507–E515, 1985. [DOI] [PubMed] [Google Scholar]
- 7. Bao S, Kennedy A, Wojciechowski B, Wallace P, Ganaway E, Garvey WT. Expression of mRNAs encoding uncoupling proteins in human skeletal muscle: effects of obesity and diabetes. Diabetes 47: 1935–1940, 1998. [DOI] [PubMed] [Google Scholar]
- 8. Bell C, Day DS, Jones PP, Christou DD, Petitt DS, Osterberg K, Melby CL, Seals DR. High energy flux mediates the tonically augmented beta-adrenergic support of resting metabolic rate in habitually exercising older adults. J Clin Endocrinol Metab 89: 3573–3578, 2004. [DOI] [PubMed] [Google Scholar]
- 9. Berriz GF, King OD, Bryant B, Sander C, Roth FP. Characterizing gene sets with FuncAssociate. Bioinformatics 19: 2502–2504, 2003. [DOI] [PubMed] [Google Scholar]
- 10. Bogardus C, Lillioja S, Ravussin E, Abbott W, Zawadzki JK, Young A, Knowler WC, Jacobowitz R, Moll PP. Familial dependence of the resting metabolic rate. N Engl J Med 315: 96–100, 1986. [DOI] [PubMed] [Google Scholar]
- 11. Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193, 2003. [DOI] [PubMed] [Google Scholar]
- 12. Boschmann M, Schroeder C, Christensen NJ, Tank J, Krupp G, Biaggioni I, Klaus S, Sharma AM, Luft FC, Jordan J. Norepinephrine transporter function and autonomic control of metabolism. J Clin Endocrinol Metab 87: 5130–5137, 2002. [DOI] [PubMed] [Google Scholar]
- 13. Bouchard C, Tremblay A, Nadeau A, Despres JP, Theriault G, Boulay MR, Lortie G, Leblanc C, Fournier G. Genetic effect in resting and exercise metabolic rates. Metabolism 38: 364–370, 1989. [DOI] [PubMed] [Google Scholar]
- 14. Cahill GF, Jr, Herrera MG, Morgan AP, Soeldner JS, Steinke J, Levy PL, Reichard GA, Jr, Kipnis DM. Hormone-fuel interrelationships during fasting. J Clin Invest 45: 1751–1769, 1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cameron-Smith D, Burke LM, Angus DJ, Tunstall RJ, Cox GR, Bonen A, Hawley JA, Hargreaves M. A short-term, high-fat diet up-regulates lipid metabolism and gene expression in human skeletal muscle. Am J Clin Nutr 77: 313–318, 2003. [DOI] [PubMed] [Google Scholar]
- 16. Christin L, O'Connell M, Bogardus C, Danforth E, Jr, Ravussin E. Norepinephrine turnover and energy expenditure in Pima Indian and white men. Metabolism 42: 723–729, 1993. [DOI] [PubMed] [Google Scholar]
- 17. Cunningham JJ. Body composition as a determinant of energy expenditure: a synthetic review and a proposed general prediction equation. Am J Clin Nutr 54: 963–969, 1991. [DOI] [PubMed] [Google Scholar]
- 18. de Lange P, Ragni M, Silvestri E, Moreno M, Schiavo L, Lombardi A, Farina P, Feola A, Goglia F, Lanni A. Combined cDNA array/RT-PCR analysis of gene expression profile in rat gastrocnemius muscle: relation to its adaptive function in energy metabolism during fasting. FASEB J 18: 350–352, 2004. [DOI] [PubMed] [Google Scholar]
- 19. DeFronzo RA. Lilly lecture 1987. The triumvirate: beta-cell, muscle, liver. A collusion responsible for NIDDM. Diabetes 37: 667–687, 1988. [DOI] [PubMed] [Google Scholar]
- 20. DeFronzo RA, Ferrannini E, Hendler R, Felig P, Wahren J. Regulation of splanchnic and peripheral glucose uptake by insulin and hyperglycemia in man. Diabetes 32: 35–45, 1983. [DOI] [PubMed] [Google Scholar]
- 21. DeFronzo RA, Jacot E, Jequier E, Maeder E, Wahren J, Felber JP. The effect of insulin on the disposal of intravenous glucose. Results from indirect calorimetry and hepatic and femoral venous catheterization. Diabetes 30: 1000–1007, 1981. [DOI] [PubMed] [Google Scholar]
- 22. Dulloo AG, Samec S. Uncoupling proteins: their roles in adaptive thermogenesis and substrate metabolism reconsidered. Br J Nutr 86: 123–139, 2001. [DOI] [PubMed] [Google Scholar]
- 23. Ferrannini E, Haffner SM, Mitchell BD, Stern MP. Hyperinsulinaemia: the key feature of a cardiovascular and metabolic syndrome. Diabetologia 34: 416–422, 1991. [DOI] [PubMed] [Google Scholar]
- 24. Ferraro R, Ravussin E. Fat mass in predicting resting metabolic rate. Am J Clin Nutr 56: 460–461, 1992. [DOI] [PubMed] [Google Scholar]
- 25. Finn PF, Dice JF. Proteolytic and lipolytic responses to starvation. Nutrition 22: 830–844, 2006. [DOI] [PubMed] [Google Scholar]
- 26. Fontvieille AM, Lillioja S, Ferraro RT, Schulz LO, Rising R, Ravussin E. Twenty-four-hour energy expenditure in Pima Indians with type 2 (non-insulin-dependent) diabetes mellitus. Diabetologia 35: 753–759, 1992. [DOI] [PubMed] [Google Scholar]
- 27. Foster DO, Frydman ML. Nonshivering thermogenesis in the rat. II. Measurements of blood flow with microspheres point to brown adipose tissue as the dominant site of the calorigenesis induced by noradrenaline. Can J Physiol Pharmacol 56: 110–122, 1978. [DOI] [PubMed] [Google Scholar]
- 28. Garvey WT, Maianu L, Hancock JA, Golichowski AM, Baron A. Gene expression of GLUT4 in skeletal muscle from insulin-resistant patients with obesity, IGT, GDM, and NIDDM. Diabetes 41: 465–475, 1992. [DOI] [PubMed] [Google Scholar]
- 29. Garvey WT, Maianu L, Zhu JH, Brechtel-Hook G, Wallace P, Baron AD. Evidence for defects in the trafficking and translocation of GLUT4 glucose transporters in skeletal muscle as a cause of human insulin resistance. J Clin Invest 101: 2377–2386, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garvey WT, Olefsky JM, Griffin J, Hamman RF, Kolterman OG. The effect of insulin treatment on insulin secretion and insulin action in type II diabetes mellitus. Diabetes 34: 222–234, 1985. [DOI] [PubMed] [Google Scholar]
- 31. Gong DW, He Y, Karas M, Reitman M. Uncoupling protein-3 is a mediator of thermogenesis regulated by thyroid hormone, beta3-adrenergic agonists, and leptin. J Biol Chem 272: 24129–24132, 1997. [DOI] [PubMed] [Google Scholar]
- 32. Hajduch E, Rencurel F, Balendran A, Batty IH, Downes CP, Hundal HS. Serotonin (5-hydroxytryptamine), a novel regulator of glucose transport in rat skeletal muscle. J Biol Chem 274: 13563–13568, 1999. [DOI] [PubMed] [Google Scholar]
- 33. Havel RJ, Carlson LA, Ekelund LG, Holmgren A. Studies on the relation between mobilization of free fatty acids and energy metabolism in man: effects of norepinephrine and nicotinic acid. Metabolism 13: 1402–1412, 1964. [DOI] [PubMed] [Google Scholar]
- 34. Henriksson J. The possible role of skeletal muscle in the adaptation to periods of energy deficiency. Eur J Clin Nutr 44, Suppl 1: 55–64, 1990. [PubMed] [Google Scholar]
- 35. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264, 2003. [DOI] [PubMed] [Google Scholar]
- 36. Jarosz PA. The effect of kappa opioid receptor antagonism on energy expenditure in the obese Zucker rat. Biol Res Nurs 8: 294–299, 2007. [DOI] [PubMed] [Google Scholar]
- 37. Jequier E, Schutz Y. Energy expenditure in obesity and diabetes. Diabetes Metab Rev 4: 583–593, 1988. [DOI] [PubMed] [Google Scholar]
- 38. Kennedy A, Gettys TW, Watson P, Wallace P, Ganaway E, Pan Q, Garvey WT. The metabolic significance of leptin in humans: gender-based differences in relationship to adiposity, insulin sensitivity, and energy expenditure. J Clin Endocrinol Metab 82: 1293–1300, 1997. [DOI] [PubMed] [Google Scholar]
- 39. Kibriya MG, Jasmine F, Argos M, Verret WJ, Rakibuz-Zaman M, Ahmed A, Parvez F, Ahsan H. Changes in gene expression profiles in response to selenium supplementation among individuals with arsenic-induced pre-malignant skin lesions. Toxicol Lett 169: 162–176, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kok P, Roelfsema F, Frolich M, van Pelt J, Stokkel MP, Meinders AE, Pijl H. Activation of dopamine D2 receptors simultaneously ameliorates various metabolic features of obese women. Am J Physiol Endocrinol Metab 291: E1038–E1043, 2006. [DOI] [PubMed] [Google Scholar]
- 41. Laakso M. Insulin resistance and coronary heart disease. Curr Opin Lipidol 7: 217–226, 1996. [DOI] [PubMed] [Google Scholar]
- 42. Lamont LS, Patel DG, Kalhan SC. Beta-adrenergic blockade alters whole-body leucine metabolism in humans. J Appl Physiol 67: 221–225, 1989. [DOI] [PubMed] [Google Scholar]
- 43. Lusk G, Du Bois EF. On the constancy of the basal metabolism. J Physiol 59: 213–216, 1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mansour A, Fox CA, Akil H, Watson SJ. Opioid-receptor mRNA expression in the rat CNS: anatomical and functional implications. Trends Neurosci 18: 22–29, 1995. [DOI] [PubMed] [Google Scholar]
- 45. McClintick JN, Edenberg HJ. Effects of filtering by Present call on analysis of microarray experiments. BMC Bioinformatics 7: 49, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Millenaar FF, Okyere J, May ST, van Zanten M, Voesenek LA, Peeters AJ. How to decide? Different methods of calculating gene expression from short oligonucleotide array data will give different results. BMC Bioinformatics 7: 137, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Monroe MB, Seals DR, Shapiro LF, Bell C, Johnson D, Parker Jones P. Direct evidence for tonic sympathetic support of resting metabolic rate in healthy adult humans. Am J Physiol Endocrinol Metab 280: E740–E744, 2001. [DOI] [PubMed] [Google Scholar]
- 48. Nair KS, Woolf PD, Welle SL, Matthews DE. Leucine, glucose, and energy metabolism after 3 days of fasting in healthy human subjects. Am J Clin Nutr 46: 557–562, 1987. [DOI] [PubMed] [Google Scholar]
- 49. Nakai Y, Hashida H, Kadota K, Minami M, Shimizu K, Matsumoto I, Kato H, Abe K. Up-regulation of genes related to the ubiquitin-proteasome system in the brown adipose tissue of 24-h-fasted rats. Biosci Biotechnol Biochem 72: 139–148, 2008. [DOI] [PubMed] [Google Scholar]
- 50. Nishizawa H, Matsuda M, Yamada Y, Kawai K, Suzuki E, Makishima M, Kitamura T, Shimomura I. Musclin, a novel skeletal muscle-derived secretory factor. J Biol Chem 279: 19391–19395, 2004. [DOI] [PubMed] [Google Scholar]
- 51. Nonogaki K. New insights into sympathetic regulation of glucose and fat metabolism. Diabetologia 43: 533–549, 2000. [DOI] [PubMed] [Google Scholar]
- 52. Osier MV, Zhao H, Cheung KH. Handling multiple testing while interpreting microarrays with the Gene Ontology Database. BMC Bioinformatics 5: 124, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pascual RM, Billington CK, Hall IP, Panettieri RA, Jr, Fish JE, Peters SP, Penn RB. Mechanisms of cytokine effects on G protein-coupled receptor-mediated signaling in airway smooth muscle. Am J Physiol Lung Cell Mol Physiol 281: L1425–L1435, 2001. [DOI] [PubMed] [Google Scholar]
- 54. Pecqueur C, Couplan E, Bouillaud F, Ricquier D. Genetic and physiological analysis of the role of uncoupling proteins in human energy homeostasis. J Mol Med 79: 48–56, 2001. [DOI] [PubMed] [Google Scholar]
- 55. Peters SJ, Harris RA, Wu P, Pehleman TL, Heigenhauser GJ, Spriet LL. Human skeletal muscle PDH kinase activity and isoform expression during a 3-day high-fat/low-carbohydrate diet. Am J Physiol Endocrinol Metab 281: E1151–E1158, 2001. [DOI] [PubMed] [Google Scholar]
- 56. Ravussin E, Lillioja S, Knowler WC, Christin L, Freymond D, Abbott WG, Boyce V, Howard BV, Bogardus C. Reduced rate of energy expenditure as a risk factor for body-weight gain. N Engl J Med 318: 467–472, 1988. [DOI] [PubMed] [Google Scholar]
- 57. Rehnmark S, Bianco AC, Kieffer JD, Silva JE. Transcriptional and posttranscriptional mechanisms in uncoupling protein mRNA response to cold. Am J Physiol Endocrinol Metab 262: E58–E67, 1992. [DOI] [PubMed] [Google Scholar]
- 58. Ricquier D, Bouillaud F, Toumelin P, Mory G, Bazin R, Arch J, Penicaud L. Expression of uncoupling protein mRNA in thermogenic or weakly thermogenic brown adipose tissue. Evidence for a rapid beta-adrenoreceptor-mediated and transcriptionally regulated step during activation of thermogenesis. J Biol Chem 261: 13905–13910, 1986. [PubMed] [Google Scholar]
- 59. Roberts SB, Savage J, Coward WA, Chew B, Lucas A. Energy expenditure and intake in infants born to lean and overweight mothers. N Engl J Med 318: 461–466, 1988. [DOI] [PubMed] [Google Scholar]
- 60. Schiffelers SL, Brouwer EM, Saris WH, van Baak MA. Inhibition of lipolysis reduces beta1-adrenoceptor-mediated thermogenesis in man. Metabolism 47: 1462–1467, 1998. [DOI] [PubMed] [Google Scholar]
- 61. Schwartz RS, Jaeger LF, Veith RC. Effect of clonidine on the thermic effect of feeding in humans. Am J Physiol Regul Integr Comp Physiol 254: R90–R94, 1988. [DOI] [PubMed] [Google Scholar]
- 62. Seo J, Hoffman EP. Probe set algorithms: is there a rational best bet? BMC Bioinformatics 7: 395, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Silva JE. Full expression of uncoupling protein gene requires the concurrence of norepinephrine and triiodothyronine. Mol Endocrinol 2: 706–713, 1988. [DOI] [PubMed] [Google Scholar]
- 64. Snitker S, Tataranni PA, Ravussin E. Respiratory quotient is inversely associated with muscle sympathetic nerve activity. J Clin Endocrinol Metab 83: 3977–3979, 1998. [DOI] [PubMed] [Google Scholar]
- 65. Spraul M, Ravussin E, Fontvieille AM, Rising R, Larson DE, Anderson EA. Reduced sympathetic nervous activity. A potential mechanism predisposing to body weight gain. J Clin Invest 92: 1730–1735, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tataranni PA, Baier L, Jenkinson C, Harper I, Del Parigi A, Bogardus C. A Ser311Cys mutation in the human dopamine receptor D2 gene is associated with reduced energy expenditure. Diabetes 50: 901–904, 2001. [DOI] [PubMed] [Google Scholar]
- 67. Tataranni PA, Harper IT, Snitker S, Del Parigi A, Vozarova B, Bunt J, Bogardus C, Ravussin E. Body weight gain in free-living Pima Indians: effect of energy intake vs. expenditure. Int J Obes Relat Metab Disord 27: 1578–1583, 2003. [DOI] [PubMed] [Google Scholar]
- 68. Tataranni PA, Young JB, Bogardus C, Ravussin E. A low sympathoadrenal activity is associated with body weight gain and development of central adiposity in Pima Indian men. Obes Res 5: 341–347, 1997. [DOI] [PubMed] [Google Scholar]
- 69. Tentolouris N, Liatis S, Katsilambros N. Sympathetic system activity in obesity and metabolic syndrome. Ann NY Acad Sci 1083: 129–152, 2006. [DOI] [PubMed] [Google Scholar]
- 70. Toubro S, Sorensen TI, Hindsberger C, Christensen NJ, Astrup A. Twenty-four-hour respiratory quotient: the role of diet and familial resemblance. J Clin Endocrinol Metab 83: 2758–2764, 1998. [DOI] [PubMed] [Google Scholar]
- 71. Udden J, Folkesson R, Hoffstedt J. Downregulation of uncoupling protein 2 mRNA in women treated with glucocorticoids. Int J Obes Relat Metab Disord 25: 1615–1618, 2001. [DOI] [PubMed] [Google Scholar]
- 72. Vazeille E, Codran A, Claustre A, Averous J, Listrat A, Bechet D, Taillandier D, Dardevet D, Attaix D, Combaret L. The ubiquitin-proteasome and the mitochondria-associated apoptotic pathways are sequentially downregulated during recovery after immobilization-induced muscle atrophy. Am J Physiol Endocrinol Metab 295: E1181–E1190, 2008. [DOI] [PubMed] [Google Scholar]
- 73. Wade AJ, Marbut MM, Round JM. Muscle fibre type and aetiology of obesity. Lancet 335: 805–808, 1990. [DOI] [PubMed] [Google Scholar]
- 74. Welle S, Schwartz RG, Statt M. Reduced metabolic rate during beta-adrenergic blockade in humans. Metabolism 40: 619–622, 1991. [DOI] [PubMed] [Google Scholar]
- 75. Widdowson EM, Edholm OG, McCance RA. The food intake and energy expenditure of cadets in training. Br J Nutr 8: 147–155, 1954. [DOI] [PubMed] [Google Scholar]
- 76. Wu X, Wang J, Cui X, Maianu L, Rhees B, Rosinski J, So WV, Willi SM, Osier MV, Hill HS, Page GP, Allison DB, Martin M, Garvey WT. The effect of insulin on expression of genes and biochemical pathways in human skeletal muscle. Endocrine 31: 5–17, 2007. [DOI] [PubMed] [Google Scholar]
- 77. Yang Z, Harris LE, Palmer-Toy DE, Hancock WS. Multilectin affinity chromatography for characterization of multiple glycoprotein biomarker candidates in serum from breast cancer patients. Clin Chem 52: 1897–1905, 2006. [DOI] [PubMed] [Google Scholar]
- 78. Zurlo F, Larson K, Bogardus C, Ravussin E. Skeletal muscle metabolism is a major determinant of resting energy expenditure. J Clin Invest 86: 1423–1427, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zurlo F, Lillioja S, Esposito-Del Puente A, Nyomba BL, Raz I, Saad MF, Swinburn BA, Knowler WC, Bogardus C, Ravussin E. Low ratio of fat to carbohydrate oxidation as predictor of weight gain: study of 24-h RQ. Am J Physiol Endocrinol Metab 259: E650–E657, 1990. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.