Abstract
Mechanisms controlling dendritic arbor formation affect the establishment of neuronal circuits. Candidate plasticity gene 15 (CPG15) is a glycosylphosphatidyl inositol (GPI)-linked activity-induced protein that has been shown to function as an intercellular signaling molecule that can promote the morphological and physiological development of the Xenopus retinotectal system. A thorough understanding of CPG15 function requires knowledge of the spatiotemporal expression of the endogenous protein. We therefore cloned Xenopus cpg15 and used RNA in situ hybridization and immunohistochemistry to determine the pattern of CPG15 expression. cpg15 mRNA and CPG15 protein are first detectable in the developing spinal cord and become widespread as development proceeds. CPG15 is expressed in sensory regions of the brain, including the visual, auditory, and olfactory systems. Within the retina, CPG15 is only expressed in retinal ganglion cells. CPG15 protein is concentrated in axon tracts, including retinal axons. These data support a model in which CPG15 expressed in retinal ganglion cells is trafficked to retinal axons, where it modulates postsynaptic dendritic arbor elaboration, and synaptic maturation.
Indexing terms: neuritin, ligand, retinotectal, GPI, spinal cord, amphibian, plasticity, visual system
Central nervous system (CNS) development is characterized by the coordinate development of presynaptic axonal arbors, postsynaptic dendritic arbors, and synaptic connections between them. Activity-dependent mechanisms regulate multiple aspects of nervous system connectivity during development. One way that activity can influence the development of neuronal connections is through the induction of activity-regulated genes, or candidate plasticity genes (CPGs). Numerous activity-induced genes have been identified through the use of highly selective screens (Nedivi et al., 1993; Qian et al., 1993; Yamagata et al., 1993). These screens have identified genes previously implicated in plasticity such as brain-derived neurotrophic factor (BDNF) and the transcription factors zif268, c-fos, and jun (Nedivi et al., 1993; Qian et al., 1993). They have also identified new functions for previously discovered genes such as the class 1 major histocompatibility (MHC1) molecule (Corriveau et al., 1998) and tissue plasminogen activator (tPA) (Qian et al., 1993), as well as many novel genes with unknown functions such as Homer, Arc, Narp, and cpg15 (Nedivi et al., 1993; Lyford et al., 1995; Tsui et al., 1996; Brakeman et al., 1997) Subsequent analysis has demonstrated that these novel activity-induced genes play a variety of roles in neuronal plasticity (Nedivi, 1999).
CPG15, also known as neuritin (Naeve et al., 1997), is a small secreted protein, anchored to the extracellular surface of neurons by a glycosylphosphatidyl inositol (GPI) link. cpg15 was first identified from rat (Nedivi et al., 1996) and has subsequently been cloned from cat (Corriveau et al., 1999) and human (Naeve et al., 1997). In these three mammalian species, the primary amino acid sequence of CPG15 consists of 142 amino acids and shows no homology to other proteins (Nedivi et al., 1996, 1998; Naeve et al., 1997). In situ hybridization studies in mammals show that cpg15 is expressed at peak levels during development, specifically at times of afferent axonal and dendritic growth and synaptogenesis (Nedivi et al., 1996; Naeve et al., 1997; Corriveau et al., 1999). Furthermore, cpg15 is upregulated by visual activity in adult rat visual cortex (Nedivi et al., 1996).
Viral expression of cpg15 in Xenopus optic tectal neurons promotes the elaboration of tectal cell dendritic arbors and presynaptic retinal axon arbors in a non-cell autonomous fashion (Nedivi et al., 1998; Cantallops et al., 2000). Electrophysiological recordings indicate that cpg15 promotes synaptic maturation by the insertion of AMPA-type glutamate receptors into synapses (Cantallops et al., 2000). The GPI link is required for these effects of CPG15, because expression of a truncated form of CPG15, lacking the GPI consensus sequence, does not promote axonal or dendritic arbor elaboration or synaptic maturation.
A thorough understanding of CPG15 function requires knowledge of the spatiotemporal expression pattern of cpg15 mRNA and protein. To address these issues, we cloned Xenopus cpg15 and mapped the spatiotemporal distribution of cpg15 mRNA and protein during nervous system development in Xenopus. cpg15 mRNA is detectable in Xenopus retina, brain, and spinal cord. Expression is detected early during nervous system development, corresponding to the time of initial morphological development and synaptogenesis. Interestingly, the protein is targeted to projection axons, where it might act as an intercellular signal to promote the development of dendritic arbors of postsynaptic neurons.
Materials and Methods
All experiments were carried out in accordance with animal care protocols approved by Cold Spring Harbor Labs.
Isolation and sequence analysis of cDNA clones
Poly A+ RNA was isolated from whole stage 47 tadpoles (Qiagen direct mRNA isolation, Qiagen, Santa Clarita, CA) and reverse transcribed into cDNA (Stratagene, La Jolla, CA). This cDNA pool was used as a template in a polymerase chain reaction (PCR) reaction, with the primers complementary to conserved mammalian cpg15 sequences (accession no. U88958) within the coding region (5′ primer: 225–247; 3′ primer: 500–524,) for 34 cycles of 95°C for 40 seconds, 40°C for 40 seconds, and 72°C for 40 seconds. A 299-bp fragment was amplified, sequenced, and cloned into pBluescript (Stratagene) and is referred to as pXcpg15-pcr. The sequence was highly homologous to mammalian cpg15 (data not shown). This fragment was excised, gel purified, and labeled with α32P dCTP (Amersham, Piscataway, NJ) by random primed DNA labeling (Boehringer Mannheim, Germany) and used as a probe to screen a Xenopus stage 55 tadpole brain cDNA Lambda Zap II library. Approximately 400,000 plaques were screened at high stringency (50% formamide at 42°C). Two independent clones of 0.8 and 1.3 kb were identified and sequenced from both directions. The complete cDNA sequence of the longer clone was submitted to GenBank (accession no. AF378092). Sequence analysis was carried out by using Sequencher software (Gene Codes, Ann Arbor, MI) and MegAlign software (DNASTAR, Madison, WI) on a Macintosh computer.
Northern blot
Poly A+ RNA was isolated from whole stage 47 tadpoles (Qiagen direct mRNA), and 4 μg was electrophoresed on a 1% agarose/formaldehyde gel and transferred to nylon membrane. The pXcpg15-pcr fragment used to screen the cDNA library was used as a probe prepared as described above. Hybridization was carried out under high stringency conditions (50% formamide, 50°C). Detection was carried out by autoradiography for 7 hours at −80° (Biomax film, Kodak, Rochester, NY) in the presence of enhancer screens (Kodak).
In situ hybridization
For whole-mount in situ hybridizations, the pXcpg15-pcr fragment was used for in vitro synthesis of sense and antisense digoxigenin-labeled riboprobes (MAXIscript, Ambion, Austin, TX). Whole-mount in situ hybridizations were carried out as described (Harland, 1991). Briefly, embryos were hybridized overnight at 50°C in 50% formamide, and detection was carried out by using the DIG Nucleic Acid Detection Kit (Boehringer Mannheim).
In situ hybridizations on frozen sections were carried out as described previously (Nedivi et al., 1996). Animals were fixed in 4% paraformaldehyde, frozen in OCT, and sectioned on a cryostat at 20 μm either as whole animals or as dissected retinae and brains. The sections were mounted on slides under RNase-free conditions, dehydrated through graded ethanol, and fixed again in 4% paraformaldehyde. αUTP 35S (Amersham) labeled pXcpg15-pcr sense and antisense riboprobes were synthesized in vitro (MAXIscript) and hybridized on sections. 35S-labeled probes were hybridized to sections under high stringency conditions (50% formamide at 50°C) overnight, washed, and dipped in emulsion (Kodak). Slides were exposed for 3 days at 4°C and developed in developer (Kodak). Sections were counterstained with toludine blue and viewed by darkfield and brightfield microscopy.
Quantification of in situ hybridization
mRNA levels were quantified as described (Corriveau et al., 1999) by using NIH Image (version 1.6). Briefly, brightfield and darkfield images were collected with a SPOT digital camera system, by using uniform settings for the entire series of images in an experiment. A single section through the dorsal optic tectum was analyzed for each of four stage 47 animals. The rostrolateral and caudomedial regions of tectum were identified from and outlined on the brightfield images. The mean pixel density of the rostrolateral and caudomedial regions of the tectum were measured in the darkfield images on a 0–255 scale where 255 is white. The mean pixel density of background was measured from a portion of the slide next to the tissue section and subtracted from each value separately for each section for normalization. No significant difference was found in mean pixel density of backgrounds. Statistical significance was determined by Student's t-test. The experiment was repeated twice with animals from the same clutch, and similar results were obtained. Data are shown for one experiment only.
Immunocytochemistry
Tissue was fixed in 4% paraformaldehyde overnight at 4°C and cut into 20-μm cryostat sections. Sections were incubated with preimmune serum or polyclonal antiserum to CPG15 at a 1:200 dilution in phosphate-buffered saline, pH 7.4, 5% goat serum, and 0.05% Triton X-100 as previously described (Nedivi et al., 1998). Sections were rinsed and then incubated with a fluorescein isothiocyanate (FITC)-tagged goat anti-rabbit secondary antibody (Sigma, St. Louis, MO). Immunoreactivity was imaged with a laser scanning confocal microscope (Noran Instruments, Madison, Wisconsin).
Western blot
Whole brains or eyes were homogenized in a glass Dounce homogenizer in buffer containing 10 mM Tris-HCl, pH 7.4, 1 mM EGTA, 0.5 mM dithiothreitol (DTT), 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 5 μg/ml leupeptin, 20 μg/ml soybean trypsin inhibitor, and 30 mM octyl glucosid. The samples were centrifuged at 20,000g for 2 minutes, and supernatants were collected for analysis. Samples were separated in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) 15% acrylamide and transferred to Immobilon-P nylon membrane (Millipore, MA). Blots were blocked by using standard procedures and incubated with the same rabbit anti-CPG15 antibody (1:250) as that used for immunocytochemistry (Nedivi et al., 1998). Blots were incubated with horseradish peroxidase (HRP)-tagged goat anti-rabbit (Sigma), and detection was carried out by using standard chemiluminescence technique (ECL, Amersham). All blots were exposed for the same amount of time.
Retinal explant cultures
Stage 53–56 albino Xenopus laevis tadpoles were anesthetized with 0.05% MS222, and both optic nerves were crushed 4 days prior to retinal explant preparation. Tadpoles were anesthetized as before, and then both eyes were removed and kept on cold HBST buffer (5.8 mM NaCl, 0.6 mM KCl, 0.3 mM CaCl2•2H2O, 0.83 mM MgSO4•7H2O, and 5 mM HEPES, pH 7.7). The neural retina was cut diagonally into eight pieces, which were plated in pairs onto laminin-coated (5% in HBST) glass coverslips. Two to 4 hours later, culture medium was added to the explants (5 parts of L-15, 4 parts of a solution containing 95 mM NaCl, 1 mM KCl, 0.6 mM CaCl2•2H2O, 3.9 mM MgSO4•7H2O, 4 mM glutamine, and 9.4 mM HEPES, and 1 part fetal calf serum with 5 ng/ml sodium selenite, 5 μg/ml transferrin, 5 μg/ml insulin, and 2,500 units of 1% penn strep). Cultures were maintained at room temperature for 5–7 days before use. Explants were fixed and processed for immunocytochemistry as described above except following primary antibody, explants were incubated with phalloidin conjugated to rhodamine (Molecular Probes, Eugene, OR) (1:800) for 1 hour.
Results
cpg15 is highly conserved across species
We cloned Xenopus cpg15 from a tadpole brain cDNA library. The deduced amino acid sequence aligned with the rat CPG15 sequence is shown in Figure 1A. A probe generated from this cDNA consistently detects a single band of approximately 1.7 kb on a Northern blot of stage 47 tadpole mRNA, similar to the size of mammalian cpg15 mRNA (Naeve et al., 1997) (Fig. 1B). This clone contains an open reading frame encoding a predicted protein of 144 amino acids, 2 amino acids longer than mammalian CPG15. The complete open reading frame is 67% identical to that of Rattus norvegicus suggesting that it is Xenopus laevis CPG15. Xenopus CPG15 contains a conserved N-terminal secretion signal, characteristic of extracellular proteins. In addition, the six cysteine residues, which are postulated to be critical for the correct folding of this extracellular protein, are conserved from Xenopus to rat and human (Naeve et al., 1997; Nedivi et al., 1998). Residues 29–33 are a consensus for S-nitrosylation (Stamler et al., 1997), and their conservation between Xenopus and rat suggests that this may be essential for CPG15 function. The C-terminal of Xenopus CPG15 is comprised of hydrophobic residues diagnostic of a GPI consensus sequence (Low, 1987). Both the N- and C-terminal signal sequences are cleaved during post-translational modification. The predicted mature protein (residues 28–115) bears a high level of homology to rat CPG15, with 73% of amino acids identical and 88% similar between the two species. No other proteins with significant similarity to CPG15 have yet been reported.
Fig. 1.
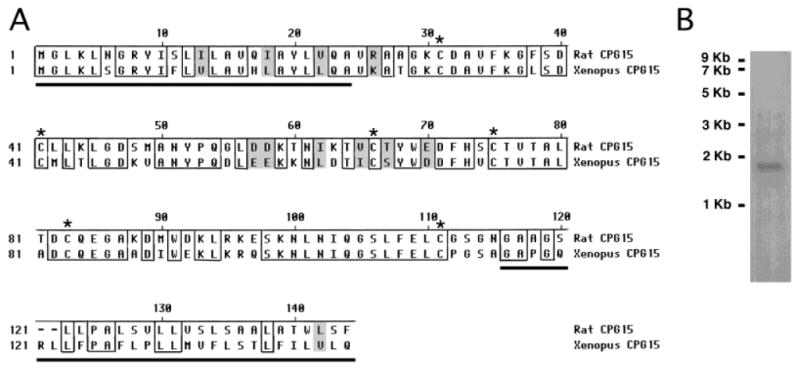
Sequence alignment of CPG15. A: Sequence alignment of rat and Xenopus CPG15. Identical amino acids are boxed and similar amino acids are shaded gray. Conserved cysteine residues are marked with asterisks. The N-terminal and C-terminal signal sequences for secretion and GPI linkage, respectively, are underlined. Sequence analysis shows an overall 67% identity between the two species and 73% identity in the mature protein. B: Northern blot analysis from whole tadpole Poly A+ RNA results in the detection of a single band of approximately 1.7 kb, similar to the size of mammalian cpg15.
Spatiotemporal expression of cpg15 in the CNS
cpg15 is detected in stage 24 embryos in dorsal and ventral regions within the spinal cord (Fig. 2A). This is a stage at which the neurons begin to send out their axons (Hartenstein, 1993). At stage 33 (Figs. 2B,C, 3A,B), there is an increase in relative numbers of cells expressing cpg15, probably reflecting an increase in the number of differentiated neurons. At this stage, cpg15 expression is prominent throughout the developing CNS and can be detected in the forebrain, midbrain, and hindbrain including the olfactory bulb and the optic tectum. cpg15 can also be detected within the olfactory sensory neurons, the olfactory pit, auditory ganglion cells, and spinal cord (Figs. 3, 4). Little mRNA is detectable in the differentiating retina at this stage (Fig. 3A,B). cpg15 expression is detectable within the tectum at stage 38 at the onset of retinotectal synaptogenesis (Holt and Harris, 1993) and continues to be expressed in this structure into adulthood (Figs. 3C–H, 4A–F).
Fig. 2.
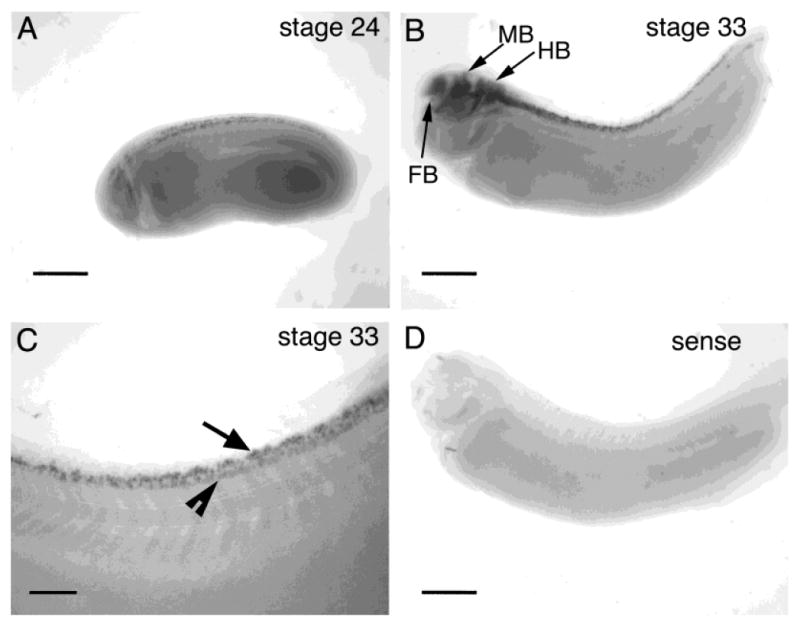
Whole-mount in situ hybridizations on albino Xenopus embryos. A: cpg15 can be detected in dorsal spinal cord at stage 24. B: cpg15 expression increases in the spinal cord and can be detected in the forebrain (FB), midbrain (MB), and hindbrain (HB) at stage 33. C: Higher magnification of spinal cord of stage 33 embryo. cpg15 expression is detectable in both dorsal (arrow) and ventral cells (arrowhead) within the spinal cord. D: Sense probe. Rostral is to the left in all images. Scale bar = 500 μm in A,B,D; 250 μm in C.
Fig. 3.
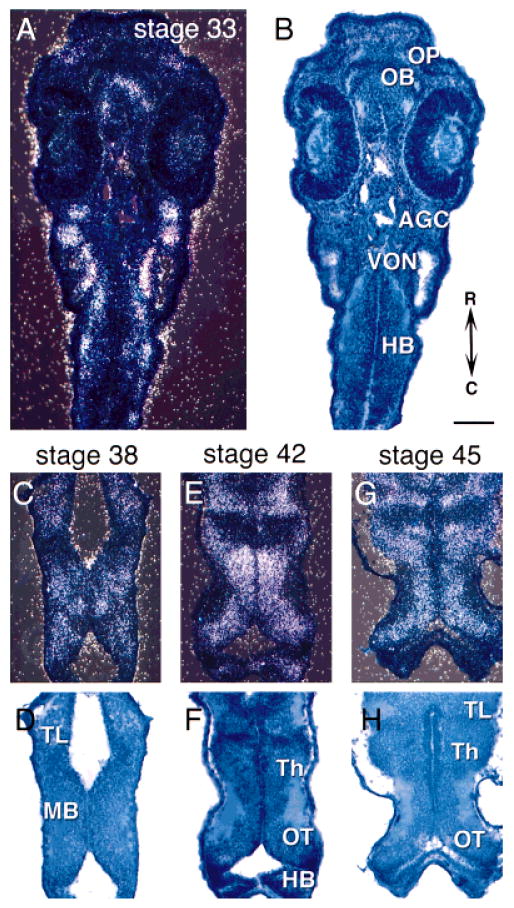
In situ hybridization of cpg15 in early stages of tadpole development. Darkfield images show mRNA (A,C,E,G) and brightfield images show toludine blue counterstain (B,D,F,H). A,B: Horizontal sections through stage 33 tadpoles. Horizontal sections through brains from stage 38 (C,D), stage 42 (E,F), and stage 45 (G,H). cpg15 is expressed in the optic tectum (OT), olfactory pits (OP), olfactory bulb (OB), telencephalon (TL), midbrain (MB), thalamus (Th), and auditory ganglion cells (AGC), as well as vestibulo-ocular nuclei (VON) and hindbrain (HB). Rostral is up in all sections. Scale bar = 100 μm.
Fig. 4.
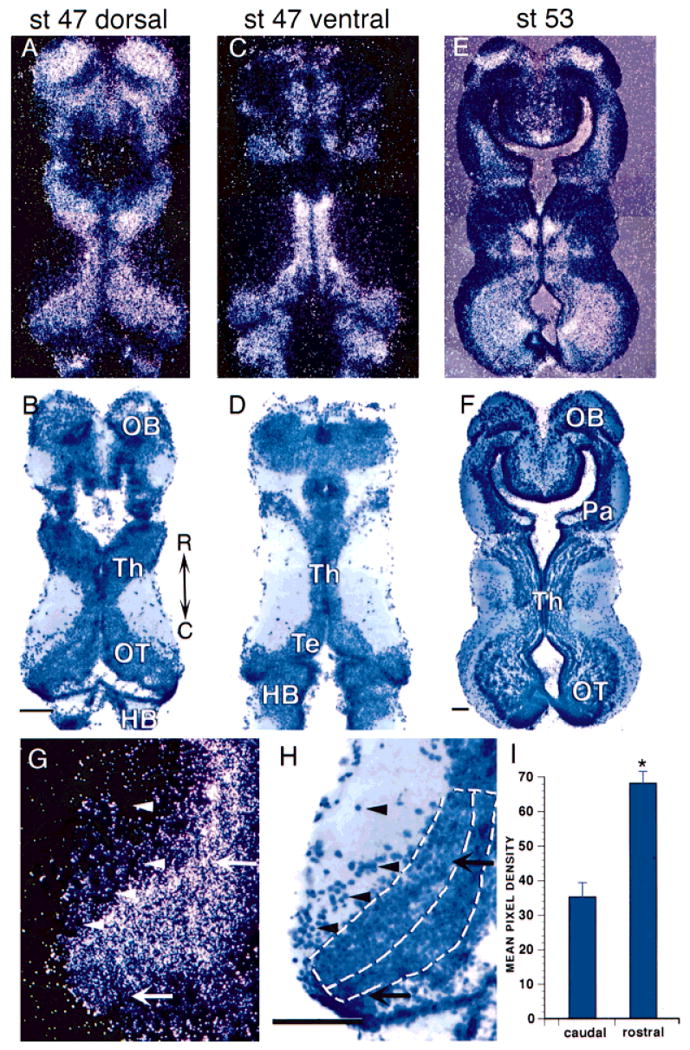
In situ hybridization of cpg15 in brains of midstage Xenopus tadpoles. Darkfield images show mRNA (A,C,E,G) and brightfield images show toludine blue counterstain (B,D,F,H). A,B: Horizontal brain sections through dorsal brain of stage 47 tadpoles show cpg15 expression in the olfactory bulb (OB), dorsal thalamus (Th), optic tectum (OT), and hindbrain (HB). C,D: Sections through ventral brain show labeling in thalamus (Th), tegmentum (Te), and hindbrain. E,F: Stage 53 tadpoles continue to express cpg15 in the olfactory bulb, thalamus, and optic tectum, as well as the pallium (Pa). G,H: High magnification of stage 47 optic tectum. Arrows show rostrolateral and caudomedial cells indicating the higher level of mRNA in rostrolateral tectum. Arrowheads indicate cells within the neuropil that have no detectable levels of cpg15. Dorsal lines in H show rostral and caudal regions of tectum used for quantification. Rostral is up in all sections. I: Quantification of cpg15 expression as mean pixel density in rostrolateral and caudomedial tectum on a 0–255 scale where 255 is white (mean ± standard error). *, P < 0.001. Scale bar = 100 μm.
cpg15 mRNA expression is high within the telencephalon, including the olfactory bulb, parts of the thalamus, and tegmentum (Fig. 4). cpg15 expression is maintained in these regions at stage 53 (Fig. 4E,F) and in postmetamorphic frogs (data not shown). cpg15 appears to be expressed at higher levels in more mature neurons in rostrolateral tectum compared with recently differentiated cells in the caudomedial tectum (Fig. 4G,H). Some neurons near the border of the neuropil are very densely labeled with silver grains. We quantified the cpg15 mRNA expression in rostrolateral and caudomedial regions of the tectum (see Materials and Methods). This analysis indicates that more mature neurons in the rostrolateral optic tectum express significantly higher levels of cpg15 than recently differentiated neurons in the caudomedial optic tectum (Fig. 4I). This gradient of mRNA level is consistent with the rostrocaudal gradient of neuronal development (Straznicky and Gaze, 1972).
cpg15 is not expressed by all cells. It is absent from cells in proliferative zones throughout the brain. cpg15 mRNA is also absent from the ventral telecephalon and some thalamic regions (Fig. 4C,D). Neurons in the optic tectal neuropil, which have been identified as γ-aminobutyric acid (GABA)-ergic interneurons (Antal, 1991; Rybicka and Udin, 1994; Wu and Cline, unpublished data), do not express cpg15 (Fig. 4G,H). cpg15 mRNA is also absent from the nucleus isthmi (data not shown), a cholinergic midbrain nucleus involved in visual system plasticity (Udin, 1999).
In the retina, cpg15 is expressed exclusively in the RGC layer (Fig. 5). It becomes detectable around stage 38 and continues to be expressed only in the RGC into adulthood (Fig. 5E,F). No expression was detected in the proliferative zone (PZ) of the retina, as was also the case in the CNS.
Fig. 5.
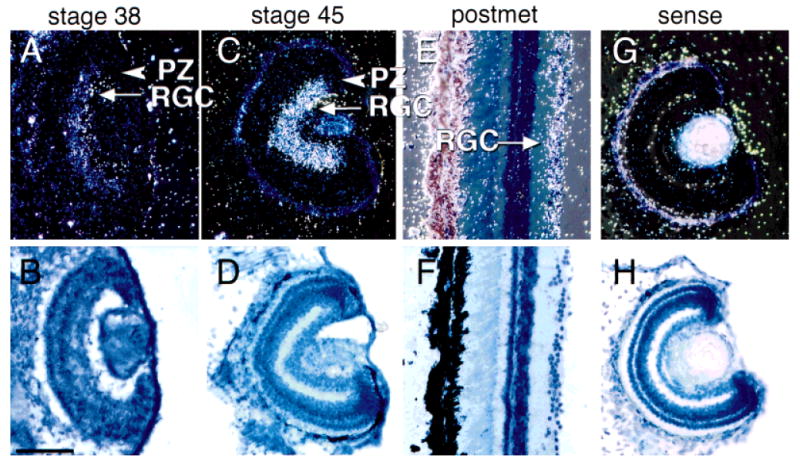
In situ hybridization of cpg15 in the retina. Darkfield images (A,C,E,G) show mRNA detection and brightfield images (B,D,F,H) show toludine blue counterstain for the same sections. cpg15 is expressed in retinal ganglion cells (RGC) but not in the proliferative zone (PZ) of retinae from stage 38 tadpoles and at all later stages examined. In G and H, sense probe did not label any cells. Scale bar = 100 μm.
Spatiotemporal expression of CPG15 in the CNS
Western blots indicate that CPG15 protein is detectable in brain and eye homogenates of tadpoles at stages 47, 55, and 60 and juvenile frogs (Fig. 6). CPG15 protein expression increases from stage 47 to 55 and remains high after metamorphosis.
Fig. 6.
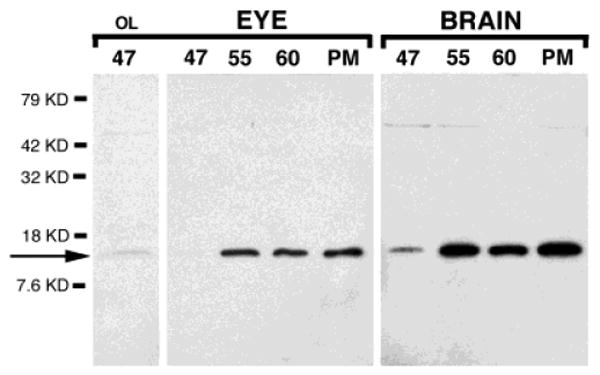
Western blot analysis of Xenopus eye and brain. CPG15 antiserum detects a band of approximately 12 kD in eye and brain homogenates from stages 47, 55, and 60 and postmetamorphic (PM) Xenopus. Proteins were loaded in equal amounts (25 μg) and overloaded (75μg) for stage 47 eye homogenate (first lane only, OL). Higher molecular weight band (∼50 kD) is not detected consistently and is also present in blots incubated in preimmune serum (Nedivi et al., 1998).
CPG15 expression in the retina is first detectable in the optic nerve (ON) of stage 40 tadpoles (Fig. 7A). At this time we cannot detect CPG15 immunoreactivity (IR) in RGC somata. The intensity of CPG15 IR in the RGC somata and in ON increases through stage 47 (Fig. 7B,C) and is detectable in retinal axons of stage 55 tadpoles (data not shown). Tissue sections treated with preimmune serum show no label in RGC somata or axons (Fig. 7E). Although CPG15 IR can be detected in RGC somata, it is concentrated in axons within the optic nerve. Unilateral optic nerve crush at stage 55 results in a loss of CPG15 IR in the damaged nerve and tract (data not shown). CPG15 IR returns with a time-course characteristic of axon regeneration (data not shown), indicating that the IR in the ON originates from RGC axons, rather than ON glia.
Fig. 7.
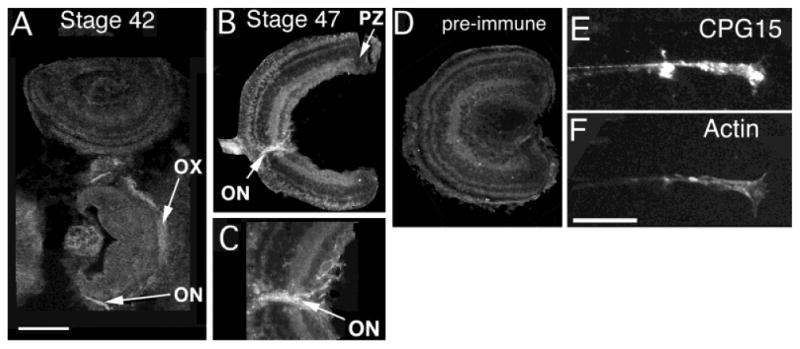
CPG15 is targeted to retinal ganglion cell axons. A: CPG15 immunoreactivity in stage 42 is detected in the optic nerve (ON) and optic chiasm (OX). B: Retinal axons within the optic nerve in stage 47. C: High-magnification image of the optic nerve head of retina from stage 47. Note the CPG15 IR around cells in the RGC layer and the concentrated immunoreactivity in the optic nerve. D: Preimmune serum does not label RGC somata or axons. E: In retinal explants, CPG15 IR can be observed all along the length of axons, including the growth cone. F: Double staining with rhodamine-phalloidin highlights actin-based axonal structure. Scale bar = 100 μm for A,B,D; 50 μm for C.
To determine whether CPG15 is targeted to axon terminals, we prepared retinal explant cultures. CPG15 IR is present throughout retinal axons including the axonal growth cones of the RGC, indicating that protein is transported to these regions (Fig. 7E,F).
CPG15 IR is first detected in the brain at about stage 42/43. By stage 45 CPG15 IR is widespread in differentiated neurons throughout the CNS (Fig. 8). CPG15 antibody labels cells in a honeycomb pattern, as previously described (Nedivi et al., 1998). In dorsal sections through the brain, CPG15 IR is highest in neurons in the olfactory region of the dorsal telencephalon and in optic tectal neurons (OB and OT in Fig. 8A,B). In more ventral sections through the brain, CPG15 IR is present in neurons throughout the ventral telencephalon (TL in Fig. 8D,E). Neurons in the tegmentum and hindbrain are immunoreactive for CPG15 (Te and HB in Fig. 8C,D).
Fig. 8.
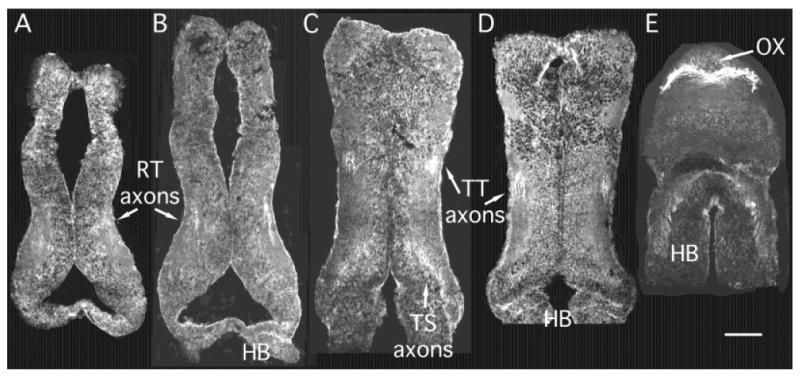
CPG15 expression in tadpole brain. A–E: Horizontal cryostat sections from a stage 45 tadpole shown in sequence from dorsal through ventral. CPG15 immunohistochemistry shows cell bodies labeled in a honeycomb pattern throughout the brain. Cells are labeled in the olfactory bulb (OB), optic tectum (OT), telencephalon (TL), and tegmentum (Te). Intense labeling occurs in axon tracts, notably the optic chiasm (OX), retinotectal axons (RT) in the optic tectal neuropil, tectotegmental axons (TT), and tectospinal (TS) axons coursing toward hindbrain (HB) and spinal cord. Scale bar = 100 μm.
In horizontal sections through the ventral brain, CPG15 IR is most intense in retinal axons in the optic chiasm and in axons within the hindbrain and spinal cord (Figs. 7A, 8). Retinal axons terminating in the optic tectal neuropil are brightly labeled (RT axons in Fig. 8A,B). Biocytin labeling of tectal neurons demonstrates tectal projections to the contralateral tegmentum and ipsilateral spinal cord (Rajan and Cline, unpublished data). Axons in these tracts are labeled by CPG15 antibody, as are tegmental axons projecting to the spinal cord (Fig. 8C,D)
CPG15 IR in the CNS remains intense through stages 49; however, by stage 55/56 CPG15 IR is decreased in the brain and was undetectable in most neurons in the CNS of postmetamorphic animals (data not shown). Labeling in the optic nerve and other axon tracts was also very low to undetectable in postmetamorphic animals (data not shown). Despite the decreased immunodetection of CPG15 in sections of older animals, Western blots indicate that CPG15 is still expressed in the CNS of these animals (Fig. 6).
In the spinal cord, we first detect CPG15 IR in commissural axons of stage 34 tadpoles (Fig. 9A). Neuronal somata in the spinal cord are not stained at this stage but become intensely immunoreactive by stage 47 (Fig. 9B). The early detection of CPG15 message and protein in spinal cord neurons suggests a potential function for CPG15 in the establishment of motor circuits.
Fig. 9.
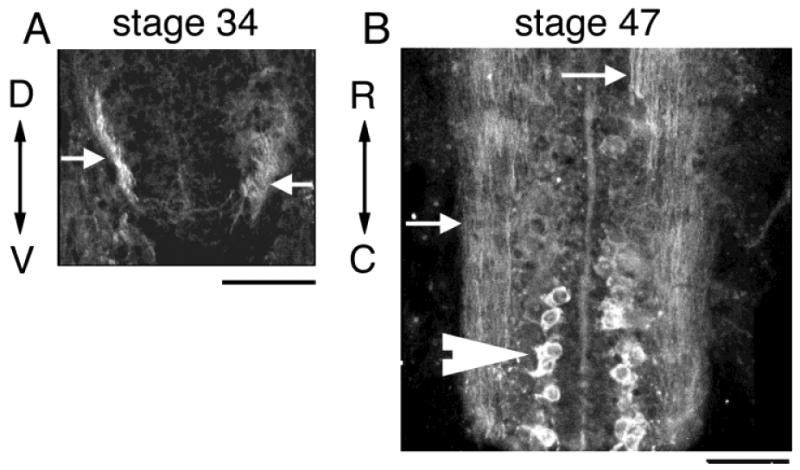
CPG15 expression in spinal cord. A: Immunohistochemistry for CPG15 labels commissural axons in the ventral spinal cord of stage 34 tadpole (arrows). B: Large somata (arrowhead) as well as axons (arrows) in spinal cord label intensely for CPG15 at stage 47. Scale bar = 30 μm in A; 50 μm in B.
Discussion
Homology and conservation of CPG15
CPG15 is a 144-amino acid protein that contains an N-terminal secretion signal sequence and a C-terminal GPI consensus sequence. CPG15 is highly conserved, with 73% of amino acid identity from Xenopus to rat and 100% identity between rat and human in the mature GPI-linked protein (Naeve et al., 1997). A recently reported zebrafish EST has a predicted open reading frame encoding for a protein of 51% identity to Xenopus and 45% identity to rat CPG15 (accession no: AI877586). The high degree of conservation across species suggests that CPG15 performs a critical function for CNS development. It is interesting to note that no CPG15 homologue has been reported in Drosophila or any invertebrate. Drosophila do not have traditional growth factors, such as nerve growth factor (NGF) or BDNF, suggesting that the molecular mechanisms underlying developmental plasticity and synaptic plasticity differ between vertebrate and invertebrate species (Jankowsky and Patterson, 1999).
CPG15 is targeted to axons
Our previous studies have shown that CPG15 functions as an intercellular signaling molecule to promote both the morphological elaboration and synaptic maturation of the retinotectal system by acting in a non-cell-autonomous fashion (Nedivi et al., 1998; Cantallops et al., 2000). Here we report that cpg15 mRNA is detectable in differentiated projection neurons within sensory systems, including the auditory, olfactory, and visual systems, and that CPG15 protein is targeted to projection axons, including the RGC axons.
The specific trafficking of CPG15 to axons is characteristic of GPI-linked proteins. The selective targeting can be most clearly seen in the retina, where cpg15 mRNA is restricted to RGCs. CPG15 IR is absent from the inner plexiform layer where RGC dendrites are located, whereas the optic nerve shows the most intense CPG15 immunoreactivity in the retina. Many developmentally regulated proteins that affect axon guidance and development are GPI-linked proteins, including semaphorins, fascilins, neural cell adhesion molecule (NCAM), and Eph ligands (Walsh and Doherty, 1997). GPI-linked proteins are concentrated in lipid rafts in the membrane where they form multimeric signaling complexes (Faivre-Sarrailh and Rougon, 1997; Tansey et al., 2000).
Under conditions of viral overexpression, CPG15 immunoreactivity is detectable in tectal cell dendrites (Nedivi et al., 1998). Therefore it is possible that under these conditions, dendritically localized CPG15 signals to other tectal cells to enhance their dendritic arbor growth and synapse maturation (Nedivi et al., 1998; Cantallops et al., 2000). Alternatively, CPG15 in local tectal cell axons overexpressing CPG15 might signal to tectal cell dendrites, leading to a cascade of events culminating in enlarged tectal cell dedritic arbors and synaptic maturation. The retinal axon arbor elaboration could arise from direct signaling from CPG15-expressing tectal neurons or indirectly through a retrograde mechanism that coordinates pre-and postsynaptic morphological development, as seen in previous experiments (Zou and Cline, 1996, 1999; Wu and Cline, 1998).
Detection of CPG15 in retinal axons is consistent with a model whereby CPG15 in presynaptic axons regulates multiple aspects of circuit formation including dendritic arbor growth, presynaptic arbor elaboration, and synaptogenesis (Nedivi et al., 1998; Corriveau et al., 1999). In this model, visual activity induces cpg15 expression in RGCs. These cells in turn synthesize CPG15 protein and traffic it to axon terminals. Visual stimulation has been shown to induce cpg15 in rat visual cortical neurons (Nedivi et al., 1996). Preliminary data indicate that light stimulation induces cpg15 expression in Xenopus RGCs (Javaherian and Cline, unpublished data).
We propose that CPG15 on the surface of retinal axons signals to postsynaptic optic tectal neurons and promotes their dendritic arbor elaboration and synaptic maturation. Similarly, CPG15 expressed in the postsynaptic tectal neurons is also trafficked to their axons in the tegmentum and spinal cord, where it may affect dendritic arbor elaboration and synaptic maturation of postsynaptic partners of tectal neurons. The expression of cpg15 in axons of projection neurons, but not in local interneurons, for instance in the retina and optic tectum, supports the idea that the source of CPG15 that regulates the development of circuitry in the CNS comes from upstream sensory afferents. The expression of cpg15 in many sensory systems in tadpole brain suggests that the function of cpg15 that we propose for visual system development may be generalizable. In particular, it is interesting to speculate that the induction of cpg15 by sensory activity may regulate the coordinated development of modality-specific circuits from the sensory periphery to motor output (Purves et al., 1996).
Acknowledgments
We thank Kim Bronson and Julianya Jay for excellent technical assistance. This work was supported by grants from the NIH (to H.T.C. and E.N.), the National Down Syndrome Society (to H.T.C.), and the Ellison Medical Foundation (to E.N.).
Literature Cited
- Antal M. Distribution of GABA immunoreactivity in the optic tectum of the frog: a light and electron microscopic study. Neuroscience. 1991;42:879–891. doi: 10.1016/0306-4522(91)90051-o. [DOI] [PubMed] [Google Scholar]
- Brakeman PR, Lanahan AA, O'Brien R, Roche K, Barnes CA, Huganir RL, Worley PF. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284–288. doi: 10.1038/386284a0. [DOI] [PubMed] [Google Scholar]
- Cantallops I, Haas H, Cline HT. Postsynaptic CPG15 expression enhances presynaptic axon growth and retinotectal synapse maturation. Nat Neurosci. 2000;3:498–503. doi: 10.1038/79823. [DOI] [PubMed] [Google Scholar]
- Corriveau RA, Huh GS, Shatz CJ. Regulation of class I MHC gene expression in the developing and mature CNS by neural activity. Neuron. 1998;21:505–520. doi: 10.1016/s0896-6273(00)80562-0. [DOI] [PubMed] [Google Scholar]
- Corriveau RA, Shatz CJ, Nedivi E. Dynamic regulation of cpg15 during activity-dependent synaptic development in the mammalian visual system. J Neurosci. 1999;19:7999–8008. doi: 10.1523/JNEUROSCI.19-18-07999.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre-Sarrailh C, Rougon G. Axonal molecules of the immunoglobulin superfamily bearing a GPI anchor: their role in controlling neurite outgrowth. Mol Cell Neurosci. 1997;9:109–115. doi: 10.1006/mcne.1997.0609. [DOI] [PubMed] [Google Scholar]
- Harland R. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hartenstein V. Early pattern of neuronal differentiation in the Xenopus embryonic brainstem and spinal cord. J Comp Neurol. 1993;328:213–231. doi: 10.1002/cne.903280205. [DOI] [PubMed] [Google Scholar]
- Holt CE, Harris WA. Position, guidance, and mapping in the developing visual system. J Neurobiol. 1993;24:1400–1422. doi: 10.1002/neu.480241011. [DOI] [PubMed] [Google Scholar]
- Jankowsky JL, Patterson PH. Cytokine and growth factor involvement in long-term potentiation. Mol Cell Neurosci. 1999;14:273–286. [PubMed] [Google Scholar]
- Low M. Biochemistry of the glycosyl-phosphatidylinositol membrane protein anchors. Biochem J. 1987;244:1–13. doi: 10.1042/bj2440001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufman WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- Naeve GS, Ramakrishnan M, Rainer K, Hevroni D, Citri Y, Theill LE. Neuritin: a gene induced by neural activity and neurotrophins that promotes neuritogenesis. Proc Natl Acad Sci USA. 1997;94:2648–2653. doi: 10.1073/pnas.94.6.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedivi E. Molecular analysis of developmental plasticity in neocortex. J Neurobiol. 1999;41:135–147. [PMC free article] [PubMed] [Google Scholar]
- Nedivi E, Hevroni D, Naot D, Israeli D, Citri Y. Numerous candidate plasticity-related genes revealed by differential cDNA cloning. Nature. 1993;363:718–722. doi: 10.1038/363718a0. [DOI] [PubMed] [Google Scholar]
- Nedivi E, Fieldust S, Theill L, Hevroni D. A set of genes expressed in response to light in the adult cerebral cortex and regulated during development. Proc Natl Acad Sci USA. 1996;93:2048–2053. doi: 10.1073/pnas.93.5.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedivi E, Wu GY, Cline HT. Promotion of dendritic growth by CPG15, an activity-induced signaling molecule. Science. 1998;281:1863–1866. doi: 10.1126/science.281.5384.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purves D, White LE, Riddle DR. Is neural development darwinian? TINS. 1996;19:460–464. doi: 10.1016/s0166-2236(96)20038-4. [DOI] [PubMed] [Google Scholar]
- Qian Z, Gilbert ME, Colicos MA, Kandel ER, Kuhl D. Tissue-plasminogen activator is induced as an immediate-early gene during seizure, kindling and long-term potentiation. Nature. 1993;361:453–457. doi: 10.1038/361453a0. [DOI] [PubMed] [Google Scholar]
- Rybicka KK, Udin SB. Ultrastructure and GABA immunoreactivity in layers 8 and 9 of the optic tectum of Xenopus laevis. Eur J Neurosci. 1994;6:1567–1582. doi: 10.1111/j.1460-9568.1994.tb00547.x. [DOI] [PubMed] [Google Scholar]
- Stamler JS, Toone EJ, Lipton SA, Sucher NJ. (S)NO signals: Translocation, regulation, and consensus motif. Neuron. 1997;18:691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- Straznicky K, Gaze RM. The development of the tectum in Xenopus laevis: an autoradiographic study. J Embryol Exp Morphol. 1972;28:87–115. [PubMed] [Google Scholar]
- Tansey MG, Baloh RH, Milbrandt J, Johnson JEM. GFRα-mediated localization of RET to lipid rafts is required for effective downstream signaling, differentiation, and neuronal survival. Neuron. 2000;25:611–623. doi: 10.1016/s0896-6273(00)81064-8. [DOI] [PubMed] [Google Scholar]
- Tsui CC, Copeland NG, Gilbert DJ, Jenkins NA, Barnes CA, Worley PF. Narp, a novel member of the pentraxin family, promotes neurite outgrowth and is dynamically regulated by neuronal activity. J Neurosci. 1996;16:2463–2478. doi: 10.1523/JNEUROSCI.16-08-02463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udin SG, S Plasticity in the tectum of Xenopus laevis: binocular maps. Prog Neurobiol. 1999;59:81–106. doi: 10.1016/s0301-0082(98)00096-3. [DOI] [PubMed] [Google Scholar]
- Walsh FS, Doherty P. Neural cell adhesion molecules of the immunoglobulin superfamily: role in axon growth and guidance. Annu Rev Cell Dev Biol. 1997;13:425–456. doi: 10.1146/annurev.cellbio.13.1.425. [DOI] [PubMed] [Google Scholar]
- Wu GY, Cline HT. Stabilization of dendritic arbor structure in vivo by CaMKII. Science. 1998;279:222–226. doi: 10.1126/science.279.5348.222. [DOI] [PubMed] [Google Scholar]
- Yamagata K, Andreasson KI, Kaufmann WE, Barnes CA, Worley PF. Expression of a mitogen-inducible cyclooxygenase in brain neurons: regulation by synaptic activity and glucocorticoids. Neuron. 1993;11:371–386. doi: 10.1016/0896-6273(93)90192-t. [DOI] [PubMed] [Google Scholar]
- Zou DJ, Cline HT. Expression of constitutively active Ca2+/calmodulin-dependent protein kinase in target tissue modifies presynaptic axon arbor growth. Neuron. 1996;16:529–539. doi: 10.1016/s0896-6273(00)80072-0. [DOI] [PubMed] [Google Scholar]
- Zou DJ, Cline HT. Coordinated regulation of retinal axon and tectal cell growth by endogenous CaMKII in vivo. J Neurosci. 1999;19:8909–8918. doi: 10.1523/JNEUROSCI.19-20-08909.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]


