Abstract
The stratification score for a case-control study is the probability of disease modeled as a function of potential confounders. The authors show that the stratification score is a retrospective balancing score and thus plays a similar role in case-control studies as the propensity score plays in prospective studies. The authors further show how standardization using the stratification score can be used to compare the distributions of exposures that would be found among case and control participants if both groups had the same distribution of confounding covariables. The authors illustrate these results using data from a genome-wide association study, the GAIN (Genetic Association Information Network) study of schizophrenia among African Americans (2006–2008).
Keywords: case-control studies, confounding factors (epidemiology), epidemiologic methods, genome-wide association study, propensity score, schizophrenia, standardization, stratification
The propensity score is a popular method for controlling confounding in prospective observational studies. The propensity score, the probability of exposure given confounding covariates, is a balancing score (1, 2); the distribution of potential confounders is independent of exposure status, conditional on the propensity score. Thus, for persons with the same propensity score, any association between exposure and outcome does not reflect a difference in potential confounders. Further, the difference in the prevalence of disease among exposed and unexposed persons, after propensity-score-based stratification, can be used to estimate the difference in the proportion of persons with disease by exposure status that would be observed in a randomized study. Of course, these statements assume that there are no unmeasured confounders and that the propensity model (or rankings based on it) is correct.
Although the propensity score is occasionally applied to case-control studies, its use is properly limited to prospective studies, for 2 reasons. First, exposure probabilities in a case-control study are not representative of the target population, so the estimated propensity score does not correspond to that in the target population. Second, comparing the difference in proportions of persons with disease in the exposed and the unexposed (the typical effect measure for a propensity score analysis) is problematic with case-control sampling, since the proportion of persons with disease in the study population is fixed by design.
We recently introduced the stratification score to control for confounding when testing hypotheses (3). Here we further develop the theory underlying the stratification score and show that it has many of the properties of a propensity score but for a retrospective study. In particular, the stratification score is a retrospective balancing score (defined below) for a case-control study. Thus, conditional on the stratification score and absent residual confounding, the distributions of exposures among case and control participants can be directly compared. Further, the stratification score can be used to estimate the exposure distribution that arises if, contrary to fact, case and control participants had been sampled with the same distribution of confounding variables. In particular, we can estimate the exposure distribution among case participants if their distribution of confounding variables matches that in control participants, which, under the rare disease approximation, approximates that in the target population. Note that the stratification score differs from Miettinen's confounder score (4), since exposure does not enter the stratification score model.
METHODS
The stratification score as a retrospective balancing score
Let D = 1 (0) denote the fact that a person is a case (control) participant. We study the association between D and exposure E, possibly distorted by confounding variables Z. The stratification score (3) is the estimated probability of case status (D = 1) given confounding variables Z. To construct the stratification score, we first model , typically by logistic regression, with parameters γ, and obtain estimates 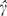 ; then the ith study participant's stratification score S(Z) is given by
; then the ith study participant's stratification score S(Z) is given by  . Note that it is not necessary that the stratification score correspond to any population quantity so long as it correctly describes the relation between disease and confounding variables in the study population.
. Note that it is not necessary that the stratification score correspond to any population quantity so long as it correctly describes the relation between disease and confounding variables in the study population.
Because a case-control study is comprised of 2 separate samples, the distribution of covariates may differ between case and control participants. If these covariates are confounders, this difference may cause a spurious association. A correctly specified stratification score is a retrospective balancing score for a case-control study, meaning that
a simple proof can be found in the Appendix. In words, the distribution of potential confounders Z is independent of case/control status, conditional on the stratification score. Recall that for a prospective study, the distribution of potential confounders, conditional on a balancing score, is independent of exposure status.
Because the stratification score is a retrospective balancing score, any observed association between disease and exposure, among persons with the same value of the stratification score, cannot be caused by differences in the distributions of confounders among cases and controls. Thus, assuming no unmeasured confounders and a properly specified stratification score, conditioning on the stratification score yields a true measure of the association between disease and exposure. This observation suggests that the stratification score be used for poststratification. Like analyses that use the propensity score, participants are assigned to one of a fixed number of strata defined by quantiles of the stratification scores in the study population. Frequently, 5 strata are used (2), although for large studies more strata can be used to better control residual confounding.
After stratification, we can test for an association between exposure and disease using standard tests such as the extended Mantel-Haenszel test. Unfortunately, odds ratio estimates may be difficult to interpret. The difficulty arises because, if we assume that a (prospective) logistic model for disease given exposure and confounding covariates holds, then the (marginal) model that only conditions on exposure and stratification score is not necessarily logistic. A similar phenomenon occurs in prospective studies that are analyzed by poststratification using the propensity score (5).
Estimation of marginal associations in the presence of confounding
Although odds ratio estimates after poststratification using the stratification score do not correspond to association parameters of interest, there are quantities of potential interest that are estimable using the stratification score. In particular, the stratification score enables comparison of the exposure distributions in the case and control populations.
A case-control study compares the difference in exposure between cases and controls. For example, we may compare allele frequencies among persons with and without a disease of interest. However, exposure may appear to vary by disease status if confounders have different distributions in persons with and without disease. Recall that potential confounders that have the same distribution by disease status cannot lead to a spurious association between disease and exposure.
The stratification score can be used to standardize data from case or control participants, so that the distribution of confounding variables Z is the same among case and control participants. For convenience, we initially assume that the data have been stratified into J strata based on the stratification score, and within each stratum S(Z) takes the fixed and distinct value sj. Then we can write
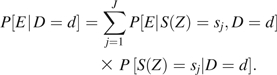 |
(1) |
The first term on the right, , can be estimated by the empirical distribution of E among cases (for D = 1) or controls (for D = 0) in the jth stratum. Because the retrospective balancing score property assures that the distribution of confounding covariates is independent of disease status among persons with the same value of the stratification score, the empirical distributions of exposure among case and control participants are directly comparable without adjustment for confounding. The second term, , can be estimated by the empirical proportions of case (for D = 1) or control (for D = 0) participants assigned to each stratum.
In equation 1, differences in the distribution of confounders Z between cases and controls have been isolated to differences in the proportions of case and control participants found in each stratum. If the same proportions were used for both case and control participants when calculating the exposure distribution in equation 1, the resulting exposure distributions could be properly compared. To this end, let denote the distribution of exposure given disease status that arises if both case and control participants have the same distribution of strata. Then we have
Note that can be compared with , with the assurance that any differences seen are not due to the effect of confounding covariates (assuming no unmeasured confounders and a correct model for the stratification score).
The distribution corresponds to standardizing the exposure distribution among case and control participants to the same distribution of stratification scores . A natural choice for the standardization distribution is , which, for a rare disease, approximates the distribution of confounding covariates in the target population. We let denote the distribution of E among case participants after this standardization. For this choice, is the actual distribution of E among controls. A second choice, standardizing to the distribution of strata among cases, can be achieved by exchanging the roles of cases and controls, and this is appropriate when the goal of the analysis is to frequency-match controls to cases. A third option is to use
where n0 (n1) is the number of control (case) participants, corresponding to the distribution of S(Z) in the (artificial) case-control study population. We let denote the distribution of general exposures E among case and control participants after this standardization.
Stratified and individually weighted estimators of the standardized exposure distributions
To develop estimators of the standardized exposure distributions, we initially restrict our attention to stratified data and a categorical (or binned) exposure E. Then, the observed data can be expressed as cell counts nedj, where e indexes exposure levels, d indicates case/control status, and j indicates stratum. Then, is estimated by
where is the empirical probability that E = e within stratum j among persons with D = d. For standardizing to the control population, 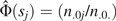 (e.g., see Table 2), while
(e.g., see Table 2), while  when standardizing to the study population.
when standardizing to the study population.
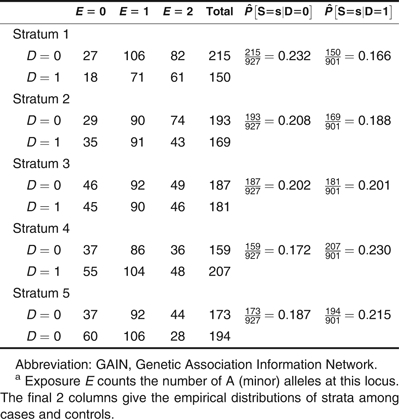
Table 2.
Distribution of Genotypes at Single Nucleotide Polymorphism rs4322256, Poststratified Using the Stratification Score, GAIN Study of Schizophrenia in African Americans, 2006–2008a
 |
To estimate , write
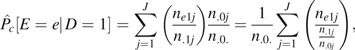 |
which can be rewritten as a sum over contributions from each person as
| (2) |
where ji is the stratum assignment for the ith individual and where
is the empirical odds of disease in stratum j. The form of equation 2 suggests the individually weighted estimator
where 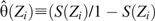 is the odds of disease given covariates Z. Note that for the individually weighted estimator, we need not assume that E is categorical or that the stratification score takes only discrete values sj..
is the odds of disease given covariates Z. Note that for the individually weighted estimator, we need not assume that E is categorical or that the stratification score takes only discrete values sj..
If the stratification score is logistic, then
and we obtain
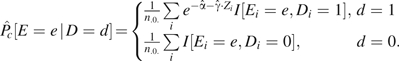 |
It is possible to prove that  estimates . Note that the intercept
estimates . Note that the intercept 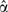 is the value obtained by fitting the stratification score model to the case-control data, not the intercept that would be obtained in a prospective study.
is the value obtained by fitting the stratification score model to the case-control data, not the intercept that would be obtained in a prospective study.
The expression for  has the form of a weighted estimator. Because the sum of the weights has the expected value 1 but the weights may not sum exactly to 1 in finite samples, we prefer the estimator
has the form of a weighted estimator. Because the sum of the weights has the expected value 1 but the weights may not sum exactly to 1 in finite samples, we prefer the estimator
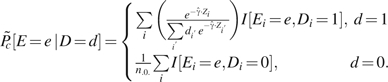 |
Because the weights used to calculate  sum exactly to 1, it has the advantage that
sum exactly to 1, it has the advantage that 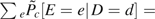 . Extrapolating the results of Lunceford and Davidian (6) to the stratification score, we would also expect
. Extrapolating the results of Lunceford and Davidian (6) to the stratification score, we would also expect  to have lower sampling variability than
to have lower sampling variability than  . For these reasons, we recommend
. For these reasons, we recommend  over
over  .
.
When standardizing the distribution of exposure given disease status to the study population, arguments that parallel those just given lead to
 |
As before, we advise normalizing the weights in 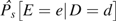 to sum to 1 in finite samples to obtain
to sum to 1 in finite samples to obtain
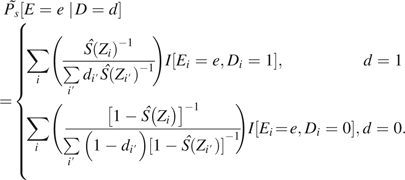 |
Simple estimators of the sampling variance of 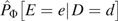 or its moments can be obtained using standard M-estimator theory (7), following the approach used by Lunceford and Davidian (6); see the Appendix for details. Finally, note that stratified estimators are special cases of the individually weighted estimators obtained by taking , where J is the number of strata used, and then estimating the stratification score model without an intercept.
or its moments can be obtained using standard M-estimator theory (7), following the approach used by Lunceford and Davidian (6); see the Appendix for details. Finally, note that stratified estimators are special cases of the individually weighted estimators obtained by taking , where J is the number of strata used, and then estimating the stratification score model without an intercept.
Often interest centers on the average exposure (especially when E is continuous) rather than the full distribution of E. For example, we may wish to study risk allele frequencies, not genotype distributions. When E is discrete and assumes levels  , the mean exposure level in the standardized population, , can be estimated by
, the mean exposure level in the standardized population, , can be estimated by 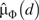 , given by
, given by
An estimate of the sampling variance of 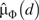 can be easily obtained from the estimated sampling variance of
can be easily obtained from the estimated sampling variance of 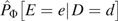 . For a continuous exposure, the sum is replaced by an integral; estimators of the sampling variance of
. For a continuous exposure, the sum is replaced by an integral; estimators of the sampling variance of 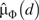 when E is continuous are discussed in the Appendix.
when E is continuous are discussed in the Appendix.
RESULTS
Association between schizophrenia and SNP rs4322256 in a genome-wide association study of African Americans
We illustrate our methods using data from the Genetic Association Information Network (GAIN) study of schizophrenia among African Americans (8). Rates of schizophrenia among African Americans are higher than those among persons with purely European ancestry (9). Here we analyze the association between disease and the single nucleotide polymorphism (SNP) rs4322256, located in the netrin G1 gene (NTNG1), a gene previously linked to schizophrenia in a Japanese sample (10). GAIN study data are available in the Database of Genotypes and Phenotypes (http://www.ncbi.nlm.nih.gov/gap) through accession number phs000021.v2.p1 (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000021.v2.p1)); in our analysis, we used data on 845,814 available SNP genotypes from the period 2006–2008. Here we show results obtained for 927 case participants and 901 control participants who had nonmissing data on genotype at SNP rs4322256. Additional information can be found in the Web Appendix, which is posted on the Journal’s Web site (http://aje.oxfordjournals.org/).
Differences in the proportion of African ancestry between cases and controls may confound the association between schizophrenia and markers that have different allele frequencies in Africans and Europeans, like SNP rs4322256, which has an A allele frequency of 0.425 in Africans and 0.950 in Europeans (11). Such confounding would manifest itself in correlated genotypes genome-wide; for example, persons with a high proportion of African ancestry would be more likely to have a pattern of genotypes characteristic of an African population, while persons with a high proportion of European ancestry would be more likely to have a pattern of genotypes characteristic of a European population. Because these correlations would occur genome-wide, not just among adjacent SNPs as would be expected due to linkage disequilibrium, this type of confounding can typically be resolved by using principal components, or related techniques, applied to the variance-covariance matrix of SNP genotypes genome-wide (12–14). We found that 3 linear combinations of SNP genotypes were adequate (14) to describe the genome-wide correlations due to the admixture of European and African ancestries in this population (see Web Appendix for additional details). We then used these linear combinations of SNP genotypes, calculated for each person, as covariates in a logistic regression model to calculate the stratification score.
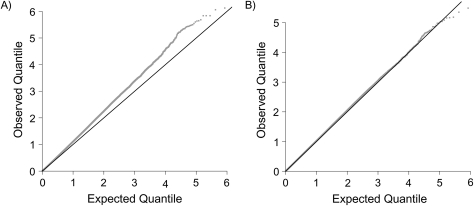
In Figure 1, we show Q-Q plots for tests of association between disease status and each of the 845,814 SNP genotypes available in these data, calculated using the Cochran-Mantel-Haenszel test for association. The extent of confounding in these data is evident in the first Q-Q plot, which has not been adjusted for confounding and which shows systematic differences between quantiles of the observed test statistics and what we would expect under the (reasonable) assumption that most loci are not associated with schizophrenia. The second Q-Q plot uses stratified Cochran-Mantel-Haenszel tests that are based on 5 nearly equally populated strata based on the quantiles of the stratification score; the close agreement between observed and expected quantiles indicates that confounding has been controlled in these data. Additionally, we show in Table 1 that the stratification score balances the potential confounders. We show the mean value of each potential confounder (standardized using its overall sample mean and standard deviation) by case/control status. The association between the outcome and each covariate is reduced by stratification, most notably for the covariates that are most associated with the outcome, and there is no significant within-stratum association between disease status and any covariate.
Figure 1.
Q-Q plots for tests of association between disease status and each of 845,814 rs4322256 single nucleotide polymorphism genotypes, before (left panel) and after (right panel) adjustment for confounding, GAIN study of schizophrenia in African Americans, 2006–2008. In each panel, the observed and expected quantiles of the log10 P value for the marginal association tests are represented by gray dots. On the black line, observed and expected quantiles are equal. (GAIN, Genetic Association Information Network).
Table 1.
Covariate Balance Between Case and Control Participants, Before and After Stratification, GAIN Study of Schizophrenia in African Americans, 2006–2008
| Mean of Z1 |
P Value | Mean of Z2 |
P Value | Mean of Z3 |
P Value | ||||
| D = 0 | D = 1 | D = 0 | D = 1 | D = 0 | D = 1 | ||||
| Unadjusted | −0.043 | 0.045 | 0.056 | 0.091 | −0.094 | 5.582e-05 | −0.009 | 0.010 | 0.684 |
| Stratum 1 | −1.096 | −1.023 | 0.644 | 1.647 | 1.590 | 0.470 | −0.075 | −0.173 | 0.502 |
| Stratum 2 | 0.527 | 0.489 | 0.233 | 0.333 | 0.328 | 0.851 | −0.104 | −0.145 | 0.758 |
| Stratum 3 | 0.622 | 0.616 | 0.861 | −0.262 | −0.265 | 0.845 | −0.145 | −0.043 | 0.339 |
| Stratum 4 | 0.383 | 0.352 | 0.356 | −0.643 | −0.647 | 0.710 | 0.102 | 0.131 | 0.626 |
| Stratum 5 | −0.487 | −0.377 | 0.065 | −1.053 | −1.028 | 0.133 | 0.222 | 0.206 | 0.713 |
| Stratified | 0.547 | 0.613 | 0.932 | ||||||
Abbreviation: GAIN, Genetic Association Information Network.
In Table 2, we show the distribution of genotypes at rs4322256 in cases and controls for the 5 strata used in these analyses. Note that cases outnumber controls in strata 1 and 2 but controls outnumber cases in strata 4 and 5, indicating systematic differences between case and control participants that must be accounted for. To illustrate our approach, we use the data in Table 2 to estimate , the distribution of exposures we would see among case participants if their distribution of confounding covariates were the same as that found among control participants. To construct this estimate, we write
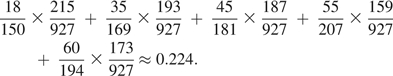 |
In writing this expression, note that we have used the empirical distributions of exposure calculated using data from case participants within each stratum (e.g., 18 of 150 case participants have E = 0 in stratum 1) but have used the empirical distribution of strata calculated using data from control participants (e.g., 215 of 927 controls are assigned to stratum 1). In contrast, the empirical proportion of cases having E = 0 is approximately 0.236 ((18 + 35 + 45 + 55 + 60)/921 ≈ 0.236), corresponding to a difference in this exposure probability of approximately 5% that is attributable to confounding.
We estimate the frequency of the A allele in case and control participants using 6 stratification-score-based estimators (Table 3). We also show the unadjusted A allele frequency. Although the differences appear small, using the analysis that standardizes to control participants implies that 27.5% (individually weighted analysis) or 26.2% (stratified analysis) of the naively observed difference in allele frequency at rs4322256 is actually explained by confounding. The test statistics obtained when standardizing to the study population are slightly larger than those obtained when standardizing to the control population, although this gain is small because the imbalance between case and control participants assigned to each of the strata in Table 2 is modest. The test statistics based on standardized allele frequency differences are comparable to the logistic regression Wald test for a gene-dose model (Table 3).
Table 3.
Estimated Frequency of the A (Minor) Allele in Cases and Controls at Single Nucleotide Polymorphism rs4322256, GAIN Study of Schizophrenia in African Americans, 2006–2008
| Method | Standard Population | Normalization |  |
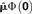 |
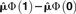 |
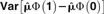 |
χ2 |
| Weighted | Controls | No | 0.5214 | 0.5588 | −0.0374 | 2.678 × 10−4 | 5.22 |
| Weighted | Controls | Yes | 0.5212 | 0.5588 | −0.0376 | 2.678 × 10−4 | 5.28 |
| Weighted | Study | No | 0.5144 | 0.5538 | −0.0394 | 2.593 × 10−4 | 5.99 |
| Weighted | Study | Yes | 0.5143 | 0.5539 | −0.0396 | 2.593 × 10−4 | 6.05 |
| Stratified | Controls | 0.5207 | 0.5588 | −0.0381 | 2.724 × 10−4 | 5.33 | |
| Stratified | Study | 0.5141 | 0.5532 | −0.0391 | 2.692 × 10−4 | 5.68 | |
| Unadjusted | 0.5072 | 0.5588 | −0.0516 | 2.726 × 10−4 | 9.77 | ||
| Logistic regression | 6.12 |
Abbreviation: GAIN, Genetic Association Information Network.
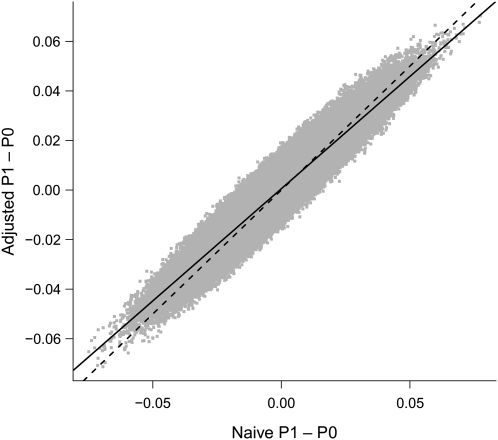
In Figure 2, we plot the unadjusted and adjusted allele frequency differences for all 845,814 loci. We also plot the 45-degree line corresponding to no adjustment, as well as the regression line. From Figure 2 we see that, on average, standardization-based adjustment for confounding in these data has resulted in shrinkage, with larger deviations being subject to larger correction.
Figure 2.
Naive difference in allele frequency versus adjusted difference in allele frequency between cases (P1) and controls (P0) for each of 845,814 rs4322256 single nucleotide polymorphism loci (gray dots), GAIN study of schizophrenia in African Americans, 2006–2008. On the dashed line, the naive and adjusted allele frequency differences are equal. The solid line is the regression line. (GAIN, Genetic Association Information Network).
DISCUSSION
The stratification score was originally proposed to control confounding when testing hypotheses in a case-control study (3). Here we have extended the stratification score approach to accommodate estimation, which is preferred by many epidemiologists over hypothesis-testing (15). By showing that the stratification score is a retrospective balancing score, we have developed a standardization-based approach to controlling confounding in case-control studies which allows us to compare the exposure distributions between case and control participants that would be observed if both groups had the same distribution of confounding covariables. This comparison is attractive, since differences in exposure frequency can be easily interpreted at the population level in a way that odds ratios from a logistic regression model cannot. Similar comparisons could also be made by stratifying on all confounders if the data were not too finely stratified. Correspondingly, matched studies are simplified by matching on stratification scores rather than matching on multiple potential confounders.
In our previous article (3), we tested whether the common odds ratio over strata was equal to 1. Here we have shown how to estimate the difference in mean exposure after standardizing the distribution of exposures, and have further described how to estimate the variance of this difference for discrete-valued exposures. As a result, we can construct confidence intervals or test hypotheses about these standardized differences. As Table 3 indicates in the context of a single analysis, these tests can be comparable in power to standard logistic regression.
We have considered both stratified and individually weighted estimators of the exposure distribution. When deriving the stratified estimators, we assumed that the stratification score had a constant value within each stratum. Violations of this assumption may lead to residual confounding and favor the individually weighted estimator. Increasing the number of strata or even fine matching based on the stratification score may be needed to resolve large-scale within-stratum variability in the stratification score. However, as Rubin (16) noted in the context of propensity score modeling, stratification is more robust to misspecification of the stratification score model. An additional advantage of stratification is that the extent of confounding can be seen. For example, our Table 2 illustrates the extent to which cases and controls are mismatched, which may be hard to ascertain when individually weighted estimators are used.
When choosing variables to include in the stratification score model, it is important to note that the goal is control of confounding, rather than prediction of case status (17). Thus, variables that predict case status but do not predict exposure should not be included in the stratification score model (18). Similarly, Brookhart et al. (19) found that variables that predict exposure but not outcome should not be included in a propensity score model. Brookhart et al. (19) further stated that variables which predict outcome but not necessarily exposure can be beneficial when modeling the propensity score. The stratification score analog to this finding would be that variables which predict exposure but not necessarily case status are salutory in a stratification score model; however, we have not evaluated this claim and hence make no recommendation at this time.
We assumed that all confounding variables were measured. In fact, we only require that unmeasured confounders U be balanced given the stratification score—that is, that . This is reasonable if, as is often assumed in epidemiologic studies, measured covariates are strongly correlated with U. For example, we may adjust for demographic covariates that may not be causal but covary with unmeasured confounders that are.
We have considered a “general exposure” without specifying its nature. Thus, levels of exposure could, for example, correspond to combinations of genotypes and environmental covariables, allowing comparison of interaction terms in case and control populations having the same distribution of potential confounders. We are also developing a modeling approach to such interaction models (unpublished data). Finally, our presentation emphasized the situation where the exposure E is categorical; this was done for ease of presentation and is not a restriction of our approach.
Supplementary Material
Acknowledgments
Author affiliations: Department of Biostatistics and Bioinformatics, School of Medicine, Duke University, Durham, North Carolina (Andrew S. Allen); Duke Clinical Research Institute, Duke University Medical Center, Durham, North Carolina (Andrew S. Allen); and Division of Reproductive Health, National Center for Chronic Disease Prevention and Health Promotion, Centers for Disease Control and Prevention, Atlanta, Georgia (Glen A. Satten).
Funding for the GAIN study of schizophrenia among African Americans was provided by the National Institutes of Health, and the genotyping of samples was supported by the Genetic Association Information Network. Dr. Andrew S. Allen received support from the National Institutes of Health through National Heart, Lung, and Blood Institute grant K25HL077663 and National Institute of Mental Health grant R01 MH084680.
The data set used for these analyses was obtained from the Database of Genotypes and Phenotypes (http://www.ncbi.nlm.nih.gov/gap) through accession number phs000021.v2.p1 (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000021.v2.p1). Samples and associated phenotype data for the GAIN study of schizophrenia among African Americans were provided by Dr. Pablo V. Gejman.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Conflict of interest: none declared.
Glossary
Abbreviations
- GAIN
Genetic Association Information Network
- SNP
single nucleotide polymorphism
APPENDIX
For this proof, we assume a “properly specified” stratification score, by which we mean that corresponds to the law that generated the study data. We wish to show
|
|
Let Ωs be the set of values of z for which S(z) = s. Since S(Z) is a coarsening of Z,
 |
Note that
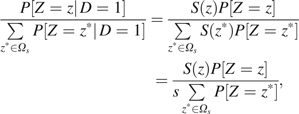 |
where P[Z = z] is the distribution of Z in the study population. Since S(z) = s when , we have
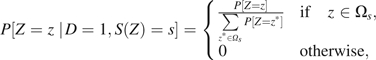 |
so that
The argument for is entirely similar. Thus, S(Z) is a retrospective balancing score.
Note that the form of given above seems to suggest that the distribution of Z among persons having S(Z) = s is the restriction of the distribution of Z in the study population to values . However, because
it is easy to see that we could just as well write
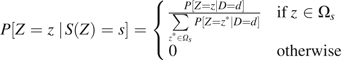 |
for either d = 0 or d = 1. This special property of the stratification score allows us to use the empirical estimate of , regardless of whether we are standardizing to the case, control, or study population.
We next outline estimation of the sampling variance of 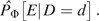 Let , and, for the ith study participant, let Let . For any standardization, the unnormalized estimators are solutions to estimating equations
Let , and, for the ith study participant, let Let . For any standardization, the unnormalized estimators are solutions to estimating equations
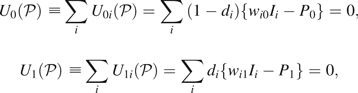 |
and
 |
For individually weighted estimators standardized to the controls, use and 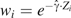 ; while standardizing to the study population, use
; while standardizing to the study population, use 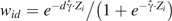 . For stratified estimators, these same equations apply but with Zi a vector of stratum-specific indicator functions in a stratification score model with no intercept. For normalized estimators, the first 2 estimating equations are modified to
. For stratified estimators, these same equations apply but with Zi a vector of stratum-specific indicator functions in a stratification score model with no intercept. For normalized estimators, the first 2 estimating equations are modified to
and
Expressing the parameters 𝒫 as solutions to estimating equations yields a sandwich estimator of their variance-covariance matrix using M-estimator theory (e.g., see Stefanski and Boos (7)). Joint estimation of all parameters addresses the concerns of Pike et al. (20) regarding the variance estimates of data poststratified using Miettinen's confounder score.
When exposure E is continuous, the sampling variance of , the mean exposure, can be calculated using a similar approach. Let the parameter vector 𝒫 be defined by , and then replace Ii by ei and Pd by in the estimating equations above. As before, standard M-estimator theory can be used to obtain a variance-covariance estimator for 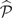 .
.
Further simplification arises assuming that is not a function of γ. Then we have
all variance-covariance estimators reported here have been calculated using this assumption.
References
- 1.Rosenbaum PR, Rubin DB. The central role of the propensity score in observational studies for causal effects. Biometrika. 1983;70(1):41–55. [Google Scholar]
- 2.Rosenbaum PR, Rubin DB. Reducing bias in observational studies using subclassification on the propensity score. J Am Stat Assoc. 1984;79(387):516–524. [Google Scholar]
- 3.Epstein MP, Allen AS, Satten GA. A simple and improved correction for population stratification in case-control studies. Am J Hum Genet. 2007;80(5):921–930. doi: 10.1086/516842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miettinen OS. Stratification by a multivariate confounder score. Am J Epidemiol. 1976;104(6):609–620. doi: 10.1093/oxfordjournals.aje.a112339. [DOI] [PubMed] [Google Scholar]
- 5.Månsson R, Joffe MM, Sun W, et al. On the estimation and use of propensity scores in case-control and case-cohort studies. Am J Epidemiol. 2007;166(3):332–339. doi: 10.1093/aje/kwm069. [DOI] [PubMed] [Google Scholar]
- 6.Lunceford JK, Davidian M. Stratification and weighting via the propensity score in estimation of causal treatment effects: a comparative study. Stat Med. 2004;23(19):2937–2960. doi: 10.1002/sim.1903. [DOI] [PubMed] [Google Scholar]
- 7.Stefanski LA, Boos DD. The calculus of M-estimation. Am Stat. 2002;56(1):29–38. [Google Scholar]
- 8.Manolio TA, Rodriguez LL, Brooks L, et al. New models of collaboration in genome-wide association studies: the Genetic Association Information Network. GAIN Collaborative Research Group. Nat Genet. 2007;39(9):1045–1051. doi: 10.1038/ng2127. [DOI] [PubMed] [Google Scholar]
- 9.Bresnahan M, Begg MD, Brown A, et al. Race and risk of schizophrenia in a US birth cohort: another example of health disparity? Int J Epidemiol. 2007;36(4):751–758. doi: 10.1093/ije/dym041. [DOI] [PubMed] [Google Scholar]
- 10.Aoki-Suzuki M, Yamada K, Meerabux J, et al. A family-based association study and gene expression analyses of netrin-G1 and -G2 genes in schizophrenia. Biol Psychiatry. 2005;57(4):382–393. doi: 10.1016/j.biopsych.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 11.International HapMap Consortium. The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 12.Zhu X, Zhang S, Zhao H, et al. Association mapping, using a mixture model for complex traits. Genet Epidemiol. 2002;23(2):181–196. doi: 10.1002/gepi.210. [DOI] [PubMed] [Google Scholar]
- 13.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 14.Lee AB, Luca D, Klei L, et al. Discovering genetic ancestry using spectral graph theory. Genet Epidemiol. 2010;34(1):51–59. doi: 10.1002/gepi.20434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothman KJ, Greenland S, Lash TL. Philadelphia PA. Lippincott Williams & Wilkins; 2008. Modern Epidemiology. 3rd ed. [Google Scholar]
- 16.Rubin DB. Estimating causal effects from large data sets using propensity scores. Ann Intern Med. 1997;127(8):757–763. doi: 10.7326/0003-4819-127-8_part_2-199710151-00064. [DOI] [PubMed] [Google Scholar]
- 17.Epstein MP, Allen AS, Satten GA. Response to Lee et al. [letter] Am J Hum Genet. 2008;82(2):526–528. [Google Scholar]
- 18.Lee S, Sullivan PF, Zou F, et al. Comment on a simple and improved correction for population stratification [letter] Am J Hum Genet. 2008;82(2):524–526. doi: 10.1016/j.ajhg.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brookhart MA, Schneeweiss S, Rothman KJ, et al. Variable selection for propensity score models. Am J Epidemiol. 2006;163(12):1149–1156. doi: 10.1093/aje/kwj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pike MC, Anderson J, Day N. Some insights into Miettinen's multivariate confounder score approach to case-control study analysis. Epidemiol Community Health. 1979;33(1):104–106. doi: 10.1136/jech.33.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.