Abstract
Sister-chromatid cohesion mediated by cohesin ensures proper chromosome segregation during cell division. Cohesin is also required for postreplicative DNA double-strand break repair and gene expression. The molecular mechanisms of these diverse cohesin functions remain to be elucidated. Here we report that the cohesin subunits Scc3 and Smc1 are both required for the production of the meiosis-specific subunit Rec8 in the budding yeast Saccharomyces cerevisiae. Using a genetic approach, we depleted Scc3 and Smc1 independently in cells that were undergoing meiosis. Both Scc3- and Smc1-depleted cells were inducible for meiosis, but the REC8 promoter was only marginally activated, leading to reduced levels of REC8 transcription and protein production. In contrast, the expression of MCD1, the mitotic counterpart of REC8, was not subject to Scc3 regulation in vegetative cells. We provide genetic evidence to show that sister-chromatid cohesion is not necessary for activation of REC8 gene expression. Cohesin appears to positively regulate the expression of a variety of genes during yeast meiosis. Our results suggest that the cohesin complex plays a dual role in gene regulation and sister-chromatid cohesion during meiotic differentiation in yeast.
MEIOSIS is a developmentally regulated cell division required for sexual reproduction in eukaryotes. In the single-celled organism Saccharomyces cerevisiae, vegetative a/α diploid cells switch to the meiotic program in response to starvation. A signal transduction cascade, which leads to changes in gene expression, initiates meiosis (Mitchell 1994; Kupiec et al. 1997). Consequently, the expression of meiosis-activating genes is increased and that of meiosis-repressing genes is decreased. The positive regulators of meiosis, of which many are transcriptional factors, then activate the expression of early, middle, and late genes that are required for recombination, chromosome segregation, and spore formation. Regulation of meiotic differentiation is facilitated by chromosome structural reorganization, which can be achieved by the actions of histone modifiers and ATP-dependent chromatin-remodeling complexes (Kassir et al. 2003). Additional chromosomal factors might be required for activating meiotic gene expression.
The evolutionarily conserved protein complex cohesin, which is composed of Smc1, Smc3, Mcd1/Scc1, and Irr1/Scc3 in the budding yeast, mediates sister-chromatid cohesion (Onn et al. 2008; Nasmyth and Haering 2009). Rec8 largely replaces Mcd1 and is the only meiosis-specific cohesin subunit in yeast of which the encoding gene is expressed early in meiosis (Chu et al. 1998). Cohesin binds to the yeast chromosome at discrete loci (Blat and Kleckner 1999; Laloraya et al. 2000; Glynn et al. 2004; Lengronne et al. 2004), and the purified cohesin complex forms a ring-shaped structure (Gruber et al. 2003). The tripartite cohesin ring made of Smc1, Smc3, and Mcd1 (probably Rec8) is sufficient for topologically entrapping a pair of sister chromatids to generate cohesion in yeast (Haering et al. 2008). Meanwhile, Scc3, which is called SA/STAG in animals, has been implicated in cohesin oligomerization (Zhang et al. 2008) and is critical for cohesin release from the chromosome (Hauf et al. 2005). Cohesin is important for establishing both the mitotic and meiotic chromosome architecture (Hirano 2006; Onn et al. 2008; Nasmyth and Haering 2009).
In addition to mediating sister-chromatid cohesion, cohesin appears to have a broad influence on chromosome metabolism that includes postreplicative DNA double-strand break repair and gene expression (Strom et al. 2004; Unal et al. 2004; Dorsett et al. 2005; Horsfield et al. 2007). Functional analysis of cohesin and its loading factor, the Scc2 and Scc4 complex, demonstrates that chromosomal binding of cohesin can generate a chromatin boundary that insulates the transcriptional activity of surrounding genes in yeast and fly (Donze et al. 1999; Rollins et al. 1999; Dorsett et al. 2005). Cohesin also plays a role in cell differentiation by modulating gene expression as demonstrated in neuron morphogenesis in flies (Pauli et al. 2008; Schuldiner et al. 2008). These studies provide insights into the understanding of the noncanonical role of cohesin in the regulation of gene expression. Cohesin function in gene expression is further supported by recent findings in vertebrates that cohesin subunits physically interact with the transcriptional factor CTCF and that they colocalize with CTCF on chromosomes (Parelho et al. 2008; Rubio et al. 2008; Wendt et al. 2008). The above observations also raise more questions yet to be answered. For example, how does cohesin regulate gene expression during cell differentiation? Is this regulatory mechanism conserved in eukaryotes? Is the cohesin holocomplex or individual subunit required for gene regulation? Is the primary role of cohesin in sister-chromatid cohesion separable from that of gene regulation?
Because cohesin subunits are essential for cell growth, genetic analysis of cohesin function in many model organisms is limited to thermosensitive or partially functional mutant alleles. Using a previously proven genetic approach (Lee and Amon 2003), we have created conditional alleles of SCC3 and SMC1 that specifically deplete Scc3 and Smc1 in yeast meiotic cells. In both Scc3- and Smc1-depleted cells, the level of the meiosis-specific subunit Rec8 is significantly lowered by a reduction of REC8 gene transcription. Our work suggests that the cohesin complex plays an important role in positively regulating the REC8 promoter when vegetative yeast cells differentiate into meiosis.
MATERIALS AND METHODS
Yeast strains and culture conditions:
Yeast strains used in this study are listed in supporting information, Table S1. We used the CLB2 promoter to replace the endogenous promoters of SCC3 and SMC1 by a PCR-based method as previously described (Jin et al. 2009). The PMET1-DEGRON-SCC3 was generated by a similar PCR method with the plasmid p378. We used plasmids pHG40 (Jin et al. 2009) and pHG105 to create PCUP1REC8 and PREC8GFP alleles by standard yeast transformation. We cloned a 1900-bp DNA sequence upstream of the REC8 start codon, which included the 5′ UTR, by PCR and placed it in front of the GFP open reading frame to create pHG105. We used the DMC1 promoter to replace the REC8 endogenous promoter to generate PDMC1REC8 using a similar method that we described previously (Yu and Koshland 2005; Jin et al. 2009). The rec8Δ, spo11-Y135F, and ndt80Δ alleles have been reported previously (Xu et al. 1995; Keeney et al. 1997; Klein et al. 1999). The tetO array was inserted into the URA3 locus on chromosome V, and tetR-GFP at the LEU2 locus on chromosome III, as previously described (Michaelis et al. 1997). A PCR-based strategy (Longtine et al. 1998) was used to create C-terminal tags of the following alleles: SCC3-3HA, SMC3-3HA, SMC3-V5, and REC8-3HA. Positive transformations were confirmed by colony PCR. PCR primer information appears in Table S2.
Synchronous meiosis was performed as previously described (Yu and Koshland 2005). Briefly, yeast cells were grown in yeast extract, peptone, acetic acid (YPA) overnight at 30° to an optical density (λ = 600 nm) of ∼1.6, washed once with H2O, and resuspended in 2% KoAC for induction of meiosis. To induce PCUP1REC8, 60 μm CuSO4 was added to the sporulation medium. The PMET1-DEGRON-SCC3 strain was grown in methionine-dropout synthetic medium at 25°. Cells were treated with α-factor (10 ng/ml) for 2 hr at 25° and washed twice with H2O. This culture was split into two equal halves; one was incubated at 25° in methionine-dropout medium and the other at 37° in complete medium.
Meiotic nuclear spreads and fluorescence microscopy:
Yeast surface nuclear spreads were performed as previously described (Jin et al. 2009). Rec8-3HA, Scc3-3HA, and Smc3-3HA were detected by an anti-HA antibody (12CA5, Roche). FITC-conjugated goat anti-mouse was used as secondary antibody (Jackson ImmunoResearch Laboratories). Chromosomal DNA was stained with DAPI. Fluorescence images were acquired with a ×100 objective lens (NA = 1.40) mounted on a motorized microscope (AxioImager, Zeiss). Acquired monotone images were merged by AxioVision software (Zeiss). For assay of sister-chromatid cohesion, yeast aliquots were withdrawn at 2-hr intervals and fixed with 1% formaldehyde. Green fluorescent protein (GFP) foci were visualized by fluorescence microscopy. At least 100 cells were counted at each time point.
Western blot:
Yeast aliquots were collected at 2-hr intervals and processed by the trichloroacetic acid (TCA) method for total protein extraction as previously described (Jin et al. 2009). Standard SDS-PAGE and Western blot procedures were followed (Sambrook and Russell 2001). Rec8-3HA and Scc3-3HA were detected by an anti-HA antibody (12CA5, Roche). Smc3-V5 was detected by an anti-V5 antibody (Invitrogen). Dmc1 and Mcd1 were detected by protein-specific antibodies (gifts of D. Bishop, University of Chicago, and V. Guacci, Carnegie Institution). GFP was detected by a GFP-specific antibody (Ab290, Abcam). The level of Tub2 (β-tubulin) served as a loading control.
Northern blot and RT-PCR:
Yeast aliquots were collected at intervals after induction of meiosis or after G1-phase release. We extracted total RNA and performed standard Northern blots (Sambrook and Russell 2001). Gene-specific probes were used to detect the mRNA of genes of interest. Labeled blots were scanned with the Storm PhosphorImager (GE). Signal intensity was quantified with the IPLab software (Scanalytics). We used the RNeasy kit (Qiagen) to extract and purify mRNA. Purified mRNA was reverse-transcribed to cDNA (Invitrogen), and a semiquantitative PCR method was used to determine the concentration of target cDNA with gene-specific primers (primer information appears in Table S2).
Chromatin immunoprecipitation:
Yeast cells were induced to undergo synchronous meiosis, fixed with 1% formaldehyde for 2 hr at room temperature, and then subjected to a chromatin immunoprecipitation (ChIP) procedure as described previously (Yu and Koshland 2005). We used an anti-V5 antibody (Invitrogen) for ChIP of Rpb3-V5-tagged yeast strains. A semiquantitative PCR-based method was used to detect the enrichment of Rpb3 at the REC8 and DMC1 genes.
RESULTS
Scc3 is required for sister-chromatid cohesion and nuclear division during yeast meiosis:
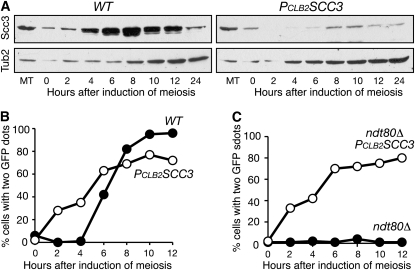
To deplete Scc3 in meiosis, we replaced the endogenous SCC3 promoter with the CLB2 promoter, which is expressed only in vegetative yeast cells (Lee and Amon 2003). Semiquantitative analysis of Scc3 by Western blot showed ∼85% depletion of Scc3 in PCLB2SCC3 cells during meiosis (Figure 1A; t = 8 hr). This conditional scc3 mutant allele was competent for meiotic DNA replication (Figure S1) and permitted us to determine whether Scc3 is required for sister-chromatid cohesion in yeast meiosis (Figure 1, B and C). We marked the centromere of one homolog of chromosome V with GFP to assay sister-chromatid cohesion (Michaelis et al. 1997). In wild-type cells, sister chromatids were cohesive and formed one GFP spot before meiosis I, which occurred ∼5 hr after induction of meiosis (Figure 1B). In contrast, in PCLB2SCC3 cells, sister chromatids were not associated after DNA replication, forming two GFP spots (Figure 1B). We incorporated an ndt80Δ mutation to arrest the cells at prophase I (Figure 1C; Xu et al. 1995). Less than 4% of ndt80Δ cells showed two GFP spots because chromosomes did not segregate and sister chromatids remained cohesive. In contrast, 86% of PCLB2SCC3 ndt80Δ cells formed two GFP spots 12 hr after induction of meiosis (Figure 1C). We therefore conclude that Scc3 is required for sister-chromatid cohesion during yeast meiosis.
Figure 1.—
Requirement for Scc3 in sister-chromatid cohesion during yeast meiosis. (A) Protein levels of Scc3 during yeast meiosis. Yeast cells were induced for synchronous meiosis, and aliquots were withdrawn at indicated times. Total protein extracts were prepared by the TCA method for Western blots, which were probed by anti-HA (12CA5) and anti-β-tubulin antibodies. The level of Tub2 (β-tubulin) served as a loading control. Note that Scc3 was largely depleted in meiosis in PCLB2SCC3 cells. MT, mitosis. Protein extracts were prepared from cells grown asynchronously in YPD medium. Wild-type (WT), strain 3072; PCLB2SCC3, strain 3200. (B and C) Assay of sister-chromatid cohesion in strains 3078C, 3206, HY2130, and HY1472. Yeast aliquots were withdrawn at indicted time points and fixed for fluorescence microscopy. An array of tetO was inserted at the URA3 locus, ∼35 kb from centromere V. Expression of tetR-GFP generated a GFP signal that could be visualized as a dot by fluorescence microscopy. Cohesed sister chromatids formed only one GFP dot. At least 100 cells were counted at each time point.
Next, to determine whether Scc3 is required for chromosome segregation, we monitored meiotic nuclear divisions (Figure S2). In wild-type cells, 12 hr after induction of meiosis, 80% of cells had finished both meiosis I and meiosis II nuclear divisions (Figure S2A). In contrast, less than 5% of PCLB2SCC3 cells were able to complete either division (Figure S2B). To determine whether PCLB2SCC3 cells are blocked by the recombination checkpoint, we introduced a spo11 mutation (spo11-Y135F; Keeney et al. 1997) to bypass the checkpoint (Figure S2, C and D). More than 55% of PCLB2SCC3spo11-Y135F cells were able to complete at least one nuclear division when double-strand break formation was eliminated (Figure S2D). Together, these data suggest that Scc3-depleted cells are competent for meiosis initiation but are arrested primarily by the recombination checkpoint.
Reduced Rec8 protein level in Scc3-depleted meiotic cells:
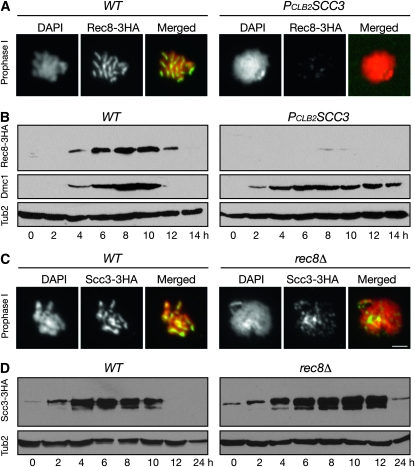
In loss of sister-chromatid cohesion and failure to complete nuclear division, the mutant phenotypes of PCLB2SCC3 resemble those of rec8Δ (Klein et al. 1999). We therefore localized Rec8 in Scc3-depleted cells by immunofluorescence (Figure 2A). As previously shown, in wild-type cells at pachytene of prophase I, Rec8 was localized along the length of chromosomes that revealed well-defined rod-shaped structures (Figure 2A, left panels for both WT and PCLB2SCC3). In Scc3-depleted cells, chromosomes became amorphous, and only traces of chromosome-associated Rec8 were observed above the background noise (Figure 2A, right panels for both WT and PCLB2SCC3). Consistent with this observation, we found very low levels of total Rec8 protein in Scc3-depleted cells by Western blot (Figure 2B). In contrast, the meiosis-specific protein Dmc1 was produced on time and in quantities similar to those in wild-type and PCLB2SCC3 cells, although its degradation was delayed in PCLB2SCC3 cells because these cells were blocked at prophase I (Figure 2B). Thus, by two different means, immunofluorescence and Western blot, we showed that the Rec8 protein level is dramatically lowered when Scc3 is absent in meiosis.
Figure 2.—
Reduced Rec8 protein level in Scc3-depleted cells. Yeast cells were induced to undergo synchronous meiosis as in Figure 1. (A) Chromosome association of Rec8 in wild-type (2824) and PCLB2SCC3 (HY2294) cells. Yeast aliquots were collected 6 hr after induction of meiosis, and surface nuclear spreads were prepared for immunofluorescence with an anti-HA antibody. Red, DNA; green, Rec8. (B) Rec8 protein level in wild-type and PCLB2SCC3 cells in meiosis. Yeast aliquots were collected at indicated times and prepared for Western blot as in A. An anti-Dmc1-specific antibody was used to detect the level of Dmc1. (C) Chromosome association of Scc3 in wild-type (3072) and rec8Δ (HY1495) cells. Surface yeast nuclear spreads were prepared as in A. Note that Scc3 remains chromosome-bound in rec8Δ cells. Red, DNA; green, Scc3. Bar, 2 μm. (D) Scc3 protein level in wild-type and rec8Δ cells. Western blots were prepared as in B. Note that the level of Scc3 remains normal in rec8Δ cells.
Next, we determined whether Rec8 is required for maintaining the Scc3 protein level. By immunofluorescence, we found that Scc3 remains to be chromosome associated in the absence of Rec8 (Figure 2C). By Western blot, we found that the total amount of Scc3 was present at a wild-type level in rec8Δ cells during meiosis (Figure 2, C and D). Therefore, Scc3 is required for mediating a normal level of Rec8 protein in meiosis, but not the reverse. Our data suggest that Scc3 can bind to the chromosome without formation of the meiotic cohesin complex during meiosis. Alternatively, a residual level of the mitotic cohesin complex remained in these cells.
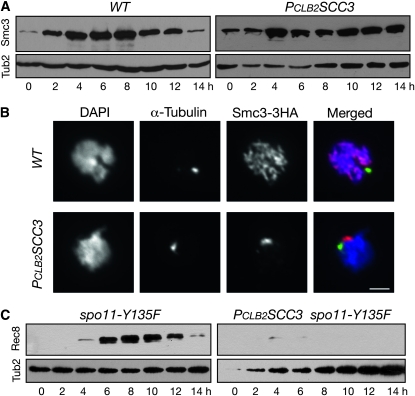
Smc1 and Smc3 are the other subunits of the meiotic cohesin, and as an example, we show that Smc3 was present at similar levels in wild-type and Scc3-depleted meiotic cells (Figure 3A). Therefore, Scc3 plays a specific role in maintaining a normal level of the meiosis-specific cohesin subunit Rec8. In Scc3-depleted cells, however, Smc3 failed to bind to meiotic chromosomes (Figure 3B), suggesting that Scc3 is required for Smc3 chromosome association.
Figure 3.—
Requirement for Scc3 for Rec8 but not for Smc3 production in yeast meiosis. Yeast cells were induced for synchronous meiosis, aliquots were withdrawn at indicated times, and protein extracts were prepared for Western blots probed by anti-V5, anti-HA, and anti-β-tubulin antibodies. (A) Protein level of Smc3 in wild-type (HY1510C) and PCLB2SCC3 (HY1566) cells. (B) Chromosome localization of Smc3 during yeast meiosis. Yeast cells were collected 6 hr after induction of meiosis and prepared for surface nuclear spread as in Figure 2A. Tub1 (α-Tubulin) was detected by a specific antibody (YOL135). Red, Smc3; green, Tub1; blue, DNA. (C) Protein levels of Rec8 in spo11-Y135F (HY1499) and PCLB2SCC3 spo11-Y135F (HY1483) cells.
To determine whether the prophase block of Scc3-depleted cells led to lowered levels of Rec8, we assayed the total protein level of Rec8 in spo11-Y135F and PCLB2SCC3spo11-Y135F double-mutant cells by Western blot (Figure 3C). Wild-type and spo11-Y135 cells did not differ in the production and degradation of Rec8 (Figure 2B and Figure 3C, left panels). In contrast, Rec8 protein level remained low in PCLB2SCC3spo11-Y135F cells (Figure 3C, right panels). Therefore, reduced Rec8 level in Scc3-depleted cells is not caused by prophase I block.
Scc3 regulates REC8 gene expression by increasing REC8 promoter activity:
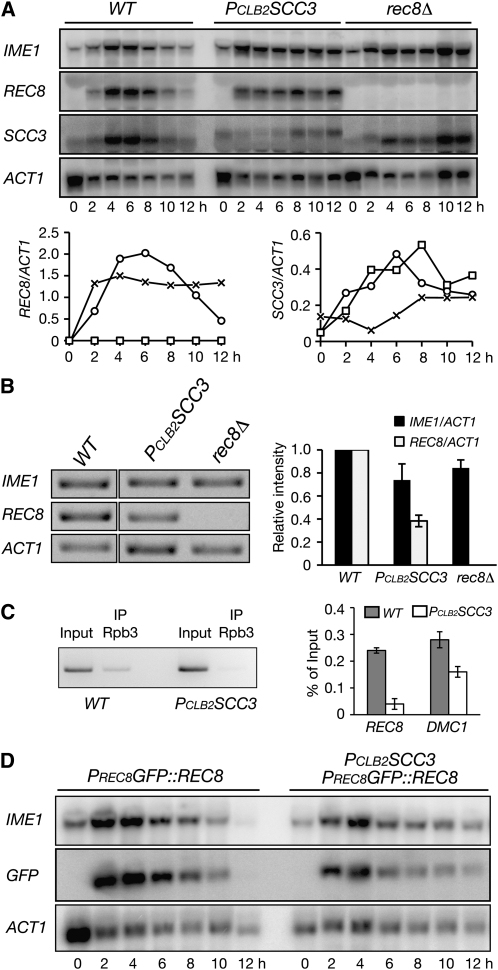
We hypothesized that Scc3 regulates REC8 gene expression in yeast meiosis. To determine the level of REC8 mRNA, we harvested yeast cells undergoing synchronous meiosis and performed Northern blots (Figure 4A). In wild-type cells, REC8 transcripts appeared after 2 hr, peaked at ∼4–6 hr, and diminished 12 hr after induction of meiosis (Figure 4A). The REC8 transcripts emerged on a similar time schedule in Scc3-depleted cells, but their levels never reached those of the wild type (Figure 4A). Quantitative analysis by RT-PCR revealed that REC8 mRNA in Scc3-depleted cells was 65% lower than that of the wild type 6 hr after induction of meiosis (Figure 4B). In contrast, the expression of the meiosis-initiating gene IME1 was reduced by only ∼20% in PCLB2SCC3 cells (Figure 4B). Scc3 therefore plays a role in REC8 gene expression in yeast meiosis.
Figure 4.—
Scc3 regulates REC8 promoter activity during yeast meiosis. (A) mRNA levels of IME1, REC8, SCC3, and ACT1 in wild-type (NH144), PCLB2SCC3 (3200), and rec8Δ (HY1495) cells. Yeast cells were induced to undergo synchronous meiosis, and aliquots were withdrawn at the indicated times and prepared for Northern blots probed by gene-specific probes. (B) RT-PCR analysis of IME1, REC8, and ACT1 transcripts. Yeast aliquots were withdrawn 6 hr after induction of meiosis; total mRNA was extracted, reversed to cDNA, and amplified by gene-specific primers. (Right) Quantitative analysis with an average of two independent experiments shown. Error bars show standard deviation. (C) ChIP of Rbp3 in wild-type (HY3000) and PCLB2SCC3 (HY3003) cells during yeast meiosis. Yeast cells were induced to undergo synchronous meiosis; aliquots were withdrawn 6 hr after induction and prepared for ChIP analysis. (Left) A representative gel image. (Right) Quantitative analysis of Rpb3 binding at the REC8 and DMC1 genes from two independent experiments. (D) A heterologous reporter assay of REC8 promoter activity (HY2106 and HY2108). Plasmid pHG105 was digested with MluI and transformed into the REC8 locus. Yeast cells were induced to undergo synchronous meiosis, and aliquots were withdrawn for Northern blots as shown in A. Gene-specific probes were used to detect the mRNA levels of IME1, GFP, and ACT1.
Our Northern blots also showed that the expression of SCC3 was largely abolished in PCLB2SCC3 cells during meiosis (Figure 4A). On the other hand, the level of SCC3 transcripts in rec8Δ remained comparable to that of wild type (Figure 4A), which is consistent with the observation that Scc3 protein levels remained normal in rec8Δ cells (Figure 2D). Therefore, Rec8 is not required for meiotic expression of SCC3.
To determine whether Scc3 is responsible for REC8 gene transcription during yeast meiosis, we assayed the density of RNA Pol II binding to the REC8 gene by ChIP (Figure 4C). Using the Pol II subunit Rpb3 as a readout, we found that the association of Rpb3 with the REC8 gene was reduced by ∼70% in PCLB2SCC3 cells after normalization of Rpb3's binding to the DMC1 gene (Figure 4C). Our data therefore suggest that the decrease in REC8 mRNA level in Scc3-depleted meiotic cells is a result of transcriptional inactivation of the REC8 promoter during yeast meiosis.
To determine further how Scc3 regulates REC8 gene transcription, we developed a heterologous reporter assay by using the REC8 promoter to drive the expression of GFP (Figure 4D and our unpublished data). The expression of PREC8GFP, which was inserted at the REC8 locus, essentially mirrored that of the REC8 gene in wild-type cells (Figure 4, A and D). In contrast, the level of GFP transcripts from PREC8GFP was low in Scc3-depleted cells during meiosis, ∼60% lower than in the wild type (Figure 4C; t = 4 hr). Because a low level of activity of the REC8 promoter still occurred in Scc3-depleted cells (Figure 4), our data suggest that Scc3 is required for increasing the REC8 promoter activity but not for its initiation. Alternatively, the low level of REC8 expression results from residual Scc3 activity in PCLB2SCC3 cells.
Scc3 is not necessary for REC8 translation but is required for Rec8 chromosome association:
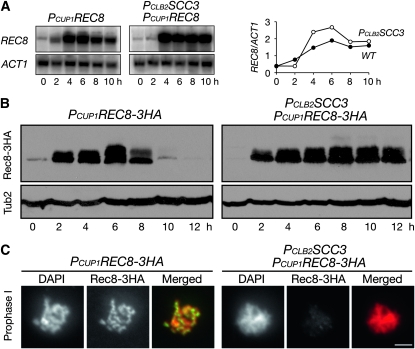
To produce Rec8 in Scc3-depleted cells, we constructed an inducible allele of REC8 (PCUP1REC8), which served as the only source of REC8 in meiosis (Figure 5). Upon the addition of copper ion at the time of induction of meiosis, PCUP1REC8 was expressed in wild-type and Scc3-depleted cells at comparable levels (Figure 5A). These REC8 transcripts produced by the CUP1 promoter appeared to be efficiently translated to produce Rec8 protein (Figure 5B). In wild-type SCC3 cells, ectopically produced Rec8 was subject to the same regulation as the endogenous Rec8; it peaked at ∼6 hr and was degraded by the end of meiosis (Figure 2B and Figure 5B). Furthermore, these Rec8 proteins localized to the meiotic chromosome along its entire length just as the endogenous Rec8 did (Figure 2A and Figure 5C). In PCLB2SCC3 cells, PCUP1REC8 was expressed, and these cells produced amounts of Rec8 similar to those seen in wild-type cells (Figure 5B). Because PCLB2SCC3 cells were arrested by the recombination checkpoint at prophase I, the degradation of Rec8 was delayed in these cells (Figure 5B). Therefore, in the absence of Scc3, Rec8 can be produced and remains relatively stable in meiosis, but ectopically produced Rec8 failed to bind to the chromosome in PCLB2SCC3 cells (Figure 5C), suggesting that Scc3 is required for Rec8 association with the chromosome.
Figure 5.—
Ectopic expression of REC8 in meiotic cells. (A) The expression level of PCUP1REC8 in wild-type (HY1417C) and PCLB2SCC3 (HY1417) cells during meiosis. Yeast cells were induced for synchronous meiosis, and aliquots were withdrawn at indicated times for Northern blots as shown in Figure 4A. Note that PCUP1REC8 is expressed in PCLB2SCC3 cells. (Right) A semiquantitative measurement of REC8 transcripts over those of ACT1. (B) Protein level of Rec8 in wild-type and PCLB2SCC3 cells. Western blots were prepared as in Figure 1A to reveal the levels of Rec8-3HA and β-tubulin. (C) Chromosome association of Rec8 in wild-type and PCLB2SCC3 cells. Yeast surface nuclear spreads were prepared for immunofluorescence as in Figure 2A. Note that Rec8 is produced but does not bind to chromosomes in PCLB2SCC3 cells. Red, DNA; green, Rec8. Bar, 2 μm.
One concern was that the CUP1 promoter perhaps overexpressed REC8 during meiosis and could obscure our interpretation. We therefore constructed a PDMC1REC8 allele, which was incorporated at the endogenous REC8 locus and produced Rec8 at a level similar to that of Dmc1 during meiosis (Figure S3A). The expression levels of PDMC1REC8 in wild-type and Scc3-depleted meiotic cells appeared comparable because the two strains produced similar amounts of Rec8 (Figure S3A). To support further a specific role of Scc3 in activating the REC8 promoter, we constructed a heterologous GFP reporter, PDCM1GFP, which produced similar amounts of GFP in wild-type and Scc3-depleted meiotic cells (Figure S3B). Together, our results suggest that Scc3 is required for REC8 gene expression because it specifically increases REC8 promoter activity during meiosis, but Scc3 is not necessary for translation of REC8 mRNA.
Scc3 is not necessary for MCD1 gene expression in proliferating cells:
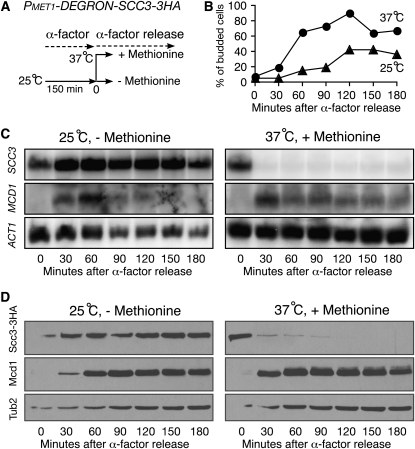
The mitotic counterpart of REC8 is MCD1, which probably arose from an ancient genome-duplication event (Kellis et al. 2004). To determine whether Scc3 plays a similar role in regulating MCD1 transcription in proliferating yeast cells, we generated a degron allele of scc3 (PMET1-DEGRON-SCC3) to deplete Scc3 in vegetative cells and observed MCD1 gene transcription and protein production with Northern and Western blots (Figure 6). To synchronize yeast culture, we used α-factor to arrest cells at the G1 phase and then released them to a nonpermissive condition to deplete Scc3 (Figure 6, A and B). The transcription of SCC3 was completely shut off in cells that were shifted to the nonpermissive condition (Figure 6C), and Scc3 became depleted in these cells after G1 phase release (Figure 6D). In contrast, MCD1 was expressed after G1 release, and its level of expression did not appear to differ greatly, except that cells expressed MCD1 earlier at the elevated temperature (Figure 6C). As a result, Mcd1 protein levels in these two treatments were comparable (Figure 6D). Scc3 is therefore required for positively regulating REC8 gene expression in meiotic cells but not for MCD1 in mitotic cells. Together, our results show that the cohesin kleisin subunit Rec8 or Mcd1 remains relatively stable when Scc3 is absent in either meiotic or mitotic cells.
Figure 6.—
Scc3 is not required for MCD1 expression in vegetative cells. (A) A diagram showing the experimental procedure. (B) Yeast budding index showing cell progression. Yeast aliquots were withdrawn at indicated times after G1 release, fixed, and examined by phase-contrast microscopy. (C) mRNA levels of SCC3, MCD1, and ACT1 after release from α-factor arrest. Yeast aliquots were withdrawn at indicated times and prepared for Northern blots probed by gene-specific probes as shown in Figure 4A. Note that MCD1 is expressed only after release from α-factor arrest. (D) Protein levels of Scc3 and Mcd1. Yeast aliquots were withdrawn at indicated times and prepared for Western blots probed by anti-HA, anti-Mcd1, and anti-β-tubulin antibodies. Note that Mcd1 remains at a normal level in Scc3-depleted vegetative cells.
Smc1 has a role similar to that of Scc3 in positively regulating REC8 gene expression during meiosis:
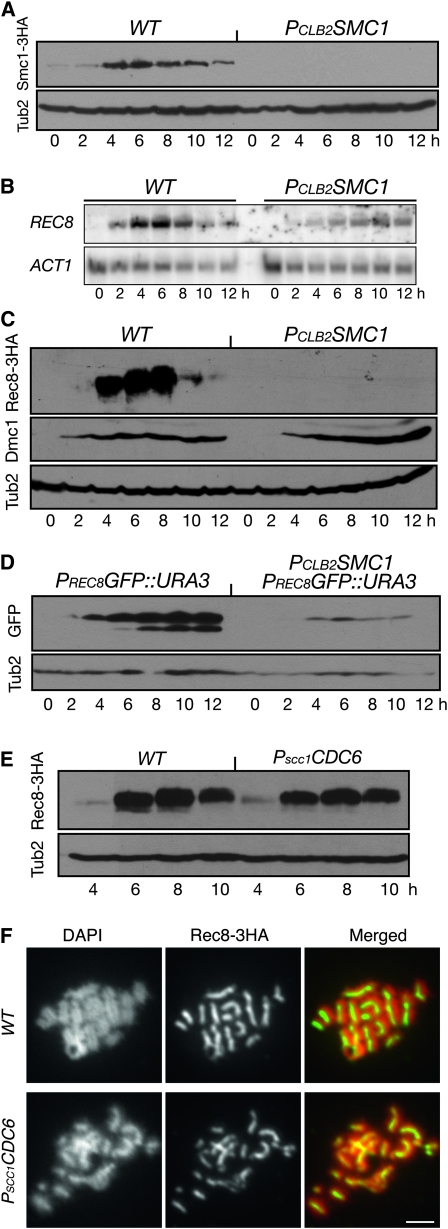
To determine whether cohesin subunits other than Scc3 have a role in positively regulating REC8 gene expression, we were able to deplete more completely Smc1 in meiosis using the same CLB2 promoter-replacement approach (Figure 7A). As in Scc3-depleted meiotic cells, the level of REC8 transcript was dramatically reduced in Smc1-depleted cells (Figure 7B). Consequently, the Rec8 protein level was very low in mutant cells (Figure 7C). In contrast, the meiosis-specific protein Dmc1 was produced at comparable levels in wild-type and PCLB2SMC1 cells (Figure 7C). These data suggest that Smc1 is also required for Rec8 production in meiosis. Using the PREC8GFP reporter assay, we found that the production of GFP was dramatically reduced in Smc1-depleted meiotic cells (Figure 7D). Taken together, our results suggest that the cohesin complex is required for positively regulating the REC8 gene transcription during yeast meiosis.
Figure 7.—
Activation of REC8 promoter requires Smc1 but not sister-chromatid cohesion. (A) Depletion of Smc1 during meiosis (2821 and HY1875). Yeast cells were induced to undergo synchronous meiosis, and aliquots were withdrawn at the indicated times for Western blots as in Figure 1A. (B) Transcriptional level of REC8 during meiosis (NH144 and HY1875). Total RNA was extracted and probed with gene-specific probes as in Figure 4A. (C) Protein level of Rec8 in wild-type (HY1503C) and PCLB2SMC1 (HY1868) cells in meiosis. Yeast protein extracts were prepared for Western blots, which detected the levels of Rec8 and Dmc1 as shown in Figure 2B. The level of β-tubulin served as a loading control. (D) A heterologous reporter assay of REC8 promoter activity in wild-type (HY2460) and PCLB2SMC1 (HY2460-1) cells in meiosis. PREC8GFP was placed at the URA3 locus by transformation. Yeast cells were induced to undergo synchronous meiosis, and aliquots were withdrawn at the indicated times and prepared for Western blots probed by anti-GFP (Ab290) and anti-β-tubulin antibodies. (E) Rec8 protein level in wild-type (HY2740) and PSCC1CDC6 (HY2741) cells during meiosis. Representative time points are shown. (F) Chromosome localization of Rec8 in wild-type and PSCC1CDC6 cells during meiosis. Yeast cells were collected 6 hr after induction of meiosis and prepared for surface nuclear spread as in Figure 2A. Note that chromosomes still formed rod-shaped structures in the absence of sister chromatids. Red, DNA; green, Rec8. Bar, 2 μm.
The presence of sister chromatids is not necessary for REC8 gene activation:
To determine whether sister-chromatid cohesion is required for activating REC8 gene expression, we used a genetic approach to abolish meiotic DNA replication with the PSCC1CDC6 allele (Hochwagen et al. 2005). In Cdc6-depleted meiotic cells, sister chromatids from chromosome V were largely absent (data not shown), but Rec8 was produced efficiently in PSCC1CDC6 cells because its protein level was comparable to that of the wild type during meiosis (Figure 7E). In addition, immunofluorescence microscopy revealed that Rec8 was localized to the chromosomes in Cdc6-depleted cells (Figure 7F). Chromosome axes in the PSCC1CDC6 cells resembled those from the wild-type cells even though Cdc6-depleted cells lacked sister chromatids in meiosis (Figure 7F). These results suggest that REC8 can be efficiently transcribed in the absence of sister chromatids, so the presence of sister chromatids is not necessary for REC8 gene expression.
Additional meiotic genes are subject to cohesin regulation:
To determine whether cohesin globally regulates gene expression during meiotic differentiation in yeast, we surveyed the gene-expression pattern using the expression microarray. Expression of 27 genes was reduced by >75% in Smc1-depleted meiotic cells 6 hr after induction of meiosis; only 8 genes showed more than a fourfold increase (data not shown). We focused on the expression pattern of ∼52 meiotic genes such as REC8 that belonged to the category of “early genes” in meiosis (Chu et al. 1998). Among them, we found by microarray analysis that the expression level of two genes (MRD1 and PAD1) was lowered by ∼50% in the PCLB2SMC1 mutant (Figure S4). The expression level of REC8 was only slightly reduced in comparison to that of DMC1 (Figure S4), demonstrating that our microarray analysis of meiotic gene expression is qualitative at best. Whether the cohesin target genes share common features is currently unknown, but our preliminary analysis supports the idea that cohesin has a positive role in meiotic gene expression.
DISCUSSION
Using a genetic approach, we have shown that cohesin subunits Scc3 and Smc1 are required for efficient transcription of a target gene, REC8, because they increase its promoter activity during yeast meiosis. Cohesin is a major chromosomal factor required for sister-chromatid cohesion (Guacci et al. 1997; Michaelis et al. 1997), but its emerging role in regulation of gene expression is best known in animal development (Dorsett et al. 2005; Horsfield et al. 2007; Wendt et al. 2008). Nonlethal mutation in genes that encode cohesin and cohesin-associated factors in humans is directly linked to developmental disorders collectively called cohesinopathies, which include Cornelia de Lange syndrome and Roberts syndrome (Liu and Krantz 2009). The etiology of these human diseases remains to be elucidated. Our work in yeast meiosis using the REC8 promoter activity as a readout of cohesin function in gene regulation lends support to the notion that this noncanonical cohesin activity is evolutionarily conserved; it also provides molecular insights into cohesin's role in cell differentiation and development.
Four lines of evidence support the idea that the cohesin complex increases REC8 gene expression by modulating the REC8 promoter activity during meiosis. First, Scc3 and Smc1 have similar effects on regulation of REC8 gene expression; second, Scc3 modulates the density of Pol II binding to the REC8 gene; third, a heterologous reporter assay using the REC8 promoter shows that it is under the influence of cohesin; and finally, the REC8 open reading frame driven by the inducible CUP1 or the meiosis-specific DMC1 promoter can be transcribed and translated at comparable levels in wild-type and cohesin mutants. Because Rec8 is a meiosis-specific cohesin subunit, feedback control by meiotic cohesin of REC8 promoter activation is not surprising (W. Lin, H. Jin and H. Yu, unpublished data). In addition, one prediction is that, if the cohesin holocomplex formation and its association with the chromosome were important, the cohesin loader, the Scc2/Scc4 complex, would have a similar role in meiotic gene activation. Indeed, our analysis of Scc2 in yeast meiosis shows that it is required for recruiting cohesin to the chromosome to activate the cohesin-regulated promoter REC8 (W. Lin, H. Jin and H. Yu, unpublished data), but our observation differs from those in the fly, where cohesin and its loader Scc2 (called Nipped B) apparently have opposite effects on gene regulation (Rollins et al. 2004). The reason for this discrepancy is currently unknown.
How, then, does cohesin activate gene transcription in yeast? In vertebrates, direct binding of cohesin to the transcriptional factor CTCF, which has been implicated in insulating gene transcription, may explain cohesin's role in gene regulation (Parelho et al. 2008; Rubio et al. 2008; Stedman et al. 2008; Wendt et al. 2008). In yeast, no equivalent of CTCF is yet known, but currently no evidence indicates that cohesin binds directly to the transcriptional machinery. Upon meiotic differentiation, yeast cells reorganize the higher-order chromosome structure that necessitates the change of gene expression pattern (Kassir et al. 2003), of which cohesin could act as an important chromosomal factor. Furthermore, cohesin binds to the chromatin-remodeling complex RSC and also interacts with modified histones during double-strand break repair (Unal et al. 2004; Chai et al. 2005). Finally, a recent study in yeast showed that scc2 and eco1 mutations that mimic human diseases lead to altered chromosome organization (Gard et al. 2009). Therefore, cohesin-mediated chromosome organization may facilitate the recruitment of transcriptional factors to the 5′ upstream sequences of cohesin-regulated genes to activate or repress gene expression. Alternatively, cohesin might directly interact with the transcriptional factors, for example, with the mediator (Kagey et al. 2010), to regulate gene expression. These two possibilities are not mutually exclusive, but this study does not distinguish between them.
Cohesin is required for REC8 gene expression during meiotic differentiation but not for that of its duplicated gene MCD1 in vegetative cells. In addition, cohesin does not seem to regulate meiotic genes universally because the expression of the meiosis-specific genes IME1, DMC1, and others is largely unaffected in Scc3- or Smc1-depleted cells (this report and data not shown). These observations imply that a complex interplay takes place between trans-acting factors and cis-acting DNA sequences in regulating the expression of cohesin-target genes during meiotic differentiation. Cohesin associates with the chromosome at specific loci of the yeast genome, which are predominately located at regions of convergent transcription (Glynn et al. 2004; Lengronne et al. 2004). These binding sites would position cohesin toward the 3′-end of the transcribed genes, rather that at promoter-proximal sequences, which might explain why only a subset of meiotic genes is subject to cohesin regulation (this study and our unpublished data). In this regard, our study is consistent with a recent observation of cohesin activity in G1-arrested vegetative cells, showing that a small number of genes changed their expression pattern in response to mcd1-1 inactivation in budding yeast (Skibbens et al. 2010).
Our genetic analysis using the cdc6 mutant indicates that the primary role of cohesin in sister-chromatid cohesion is not necessary for its regulation of its target gene. In the cdc6 mutant, cohesin, revealed by Rec8, is localized to the meiotic chromosome axis in a way that is similar to that in the wild type. Because sister-chromatid cohesion is coupled to DNA replication in yeast (Uhlmann and Nasmyth 1998), our results suggest that chromosomal binding of cohesin is sufficient for carrying out cohesin's function in gene regulation. Therefore, cohesin's role in sister-chromatid cohesion appears to be separable from its role in gene expression. Our results also lend support to the notion that regulation of gene expression by cohesin is independent of sister-chromatid cohesion in postmitotic and differentiating animal cells (Horsfield et al. 2007; Pauli et al. 2008; Schuldiner et al. 2008; Nativio et al. 2009).
In summary, we have shown that cohesin plays a positive role in target gene activation during yeast meiotic differentiation. Lack of cohesin is detrimental to yeast meiosis in many aspects, including gene transcription, recombination, and chromosome segregation. The identification of cohesin target genes in yeast provides a valuable tool for further elucidation of the biological significance and mechanism of cohesin function in gene regulation during cell differentiation in a model eukaryote.
Acknowledgments
We thank A. Amon, V. Guacci, and D. Bishop for sharing yeast strains and antibodies. S. Miller provided technical assistance. A. B. Thistle assisted with text editing. This work was supported in part by the National Science Foundation (MCB-0718384) and the Florida Biomedical Research Program (08BN-08).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.122358/DC1.
References
- Blat, Y., and N. Kleckner, 1999. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98 249–259. [DOI] [PubMed] [Google Scholar]
- Chai, B., J. Huang, B. R. Cairns and B. C. Laurent, 2005. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 19 1656–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein et al., 1998. The transcriptional program of sporulation in budding yeast. Science 282 699–705. [DOI] [PubMed] [Google Scholar]
- Donze, D., C. R. Adams, J. Rine and R. T. Kamakaka, 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 13 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett, D., J. C. Eissenberg, Z. Misulovin, A. Martens, B. Redding et al., 2005. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development 132 4743–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard, S., W. Light, B. Xiong, T. Bose, A. J. McNairn et al., 2009. Cohesinopathy mutations disrupt the subnuclear organization of chromatin. J. Cell Biol. 187 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn, E. F., P. C. Megee, H. G. Yu, C. Mistrot, E. Unal et al., 2004. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2 E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber, S., C. H. Haering and K. Nasmyth, 2003. Chromosomal cohesin forms a ring. Cell 112 765–777. [DOI] [PubMed] [Google Scholar]
- Guacci, V., D. Koshland and A. Strunnikov, 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering, C. H., A. M. Farcas, P. Arumugam, J. Metson and K. Nasmyth, 2008. The cohesin ring concatenates sister DNA molecules. Nature 454 297–301. [DOI] [PubMed] [Google Scholar]
- Hauf, S., E. Roitinger, B. Koch, C. M. Dittrich, K. Mechtler et al., 2005. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 3 e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, T., 2006. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 7 311–322. [DOI] [PubMed] [Google Scholar]
- Hochwagen, A., W. H. Tham, G. A. Brar and A. Amon, 2005. The FK506 binding protein Fpr3 counteracts protein phosphatase 1 to maintain meiotic recombination checkpoint activity. Cell 122 861–873. [DOI] [PubMed] [Google Scholar]
- Horsfield, J. A., S. H. Anagnostou, J. K. Hu, K. H. Cho, R. Geisler et al., 2007. Cohesin-dependent regulation of Runx genes. Development 134 2639–2649. [DOI] [PubMed] [Google Scholar]
- Jin, H., V. Guacci and H. G. Yu, 2009. Pds5 is required for homologue pairing and inhibits synapsis of sister chromatids during yeast meiosis. J. Cell Biol. 186 713–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey, M. H., J. J. Newman, S. Bilodeau, Y. Zhan, D. A. Orlando et al., 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassir, Y., N. Adir, E. Boger-Nadjar, N. G. Raviv, I. Rubin-Bejerano et al., 2003. Transcriptional regulation of meiosis in budding yeast. Int. Rev. Cytol. 224 111–171. [DOI] [PubMed] [Google Scholar]
- Keeney, S., C. N. Giroux and N. Kleckner, 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 375–384. [DOI] [PubMed] [Google Scholar]
- Kellis, M., B. W. Birren and E. S. Lander, 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428 617–624. [DOI] [PubMed] [Google Scholar]
- Klein, F., P. Mahr, M. Galova, S. B. Buonomo, C. Michaelis et al., 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98 91–103. [DOI] [PubMed] [Google Scholar]
- Kupiec, M., B. Byers, R. E. Esposito and A. Mitchell, 1997. Meiosis and sporulation in Saccharomyces cerevisiae, pp. 889–1036 in The Molecular and Cellular Biology of the Yeast Saccharomyces, edited by J. R. Broach, J. R. Pringle, and E. W. Jones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Laloraya, S., V. Guacci and D. Koshland, 2000. Chromosomal addresses of the cohesin component Mcd1p. J. Cell Biol. 151 1047–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B. H., and A. Amon, 2003. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science 300 482–486. [DOI] [PubMed] [Google Scholar]
- Lengronne, A., Y. Katou, S. Mori, S. Yokobayashi, G. P. Kelly et al., 2004. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430 573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., and I. D. Krantz, 2009. Cornelia de Lange syndrome, cohesin, and beyond. Clin. Genet. 76 303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953–961. [DOI] [PubMed] [Google Scholar]
- Michaelis, C., R. Ciosk and K. Nasmyth, 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91 35–45. [DOI] [PubMed] [Google Scholar]
- Mitchell, A. P., 1994. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol. Rev. 58 56–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth, K., and C. H. Haering, 2009. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43 525–558. [DOI] [PubMed] [Google Scholar]
- Nativio, R., K. S. Wendt, Y. Ito, J. E. Huddleston, S. Uribe-Lewis et al., 2009. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2–H19 locus. PLoS Genet. 5 e1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onn, I., J. M. Heidinger-Pauli, V. Guacci, E. Unal and D. E. Koshland, 2008. Sister chromatid cohesion: a simple concept with a complex reality. Annu. Rev. Cell Dev. Biol. 24 105–109. [DOI] [PubMed] [Google Scholar]
- Parelho, V., S. Hadjur, M. Spivakov, M. Leleu, S. Sauer et al., 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132 422–433. [DOI] [PubMed] [Google Scholar]
- Pauli, A., F. Althoff, R. A. Oliveira, S. Heidmann, O. Schuldiner et al., 2008. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev. Cell 14 239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins, R. A., P. Morcillo and D. Dorsett, 1999. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics 152 577–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins, R. A., M. Korom, N. Aulner, A. Martens and D. Dorsett, 2004. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol. Cell. Biol. 24 3100–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio, E. D., D. J. Reiss, P. L. Welcsh, C. M. Disteche, G. N. Filippova et al., 2008. CTCF physically links cohesin to chromatin. Proc. Natl. Acad. Sci. USA 105 8309–8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and D. W. Russell, 2001. Molecular Cloning: A Laboratory Manual. Cold Sping Harbor Laboratory Press, Cold Sping Harbor, NY.
- Schuldiner, O., D. Berdnik, J. M. Levy, J. S. Wu, D. Luginbuhl et al., 2008. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev. Cell 14 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens, R. V., J. Marzillier and L. Eastman, 2010. Cohesins coordinate gene transcriptions of related function within Saccharomyces cerevisiae. Cell Cycle 9 1601–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman, W., H. Kang, S. Lin, J. L. Kissil, M. S. Bartolomei et al., 2008. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 27 654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom, L., H. B. Lindroos, K. Shirahige and C. Sjogren, 2004. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16 1003–1015. [DOI] [PubMed] [Google Scholar]
- Uhlmann, F., and K. Nasmyth, 1998. Cohesion between sister chromatids must be established during DNA replication. Curr. Biol. 8 1095–1101. [DOI] [PubMed] [Google Scholar]
- Unal, E., A. Arbel-Eden, U. Sattler, R. Shroff, M. Lichten et al., 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16 991–1002. [DOI] [PubMed] [Google Scholar]
- Wendt, K. S., K. Yoshida, T. Itoh, M. Bando, B. Koch et al., 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451 796–801. [DOI] [PubMed] [Google Scholar]
- Xu, L., M. Ajimura, R. Padmore, C. Klein and N. Kleckner, 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15 6572–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H. G., and D. E. Koshland, 2005. Chromosome morphogenesis: condensin-dependent cohesin removal during meiosis. Cell 123 397–407. [DOI] [PubMed] [Google Scholar]
- Zhang, N., S. G. Kuznetsov, S. K. Sharan, K. Li, P. H. Rao et al., 2008. A handcuff model for the cohesin complex. J. Cell Biol. 183 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]