Abstract
Histone variants and histone modification complexes act to regulate the functions of chromatin. In Saccharomyces cerevisiae the histone variant H2A.Z is encoded by HTZ1. Htz1 is dispensable for viability in budding yeast, but htz1Δ is synthetic sick or lethal with the null alleles of about 200 nonessential genes. One of the strongest of these interactions is with the deletion of SET3, which encodes a subunit of the Set3/Hos2 histone deacetylase complex. Little is known about the functions of Set3, and interpreting these genetic interactions remains a highly challenging task. Here we report the results of a forward genetic screen to identify bypass suppressors of the synthetic slow-growth phenotype of htz1Δ set3Δ. Among the identified loss-of-function suppressors are genes encoding subunits of the HDA1 deacetylase complex, the SWR1 complex, the H2B deubiquitination module of SAGA, the proteasome, Set1, and Sir3. This constellation of suppressor genes is uncommon among the global set of htz1Δ synthetic interactions. BDF1, AHC1, RMR1, and CYC8 were identified as high-copy suppressors. We also identified interactions with SLX5 and SLX8, encoding the sumoylation-targeted ubiquitin ligase complex. In the context of htz1Δ set3Δ, suppressors in the SWR1 and the H2B deubiquitination complexes show strong functional similarity, as do suppressors in the silencing genes and the proteasome. Surprisingly, while both htz1Δ set3Δ and swr1Δ set3Δ have severe slow-growth phenotypes, the htz1Δ swr1Δ set3Δ triple mutant grows relatively well. We propose that Set3 has previously unrecognized functions in the dynamic deposition and remodeling of nucleosomes containing H2A.Z.
HIGHLY sophisticated coordination of distinct sets of chromatin regulators is required for the information to flow correctly from DNA to RNA in eukaryotic cells. Reconstructing the temporal and spatial structure of this coordination is the central task of studying eukaryotic gene expression. The direct entry points are physical interactions among the chromatin regulators but many functional relationships are beyond immediate physical interactions and only visible through genetic interactions. Interpreting the genetic interactions in the context of physical process remains extremely challenging. In recent years, functional genomics studies utilizing yeast deletion collection (Tong et al. 2001; Ooi et al. 2003; Pan et al. 2006; Collins et al. 2007; Lin et al. 2008; Fiedler et al. 2009; Costanzo et al. 2010) have generated large data sets of genetic interactions but most of these await prioritization and investigation. In this report, we chose to study the genetic interactions between the gene for histone H2A variant HTZ1 and the gene for SET3 because of the critical roles of their functional homologs in metazoan development.
The results of early chromatin immunoprecipitation experiments in the budding yeast Saccharomyces cerevisiae showed that Htz1 preferentially occupies the promoter regions of two transcriptionally inactive but inducible genes, GAL1 and PHO5 (Santisteban et al. 2000). This promoter enrichment was later revealed to be a global pattern in budding yeast by several independent studies (Guillemette et al. 2005; Li et al. 2005; Raisner et al. 2005; Zhang et al. 2005; Albert et al. 2007). Both the protein sequence of H2A.Z and its distinct genomic geography are highly conserved from budding yeast to mammalian cells as the promoter enrichment pattern of H2A.Z has been reported in worm (Whittle et al. 2008), fly (Mavrich et al. 2008), plant (Zilberman et al. 2008), murine (Creyghton et al. 2008), and human cells (Barski et al. 2007). It is well established in yeast that the ATP-dependent SWR1 complex deposits H2A.Z onto chromatin (Krogan et al. 2003; Kobor et al. 2004; Mizuguchi et al. 2004). Although the specific targeting mechanism remains to be determined, NuA4-mediated histone acetylation and the double bromodomain subunit of SWR1 complex, Bdf1, have been suggested to be the contributing factors of the selective deposition (Altaf et al. 2010). In budding yeast, the Htz1 molecules are removed from the promoters during the activation process of inducible genes (Santisteban et al. 2000), which may be partially facilitated by the intrinsic fragility of Htz1–H2B dimer compared to the canonical H2A–H2B dimer (Zhang et al. 2005). The detailed mechanism and the relationships between this dynamic process and other transcriptional initiation events are largely unclear. The 5′-end enrichment pattern of H2A.Z appears to be dynamic in metazoan cells as well, observed most prominently during development (Creyghton et al. 2008; Whittle et al. 2008; Cui et al. 2009) and regulated gene expression responding to environmental signals (John et al. 2008; Sutcliffe et al. 2009). One striking example is in murine embryonic stem cells. H2A.Z molecules occupy the promoters of genes critical for development and this genomic distribution shows remarkable correlations with the patterns of Polycomb group (PcG) proteins. In lineage-committed cells, H2A.Z molecules redistribute to the promoters of different sets of genes (Creyghton et al. 2008). The link between H2A.Z and PcG group proteins was revealed earlier by genetic studies in Drosophila (Swaminathan et al. 2005). It was reported that the developmental abnormalities of Pc mutants could be enhanced by additional mutation in His2AV, the H2A.Z gene in Drosophila. Consistently, the mutant phenotypes of Trithorax group (TrxG) genes, encoding the development regulators antagonizing PcG proteins, could be suppressed by His2AV mutation.
In budding yeast, Set3 was found by TAP-tag affinity purification to be a component of a 7-subunit HDAC complex (Pijnappel et al. 2001). Based on the complex composition, Set3 complex was proposed to be a functional homolog of metazoan NCoR/SMRT complexes, which are transcriptional corepressors directly interacting with unliganded nuclear receptors to shape the chronology of gene expression through mediating active repression. They play critical roles during development, tissue differentiation and metabolism (Mckenna and O'malley 2002). As a platform, NCoR/SMRT forms various complexes with different sets of subunits to achieve cell-type and promoter specificities. NCoR/SMRT forms a stable ternary complex with histone deacetylase HDAC3 (Guenther et al. 2000; Li et al. 2000) and TBL1 (Guenther et al. 2000), a WD-repeat-containing protein. HDAC3 and TBL1 are mammalian homologs of Hos2 and Sif1, both of which are core subunits of the budding yeast Set3 complex. The functional homology between Set3 complex and NCoR/SMRT extends further to the SANT domain shared by NCoR/SMRT and another core subunit of Set3 complex, Snt1 (Yu et al. 2003). Histone methyltransferase activity has not been associated with NCoR/SMRT and no methyltransferase activity has been reported for Set3 complex either. The main enzymatic activity of Set3 complex appears to be the HDAC subunit Hos2, which deacetylates histone H3 and H4 at the 5′ ends of actively transcribing genes. The recruitment of the Set3 complex to the 5′ end requires both the H3K4 methylation and the plant homeo domain of Set3 (Kim and Buratowski 2009). Interestingly, in addition to the different compositions of the NCoR/SMRT complex, another level of specificity of nuclear receptor responses can be achieved by cell-specific localization of H2A.Z. It is reported that H2A.Z plays a critical role in shaping the chromatin signature at the nuclear receptor interaction sites (John et al. 2008).
Although it is dispensable for budding yeast to grow under optimal conditions, htz1Δ is synthetic sick or lethal with null alleles of about 200 genes, together covering a wide spectrum of cellular functions. A major subgroup of genes having genetic interactions with htz1Δ consists of chromatin regulators, among which are the genes encoding the four core subunits of Set3 complex, SET3, HOS2, SIF2, and SNT1. The synthetic interaction between Htz1 and Set3 complex suggests overlapping roles in at least one essential biological process. While Htz1 is convincingly involved in a variety of cellular functions such as transcription (Santisteban et al. 2000), chromatin boundary maintenance (Meneghini et al. 2003), chromosome segregation (Rangasamy et al. 2004), and cell-cycle control (Dhillon et al. 2006), the function of Set3 complex is relatively poorly examined. Disruption of Set3 complex results in early induction of meiotic gene program upon nitrogen starvation (Pijnappel et al. 2001), indicating a repressive role of Set3 complex in transcription, which is reminiscent of its metazoan corepressor homologs. This notion is complicated by another report (Wang et al. 2002), which showed that Set3 and Hos2 are required for efficient transcription of GAL1, and HOS2 physically associates with actively transcribing genes. This result has been recently reinforced and the mechanism behind this active role is linked to H3K4 methylation-dependent recruitment of Set3 complex to the 5′ of the gene and Hos2-dependent deacetylation of the region (Kim and Buratowski 2009). The association of Set3 with actively transcribed genes can also be methylation independent, as phosporylation of the Pol II C-terminal domain by Kin28 has been found to stimulate the cotranscriptional recruitment of the Set3 complex to coding regions (Govind et al. 2010).
The intricate involvements of both H2A.Z and NCoR/SMRT in some of the most critical biological processes invoked us to gain further insight into their relationship. We propose that the fundamental biology is conserved in the synthetic interaction between their nonessential functional homologs in budding yeast, and we can expand this genetic interaction through yeast genetics, which is of great advantage in this case since the essential requirements of H2A.Z and NCoR/SMRT in higher eukaryotes preclude the task in metazoan system.
MATERIALS AND METHODS
Yeast strains, plasmids, and methods:
Yeast strains used in this study are listed in Table 1. Plasmids used are listed in Table 2. Standard yeast manipulations were performed as described (Adams et al. 1998). 5-Fluoroortic acid (5-FOA) was added to the final concentration of 1 mg/ml. The concentrations of other drugs are described in relevant contexts. Yeast transformation is based on the lithium acetate method. Cells were grown to 2 × 107 cells/ml, and the transformation mix was subject to heat shock at 42° for 30 min. For recovery of plasmids, 5 ml overnight yeast cultures were resuspended in STET buffer (8% sucrose, 50 mm Tris HCl pH 8.0, 50 mm EDTA, 5% Triton X-100) and mechanically disrupted in the presence of glass beads. After removing the impurities, DNA was ethanol precipitated, recovered, and used to transform a leu− Escherichia coli strain JA221.
TABLE 1.
Yeast strains
| Strains | Genotypes |
|---|---|
| MSY2029 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| MSY4477 | MATaade2-1 ade3∷hisG can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1Δ set3Δ∷NatR pMSS59[HTZ1 URA3 ADE3] |
| MSY4478 | MATα ade2-1 ade3∷hisG can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1Δ set3Δ∷NatR pMSS59[HTZ1 URA3 ADE3] |
| MSY4100 | MATα can1∷Prom-STE2-Sphis5 lyp1Δ cyh2 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0htz1Δ∷NatR |
| MSY4506 | MATα can1∷Prom-STE2-Sphis5 lyp1Δ cyh2 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0htz1Δ∷NatR swr1Δ∷hphR |
| MSY4652 | MATα can1∷Prom-STE2-Sphis5 lyp1Δ cyh2 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0htz1Δ∷NatR vps72Δ∷hphR |
| MSY4653 | MATα can1∷Prom-STE2-Sphis5 lyp1Δ cyh2 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0htz1Δ∷NatR hda2Δ∷hphR |
| MSY4654 | MATα can1∷Prom-STE2-Sphis5 lyp1Δ cyh2 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0htz1Δ∷NatR hda3Δ∷hphR |
| MSY4508 | MATα can1∷Prom-STE2-Sphis5 lyp1Δ cyh2 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0htz1Δ∷NatR ubp8Δ∷hphR |
| MSY4655 | MATα can1∷Prom-STE2-Sphis5 lyp1Δ cyh2 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0htz1Δ∷NatR sgf11Δ∷hphR |
| MSY4509 | MATα can1∷Prom-STE2-Sphis5 lyp1Δ cyh2 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0htz1Δ∷NatR rpn10Δ∷hphR |
| MSY4656 | MATα can1∷Prom-STE2-Sphis5 lyp1Δ cyh2 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0htz1Δ∷NatR pre9Δ∷hphR |
| MSY4537 | MATα can1∷Prom-STE2-Sphis5 lyp1Δ cyh2 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 set3Δ∷NatR |
| MSY4657 | MATα can1∷Prom-STE2-Sphis5 lyp1Δ cyh2 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0set3Δ∷NatR hda2Δ∷hphR |
| MSY4658 | MATα can1∷Prom-STE2-Sphis5 lyp1Δ cyh2 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 set3Δ∷NatR hda3Δ∷hphR |
| MSY4659 | MATα can1∷Prom-STE2-Sphis5 lyp1Δ cyh2 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 set3Δ∷NatR ubp8Δ∷hphR |
| MSY4660 | MATα can1∷Prom-STE2-Sphis5 lyp1Δ cyh2 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 set3Δ∷NatR sgf11Δ∷hphR |
| MSY4661 | MATα can1∷Prom-STE2-Sphis5 lyp1Δ cyh2 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 set3Δ∷NatR rpn10Δ∷hphR |
| MSY4662 | MATα can1∷Prom-STE2-Sphis5 lyp1Δ cyh2 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 set3Δ∷NatR pre9Δ∷hphR |
| MSY4586 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1Δ∷NatR |
| MSY4587 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1Δ∷NatR hda1Δ∷hphR |
| MSY4588 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1Δ∷NatR hda2Δ∷hphR |
| MSY4589 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1Δ∷NatR swr1Δ∷hphR |
| MSY4590 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1Δ∷NatR arp6Δ∷hphR |
| MSY4591 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1Δ∷NatR ubp8Δ∷hphR |
| MSY4592 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1Δ∷NatR sgf11Δ∷hphR |
| MSY4593 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1Δ∷NatR set1Δ∷hphR |
| MSY4594 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1Δ∷NatR sir3Δ∷hphR |
| MSY4595 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1Δ∷NatR rpn9Δ∷hphR |
| MSY4596 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1Δ∷NatR rpn10Δ∷hphR |
| MSY4544 | MATaade2-1 ade3∷hisG can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1Δ set3Δ∷NatR pMSS59[HTZ1 URA3 ADE3] pMH02 [SLX5 TRP1 CEN] |
| MSY4605 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1Δ∷NatR pRS425 |
| MSY4606 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1Δ∷NatR pMH05[AHC1 LEU2 2μ] |
| MSY4607 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1Δ∷NatR pMH04[BDF1 LEU2 2μ] |
| MSY4608 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1Δ∷NatR pMH07[CYC8 LEU2 2μ] |
| MSY4609 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1Δ∷NatR pMH06[RMR1 LEU2 2μ] |
| MSY4603 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 set3Δ∷KanR |
| MSY4610 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 set3Δ∷KanR hda1Δ∷hphR |
| MSY4611 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 set3Δ∷KanR hda2Δ∷hphR |
| MSY4612 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 set3Δ∷KanR ubp8Δ∷hphR |
| MSY4613 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 set3Δ∷KanR sgf11Δ∷hphR |
| MSY4614 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 set3Δ∷KanR set1Δ∷hphR |
| MSY4615 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 set3Δ∷KanR sir3Δ∷hphR |
| MSY4616 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 set3Δ∷KanR rpn10Δ∷hphR |
| MSY4617 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 set3Δ∷KanR pRS425 |
| MSY4618 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 set3Δ∷KanR pMH05[AHC1 LEU2 2μ] |
| MSY4619 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 set3Δ∷KanR pMH04[BDF1 LEU2 2μ] |
| MSY4620 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 set3Δ∷KanR pMH07[CYC8 LEU2 2μ] |
| MSY4621 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 set3Δ∷KanR pMH06[RMR1 LEU2 2μ] |
| MSY4523 | MATaURA3-TEL-VIIL ppr1∷TRP1 ura3Δ0 trp1-Δ63 (1) |
| MSY4528 | MATaURA3-TEL-VIIL ppr1∷TRP1 ura3Δ0 trp1-Δ63 sir3Δ∷hphR |
| MSY4529 | MATaURA3-TEL-VIIL ppr1∷TRP1 ura3Δ0 trp1-Δ63 rpn10Δ∷hphR |
| MSY4531 | MATaURA3-TEL-VIIL ppr1∷TRP1 ura3Δ0 trp1-Δ63 ubp14Δ∷hphR |
| MSY4663 | MATaade2-1 can1-100 his3-Δ200 leu2-3,112 trp1 URA3∷TelVR (2) |
| MSY4664 | MATaade2-1 can1-100 his3-Δ200 leu2-3,112 trp1 URA3∷TelVR sir3Δ∷hphR |
| MSY4665 | MATaade2-1 can1-100 his3-Δ200 leu2-3,112 trp1 URA3∷TelVR rpn10Δ∷hphR |
| MSY4666 | MATaade2-1 can1-100 his3-Δ200 leu2-3,112 trp1 URA3∷TelVR ubp14Δ∷hphR |
| MSY4667 | MATaade2-1 can1-100 his3-Δ200 leu2-3,112 trp1 URA3∷TelVR hda1Δ∷hphR |
All strains were constructed during this study except for: (1) Kahana and Gottschling (1999) and (2) Rundlett et al. (1996).
TABLE 2.
Plasmids strains
| Plasmid name | Genes on plasmid |
|---|---|
| pMSS59 | HTZ1 URA3 ADE3 2μ |
| pMH02 | SLX5 TRP1 CEN6 ARSH4 |
| pMH03 | SLX8 TRP1 CEN6 ARSH4 |
| pMH04 | BDF1 LEU2 2μ |
| pMH05 | AHC1 LEU2 2μ |
| pMH06 | RMR1 LEU2 2μ |
| pMH07 | CYC8 LEU2 2μ |
| pMH08 | SLX5 LEU2 CEN6 ARSH4 |
| pMH10 | SIR3 TRP1 2μ |
Mutagenesis, screening, and cloning:
The mTn-LEU2/lacZ yeast genomic library was constructed and described by Ross-MacDonald et al. (1999). Plasmid DNA collected with midi-prep kit (Qiagen) from the 14 pools of the mTn3 library was digested with NotI and gel purified. MSY4477 was transformed with purified DNA and the cells were plated onto SC–Leu plates at about 1200 colonies/large petri dish (150 × 15 mm). Approximately 90,000 colonies were subjected to the subsequent screening. The transformants were microscopically examined and colonies that displayed sectoring phenotype were streaked onto SC–Ura–Leu plates to select the pMSS59[HTZ1URA3ADE3] plasmids. Nonsectoring red colonies of these candidates from SC–Ura–Leu plates were restreaked onto YPD plates to confirm their sectoring phenotype. Candidates maintaining the sectoring phenotype were crossed with MSY4478, which is isogenic to MSY4477 except for having the opposite mating type. The diploids were subjected to tetrad analysis. If a single transposon-disrupted ORF is responsible for the sectoring phenotype, the sectoring phenotype cosegregates with the LEU2 marker on the transposon and follow a 2:2 segregation pattern. The transposon insertion sites in the final candidates were determined by inverse PCR. Genomic DNA from these candidates was purified and digested with RsaI. The digested genomic DNA was treated with T4 ligase at low concentration (1 μg/ml) to promote self-ligation. The ligation products were used as PCR templates and amplified using the primer pair 5′-TAAGTTGGGTAACGCCAGGGTTTTC-3′ and 5′-TGTTGCCACTCGCTTTAATG-3′. The PCR products were gel purified and sequenced with primer 5′-CGTTGTAAAACGACGGGATCCCCC-3′. The sequencing results were subject to BLAST search and the transposon insertion sites on the chromosome were determined.
EMS mutagenesis:
After the determination of the ethyl methanesulfonate (EMS) kill curve, the actual mutagenesis screen was performed by adding 23 μl EMS (Sigma) to 3 ml fresh cell culture (grown to log phase at 1 × 107 cells/ml). After 1 hr incubation at 28°, 1 ml 5% sodium thiosulfate was added to quench the mutagen. The cell mix was sonicated and diluted, after which 6000 cells were plated onto each YPD plate (150 × 15 mm) to achieve roughly 2000 surviving colonies per plate. Approximately 60,000 cells were subjected to the visual screening as described in the transposon mutagenesis method and 14 suppressor candidates were isolated. These candidates were crossed to MSY4478 to determine whether the suppressor mutations were recessive or dominant, and the resultant diploids were subjected to tetrad analysis. To clone the recessive suppressor candidate, a p366-based yeast genomic library was used to transform the suppressor strain. The Leu+ transformants were visually screened for revertants displaying nonsectoring color pattern due to the complementation of the suppressor gene by the wild-type copy on the library plasmid.
Generating deletion mutations:
All deletion mutants were generated by one-step replacement by homologous recombination. The knockout DNA fragments consisted of the ORF-flanking sequences PCR amplified together with a drug-resistant gene using GoTaq polymerase (Promega). The natMX template was plasimd pAG25 and the hphMX template was plasmid pAG32 using primers with following features: from 5′ to 3′, forward (reverse) primers have ∼40 nucleotides homologous to the 5′ ends (3′ ends) of the genomic target loci followed by 5′-CGTACGCTGCAGGTCGAC-3′ (5′-ATCGATGAATTCGAGCTCG-3′). The PCR products were gel purified (Qiagen) and 1 μg DNA was used for each transformation. After heat shock, cells were recovered in YPD media for at least 4 hr before being plated onto YPD + drug plates (kanamycin, 400 μg/ml; clonNAT, 100 μg/ml; hygromycin B, 500 μg/ml). The transformants were screened by analytical PCR using a forward primer located upstream of the replacement DNA and a common reverse primer located inside the drug-resistant cassette (5′-GTATGGGCTAAATGTACGGGC-3′).
Highcopy suppressor screen:
The 1588 clones of the systematic yeast genomic library (Jones et al. 2008) were individually cultured overnight at 37° in 200 μl/well LB + kanamycin (50 μg/ml) in 96-well plates. The overnight cultures from all the wells were pooled and plasmids were extracted by midi-prep kit (Qiagen). MSY4477 was transformed with the library plasmids and plated onto SC–Leu in large petri plates. About 40,000 Leu+ transformants were visually screened for color-sectoring phenotype. The suppressor candidates that showed color sectoring were restreaked onto SC–Ura–Leu plates to select for the pMSS59[HTZ1URA3ADE3] plasmids and the solid red colony morphology. After this step, the candidates were restreaked again on SC–Leu plates to determine which candidates would maintain the color-sectoring phenotype. The plasmids were extracted from the yeast cells and sequenced to identify the library plasmid and the genes it carried. All individual genes with their endogenous promoters on the candidate library plasmids were subcloned into pRS425, used to transform MSY4477, and tested for their suppressor activities on the basis of color sectoring and 5-FOA sensitivity.
Synthetic genetic array analysis:
Yeast strains for the specialized miniarrays were manually selected from the systematic gene knockout collection created in the MATa BY4741 background (Giaever et al. 2002) (EUROSCARF, Institute for Molecular Biosciences, Frankfurt, Germany). Deletions having negative genetic interactions with htz1Δ and set3Δ were chosen on the basis of annotations in the Saccharomyces Genome Database (http://www.yeastgenome.org). Spreadsheets of the miniarrays are presented in supporting information, Table S1 and Table S2. The query strains were constructed in the background of MATα can1∷Prom-STE2-Sphis5 lyp1Δ cyh2 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 (Tong et al. 2004). The htz1Δ and set3Δ alleles were replaced by natMX in the single query strains. These two strains were then used to construct the double-query strains, each of which has a suppressor allele replaced by hphMX. Synthetic genetic array analysis was carried out as described previously (Tong et al. 2004) with the modification including hygromycin, in addition to kanamycin and clonNAT, for the final triple-mutant selection. After 4 days of selection, the growth of the pinned colonies was recorded by digital photography and colony sizes were determined using custom image analysis software based on the Python Imaging Library (Pythonware, http://www.pythonware.com/products/pil/). Colony sizes were normalized across different plates using the set of control strains arrayed with the test sets. Synthetic negative genetic interactions were scored using as a cutoff a reduction of at least 25% in colony spot size relative to the single mutant alone. Bypass suppression was scored using a criterion of an increase in colony spot size of at least 33% with the suppressor relative to the double-mutant strain. For htz1Δ, each interacting deletion strain was represented once on the array (Table S1) and each candidate suppressor was screened in three independent experiments. For set3Δ, each deletion strain was represented three times on the array (Table S2) and each suppressor was screened once.
RESULTS
Isolation of htz1Δ set3Δ bypass suppressor mutations:
To characterize the network of HTZ1 and SET3 genetic interactions, we sought to isolate suppressors of the htz1Δ set3Δ slow-growth phenotype using a colony color screen (Bender and Pringle 1991). Growth of the htz1Δ set3Δ strain MSY4477 is maintained by the presence of plasmid pMSS59[HTZ1URA3ADE3], which also confers 5-FOA sensitivity and nonsectoring red colony color. The introduction of a suppressor mutation that bypasses the htz1Δ set3Δ synthetic slow-growth phenotype allows the plasmid to be lost during mitotic growth, giving rise to red/white sectored colonies and the segregation of 5-FOA-resistant isolates. We exploited the sectoring pattern for visual screening and 5-FOA resistance for secondary characterization.
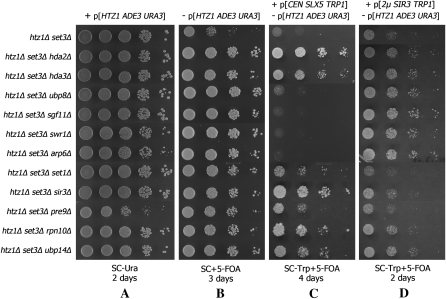
Bypass suppressors were generated by transposon mutagenesis (Ross-MacDonald et al. 1999). Pooled DNA prepared from the transposon library was used to transform MSY4477 and ∼90,000 Leu+ transformants were subjected to several steps of screening and verification (materials and methods). DNA sequence analysis of the transposon gene disruptions identified the following 12 candidate suppressor genes (and the number of times each was isolated): hda2 (7), hda3 (10), ubp8 (2), sgf11 (3), swr1 (2), vps72 (2), arp6 (3), set1 (1), sir3 (2), rpn10 (3), pre9 (2), and ubp14 (1). These candidates were crossed to MSY4478 and the diploids were subjected to tetrad analysis (sir3 was not tested due to sterility). In all cases, the sectoring phenotype segregated together with the transposon disrupted allele, marked by LEU2, and the tetrads showed 2:2 segregation. Finally, we confirmed the identities of the suppressors by deleting each of the candidate genes in MSY4477 and found that all the deletions resulted in strong suppression of the htz1Δ set3Δ synthetic slow-growth phenotype (Figure 1, A and B).
Figure 1.—
Loss-of-function suppressors of the htz1Δ set3Δ synthetic slow-growth phenotype. (A) The indicated strains were cultured in SC–Ura media in the presence of pMSS59[HTZ1 ADE3 URA3]. Serial dilutions were plated on SC–Ura plates and grown for 2 days. (B) Small portions of the overnight cell cultures from A were resuspended in YPD and grown overnight to allow the segregational loss of pMSS59. Serial dilutions were plated on SC + 5-FOA plates and grown for 3 days. (C) Differential responses of suppressors to SLX5 overexpression. The indicated strains were transformed with pMH02[SLX5 TRP1] and the transformants were grown overnight in SC–Trp media to permit the loss of pMSS59 while maintaining pMH02. Serial dilutions of the overnight cultures were plated on SC–Trp + 5-FOA plates and grown for 4 days. (D) Differential responses of suppressors to SIR3 overexpression. The indicated strains were transformed with plasmid pMH10[2μ SIR3 TRP1] and the transformants were grown in SC–Trp media overnight to permit the loss of pMSS59 while maintaining pMH10. Serial dilutions of the overnight cultures were plated on SC–Trp + 5-FOA plates and grown for 2 days.
One striking feature of this collection is that most of the genes encode subunits of protein complexes with broad functional significance, including the trimeric Hda1 histone deacetylation complex (hda2, hda3), the H2B deubiquitination submodule of the SAGA complex (ubp8, sgf11), the SWR1 complex (swr1, arp6, vps72), and the proteasome (rpn10, pre9). Thus, we next tested whether the deletion of other nonessential subunits of these complexes could also suppress htz1Δ set3Δ (data not shown). We found that deletions of the genes encoding all three subunits of the Hda1 HDAC complex, including HDA1, which encodes the enzymatic subunit, suppress htz1Δ set3Δ. In the case of the SAGA complex, only deletion of the H2B deubiquitination module genes could suppress. We tested the deletion of three genes outside the H2B deubiquitination module (gcn5, spt3, and spt8) and found that they actually exacerbate the synthetic phenotype of htz1Δ set3Δ. Deletions of all the nonessential subunits of the SWR1 complex, except YAF9, are strong suppressors, and the same is true for all the nonessential subunits of the proteasome. These results collectively indicate that it is the functions of these multisubunit protein complexes that become toxic in the absence of Htz1 and Set3.
SLX5 is a dosage inhibitor of a subset of suppressors:
We also isolated 13 recessive suppressor mutations following random mutagenesis with EMS. We attempted to clone the corresponding wild-type alleles by complementation using a wild-type yeast genomic plasmid library and screening for restoration of the nonsectoring color colony phenotype. Out of ∼60,000 transformants of one candidate strain, we repeatedly isolated two different CEN plasmids, both containing SLX5. Surprisingly, however, DNA sequence analysis showed that the the candidate suppressor strain did not contain mutations in either the ORF or the promoter region of chromosomal SLX5. To confirm this unexpected result, we cloned the chromosomal copy of SLX5 from the suppressor strain into a CEN yeast shuttle vector (pRS315), transformed the candidate suppressor strain with this plasmid, and found that it also restored the nonsectoring phenotype. Thus, while the suppressor mutation in the candidate strain remains unidentified, these results did reveal that an extra gene dose of SLX5 inhibits suppression. Transformation of htz1Δ set3Δ double mutants with a CEN plasmid copy of wild-type SLX5 also eliminates the marginal viability of the strain. Interestingly, SLX8 has a similar, although milder, effect (data not shown).
Slx5 and Slx8 comprise a SUMOylation targeted ubiquitin ligase (STUbL) complex, a functionally conserved entity from fission yeast to human cells. Slx5 binds to the SUMOylated subunit and Slx8 acts as the E3 ligase (Burgess et al. 2007; Li et al. 2007b; Mullen and Brill 2008). This heterodimer complex has been proposed to participate in DNA repair (Zhang et al. 2006; Burgess et al. 2007; Li et al. 2007a; Nagai et al. 2008), telomere silencing (Darst et al. 2008), and protein quality control (Wang and Prelich 2009). No endogenous SUMOylated substrates of the Slx5–Slx8 complex have been identified although two recent reports identified mating-type switching regulators alpha2 (Xie et al. 2010) and alpha1 (Nixon et al. 2010) to be un-SUMOylated endogenous substrates.
Phenotypic classification of htz1Δ set3Δ suppressors:
We took advantage of the serendipitous discovery that SLX5 could eliminate the activity of at least one suppressor and tested the effect of a pMH02[SLX5 CEN TRP1] plasmid on our set of known transposon-mediated suppressor mutations. Deletion of genes encoding the SWR1 complex and the H2B deubiquitination module of SAGA complex were no longer able to bypass htz1Δ set3Δ in the presence of extra copies of SLX5. However, the other gene deletions retained the ability to suppress (Figure 1, B and C).
The isolation of sir3 as a suppressor of htz1Δ set3Δ indicates that this silent information regulator becomes toxic in the absence of Htz1 and Set3, and the direct removal of this genetic toxin is sufficient to rescue the double mutant. Set1-mediated H3K4 methylation contributes to preventing Sir3 from spreading into euchromatin and set1Δ causes dilution of the limited pool of cellular Sir3 from its physiological targets (Venkatasubrahmanyam et al. 2007). We hypothesized that the suppressor activity of set1Δ might be an indirect consequence of mimicking Sir3 depletion and predicted that its suppressor activity would be reduced by overexpressing SIR3. Consistent with this hypothesis, suppression by set1Δ was impaired when SIR3 was overexpressed. Surprisingly, however, the suppression phenotypes exhibited by rpn10Δ, pre9Δ, and ubp14Δ were also notably reduced in the presence of the SIR3 2μ plasmid (Figure 1, B and D). Thus, on the basis of their distinct responses to overexpression of either SLX5 or SIR3, the bypass suppressors can be categorized into three phenotypic classes (Table 3).
TABLE 3.
Categorization of loss of function suppressors into three classes on the basis of their responses to overexpression of SLX5 or SIR3
| Suppression in the presence of |
||
|---|---|---|
| Suppressors | SLX5(CEN) | SIR3(2μ) |
| Hda1 complex | Yes | Yes |
| SWR1 complex | No | Yes |
| Ubp8–Sgf11 | No | Yes |
| Set1 | Yes | Reduced |
| Sir3 | Yes | Reduced |
| Proteolytic factors | Yes | Reduced |
Telomere silencing requires the protein degradation pathway:
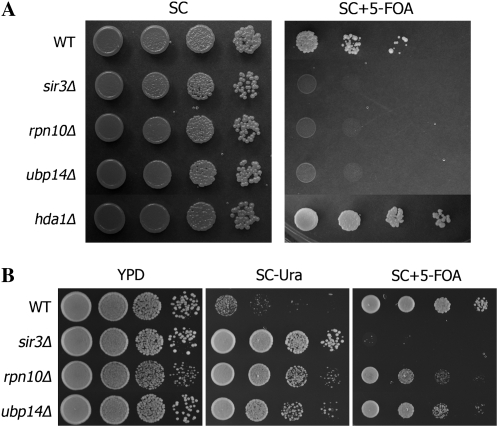
The responses of rpn10Δ, pre9Δ, and ubp14Δ to SIR3 overexpression suggested that the protein degradation pathway might have a previously unrecognized role in Sir3-dependent silencing. To test this, subtelomeric position effect variegation (PEV) was examined in these mutants. PEV was measured by monitoring the 5-FOA sensitivity of a strain in which URA3 is integrated ∼2.1 kb away from the right end of chromosome V (Rundlett et al. 1996). URA3 is randomly silenced in wild-type clonal cells (Figure 2A). Deletion of SIR3 substantially removes the clonal repression as expected. Interestingly, ubp14Δ and rpn10Δ both show strong URA3 derepression at a level comparable to sir3Δ (Figure 2A). Deletion of HDA1, whose suppression activity does not respond to SIR3 overexpression, increases subtelomeric silencing, consistent with previous results (Rundlett et al. 1996).
Figure 2.—
Sir3-dependent telomere silencing requires the protein degradation pathway. (A) The indicated strains were constructed in a reporter strain in which the URA3 gene is inserted approximately 2.1 kb away from the left end of the chromosome VII (Rundlett et al. 1996). Serial dilutions were plated on SC and SC + 5-FOA plates. (B) The indicated strains were constructed in a reporter strain in which the URA3 gene is placed at the very end of the chromosome V (kahana and gottschling 1999). Serial dilutions were plated on YPD, SC–Ura, and SC + 5-FOA plates.
To further confirm this novel relationship between SIR3 and protein degradation, we used another reporter strain in which the URA3 reporter gene is integrated at the left tip of chromosome VII (Kahana and Gottschling 1999). This strain is barely viable on plates lacking uracil because URA3 is strictly silenced. In this strain sir3Δ dramatically improves growth on Ura− plates as expected because of the derepression of URA3. Consistent with our PEV results at the telomere of chromosome V, rpn10Δ and ubp14Δ also increase URA3 expression (Figure 2B) although the derepression in rpn10Δ and ubp14Δ is at a lower level compared to sir3Δ, as indicated by the growth on 5-FOA plates.
We investigated whether the silencing defects of the protein degradation mutants are associated with a decrease of Sir3 at the telomere region. We introduced rpn10Δ and ubp14Δ into a strain carrying an allele of SIR3 encoding a triple-HA epitope tag at the C terminus of Sir3. Sir3–3HA binding at the right end of chromosome V in these deletion mutants was then compared to wild type using chromatin immunoprecipitation (ChIP). No differences between these deletion mutants and the wild-type strain were observed (data not shown), suggesting that in addition to Sir3 binding, one or more additional steps involving the protein degradation pathway are required for efficient telomere silencing.
Suppression of htz1Δ and set3Δ single defects:
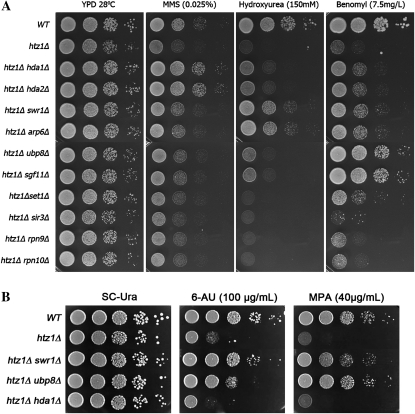
The htz1Δ set3Δ double mutant encounters genetic stress from each of the gene deletions individually as well as the synthetic effect of the double deletion. Thus, in principle, we might expect at least three classes of suppressors: those that bypass one or the other of the single deletions and those that bypass the synthetic defects. Indeed, hda1Δ has been reported to rescue the growth phenotype of htz1Δ and reduce its sensitivity to hydroxyurea (HU), methyl-methanesulfonate (MMS), and the microtubule-interfering drug benomyl (Lin et al. 2008). Therefore, we investigated the effect of the other htz1Δ set3Δ suppressors on htz1Δ single-mutant defects. A series of double mutants containing htz1Δ and each of the suppressor genes were constructed in the BY4741 strain background. Their growth conditions on YPD and in the presence of various drugs were compared to wild-type and htz1Δ cells. Each functional module was represented by two members to monitor the phenotypic consistency within the group.
In agreement with the observations of Lin et al. (2008), disruption of the Hda1 complex weakly suppresses the sensitivities of htz1Δ to HU and benomyl and strongly suppresses its MMS sensitivity (Figure 3A). Suppressors within the SWR1 complex genes also improve the growth of htz1Δ in the presence of these drugs, but with a more prominent effect on HU sensitivity (Figure 3A). Deletions of genes encoding the SAGA H2B deubiquitination module strongly rescue benomyl sensitivity but only display subtle effects on the sensitivities to the DNA-damaging reagents (Figure 3A). In the case of 6-azauracil (6-AU) and mycophenolic acid (MPA), two drugs that affect transcription elongation by reducing the cellular pool of ribonucleotides, both swr1Δ and ubp8Δ but not hda2Δ, reduce the sensitivity of htz1Δ (Figure 3B).
Figure 3.—
Suppression of htz1Δ drug phenotypes by gene deletions. (A) Serial dilutions were plated on YPD, YPD + MMS (0.025%), YPD + hydroxyurea (150 mm), and YPD + benomyl (7.5 μg/ml). (B) The indicated strains were transformed with the control plasmid pRS316 [URA3 CEN] and serial dilutions were plated on SC–Ura, SC–Ura + 6-AU (100 μg/ml) and SC–Ura + MPA (40 μg/ml).
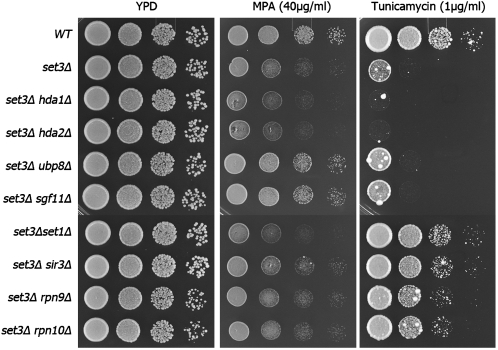
We carried out a similar set of tests for set3Δ. Fewer phenotypes are known for set3; however, the deletion strain is hypersensitive to MPA (Kim and Buratowski 2009) and tunicamycin (Cohen et al. 2008), which induces secretory stress response. We again constructed a set of double mutants containing set3Δ and each of the suppressor genes and assayed their growth in the presence of MPA and tunicamycin. The set3Δ swr1Δ and set3Δ arp6Δ double mutants could not be made due to synthetic lethality between set3Δ and loss of SWR1 complex function in the background of BY4741. As expected, MPA markedly inhibited the growth of set3Δ. Among the tested suppressors, only those in the H2B deubiquitination module show an ability to rescue the drug sensitivities (Figure 4). Interestingly, set1Δ, sir3Δ, and the deletion of genes encoding proteasome subunits all rescue the tunicamycin sensitivity of set3Δ cells (Figure 4), providing additional evidence for the functional relationship between Sir3 and the protein degradation pathway.
Figure 4.—
Suppression of set3Δ drug phenotypes by gene deletions. Serial dilutions were plated on YPD, YPD + MPA (40 μg/ml), and YPD + tunicamycin (1 μg/ml).
Bypass suppressors ubp8Δ and sgf11Δ can act at the level of Htz1- and Set3-dependent gene expression:
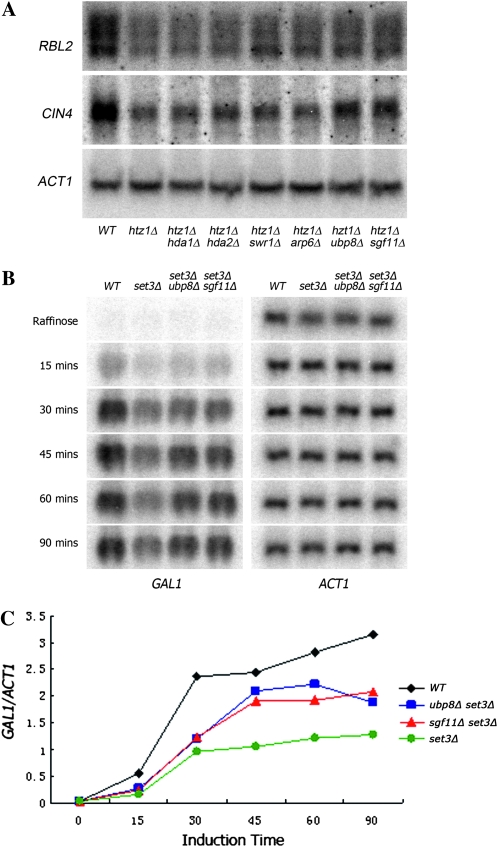
We next sought to determine if any of the htz1Δ set3Δ bypass suppressors might be acting at the level of Htz1- and Set3-dependent gene transcription. The hypersensitivity of htz1Δ to benomyl is due, in part, to reduced transcription of RBL2 and CIN4, two genes encoding factors required for β-tubulin folding (A. Allison, M. Santisteban, and M.M.S., unpublished results). Since suppressors in genes encoding the Hda1 complex, SWR1 complex, and H2B deubiquitination subcomplex suppress the benomyl sensitivity of htz1Δ, we assayed the levels of RBL2 and CIN4 mRNAs in these suppressor strains. As expected, the mRNA levels of RBL2 and CIN4 in htz1Δ are significantly lower than in wild-type cells (Figure 5A). In the case of the tested suppressors, the pattern of mRNA expression matched that of the benomyl sensitivity (Figure 3A). The most potent suppressors of htz1Δ benomyl sensitivity are ubp8 and sgf11. Similarly, CIN4 transcription is markedly increased in ubp8 and sgf11 (Figure 5A). Lesser effects are observed for hda1Δ, hda2Δ, swr1Δ, and arp6Δ.
Figure 5.—
Suppression of htz1Δ and set3Δ transcriptional phenotypes by ubp8Δ and sgf11Δ. (A) Total RNA (20 μg) isolated from each strain was subjected to Northern analysis. The membrane was blotted sequentially with probes that detect RBL2, CIN4, and ACT1. The membrane was stripped with boiling 0.1% SDS between each probing. (B) Northern analysis of 20 μg of total RNA from each strain is shown following galactose addition. The membrane was assayed sequentially with probes detecting GAL1 and ACT1. (C) The ratios of GAL1 mRNA to ACT1 mRNA are shown across the course of induction of each strain.
The induction of GAL1 transcription is known to be impaired in set3Δ (Wang et al. 2002; Kim and Buratowski 2009). Therefore, since ubp8Δ and sgf11Δ are strong suppressors of the MPA phenotype of set3Δ, we compared the induction of GAL1 transcription in wild type, set3Δ, set3Δ ubp8Δ, and set3Δ sgf11Δ. Consistent with the previous reports, induction of GAL1 transcription is notably slower in set3Δ cells. Interestingly, GAL1 transcription is partially restored in set3Δ ubp8Δ and set3Δ sgf11Δ (Figure 5B). Together, these results define examples in which bypass suppressors ubp8Δ and sgf11Δ affect Htz1- and Set3-dependent gene transcription.
Suppression of the global set of htz1Δ and set3Δ negative genetic interactions:
All of the suppressors isolated in our genetic screen are general factors with broad roles in gene expression, rather than regulators of a specific gene or pathway. Consequently, we reasoned that this set of suppressors might be rather general and bypass most synthetic defects involving htz1Δ or set3Δ. To test this prediction, we carried out a modified synthetic genetic array (SGA) analysis to assess the genome-wide suppression of htz1Δ and set3Δ negative genetics interactions (Figure 6). First, for each of the htz1Δ set3Δ bypass suppressor genes, we created a pair of query strains carrying both the suppressor deletion and either htz1Δ or set3Δ. These double-mutant query strains were then crossed against specialized miniarrays that included strains with gene deletions that have negative genetic interactions with either htz1Δ or set3Δ (materials and methods). Control strains that show no genetic interactions with htz1Δ and set3Δ were also included in the arrays to normalize colonies sizes across replicate plates. After mating with the query strain and sporulation, the triple mutants were selected and their relative colony sizes were quantified.
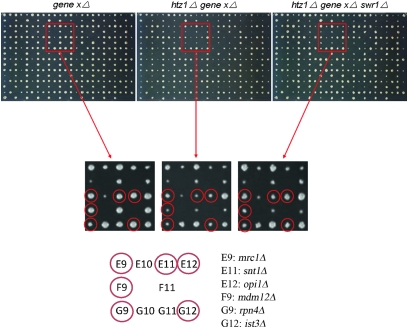
Figure 6.—
SGA miniarray analysis. Three final ordered arrays from one set of the suppressor SGA experiments are shown. Crosses incorporating swr1Δ are used in this example, but similar assays were carried out for all of the suppressors tested. After colony sizes were first normalized across the plates using a set of control strains, synthetic negative gene interactions were scored by comparing plate A to plate B (materials and methods). Bypass suppression was then scored as an increase in colony size of at least 33% comparing plate C to plate B. Reductions or increases in colony sizes can be visualized for the selected deletions showing interactions with htz1Δ. The enlarged areas show examples of suppression with the genes and their locations in the miniarray indicated. Note that snt1 (E11) is one of the core subunits of the Set3 HDAC complex.
The htz1Δ-based miniarray was composed of 241 strains with gene deletions known or suspected to produce negative genetic interactions. In control screens where bypass suppressors were not included, 164 gene deletions in the miniarray scored as having negative genetic interactions with htz1Δ. We next analyzed the ability of hda2Δ, hda3Δ, swr1Δ, vps72Δ, ubp8Δ, sgf11Δ, pre9Δ, and rpn10Δ to suppress these negative synthetic genetic interactions. Contrary to expectations, each suppressor only bypassed a limited number of interactions (Table 4 and Table S3). Furthermore, the htz1Δ set3Δ suppressors, as a group, did not function coordinately to suppress other htz1Δ synthetic interactions. Only 14 of the 164 negative genetic interactions were bypassed by at least 7 of the suppressors, and growth of the average double mutant was improved by less than 3 of the htz1Δ set3Δ suppressors.
TABLE 4.
Suppression of genome-wide interactions of htz1 and set3
| Number suppresseda |
||
|---|---|---|
| Suppressors | htz1 (164) | set3 (25) |
| hda2 | 53 | 8 |
| hda3 | 62 | 7 |
| pre9 | 32 | 16 |
| rpn10 | 40 | 7 |
| sgf11 | 67 | 17 |
| ubp8 | 58 | 20 |
| swr1 | 55 | N/Ab |
| vps72 | 57 | N/Ab |
The total numbers of scored interactions are shown in parentheses.
swr1 and vps72 are synthetic lethal in combination with set3 in the background of BY4741.
The set3Δ miniarray was composed of 53 gene deletions. Of these, 25 deletions were scored as having a synthetic negative growth phenotype in combination with set3Δ. Since swr1Δ and vps72Δ are themselves synthetic lethal with set3Δ in the SGA strain background, we could create only double-mutant query strains with hda2Δ, hda3Δ, ubp8Δ, sgf11Δ, pre9Δ, and rpn10Δ. As with the htz1Δ analysis, most of the 25 synthetic negative interactions with set3Δ were suppressed by relatively few of the query genes; only 7 of the 25 interactions were suppressed by more than 4 of the 6 suppressors. However, unlike the htz1Δ case, the sgf11Δ, and ubp8Δ suppressors in the H2B ubiquitination module stand out as interacting with a majority of the scored set3Δ interactions (Table 4 and Table S4). Taken together, these results argue that htz1Δ set3Δ has defects in chromatin function that are relatively restricted to that double mutant. These defects are bypassed by a set of suppressors that is also an uncommon constellation of genes suggesting a special functional relationship among htz1Δ, set3Δ, and their bypass suppressors.
High-copy suppressors of htz1Δ set3Δ genetic interaction:
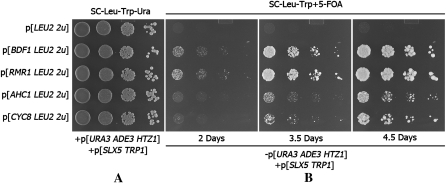
To gain further insights into the genetic interaction between htz1Δ and set3Δ, we performed a genome-wide screen to isolate high-copy suppressors of the htz1Δ set3Δ synthetic growth phenotype using a 2μ-plasmid-based systematic genomic library (Jones et al. 2008). Each clone of this library contains a sequenced fragment of the yeast chromosome and together they cover the complete S. cerevisiae genome. The library plasmids were used to transform MSY4544, which is MSY4474 containing a low-copy SLX5 plasmid to enhance sensitivity. Leu+ transformants were screened for color sectored colonies (materials and methods) and five library plasmids showing strong suppression of htz1Δ set3Δ were recovered (data not shown). The promoter and ORF of each individual gene on these library plasmids were subcloned into a 2μ plasmid shuttle vector and tested individually in MSY4544. These assays identified the following high-copy suppressors (and the number of times they were isolated): BDF1 (6), AHC1 (2), RMR1 (3), and CYC8 (2). Suppression was independent of low-copy SLX5 overexpression (Figure 7). The suppression of a fifth library plasmid could not be assigned to any single gene on the plasmid. Interestingly, this plasmid contains a copy of the HMR locus, which could theoretically serve as a titrator of Sir3 protein and mimic the suppression by sir3Δ. Since BDF1 provided strong suppression, we also tested overexpression of BDF2, but it failed to confer suppression of the htz1Δ set3Δ synthetic phenotype (data not shown).
Figure 7.—
(A) An htz1Δ set3Δ strain carrying pMSS59[HTZ1 ADE3 URA3] and pMH02[SLX5 TRP1] was transformed with the indicated plasmids. The transformants were cultured in SC–Ura–Leu–Trp media and serial dilutions were plated on SC–Ura–Leu–Trp plates to maintain the three plasmids. (B) Strains from A were then grown in SC–Leu–Trp media overnight to permit loss of pMSS59. Serial dilutions of the overnight cultures were plated on SC–Leu–Trp + 5-FOA plates. Growth at different times is shown.
Suppression of htz1Δ and set3Δ single defects by high-copy suppressors:
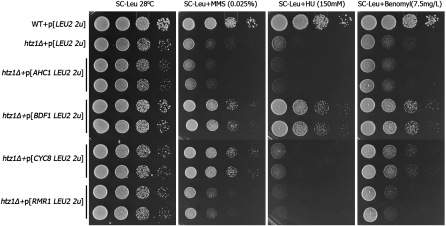
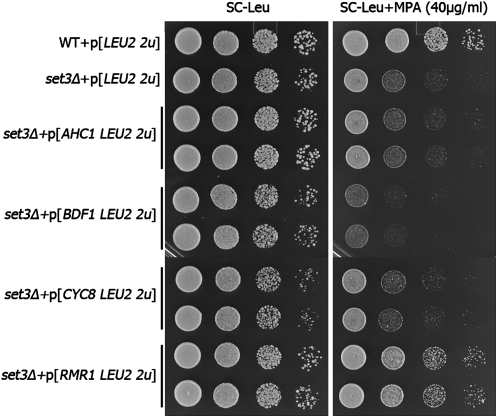
Loss-of-function suppressors differentially alleviate the single defects of htz1Δ or set3Δ cells and we attempted to characterize the high-copy suppressors using the same approach. Derivatives of the 2μ plasmid pRS425 containing BDF1, AHC1, RMR1 and CYC8 were introduced into htz1Δ cells and pRS425 empty vector was transformed as control. We then compared the sensitivities of these transformants to MMS, HU, and benomyl (Figure 8). CYC8 overexpression notably suppresses the MMS sensitivity of htz1Δ but it has very subtle, if any, effects on the HU and benomyl sensitivities. This profile is similar to that observed earlier for deletions of genes encoding the Hda1 complex. Interestingly, Cyc8 forms a transcriptional corepressor complex with Tup1 and together they recruit the Hda1 complex to its targets (Wu et al. 2001). Thus, CYC8 overexpression could exert its dominant suppression by sequestering the Hda1 complex away from its physiological targets mimicking the disruption of the complex by the Hda1 complex gene deletions. Overexpression of BDF1 exhibits a wide spectrum suppression of all of the htz1Δ phenotypes tested, while overexpression of AHC1 and RMR1 does not suppress any (Figure 8). In the case of set3Δ, the overexpression of RMR1 markedly improves growth in the presence of MPA (Figure 9), suggesting a regulatory role of Rmr1 in transcription. We also observed that BDF1 overexpression exacerbates the set3Δ MPA sensitivity.
Figure 8.—
Suppression of htz1Δ drug sensitivities by high-copy suppressors. Serial dilutions of the cell cultures of htz1Δ cells containing the indicated plasmids were plated on SC–Leu, SC–Leu + MMS (0.025%), SC–Leu + hydroxyurea (150 mm) and SC–Leu + benomyl (7.5 μg/ml).
Figure 9.—
Suppression of set3Δ drug sensitivities by high-copy suppressors. Serial dilutions of set3Δ cells containing the indicated plasmids were plated on SC–Leu, SC–Leu + MPA (40 μg/ml).
DISCUSSION
The results reported here reveal a surprisingly close functional relationship between Htz1 and Set3 and suggest a model for Set3 activity. Synthetic growth defects for the binary combinations of both htz1Δ set3Δ and swr1Δ set3Δ are well known (Krogan et al. 2003; Collins et al. 2007; Wilmes et al. 2008; Costanzo et al. 2010). Furthermore, Swr1 is detrimental to cells in the absence of its normal Htz1–H2B substrate (Halley et al. 2010; Morillo-Huesca et al. 2010). What is surprising, then, is the striking finding that htz1Δ swr1Δ set3Δ triple mutants grow relatively well. This implies that not only is Swr1 severely detrimental in the absence of Htz1 and Set3, but Htz1 is also severely detrimental in the absence of Swr1 and Set3, a previously unrecognized activity of Htz1. Furthermore, the partial redundancy of these three genes is likely to be in a specific pathway of chromatin function since we find that the particular combination of suppressor genes that can bypass htz1Δ set3Δ is uncommon among the global set of htz1Δ synthetic interactions. This interlocking genetic relationship between HTZ1, SWR1, and SET3 argues that Set3 may participate in the dynamic deposition and remodeling of Htz1-containing nucleosomes. In the absence of Htz1, we propose that the Set3 HDAC complex can partially overcome the effect of SWR1 complex either by decreasing its recruitment or modifying its target nucleosomes. In the absence of Swr1, we reason that Set3 activity may partially overcome either the negative effects of Htz1 nucleosomes no longer remodeled by the SWR1 complex or the stochastic misincorporation of Htz1 into abnormal sites of deposition.
The phenotypic characterization of our suppressor collection also revealed a surprising functional similarity between the SWR1 complex and the SAGA H2B deubiquitination subcomplex with respect to htz1Δ set3Δ. The simplest interpretation of these results is that the activities of these two complexes are partially redundant for achieving dynamic regulation of chromatin by Htz1. In the context of our experiments, the toxic effects of Swr1 in the absence of Htz1 might be relieved by the increased H2B ubiquitination in ubp8Δ or sgf11Δ mutants. This hypothesis is supported by the observation that H2B ubiquitination increases nucleosome stability over the promoters of repressed genes (Chandrasekharan et al. 2009). We carried out initial tests of this model by examining the steady-state levels of Htz1 at the promoters of repressed GAL1 and PHO5 by chromatin immunoprecipitation in UBP8 and ubp8Δ cells. We also examined the rate of Htz1 deposition at the PHO5 promoter following repression. Neither of these experiments revealed any effect of ubp8Δ on Htz1 occupancy (data not shown). However, both studies were carried out in the context of HTZ1 and SWR1 and it remains possible that ubp8Δ and sgf11Δ can relieve the detrimental Swr1-dependent remodeling of nucleosomes in the absence of Htz1. Alternatively, the Swr1 complex and the Ubp8–Sgf11 H2B deubiquitination subcomplex might function sequentially in a common pathway, although the downstream effect of this hypothetical pathway is unlikely to be Htz1 deposition.
Our suppressor analysis also revealed a second functional similarity between Sir3 and the proteasome protein degradation pathway. Overexpression of SIR3 not only reduced htz1Δ set3Δ suppression by set1, but also antagonized suppression by rpn10Δ, pre9Δ, and ubp14Δ. Furthermore, we also found that telomere silencing is impaired in rpn10Δ, pre9Δ, and ubp14Δ, just as it is in sir3Δ. A number of links between protein degradation and silencing are known. Physical interactions have been reported between the ubiquitin-specific protease Ubp3 and Sir4, and increased telomere silencing was observed in ubp3Δ (Moazed and Johnson 1996). Another ubiquitin protease, Ubp6, and the proteasome subunit Sem1 are also involved in telomere silencing. Silencing is reduced in sem1Δ, suggesting a positive role in silencing, while defective silencing in ubp10Δ is rescued by ubp6Δ, suggesting a negative role for Ubp6 (Qin et al. 2009). Our results suggest additional functional overlaps between the proteasome, ubiquitin recycling, and Sir3-dependent silencing.
It was curious that the deletion of genes encoding the HDA1 deacetylase complex would bypass defects involving the deletion set3Δ, which is a subunit of a deacetylase complex. However, a similar antagonism between the HDA1 and HOS2 deacetylases has recently been observed in Candida albicans (Zacchi et al. 2010). HDA1 specifically deacetylates H2B and H3 and, in its absence, intergenic promoter regions and so-called HDA1-affected subtelomeric regions (HAST) become hyperacetylated (Robyr et al. 2002). Interestingly, HDA1 is a downstream effector of the Tup1–Cyc8 repressor complex, establishing transcriptionally repressive chromatin at promoter regions (Wu et al. 2001; Green and Johnson 2004). Furthermore, Tup1 likely participates in specifying some of the sites of Htz1 deposition by SWR1 (Gligoris et al. 2007). Although we did not recover tup1Δ in our suppressor screens, the fact that CYC8 is a high-copy suppressor suggests that reducing HDA1 activity at its Tup1-independent targets might contribute to the suppression of htz1Δ set3Δ.
The functional involvement of Slx5–Slx8 in the Htz1 and Set3 network is provocative. SLX5 was previously recovered in a systematic study of overexpression toxicity (Sopko et al. 2006). Interestingly, SLX5 overexpression specifically blocks the ability of deletions in genes encoding SWR1 and the SAGA H2B deubiquitination subcomplex to suppress the growth phenotype of htz1Δ set3Δ. We reason that overexpression of SLX5 increases the polyubiquitination and degradation of one or more target proteins critical for growth in the suppression genotypes. Given the DNA damage sensitivity of htz1Δ, and the roles of H2B ubiquitination in repair (Lis and Romesberg 2006; Game and Chernikova 2009), one potential pathway for these targets is DNA damage repair. Nagai et al. (2008) have proposed that Slx5–Slx8 is required to target degradation of an unknown SUMOylated factor that must be removed for repair to proceed. Thus, excess degradation could be lethal in htz1Δ set3Δ regardless of the altered chromatin structure provided by swr1Δ or ubp8Δ. Transcription initiation is a second candidate pathway. Temperature-sensitive mot1-301, which cannot efficiently dissociate TATA-binding protein from the promoter DNA, is suppressed by slx5Δ (Wang et al. 2006). This model suggests that Slx5–Slx8 polyubiquitinates an unknown SUMOylated protein that enhances transcription initiation and that the increased concentration of this factor in the slx5Δ can suppress the mot1-301 defect. Because SLX5 overexpression diminishes the beneficial effects of disrupting either SWR1 complex or Ubp8–Sgf11 in htz1Δ set3Δ and because both Htz1-containing nucleosomes and Set3 HDAC modified nucleosomes are promoter proximal (Albert et al. 2007; Kim and Buratowski 2009), the transcription initiation pathway is an attractive model for the targets of Slx5–Slx8 in our network.
Both models predict that overexpression of genes encoding Slx5–Slx8 substrates might relieve the negative effect of increased SLX5 dosage and that the proteins expressed should be SUMOylated. Both Bdf1 and Rmr1 have been isolated in proteomics screens for SUMOylated proteins (Wohlschlegel et al. 2004; Hannich et al. 2005; Yu et al. 2008). As a component of SWR1, increased Slx5–Slx8 targeted degradation of Bdf1 might further impair transcription in htz1Δ set3Δ. Overexpression of BDF1 would compensate for this increased degradation. Furthermore, high-copy BDF1 might confer additional suppression by diluting SWR1 complex and mimicking suppression by swr1Δ. However, the mechanism is likely to be more complicated since the suppression of htz1Δ phenotypes by BDF1 is more extensive than that achieved by swr1Δ. Indeed, Bdf1 is more widely distributed across chromosomes than would be expected on the basis of its association with TFIID and SWR1 (Chua and Roeder 1995; Matangkasombut and Buratowski 2003). In the case of RMR1, relatively little is known about its functions, although our results suggest a potential role in transcription. Interestingly, RMR1 is a weak high-copy suppressor of the null allele of ulp2, one of two essential SUMO proteases in budding yeast (Hannich et al. 2005).
Expanding the binary synthetic htz1Δ set3Δ interaction to a network of ternary gene interactions has revealed strong functional links among a small set of protein complexes with general regulatory functions. Importantly, these results suggest an important new role for the Set3 HDAC complex in Htz1–nucleosome dynamics. Further genetic, biochemical, and molecular studies will help to dissect these interactions at a mechanistic level, providing new insights into the epigenetic regulation of chromatin function.
Acknowledgments
This work was supported by a Robert R. Wagner Fellowship to M.H. and grants GM28920 and GM60444 from the National Institutes of Health to M.M.S.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.125419/DC1.
References
- Adams, A., C. Kaiser and C. S. H. Laboratory, 1998. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Albert, I., T. N. Mavrich, L. P. Tomsho, J. Qi, S. J. Zanton et al., 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446 572–576. [DOI] [PubMed] [Google Scholar]
- Altaf, M., A. Auger, J. Monnet-Saksouk, J. Brodeur, S. Piquet et al., 2010. NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J. Biol. Chem. 285 15966–15977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones et al., 2007. High-resolution profiling of histone methylations in the human genome. Cell 129 823–837. [DOI] [PubMed] [Google Scholar]
- Bender, A., and J. R. Pringle, 1991. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell Biol. 11 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, R. C., S. Rahman, M. Lisby, R. Rothstein and X. Zhao, 2007. The Slx5–Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol. Cell Biol. 27 6153–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan, M. B., F. Huang and Z. W. Sun, 2009. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc. Natl. Acad. Sci. USA 106 16686–16691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, P., and G. S. Roeder, 1995. Bdf1, a yeast chromosomal protein required for sporulation. Mol. Cell Biol. 15 3685–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, T. J., M. J. Mallory, R. Strich and T. P. Yao, 2008. Hos2p/Set3p deacetylase complex signals secretory stress through the mpk1p cell integrity pathway. Eukaryot. Cell 7 1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, S. R., K. M. Miller, N. L. Maas, A. Roguev, J. Fillingham et al., 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446 806–810. [DOI] [PubMed] [Google Scholar]
- Costanzo, M., A. Baryshnikova, J. Bellay, Y. Kim, E. D. Spear et al., 2010. The genetic landscape of a cell. Science 327 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton, M. P., S. Markoulaki, S. S. Levine, J. Hanna, M. A. Lodato et al., 2008. H2AZ is enriched at polycomb complex target genes in es cells and is necessary for lineage commitment. Cell 135 649–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, K., C. Zang, T. Y. Roh, D. E. Schones, R. W. Childs et al., 2009. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell. Stem Cell 4 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darst, R. P., S. N. Garcia, M. R. Koch and L. Pillus, 2008. Slx5 promotes transcriptional silencing and is required for robust growth in the absence of Sir2. Mol. Cell Biol. 28 1361–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon, N., M. Oki, S. J. Szyjka, O. M. Aparicio and R. T. Kamakaka, 2006. H2A.Z functions to regulate progression through the cell cycle. Mol. Cell Biol. 26 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler, D., H. Braberg, M. Mehta, G. Chechik, G. Cagney et al., 2009. Functional organization of the S. cerevisiae phosphorylation network. Cell 136 952–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game, J. C., and S. B. Chernikova, 2009. The role of RAD6 in recombinational repair, checkpoints and meiosis via histone modification. DNA Repair 8 470–482. [DOI] [PubMed] [Google Scholar]
- Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles et al., 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418 387–391. [DOI] [PubMed] [Google Scholar]
- Gligoris, T., G. Thireos and D. Tzamarias, 2007. The Tup1 corepressor directs Htz1 deposition at a specific promoter nucleosome marking the GAL1 gene for rapid activation. Mol. Cell Biol. 27 4198–4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind, C. K., H. Qiu, D. S. Ginsburg, C. Ruan, K. Hofmeyer et al., 2010. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol. Cell 39 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, S. R., and A. D. Johnson, 2004. Promoter-dependent roles for the Srb10 cyclin-dependent kinase and the Hda1 deacetylase in Tup1-mediated repression in Saccharomyces cerevisiae. Mol. Biol. Cell 15 4191–4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther, M. G., W. S. Lane, W. Fischle, E. Verdin, M. A. Lazar et al., 2000. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 14 1048–1057. [PMC free article] [PubMed] [Google Scholar]
- Guillemette, B., A. R. Bataille, N. Gevry, M. Adam, M. Blanchette et al., 2005. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3 e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halley, J. E., T. Kaplan, A. Y. Wang, M. S. Kobor and J. Rine, 2010. Roles for H2A.Z and its acetylation in GAL1 transcription and gene induction, but not GAL1-transcriptional memory. PLoS Biol. 8 e1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannich, J. T., A. Lewis, M. B. Kroetz, S. J. Li, H. Heide et al., 2005. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 280 4102–4110. [DOI] [PubMed] [Google Scholar]
- John, S., P. J. Sabo, T. A. Johnson, M. H. Sung, S. C. Biddie et al., 2008. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol. Cell 29 611–624. [DOI] [PubMed] [Google Scholar]
- Jones, G. M., J. Stalker, S. Humphray, A. West, T. Cox et al., 2008. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nat. Methods 5 239–241. [DOI] [PubMed] [Google Scholar]
- Kahana, A., and D. E. Gottschling, 1999. DOT4 links silencing and cell growth in Saccharomyces cerevisiae. Mol. Cell Biol. 19 6608–6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T., and S. Buratowski, 2009. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor, M. S., S. Venkatasubrahmanyam, M. D. Meneghini, J. W. Gin, J. L. Jennings et al., 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2 E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan et al., 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12 1565–1576. [DOI] [PubMed] [Google Scholar]
- Li, B., S. G. Pattenden, D. Lee, J. Gutierrez, J. Chen et al., 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. USA 102 18385–18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., J. Wang, Z. Nawaz, J. M. Liu, J. Qin et al., 2000. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 19 4342–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T., J. Fung, J. R. Mullen and S. J. Brill, 2007. a The yeast Slx5-Slx8 DNA integrity complex displays ubiquitin ligase activity. Cell Cycle 6 2800–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T., J. R. Mullen, C. E. Slagle and S. J. Brill, 2007. b Stimulation of in vitro sumoylation by Slx5-Slx8: evidence for a functional interaction with the SUMO pathway. DNA Repair 6 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y. Y., Y. Qi, J. Y. Lu, X. Pan, D. S. Yuan et al., 2008. A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev. 22 2062–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis, E. T., and F. E. Romesberg, 2006. Role of Doa1 in the Saccharomyces cerevisiae DNA damage response. Mol. Cell Biol. 26 4122–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matangkasombut, O., and S. Buratowski, 2003. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol. Cell 11 353–363. [DOI] [PubMed] [Google Scholar]
- Mavrich, T. N., C. Jiang, I. P. Ioshikhes, X. Li, B. J. Venters et al., 2008. Nucleosome organization in the Drosophila genome. Nature 453 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, N. J., and B. W. O'Malley, 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108 465–474. [DOI] [PubMed] [Google Scholar]
- Meneghini, M. D., M. Wu and H. D. Madhani, 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112 725–736. [DOI] [PubMed] [Google Scholar]
- Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen et al., 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303 343–348. [DOI] [PubMed] [Google Scholar]
- Moazed, D., and D. Johnson, 1996. A deubiquitinating enzyme interacts with Sir4 and regulates silencing in S. cerevisiae. Cell 86 667–677. [DOI] [PubMed] [Google Scholar]
- Morillo-Huesca, M., M. Clemente-Ruiz, E. Andujar and F. Prado, 2010. The SWR1 histone replacement complex causes genetic instability and genome-wide transcription misregulation in the absence of H2A. Z. PLoS One 5 e12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen, J. R., and S. J. Brill, 2008. Activation of the Slx5-Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates. J. Biol. Chem. 283 19912–19921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai, S., K. Dubrana, M. Tsai-Pflugfelder, M. B. Davidson, T. M. Roberts et al., 2008. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon, C. E., A. J. Wilcox and J. D. Laney, 2010. Degradation of the Saccharomyces cerevisiae mating-type regulator alpha1: genetic dissection of cis-determinants and trans-acting pathways. Genetics 185 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi, S. L., D. D. Shoemaker and J. D. Boeke, 2003. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat. Genet. 35 277–286. [DOI] [PubMed] [Google Scholar]
- Pan, X., P. Ye, D. S. Yuan, X. Wang, J. S. Bader et al., 2006. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124 1069–1081. [DOI] [PubMed] [Google Scholar]
- Pijnappel, W. W., D. Schaft, A. Roguev, A. Shevchenko, H. Tekotte et al., 2001. The S. cerevisiae Set3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 15 2991–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, S., Q. Wang, A. Ray, G. Wani, Q. Zhao et al., 2009. Sem1p and Ubp6p orchestrate telomeric silencing by modulating histone H2B ubiquitination and H3 acetylation. Nucleic Acids Res. 37 1843–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner, R. M., P. D. Hartley, M. D. Meneghini, M. Z. Bao, C. L. Liu et al., 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123 233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangasamy, D., I. Greaves and D. J. Tremethick, 2004. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat. Struct. Mol. Biol. 11 650–655. [DOI] [PubMed] [Google Scholar]
- Robyr, D., Y. Suka, I. Xenarios, S. K. Kurdistani, A. Wang et al., 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109 437–446. [DOI] [PubMed] [Google Scholar]
- Ross-MacDonald, P., A. Sheehan, C. Friddle, G. S. Roeder and M. Snyder, 1999. Transposon mutagenesis for the analysis of protein production, function, and localization. Methods Enzymol. 303 512–532. [DOI] [PubMed] [Google Scholar]
- Rundlett, S. E., A. A. Carmen, R. Kobayashi, S. Bavykin, B. M. Turner et al., 1996. Hda1 and Rpd3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93 14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santisteban, M. S., T. Kalashnikova and M. M. Smith, 2000. Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell 103 411–422. [DOI] [PubMed] [Google Scholar]
- Sopko, R., D. Huang, N. Preston, G. Chua, B. Papp et al., 2006. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell 21 319–330. [DOI] [PubMed] [Google Scholar]
- Sutcliffe, E. L., I. A. Parish, Y. Q. He, T. Juelich, M. L. Tierney et al., 2009. Dynamic histone variant exchange accompanies gene induction in T cells. Mol. Cell Biol. 29 1972–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan, J., E. M. Baxter and V. G. Corces, 2005. The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev. 19 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader et al., 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294 2364–2368. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu et al., 2004. Global mapping of the yeast genetic interaction network. Science 303 808–813. [DOI] [PubMed] [Google Scholar]
- Venkatasubrahmanyam, S., W. W. Hwang, M. D. Meneghini, A. H. Tong and H. D. Madhani, 2007. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A. Z. Proc. Natl. Acad. Sci. USA 104 16609–16614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, A., S. K. Kurdistani and M. Grunstein, 2002. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 298 1412–1414. [DOI] [PubMed] [Google Scholar]
- Wang, Z., and G. Prelich, 2009. Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol. Cell Biol. 29 1694–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., G. M. Jones and G. Prelich, 2006. Genetic analysis connects Slx5 and Slx8 to the SUMO pathway in Saccharomyces cerevisiae. Genetics 172 1499–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle, C. M., K. N. McClinic, S. Ercan, X. Zhang, R. D. Green et al., 2008. The genomic distribution and function of histone variant htz-1 during C. elegans embryogenesis. PLoS Genet. 4 e1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes, G. M., M. Bergkessel, S. Bandyopadhyay, M. Shales, H. Braberg et al., 2008. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol. Cell 32 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel, J. A., E. S. Johnson, S. I. Reed and J. R. Yates, 3rd, 2004. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 279 45662–45668. [DOI] [PubMed] [Google Scholar]
- Wu, J., N. Suka, M. Carlson and M. Grunstein, 2001. Tup1 utilizes histone H3/H2B-specific Hda1 deacetylase to repress gene activity in yeast. Mol. Cell 7 117–126. [DOI] [PubMed] [Google Scholar]
- Xie, Y., E. M. Rubenstein, T. Matt and M. Hochstrasser, 2010. SUMO-independent in vivo activity of a SUMO-targeted ubiquitin ligase toward a short-lived transcription factor. Genes Dev. 24 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H., P. Braun, M. A. Yildirim, I. Lemmens, K. Venkatesan et al., 2008. High-quality binary protein interaction map of the yeast interactome network. Science 322 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., Y. Li, T. Ishizuka, M. G. Guenther and M. A. Lazar, 2003. A SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation. EMBO J. 22 3403–3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacchi, L. F., W. L. Schulz and D. A. Davis, 2010. HOS2 and HDA1 encode histone deacetylases with opposing roles in Candida albicans morphogenesis. PLoS One 5 e12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C., T. M. Roberts, J. Yang, R. Desai and G. W. Brown, 2006. Suppression of genomic instability by Slx5 and Slx8 in Saccharomyces cerevisiae. DNA Repair 5 336–346. [DOI] [PubMed] [Google Scholar]
- Zhang, H., D. N. Roberts and B. R. Cairns, 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman, D., D. Coleman-Derr, T. Ballinger and S. Henikoff, 2008. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]