Abstract
The genus Drosophila has been the subject of intense comparative phylogenomics characterization to provide insights into genome evolution under diverse biological and ecological contexts and to functionally annotate the Drosophila melanogaster genome, a model system for animal and insect genetics. Recent sequencing of 11 additional Drosophila species from various divergence points of the genus is a first step in this direction. However, to fully reap the benefits of this resource, the Drosophila community is faced with two critical needs: i.e., the expansion of genomic resources from a much broader range of phylogenetic diversity and the development of additional resources to aid in finishing the existing draft genomes. To address these needs, we report the first synthesis of a comprehensive set of bacterial artificial chromosome (BAC) resources for 19 Drosophila species from all three subgenera. Ten libraries were derived from the exact source used to generate 10 of the 12 draft genomes, while the rest were generated from a strategically selected set of species on the basis of salient ecological and life history features and their phylogenetic positions. The majority of the new species have at least one sequenced reference genome for immediate comparative benefit. This 19-BAC library set was rigorously characterized and shown to have large insert sizes (125–168 kb), low nonrecombinant clone content (0.3–5.3%), and deep coverage (9.1–42.9×). Further, we demonstrated the utility of this BAC resource for generating physical maps of targeted loci, refining draft sequence assemblies and identifying potential genomic rearrangements across the phylogeny.
THE genus Drosophila contains ∼2000 species of diverse morphology, ecology, and behavior that are placed in three major lineages: subgenus Sophophora, subgenus Drosophila, and subgenus Dorsilopha (Markow and O'Grady 2006, 2007). The most widely studied species in the genus, Drosophila melanogaster, is firmly established as the premier model system for many biological research areas such as neurobiology, medicine, and population biology (Rubin and Lewis 2000). Several other species in this genus, such as D. pseudoobscura and D. virilis, have also been utilized as genetic model systems particularly for evolutionary studies (Orr and Coyne 1989; Anderson et al. 1991; Popadic and Anderson 1994; Charlesworth et al. 1997; Vieira et al. 1997; Sweigart 2010). Recently, the genomes of D. melanogaster and 11 other Drosophila species, whose most recent common ancestor occurred >45–50 MYA, have been sequenced, assembled, and annotated (Adams et al. 2000; Myers et al. 2000; Celniker et al. 2002; Richards et al. 2005; Drosophila 12 Genomes Consortium 2007; Gilbert 2007). Species were selected for genome sequencing partly on the basis of their relationship with D. melanogaster. Nine of the 12 sequenced genomes were sampled from one subgenus, Sophophora, to which D. melanogaster belongs, and the remaining 3 are from the Drosophila subgenus. These sequences have already greatly improved understanding of the evolution and regulation of eukaryotic genes and genomes through comparative analyses (Stark et al. 2007). However, to fully reap the benefits from this unique resource, the Drosophila community is faced with two critical needs: first, the development of additional genomics resources to aid in finishing the 11 existing draft genome sequences and, second, the generation of additional genomic resources that encompass a much broader range of phylogenetic diversity.
Toward this direction, we constructed a comprehensive set of bacterial artificial chromosome (BAC) libraries for 19 different Drosophila species representing a broad spectrum of phylogenetic diversity. BAC libraries are powerful tools for comparative genome research (Kim et al. 1996; Hoskins et al. 2000; International Human Genome Mapping Consortium 2000a,b; Locke et al. 2000; Osoegawa et al. 2000, 2001, 2004; Eichler and Dejong 2002; Gregory et al. 2002; Gibbs et al. 2003; Krzywinske et al. 2004; Gonzalez et al. 2005; Ammiraju et al. 2006; Drosophila 12 Genomes Consortium 2007; Kim et al. 2008; Murakami et al. 2008), especially in taxa containing highly repetitive genomes (Havlak et al. 2004; Ellison and Shaw 2010; Fang et al. 2010). Genome sequences are available for 10 of 19 species for which BAC libraries are constructed, some of which were instrumental in facilitating sequence assemblies (Drosophila 12 Genomes Consortium 2007), and they remain a high-priority resource for improving and finishing several of the low coverage draft genome assemblies. BAC libraries for species without sequenced genomes present an important resource for positional cloning and large-scale targeted comparative genome analyses.
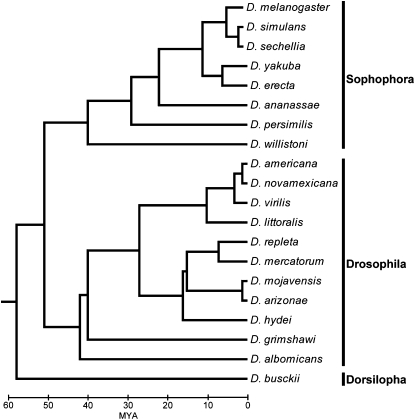
We selected 19 species within three lineages of the genus Drosophila for BAC library construction (Figure 1). These species shared a common ancestor ∼40–60 MYA (Powell 1997) and were selected because of their varied evolutionary distances from D. melanogaster and other sequenced species, their diverse ecologies and life history characters, and the fact that they can be reared in the laboratory and used in experimental work in the future. Ten BAC libraries were constructed as a resource for generating BAC end mate-pair sequence to assist in the assembly of whole-genome shotgun sequences and for enabling future genomic research (Drosophila 12 Genomes Consortium 2007). Beyond those 10 species, we are interested in generating BAC library resources for representative species of lineages not yet targeted for sequencing but that fill in large phylogenetic gaps. The majority of these species have at least one previously sequenced reference genome for immediate comparative benefit. In addition, this new set of species facilitates the “ladder and constellation” approach of modified phylogenetic shadowing proposed by Clark et al. (http://flybase.org/static_pages/news/whitepapers/GenomesWP2003.pdf) for annotating genome data. In this approach ladder rungs constitute successively increasing divergence points and constellations are clusters of species attaching to these divergence points. This set of 19 BAC libraries documented here will further advance the genus Drosophila as an ideal eukaryotic comparative genomics system designed to (1) provide sequencing resources for comparative annotation of the D. melanogaster genome and (2) provide genomic resources for experimental investigation of gene function throughout the genus Drosophila.
Figure 1.—
Phylogenetic tree of 19 species and D. melanogaster selected for the Drosophila BAC resource project. The phylogenetic relationships and approximate divergence times among the Drosophila species in our study were determined from a compilation of prior analyses (Pitnick et al. 1995; Markow and O'Grady 2006; Drosophila 12 Genomes Consortium 2007).
MATERIALS AND METHODS
Fly culturing and embryo collection:
Fly cultures were expanded on banana/opuntia medium (http://flyfood.arl.arizona.edu/opuntia.php3) and healthy sexually mature adult flies were introduced into plexiglass oviposition chambers kept on a 16/8 light/dark cycle at 24°–25° with a relative humidity of 60–80%. Exceptions to this procedure were D. littoralis, D. novamexicana, D. americana, D. grimshawi, and D. persimilis cultures, which were oviposited at 20°–22°, whereas D. albomicans was oviposited at 17°. Medium for D. sechellia was supplemented with 0.5% (v/v) hexanoic acid and 0.5% (v/v) octanoic acid to stimulate oviposition. Oviposition medium for D. grimshawi was supplemented with 2% (w/v) methylparaben to prevent overgrowth of fungus. D. busckii and D. grimshawi cultures were grown on Wheeler–Clayton medium (http://flyfood.arl.arizona.edu/wheeler.php3). D. grimshawi adults were separated by sex until day of placement in the oviposition chamber to enhance embryo production. Adult flies were allowed to oviposit on a given plate for as long as possible without larval hatch. This interval varied between 4 and 48 hr depending on the species. About 1.2–1.5 g wet weight embryos were pooled in batches and stored at −80° at the end of each oviposition session.
Nuclei preparation and BAC library construction:
Embryos were gently homogenized in PBS buffer (0.76% NaCl, 4 mm NaH2PO4, 9 mm Na2HPO4, pH 7.0) using a Dounce Tissue Grinder (Wheaton Science), centrifuged at 4° at 1430 × g for 15 min, and resuspended in PBS buffer. The suspension was then mixed with an equal volume of 1% InCert Agarose (CAMBREX) (in PBS buffer) at 45° and transferred into plug molds. Treatment of plugs to produce unsheared megabase-size DNA was as described (Luo and Wing 2003). BAC libraries were constructed as previously described (Luo and Wing 2003; Ammiraju et al. 2006).
BAC library characterization:
DNA from a random sample of 260–480 BAC clones from each library was isolated, restriction digested with NotI, and run on CHEF gels for insert size determination as previously described (Luo and Wing 2003; Ammiraju et al. 2006).
High colony density hybridization filters for each library were prepared using Genetix Q-bots (Genetix) as described previously (Ammiraju et al. 2006; Luo et al. 2006). Nine gene-specific probes were chosen that represented all chromosomes of D. melanogaster (supporting information, Table S1 and Table S2). All probe DNA fragments were PCR amplified from the D. mojavensis genome and gel purified using a QIAEX II (QIAGEN, Valencia, CA) kit. Table S1 lists the primer sequences used for each probe. Purified DNA fragments were sequenced and similarity searches were conducted to validate their specificity. Probes were prepared by labeling with [32P]dCTP using a DecaprimeII random prime labeling kit (Ambion), and hybridizations were carried out as described by Ammiraju et al. (2006). Positive clones were picked and rearrayed onto colony filters, followed by a secondary hybridization with individual probes.
Fingerprinting and contig assembly:
Positive hybridization clones were fingerprinted using SNaPshot (Luo et al. 2003; Kim et al. 2008) and assembled into contigs with FPC v 8.5.2 (Soderlund et al. 2000; www.agcol.arizona.edu) at a fixed tolerance value 4 and an initial Sulston score 1e−50 (Ammiraju et al. 2006).
BAC end sequencing and in silico analysis:
Fingerprinted BAC clones were end sequenced with a universal T7 primer (5′ TAA TAC GAC TCA CTA TAG GG 3′) and a custom primer BES_HR (5′ CAC TCA TTA GGC ACC CCA 3′) following previously described methods (Kim et al. 2008). BAC end sequences (BES) were submitted to GenBank with the following accession numbers: D. simulans, EI211963.1–EI212067.1; D. sechellia, CZ549016.1–CZ549204.1; D. yakuba, EI89369.1–EI189559.1; D. erecta, CZ548656.1–CZ548834.1; D. ananasseae, CZ548467.1–CZ548655.1; D. persimilis, EI188778.1–EI189177.1; D. willistoni, EI189178.1–EI189368.1; D. Americana, EI189178.1–EI189368.1; D. novamexicana, DU169152.1–DU169329.1; D. virilis, CZ549205.1–CZ549371.1; D. littoralis, EI211597.1–EI211779.1; D. replete, EI211780.1–EI211962.1; D. mercatorum, EI188452.1–EI188610.1; D. mojavensis, CZ548835.1–CZ549015.1; D. arizonae, EI211417.1–EI211231.1; D. hydei, EI188451.1–EI188450.1; D. grimshawi, EI188111.1–EI188299.1; D. albomicans, EI211043–EI211230.1; and D. busckii, EI211418.1–EI211596.1.
All BESs were masked with Repeat Masker (version 3.1.0) against a redundant repeat database with sequences obtained from FlyBase (www.FlyBase.org) and Repbase (www.girinst.org). These sequences were used to conduct BLAST analysis against the mitochondrial (NC_001709, 19,517 bp) and nuclear genome sequences of D. melanogaster (Build 5.1) and the freeze 1 genome assemblies from the remaining 11 species (http://rana.lbl.gov/drosophila/caf1.html and http://insects.eugenes.org/species/data/). To compensate for the lack of whole-genome sequences and to minimize the bias of sequence divergence, the genome sequences of D. virilis and D. mojavensis were used as pseudoreference sequences for the D. virilis and D. repleta species group, respectively. BES from D. albomicans and D. busckii was compared to the D. grimshawi sequences.
In addition, similarity searches were conducted with complete gene sequences of each probe against the 12 Drosophila whole-genome sequences (Drosophila 12 Genomes Consortium 2007). Homologs with a minimum alignment length of 100 bp and 75% of nucleotide identity were retained for further analysis and for a comparison of their presence or absence in FPC-derived contigs.
RESULTS AND DISCUSSION
Drosophila strain selection and genome sizes:
Several criteria were used for careful evaluation of the different Drosophila species strains used for BAC resource development in this study. First, all fly lines were inbred for a minimum of eight generations by sib–sib mating to reduce the extent of heterozygosity and subsequently sequenced at six nuclear loci to verify homozygosity (T. A. Markow, unpublished data). Second, to minimize endosymbiont contamination (Wolbachia spp. and Spiroplasma spp.) at least five adult fly DNA samples from each species were screened with established protocols (Mateos et al. 2006). Finally, species identity was confirmed by both morphological and molecular approaches. When a suitable nuclear or mitochondrial DNA marker was known for a species, that marker was amplified, sequenced, and validated. Additionally, salivary gland chromosomes from third instar larvae were prepared and inspected for inversion polymorphism microscopically. Only homokaryotypic lines were used. All strains (Table 1) are deposited in the University of California at San Diego Drosophila Stock Center and are publicly available as a community resource.
TABLE 1.
Characteristics of the 19 Drosophila BAC library set
| Species | Groupa | Stock no.b | Library name | Enzyme | Genome size (Mb) | Average insert size (kb) | Clone no. | Calculated genome coveragec |
|---|---|---|---|---|---|---|---|---|
| D. simulans | MEL | DSSC 14021-0251.195 | DS_ABa | HindIII | 160d | 158 | 18,432 | 18.2 |
| D. sechellia | MEL | DSSC 14021-0248.25 | DS__Ba | HindIII | 166d | 139 | 18,432 | 15.4 |
| D. yakuba | MEL | DSSC 14021-0261.01 | DY__Ba | HindIII | 188d | 148 | 11,520 | 9.1 |
| D. erecta | MEL | DSSC 14021-0224.01 | DE_TBa | HindIII | 145d | 149 | 18,432 | 18.9 |
| D. ananassae | MEL | DSSC 14024-0371.13 | DA__Ba | BamHI | 215a | 148 | 36,864 | 25.4 |
| D. persimilis | OBS | DSSC 14011-0111.49 | DP__Ba | HindIII | 183d | 151 | 18,432 | 15.2 |
| D. willistoni | WIL | DSSC 14030-0811.24 | DW__Ba | HindIII | 206d | 150 | 18,432 | 13.4 |
| D. americana | VIR | DSSC 15010-0951.15 | DA_ABa | BstYI | 275d | 136 | 11,520 | 5.7 |
| D. novamexicana | VIR | DSSC 15010-1031.14 | DN__Ba | HindIII | 244e | 155 | 13,440 | 8.5 |
| D. virilis | VIR | DSSC 15010-1051.87 | DV_VBa | BstYI | 404d | 127 | 55,296 | 17.4 |
| D. littoralis | VIR | DSSC 15010-1001.11 | DL__Ba | HindIII | 238b | 168 | 36,864 | 26 |
| D. repleta | REP | DSSC 15084-1611.10 | DR__Ba | HindIII | 167e | 143 | 36,864 | 31.6 |
| D. mercatorum | REP | DSSC 15082-1521.36 | DM__Ba | HindIII | 128d | 125 | 18,432 | 18 |
| D. mojavensis | REP | DSSC 15081-1352.22 | DM_CBa | BamHI | 152d | 143 | 30,720 | 28.9 |
| D. arizonae | REP | DSSC 15081-1271.27 | DA_CBa | HindIII | 152f | 133 | 18,432 | 16.1 |
| D. hydei | REP | DSSC 15085-1641.58 | DH__Ba | HindIII | 164d | 146 | 36,864 | 32.8 |
| D. grimshawi | HAW | DSSC 15287-2541.00 | DG__Ba | HindIII | 231d | 127 | 18,432 | 10.1 |
| D. albomicans | IMM | DSSC 15112-1751.08 | DA_BBa | HindIII | 299f | 130 | 18,432 | 8 |
| D. busckii | DOR | DSSC 13000-0081.31 | DB__Ba | HindIII | 194e | 166 | 18,432 | 15.8 |
MEL, melanogaster; OBS, obscura; WIL, willistoni; VIR, virilis; REP, repleta; HAW, Hawaiian; IMM, immigrans; DOR, subgenus Drosophila.
DSSC: Drosophila Species Stock Center.
Calculated genome coverage: by insert size, genome size, and number of clones in the library.
Genome size measured by the PI method (Bosco et al. 2007).
Genome size measured by the DAPI method (Bosco et al. 2007).
Genome sizes of D. arizonae and D. albomicans were adopted from the genome size of close relatives, D. mojavensis and D. immigrans, respectively.
Genome size of an organism is the most important factor in determining the depth of a genomic library (reviewed in Gregory 2005). Previously determined genome sizes (Bosco et al. 2007) were used in this study for estimating the coverage of the BAC libraries for different Drosophila species. Bosco et al. (2007) employed two nucleic-acid–binding fluorescent dyes, propidium iodide (PI) and 4′,6-diamidino-2-phenylindole (DAPI), in conjunction with flow cytometry to determine genome sizes of 38 species of Drosophilidae, including the 12 sequenced Drosophila species (Drosophila 12 Genomes Consortium 2007).
The genome sizes of 15 of the 19 Drosophila species used in this study were based on the PI method and the remaining species (D. novamexicana, D. littoralis, D. repleta, and D. busckii) genome sizes were based on the DAPI method alone (for which the PI data were not available) (Table 1). Nine of the Drosophila species strains were not the same as the strains analyzed by Bosco et al. (2007). An important finding to consider, as reported by Bosco et al. (2007) and Gregory and Johnston (2008), is that DAPI may overestimate genome size, which could affect the estimated genome coverage of these four libraries.
Genome sizes of two species, D. arizonae and D. albomicans, were not known, so the genome sizes of closest relatives D. mojavensis and D. immigrans, respectively, were applied to estimate the tentative genome coverages of their respective BAC libraries. The genome sizes among the 19 Drosophila species varied by ∼3.2-fold, with the smallest being D. mercatorum and the largest D. virilis (Table 1).
BAC library construction and characterization:
Three different restriction enzymes were used for BAC library construction: HindIII, BamHI, and BstYI. Fifteen of the 19 libraries were constructed from DNA partially digested with HindIII, followed by size selection and ligation into the HindIII site of pIndigoBAC536SwaI (Ammiraju et al. 2006) (Table 1). Two libraries each were generated similarly from BamHI (D. ananassae and D. mojavensis) and BstYI (D. virilis and D. americana) restriction digests. All libraries, except for the D. busckii library (two ligations), were built from single ligations. The number of clones in the 19-BAC library set ranged between 11,520 and 55,296 (Table 1), which were arrayed into 384-well microtiter plates for long-term storage in −80° freezers at the Arizona Genomics Institute's (AGI) BAC/EST Resource Center (www.genome.arizona.edu).
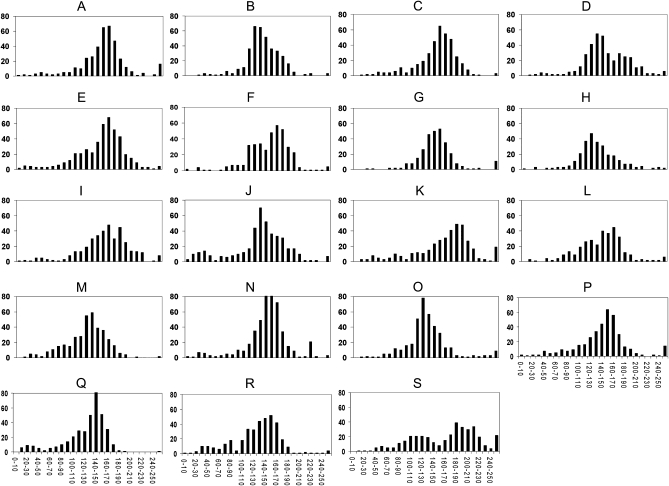
Insert sizes of individual clones in each library ranged from 10 to 371 kb, with the majority >120 kb (Figure 2). The average insert sizes of these libraries ranged from 125 to 168 kb (Table 1). Percentages of non-insert–containing clones ranged between 0.3 and 5.3%, which is typical for BAC libraries constructed at AGI (Ammiraju et al. 2006).
Figure 2.—
Insert size distribution of 19 Drosophila BAC libraries. Histograms A–S depict the insert size distribution in the 19 different libraries. For each histogram, the x-axis represents insert size (kilobases) and the y-axis represents the number of clones in a particular insert size range. (A) D. simulans (DS_ABa), average insert size 158 kb; (B) D. sechellia (DS__Ba), average insert size 139 kb; (C) D. yakuba (DY__Ba), average insert size 148 kb; (D) D. erecta (DE_TBa), average insert size 149 kb; (E) D. ananassae (DA__Ba), average insert size 148 kb; (F) D. persimilis (DP__Ba), average insert size 151 kb; (G) D. willistoni (DW__Ba), average insert size 150 kb; (H) D. americana (DA_ABa), average insert size 136 kb; (I) D. novamexicana (DN__Ba), average insert size 155 kb; (J) D. virilis (DV_VBa), average insert size 127 kb; (K) D. littoralis (DL__Ba), average insert size 168 kb; (L) D. repleta (DR__Ba), average insert size 143 kb; (M) D. mercatorum (DM__Ba), average insert size 125 kb; (N) D. mojavensis (DM_CBa), average insert size 143 kb; (O) D. arizonea (DA_CBa), average insert size 133 kb; (P) D. hydei (DH__Ba), average insert size 146 kb; (Q) D. grimshawi (DG__Ba), average insert size 127 kb; (R) D. albomicans (DA_BBa), average insert size 130 kb; (S) D. busckii (DB__Ba), average insert size 166 kb.
Genomic redundancy of the Drosophila BAC libraries:
We estimated the genomic depth of the 19 Drosophila BAC set by three different, but complementary approaches. First, we estimated the redundancy of each library empirically from the average insert size, total number of clones, and the genome size of the corresponding lineage, which ranged approximately between 5.7- and 32.8-fold (Table 1). To assess the randomness and extent of representational heterogeneity for different genomic regions, we screened the entire set of 19 Drosophila BAC libraries with nine gene-specific probes in two successive rounds of hybridizations (materials and methods, Table S1, and Table S2).
In brief, 4196 putative positive BAC clones were identified in the first round of hybridization, and 3809 (91%) were confirmed by a second hybridization. The number of positive hits per library ranged from 1 to 108 (Table S3). At least one positive hit per each probe was detected for all the libraries with the exceptions of the D. americana, D. repleta, and D. hydei libraries for probe X-CG11387 and D. ananassae for probe 3R-CG31247 (Table S3). In these four species no hits were found, even upon three rounds of library screening, with different hybridization stringencies. For D. ananassae, the whole-genome draft sequence was available (http://rana.lbl.gov/drosophila/caf1.html), and similarity searches revealed the presence of the probe sequence (3R-CG31247; Table S2) in the draft sequence assembly. Therefore, at least in the case of D. ananassae, it appears that methodological and/or library coverage issues prevented recovery of this gene via the hybridization-based approach, possibly due to use of heterologous probes, multiple usage of high-density colony filters, or cloning bias (under- and overrepresentation of genomic regions due to usage of a single restriction enzyme during library construction). More data are required to confirm the absence of the gene X-CG11387 in the other three species (D. americana, D. repleta, and D. hydei).
Hybridization-based genome coverages ranged from 9.1× (D. americana) to 42.9× (D. hydei). In only two species, D. mercatorum and D. willistoni, the hybridization-based coverage was slightly lower than expected (Table 2). The remaining 17 libraries had either nearly equal or higher coverage than predicted (Table 2 and Table S3). The D. albomicans BAC library showed an ∼3.6-fold higher than expected coverage on the basis of hybridization (Table 2), which could have resulted from not having accurate genome size estimation for this species (Table 1).
TABLE 2.
A comparison of genomic redundancies of each Drosophila BAC library as estimated by empirical, hybridization, and FPC approaches
| Species | Calculated genome coveragea | Average hyb coverageb | FPC, generalc | Ratio of a:b:c |
|---|---|---|---|---|
| D. simulans | 18.2 | 25.0 | 17 | 1:1.4:0.94 |
| D. sechellia | 15.4 | 20.2 | 14 | 1:1.3:0.88 |
| D. yakuba | 9.1 | 11.0 | 9 | 1:1.2:1.01 |
| D. erecta | 18.9 | 19.7 | 14 | 1:1.0:0.75 |
| D. ananassae | 25.4 | 25.3 | 22 | 1:1.0:0.87 |
| D. persimilis | 15.2 | 18.3 | 13 | 1:1.2:0.86 |
| D. willistoni | 13.4 | 9.6 | 7 | 1:0.7:0.52 |
| D. americana | 5.7 | 9.1 | 8 | 1:1.6:1.36 |
| D. novamexicana | 8.5 | 14.8 | 13 | 1:1.7:1.48 |
| D. virilis | 17.4 | 32.7 | 19 | 1:1.9:1.11 |
| D. littoralis | 26 | 25.1 | 18 | 1:1.0:0.71 |
| D. repleta | 31.6 | 35.7 | 14 | 1:1.1:0.44 |
| D. mercatorum | 18 | 11.7 | 10 | 1:0.6:0.54 |
| D. mojavensis | 28.9 | 31.1 | 17 | 1:1.1:0.59 |
| D. arizonae | 16.1 | 20.2 | 10 | 1:1.3:0.63 |
| D. hydei | 32.8 | 42.9 | 37 | 1:1.3:1.12 |
| D. grimshawi | 10.1 | 14.2 | 9 | 1:1.4:0.87 |
| D. albomicans | 8 | 28.4 | 10 | 1:3.6:1.22 |
| D. busckii | 15.8 | 28.2 | 9 | 1:1.8:0.58 |
A third and a more rigorous approach using fingerprinted contig (FPC)-based estimations of genomic redundancy of BAC libraries was applied, using a similar strategy to our previous analysis of a set of 11 Oryza (cultivated and wild rice) BAC libraries (Ammiraju et al. 2006). This approach can discriminate the unavoidable cloning bias from those of cross-hybridizations and genetic rearrangements such as duplications. All 3809 hybridization-derived BAC clones were fingerprinted and 3005 (79%) successful fingerprints were assembled into physical contigs (Table S4 and Table S5). Under a scenario of single-copy probes and one contig per probe for each species, the theoretically expected number of contigs is 171 (nine probes for 19 libraries). However, several exceptions were found: (a) as described above, 1 probe, X-CG11387, had no hits in the D. Americana, D. repleta, and D. hydei libraries, and another probe, 3R-CG31247, had no hits in the D. ananassae library (Table S3); (b) clones detected from six hybridizations (D. yakuba, D. persimilis, and D. willistoni with probe X-CG11387; D. mercatorum with probe 2L-CG4128; and D. mercatorum and D. grimshawi with probe 4-CG2999) resulted in the presence of singletons (Table S5) (all these instances resulted in less than three positive clones, Table S3). Taking into account the absence of these contigs in these species, 161 contigs are expected.
Our FPC analysis revealed a total of 211 contigs, 50 additional contigs than the expected number of 161 (Table S4). The number of contigs and respective coverage differed among different Drosophila libraries for the same probe (Table S5). Five probes (X-CG11387, X-CG32611, 3L-CG10948, 3R-CG31247, and 4-CG2999) essentially behaved as single-copy probes in most Drosophila libraries (Table S5). The remaining four detected on average ≥1.4 contigs per probe (Table S5). To better understand whether these deviations from expectation (50 additional contigs) were due to technical issues (cross-hybridization and assembly artifacts) and/or lineage-specific genetic changes, we gathered data from two additional experiments. First, on the basis of BES mapping information (materials and methods), we classified 142 contigs as primary (those that map to the expected genomic location) and 69 additional contigs as secondary (27 contigs that cannot be positioned in any genome and 42 contigs that map to nonorthologous locations), a good agreement between the results of FPC analyses and mapping information (Table S2 and Table S6).
Second, nucleotide and protein similarity searches of the probe (or gene) sequences revealed that several secondary sites (17/42 secondary contigs) contained small cross-hybridizing paralogous sequences (Table S6, indicated with *). It is possible that the 25 remaining secondary sites also contained very small cross-hybridizing sequences that were not easily detected through similarity searches. In addition, sequence analysis of the extended flanking sequences of the primary sites with the secondary sites revealed no evidence of synteny, suggesting cross-hybridization as the main cause for these additional contigs.
To provide a conservative estimate of genome coverage, we considered each identified contig as an independent locus and calculated a weighted FPC coverage that accounts for the presence of several loci (Table S4; Ammiraju et al. 2006). Estimated FPC coverage for the 19 libraries (Table 2 and Table S4) ranged between 7× and 37×. Only 2 libraries had coverage <9×: D. willistoni (7×) and D. americana (8×).
Twelve libraries showed a ratio close to 1:1 between the FPC and empirically estimated coverage (Table 2). The D. willistoni, D. littoralis, D. repleta, D. mercatorum, D. mojavensis, D. arizonae, and D. busckii libraries showed ratios ≤0.7:1 (Table 2 and Table S4). The difference between hybridization-based and contig-based estimates of library coverage is due to the difference in the number of loci used to calculate the coverage. While each probe is considered as a single locus in the hybridization-based approach, each secondary contig is considered as an independent locus in the FPC-based approach (Table 2, Table S3, and Table S4). Together, these results showcase the high quality and deep representational coverage of each of 19 Drosophila genomes in their respective libraries.
Utilization of BAC libraries:
Although a few Drosophila BAC libraries have already been reported in the literature (Hoskins et al. 2000; Locke et al. 2000; Gonzalez et al. 2005; Osoegawa et al. 2007; Murakami et al. 2008), this is the first synthesis and characterization of a comprehensive set of BAC library resources for the genus, which fills a critical void for the Drosophila research community. Hybridization of nine different probes to the full set of libraries demonstrates the feasibility of isolating homologous regions across the entire genus. Combined with high-throughput sequencing methods (Wicker et al. 2006), this set of libraries provides an excellent resource for comparative studies of targeted genomic regions (e.g., Leung et al. 2010).
First, BAC libraries from species that do not yet have a reference genome sequence themselves provide a source for identifying genome rearrangements in comparisons with the available genome sequences. For example, end sequences of BACs isolated with the X-linked probe CG32611 from D. novamexicana map at an unexpected position within contig 12,970 of D. virilis, indicating a putative small inversion at the base of the X chromosome that had not been previously identified (Vieira et al. 1997). Another putative inversion was also revealed in D. arizonae by the localization of end sequences of clones hybridizing to CG3139 in the genome sequence of D. mojavensis. Targeted analyses inversion breakpoints are also enabled by the availability of these BAC libraries and informed by the reference genome sequences. Evans et al. (2007) used cytological evidence on the position of an inversion in D. americana to develop probes for isolating its breakpoints from the respective BAC clones. In addition, the BAC libraries for the nine unsequenced Drosophila species provide robust templates for the whole-genome physical and sequence frameworks. In this direction, the entire D. persimilis BAC library was fingerprinted, bidirectionally end sequenced, and assembled into a whole-genome physical map. This map was aligned to the D. persimilis and D. pseudoobscura draft sequences and is currently under editing (data not shown).
An extremely important application of the BAC resources reported here is in the ability to use functional genomics to test genes underlying the differences between Drosophila species. The tool kit for functional analyses of Drosophila has taken a major leap forward with the recent establishment of the P/ΦC31 artificial chromosome manipulation (P[acman]) transgenesis platform (Venken et al. 2006, 2009; Venken and Bellen 2007). While still reliant on the P-transposable element for transformation, this BAC transgenic system significantly improves upon the size of the DNA to be carried in the vector (>130 kb) and its site-specific integration in the fly genome. An important feature of the P[acman] system is recombineering, which permits cloning/transfer of large DNA fragments from existing Drosophila P1 or BAC clones through a homologous recombination-mediated gap repair process. Therefore, a combination of the P[acman] system with the 19 Drosophila BAC libraries will provide an unprecedented opportunity to the fly community to access, transfer, and manipulate virtually any genomic region of interest (large genes or even gene clusters) covering the entire phylogenomic range of the genus Drosophila.
Finally, the BAC library set reported here can be used to further improve many of the existing Drosophila draft sequence assemblies (Drosophila 12 Genomes Consortium 2007) and aid in the characterization of lineage-specific rearrangements. For example, physical mapping of BAC contigs, or individual BAC clones, identified by hybridization probes designed from draft Drosophila genome sequences, has revealed and confirmed chromosomal location of several sequence contigs from the draft assemblies, as well as their relationship to D. melanogaster (Table S6). Conserved linkage and physical markers were used to infer the physical organization of the assembled genome assemblies relative to reference chromosome maps (Schaeffer et al. 2008), and these BAC libraries serve as an appropriate resource to isolate regions at inferred gaps between adjacent contigs (e.g., Hoskins et al. 2000). Using hybridization to recover genome regions containing target genes, combined with end sequencing of positive clones, further reveals the conserved linkage among Drosophila species. For example, scaffolds 20 and 24 map to X[A], 29 to 3L[D], and 30 to 4[F] in D. sechellia; 4512 to 4[F] in D. erecta, 12,984 to 3R[B] and 12,947 to 4(LR)[F] in D. ananassae; 48 to XR[D/A] and 103 to 5[F] in D. persimilis; 5 group M to 5[F] in D. pseudoobscura; 13,052 to 6[F] in D. virilis (Drosophila 12 Genomes Consortium 2007); 6,498 to 6[F] in D. mojavensis; and 14,822 to 6[F] in D. grimshawi (Table S6).
These libraries are likely to facilitate a wide array of comparative, evolutionary, and functional genomics studies and play a major role in advancing the Drosophila biology.
Acknowledgments
This work was supported by National Institutes of Health grant U1HG02525A.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.111.126540/DC1.
References
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287 2185–2195. [DOI] [PubMed] [Google Scholar]
- Ammiraju, J. S., M. Luo, J. L. Goicoechea, W. Wang, D. Kudrna et al., 2006. The Oryza bacterial artificial chromosome library resource: construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res. 16 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, W. W., J. Arnold, D. G. Baldwin, A. T. Beckenbach, C. J. Brown et al., 1991. Four decades of inversion polymorphism in Drosophila pseudoobscura. Proc. Natl. Acad. Sci. USA 88 10367–10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco, G., P. Campbell, J. T. Leiva-Neto and T. A. Markow, 2007. Analysis of Drosophila species genome size and satellite DNA content reveals significant differences among strains as well as between species. Genetics 177 1277–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker, S. E., D. A. Wheeler, B. Kronmiller, J. W. Carlson, A. Halpern et al., 2002. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3 research0079.1–0079.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., D. Charlesworth, J. Hnilicka, A. Yu and D. S. Guttman, 1997. Lack of degeneration of loci on the neo-Y chromosome of Drosophila americana. Genetics 145 989–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium, 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450 203–218. [DOI] [PubMed] [Google Scholar]
- Eichler, E. E., and P. J. Dejong, 2002. Biomedical applications and studies of molecular evolution: a proposal for a primate genomic library resource. Genome Res. 12 673–678. [DOI] [PubMed] [Google Scholar]
- Ellison, C. K., and K. L. Shaw, 2010. Mining non-model genomic libraries for microsatellites: BAC versus EST libraries and the generation of allelic richness. BMC Genomics 11 428–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, A. L., P. A. Mena and B. F. Mcallister, 2007. Positive selection near an inversion breakpoint on the neo-X chromosome of Drosophila americana. Genetics 177 1303–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, G., B. P. Blackmon, D. C. Henry, M. E. Staton, C. A. Saski et al., 2010. Genomic tools development for Aquilegia: construction of a BAC-based physical map. BMC Genomics 11 621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs, R. A., G. M. Weinstock, M. L. Metzker, D. M. Muzny, E. J. Sodergren et al., 2003. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428 493–521. [DOI] [PubMed] [Google Scholar]
- Gilbert, D. G., 2007. DroSpeGe: rapid access database for new Drosophila species genomes. Nucleic Acids Res. 35 D480–D485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, J., M. Nefedov, I. Bosdet, F. Casals, O. Calvete et al., 2005. A BAC-based physical map of the Drosophila buzzatii genome. Genome Res. 15 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory, S. G., M. Sekhon, J. Schein, S. Zhao, K. Osoegawa et al., 2002. A physical map of the mouse genome. Nature 418 743–750. [DOI] [PubMed] [Google Scholar]
- Gregory, T. R., 2005. Synergy between sequence and size in large-scale genomics. Nat. Rev. Genet. 6 699–708. [DOI] [PubMed] [Google Scholar]
- Gregory, T. R., and J. S. Johnston, 2008. Genome size diversity in the family Drosophilidae. Heredity 101 228–238. [DOI] [PubMed] [Google Scholar]
- Havlak, P., R. Chen, K. J. Durbin, A. Egan, Y. Ren et al., 2004. The Atlas genome assembly system. Genome Res. 14 721–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins, R. A., C. R. Nelson, B. P. Berman, T. R. Laverty, R. A. George et al., 2000. A BAC-based physical map of the major autosomes of Drosophila melanogaster. Science 287 2271–2274. [DOI] [PubMed] [Google Scholar]
- International Human Genome Mapping Consortium, 2000. a A physical map of the human genome. Nature 409 934–941. [DOI] [PubMed] [Google Scholar]
- International Human Genome Mapping Consortium, 2000. b Initial sequencing and analysis of the human genome. Nature 409 860–921. [DOI] [PubMed] [Google Scholar]
- Kim, H., B. Hurwitz, Y. Yu, K. Collura, M. Gill et al., 2008. Construction, alignment and analysis of twelve framework physical maps that represent the ten genome types of the genus Oryza. Genome Biol. 9 R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, U. J., B. W. Birren, T. Slepak, V. Mancina, C. Boysen et al., 1996. Construction and characterization of a human bacterial artificial chromosome library. Genomics 34 213–218. [DOI] [PubMed] [Google Scholar]
- Krzywinske, M., J. Wallis, C. Gosele, I. Bosdet, R. Chiu et al., 2004. Integrated and sequence-ordered BAC- and YAC-based physical maps for the rat genome. Genome Res. 14 766–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, W., C. D. Shaffer, T. Cordonnier, J. Wong, M. S. Itano et al., 2010. Evolution of a distinct genomic domain in Drosophila: comparative analysis of the dot chromosome in Drosophila melanogaster and Drosophila virilis. Genetics 185 1519–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke, J., L. Podemski, N. Aippersbach, H. Kemp and R. Hodgetts, 2000. A physical map of the polytenized region (101EF–102F) of chromosome 4 in Drosophila melanogaster. Genetics 155 1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M., and R. A. Wing, 2003. An improved method for plant BAC library construction, pp. 3–20 in Plant Functional Genomics, edited by E. Grotewold. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Luo, M., H. Kim, D. Kudrna, N. B. Sisneros, S. Lee et al., 2006. Construction of a nurse shark (Ginglymostoma cirratum) bacterial artificial chromosome (BAC) library and a preliminary genome survey. BMC Genomics 7 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M. C., C. Thomas, F. M. You, J. Hsiao, S. Ouyang et al., 2003. High-throughput fingerprinting of bacterial artificial chromosomes using the snapshot labeling kit and sizing of restriction fragments by capillary electrophoresis. Genomics 82 378–389. [DOI] [PubMed] [Google Scholar]
- Markow, T. A., and P. M. O'Grady, 2006. Evolutionary genetics of reproductive behavior in Drosophila: connecting the dots. Annu. Rev. Genet. 39 263–291. [DOI] [PubMed] [Google Scholar]
- Markow, T. A., and P. M. O'Grady, 2007. Drosophila biology in the genomic age. Genetics 177 1269–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos, M., S. J. Castrezana, B. J. Nnakivell, A. M. Estes, T. A. Markow et al., 2006. Heritable endosymbionts of Drosophila. Genetics 174 363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, K., A. Toyoda, M. Hattori, Y. Kuroki, A. Fujiyama et al., 2008. BAC library construction and BAC end sequencing of five Drosophila species: the comparative map with the D. melanogaster genome. Genes Genet. Syst. 83 245–246. [DOI] [PubMed] [Google Scholar]
- Myers, E. W., G. G. Sutton, A. L. Delcher, I. M. Dew, D. P. Fasulo et al., 2000. A whole-genome assembly of Drosophila. Science 287 2196–2204. [DOI] [PubMed] [Google Scholar]
- Osoegawa, K., A. M. Tateno, P. Y. Woon, E. Frengen, A. G. Mammoser et al., 2000. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 10 116–128. [PMC free article] [PubMed] [Google Scholar]
- Osoegawa, K., A. G. Mammoser, C. Wu, E. Frengen, C. Zeng et al., 2001. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 11 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osoegawa, K., B. Zhu, C. L. Shu, T. Ren, Q. Cao et al., 2004. BAC resources for the rat genome project. Genome Res. 14 780–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osoegawa, K., G. M. Vessere, C. L. Shu, R. A. Hoskins, J. P. Abad et al., 2007. BAC clones generated from sheared DNA. Genomics 89 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., and J. A. Coyne, 1989. The genetics of postzygotic isolation in the Drosophila virilis group. Genetics 121 527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitnick, S., T. A. Markow and G. S. Spicer, 1995. Delayed male maturity is a cost of producing large sperm in Drosophila. Proc. Natl. Acad. Sci. USA 92 10614–10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popadic, A., and W. W. Anderson, 1994. The history of a genetic system. Proc. Natl. Acad. Sci. USA 91 6819–6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, J. R., 1997. Progress and Prospects in Evolutionary Biology: The Drosophila Model. Oxford University Press, London/New York/Oxford.
- Richards, S., Y. Liu, B. R. Bettencourt, P. Hradecky, S. Letovsky et al., 2005. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, geni, and cis-element evolution. Genome Res. 15 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, G. M., and E. B. Lewis, 2000. A brief history of Drosophila's contributions to genome research. Science 287 2216–2218. [DOI] [PubMed] [Google Scholar]
- Schaeffer, S. W., A. Bhutkar, B. F. Mcallister, M. Matsuda, L. M. Matzkin et al., 2008. Polytene chromosomal maps of 11 Drosophila species: the order of genomic scaffolds inferred from genetic and physical maps. Genetics 179 1601–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund, C., S. Humphray, A. Dunham and L. French, 2000. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 10 1772–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, A., M. F. Lin, P. Kheradpour, J. S. Pedersen, L. Parts et al., 2007. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 450 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigart, A. L., 2010. Simple Y-autosomal incompatibilities cause hybrid male sterility in reciprocal crosses between Drosophila virilis and D. americana. Genetics 184 779–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken, K. J. T., and H. J. Bellen, 2007. Transgenesis upgrades for Drosophila melanogaster. Development 134 3571–3584. [DOI] [PubMed] [Google Scholar]
- Venken, K. J. T., Y. He, R. A. Hoskins and H. J. Bellen, 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314 1747–1751. [DOI] [PubMed] [Google Scholar]
- Venken, K. J. T., J. W. Carlson, K. L. Schulze, H. Pan, Y. He et al., 2009. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods 6 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira, J., C. P. Vieira, D. L. Hartl and E. R. Lozovskaya, 1997. Discordant rates of chromosome evolution in the Drosophila virilis species group. Genetics 147 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker, T., E. Schlagenhauf, A. Graner, T. J. Close, B. Keller et al., 2006. 454 sequencing put to the test using the complex genome of barley. BMC Genomics 7 275. [DOI] [PMC free article] [PubMed] [Google Scholar]