Abstract
Genome annotation is a synthesis of computational prediction and experimental evidence. Small genes are notoriously difficult to detect because the patterns used to identify them are often indistinguishable from chance occurrences, leading to an arbitrary cutoff threshold for the length of a protein-coding gene identified solely by in silico analysis. We report a systematic reappraisal of the Schizosaccharomyces pombe genome that ignores thresholds. A complete six-frame translation was compared to a proteome data set, the Pfam domain database, and the genomes of six other fungi. Thirty-nine novel loci were identified. RT-PCR and RNA-Seq confirmed transcription at 38 loci; 33 novel gene structures were delineated by 5′ and 3′ RACE. Expression levels of 14 transcripts fluctuated during meiosis. Translational evidence for 10 genes, evolutionary conservation data supporting 35 predictions, and distinct phenotypes upon ORF deletion (one essential, four slow-growth, two delayed-division phenotypes) suggest that all 39 predictions encode functional proteins. The popularity of S. pombe as a model organism suggests that this augmented annotation will be of interest in diverse areas of molecular and cellular biology, while the generality of the approach suggests widespread applicability to other genomes.
Keywords: mass spectrometry, comparative genomics, Schizosaccharomyces pombe, new, tam
THE fission yeast Schizosaccharomyces pombe is a widely exploited model organism that shares many critical pathways with Homo sapiens. It undergoes a limited, yet highly defined, differentiation program in its sexual cycle. The initial genome annotation (Wood et al. 2002) has been refined to a complement of 5027 protein-coding genes (Dutrow et al. 2008; Wilhelm et al. 2008; Quintales et al. 2010).
Since small protein-coding genes are notoriously difficult to detect against a random background of short open reading frames (ORFs), pioneering studies in Saccharomyces cerevisiae set a preliminary cutoff threshold of 100 amino acids for the annotation of a small protein-coding gene in the absence of other supporting evidence (reviewed in Fisk et al. 2006); any ORFs smaller than this 100-amino-acid threshold were ignored. The same threshold was applied in the first full annotation of the fission yeast genome (Wood et al. 2002). However, the subsequent discovery of an additional 63 small protein-coding genes combines with the realization that 94% of the genome is transcribed (Dutrow et al. 2008; Wilhelm et al. 2008; Quintales et al. 2010) to raise the distinct possibility that further small protein-coding genes await discovery.
A number of pipelines have successfully exploited high-throughput tandem mass-spectrometry (MS/MS) data to predict novel proteins in H. sapiens (Fermin et al. 2006; Tanner et al. 2007; Bitton et al. 2010), Caenorhabditis elegans (Schrimpf et al. 2009), Drosophila melanogaster (Schrimpf et al. 2009), Arabidopsis thaliana (Castellana et al. 2008), and bacteria (Gupta et al. 2007). We have therefore applied a pipeline that integrates this proteogenomics approach (Bitton et al. 2010) with comparative genomics and genome-wide domain prediction to augment the current annotation of S. pombe. Exploitation of data sets from vegetative and sexually differentiating cultures predicts an additional 0.8% expansion in the set of protein-coding genes. Complementary functional studies suggest that most genes are likely to represent bona fide protein-coding genes.
MATERIALS AND METHODS
Fission yeast techniques:
Standard fission yeast and molecular biology methods were used throughout (Moreno et al. 1991). PCR deletion was done as described in Bahler et al. (1998), using pFA6a natMX6 as a template (Hentges et al. 2005). For complementation of deletion strains, each predicted ORF was amplified from genomic DNA with an NdeI site at the initiator methionine codon and BamHI after the stop codon. This NdeI/BamHI fragment was cloned into pREP1, pREP41, and pREP81 vectors (Basi et al. 1993; Maundrell 1993) in which the LEU2+ marker had been replaced with hygR (A. Grallert and I. M. Hagan, unpublished results). Wild-type asynchronous haploid (972 h−) and pat1.114 (pat1.114/pat1.114 ade6.M210/ade6.M216 h−/h+) diploid cells were grown in EMM2 media at 25°, the pat1.114 diploids were transferred to 32° to induce meiosis, and samples were taken before (0 hr) and 3, 5, and 10 hr after the temperature shift. Standard trichloroacetic acid-precipitated samples were run on 7, 10, and 12% acrylamide gels. The gels were cut into ∼80 slices. To minimize redundancy between data sets, only the bottom 40 slices from the 7% gel, all slices from the 10% gel, and the top 40 slices from the 12% gel were subjected to liquid chromatography (LC)-MS/MS analysis. Calcofluor/DAPI staining and antitubulin/Sad1 immunofluorescence staining were as described previously (Marks and Hyams 1985; Hagan and Yanagida 1995).
Gel slice destaining and washing:
Coomassie blue-stained gel slices were destained with 3 × 20-min changes of 1 ml 200 mm ammonium bicarbonate and 40% (v/v) acetonitrile. Gel slices were dehydrated by the addition of 500 μl acetonitrile for 15 min followed by rehydration in 500 μl of water for a further 15 min. This dehydration–rehydration procedure was performed a total of three times followed by a final dehydration in acetonitrile.
In-gel tryptic digestion:
Gel slices were rehydrated in 25 μl of 50 mm ammonium bicarbonate, 9% (v/v) acetonitrile, and 20 ng/μl sequencing grade trypsin (Sigma-Aldrich) for 20 min. The slices were then covered in 100 μl of 50 mm ammonium bicarbonate and 9% (v/v) acetonitrile and incubated at 37° for 18 hr. Following digestion, samples were acidified by the addition of 10 μl of 10% (v/v) formic acid. The isolated supernatant was dried in a vacuum centrifuge at 40° for 30 min before the peptides were resuspended in 20 μl of water and 0.1% trifluoroacetic acid (Sigma-Aldrich) prior to LC-MS/MS analysis.
nano liquid chromatography-MS/MS analysis:
Peptides were separated utilizing a Nano-Acquity UPLC system (Waters) as detailed below. Each sample was loaded onto a Waters C18 Symmetry trap column [180 μm internal diameter (ID), 5 μm 5 cm] in water, 0.1% (v/v) acetonitrile, and 0.1% (v/v) formic acid at a flow rate of 7 μl/min for 5 min. Peptides were then separated using a Waters NanoAcquity BEH C18 column (75 μm ID, 1.7 μm, 25 cm) with a gradient of 1–40% (v/v) of acetonitrile, 0.1% formic acid over 15 min at a flow rate of 400 nl/min. The nano liquid chromatography (nLC) effluent was sprayed directly into the LTQ-Orbitrap XL mass spectrometer aided by the Proxeon nano source at a voltage offset of 2.5 kV. The mass spectrometer was operated in parallel data-dependent mode where the MS survey scan was performed at a nominal resolution of 60,000 [at mass/charge (m/z) 400] in the Orbitrap analyzer in an m/z range of 400–2000. The top six multiply charged precursors were selected for collision induced dissociation (CID) in the LTQ at a normalized collision energy of 35%. Dynamic exclusion was enabled to prevent the selection of a formally targeted ion for a total of 20 sec.
Proteogenomics:
The LC-MS/MS analysis of the six samples [pat1.114 diploid cultures, before (0 hr), 3, 5, and 10 hr after shift and asynchronous wild-type haploid (972 h−)] generated six independent data sets (i.e., 7%, wild type; 7%, pat1.114, etc.). The S. pombe genome was downloaded from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and translated in all possible six frames as described in Bitton et al. (2010), producing 783,342 putative protein sequences. Each acquired data set (supporting information, Table S6) was searched independently once against the S. pombe six-frame database and once against the standard GeneDB protein database. Searches were performed using ProteinPilot (Shilov et al. 2007) [version 3.0, settings: Search—Rapid; Instrument—OrbiFT MS (1–3 ppm), LTQ MS/MS; Enzyme—Trypsin; FDR Analysis—Yes] against the target database concatenated to its decoy database counterpart (Tang et al. 2008). False discovery rates (FDRs) were calculated as described (Kall et al. 2008; Jones et al. 2009; Bitton et al. 2010) (Table S7). Only peptides with confidence levels >95% were considered. Peptide sequences obtained from the six-frame and standard searches were compared using the R statistical package (Gentleman et al. 2004). A second series of database searches was then conducted (settings as before) against a modified GeneDB database that includes all GeneDB database entries (5027), putative novel sequences (219), and all decoy hits reported by ProteinPilot (17,150, regardless of their assigned confidence). The 167 six-frame peptides that were re-identified with >95% confidence (FDR < 2%) were mapped back to their genomic coordinates (Perl, GeneDB annotation data). Each of these putative novel loci was then subjected to manual curation. (Raw proteomics data are available upon request.)
Comparative genomics:
Six fungal genomes (Schizosaccharomyces japonicus, Schizosaccharomyces octosporus, Schizosaccharomyces cryophilus, S. cerevisiae, Aspergillus fumigatus, and Neurospora crassa) were downloaded from the Fungal Genome Initiative webpage at the Broad Institute (http://www.broadinstitute.org/science/projects/fungal-genome-initiative). These and the S. pombe genome were translated in all possible reading frames [translations were carried out as described in Bitton et al. (2010), with the exception that proteins not containing arginine (R) or lysine (K) were retained]. Database sizes can be found in Table S5. Each protein database was then compared to the S. pombe six-frame database using InParanoid (Remm et al. 2001) (version 3.0, default settings plus bootstrapping). Ortholog and in-paralogue lists were compared (Table S5 and Figure 1) and mapped back to their genomic coordinates. Only intergenic orthologs/in-paralogues were retained. Each fungal database was also permuted and compared to the nonpermuted S. pombe database, as before, to evaluate the false-positive rate. Each of the putative intergenic ORFs was then subjected to manual curation.
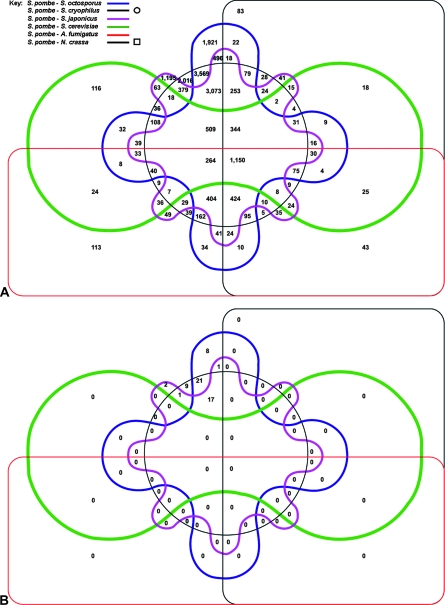
Figure 1.—
Six-way Venn diagram showing the number of conserved loci identified among the different fungi. Six-frame translations of six different fungi (key, top left) were each compared to that of S. pombe. Each shape represents the set of loci in S. pombe with significant conservation to one of the six other fungi. Intersections between shapes represent intersections between each of these sets, and the size of each intersection is indicated. For example, 1921 conserved loci were identified between S. pombe and S. octosporus (top center), and an additional 496 loci between S. pombe, S. octosporus, and S. japonicus. (A) All conserved loci. (B) Loci after filtering to remove known coding regions.
Pfam scan:
The S. pombe six-frame database was scanned using Pfam software (Finn et al. 2010) (pfam_scan.pl) installed locally (available at ftp://ftp.sanger.ac.uk/pub/databases/). Domains were mapped back to their genomic location and subjected to manual curation, as before.
RNA extraction:
Total RNA was extracted from S. pombe cells as described previously (Lyne et al. 2003), and quality was determined using an Agilent 2100 Bioanalyser.
Reverse transcription PCR:
Following extraction and purification of total RNA, genomic DNA was digested using RNase-free DNase (Qiagen). Reverse transcription was performed using Taqman reverse transcription reagents (Applied Biosystems). The reaction mixture was incubated at 25° for 10 min, 48° for 30 min, and 95° for 5 min. PCR was performed using 0.4 μm of each primer, 10 ng cDNA, 12.5 μl Jumpstart RedTaq PCR mastermix (Sigma), and RNase-free water to a volume of 25 μl. Cycling conditions included denaturation at 94° for 5 min, 30 cycles of (1) denaturation at 94° for 30 sec, (2) annealing at 60° for 30 sec, and (3) extension at 72° for 2 min, finishing with a final extension of 72° for 5 min. PCR fragments were resolved using agarose gel electrophoresis, gel-purified (Qiagen), and TA-cloned into the pGEM-T Easy vector (Promega). Amplified vectors were subsequently sequenced for identification.
5′ and 3′ RACE:
5′ and 3′ RACE were performed using the SMARTer RACE cDNA amplification kit (Clontech) as per the manufacturer's instructions. RACE PCR was performed using the Advantage 2 PCR kit (Clontech) containing a mixture of Taq and a proofreading polymerase. RACE PCR products were cloned and sequenced as described above.
RESULTS
The search for novel proteins was conducted using a modified version of a pipeline that we have previously used to search for novel human protein-coding genes (Bitton et al. 2010). The entire S. pombe genome was translated in all six reading frames and partitioned at stop codons to create a database containing 783,342 candidate sequences. To provide a coverage of the overall proteome that was as comprehensive as possible, we assessed the proteomic profiles of vegetatively growing wild-type haploid cells (972 h−) and cultures representing all stages of sexual differentiation. These differentiating cultures were generated by manipulation of the pat1.114 mutant. pat1+ encodes a protein kinase whose activity toward the meiotic inducer Mei2 during vegetative growth blocks commitment to sexual differentiation (Watanabe et al. 1997). Relief of Pat1 kinase inhibition of Mei2 is sufficient to induce sexual differentiation. Under normal circumstances, Pat1 activity is downregulated by the production of a pseudosubstrate called Mei3 (McLeod and Beach 1988; Li and McLeod 1996). However, temperature-dependent inactivation of the product of the pat1.114 gene offers an equally effective route to relieve Mei2 inhibition (Iino and Yamamoto 1985; Nurse 1985). A key feature of this temperature-dependent inactivation of Pat1.114 is the immediate induction of sexual differentiation of all cells within the culture, irrespective of factors that normally determine the potential to differentiate (cell cycle or mating-type status, nutrient level, and pheromone signaling). As progress through the different stages of sexual differentiation occurs at similar rates within each cell within these pat1.114-shifted cultures, differentiation proceeds in a largely synchronous fashion throughout the culture. The molecular changes that accompany sexual differentiation within such bulk cultures can therefore be taken as representative of the differentiation steps taken within the individual cells within the culture (Mata et al. 2002).
Samples from the pat1.114 culture were taken immediately before the temperature shift to 32° to induce meiosis (0 hr) and at 3, 5, and 10 hr after. These were then pooled to give a single pat1.114 meiotic sample for proteomic analysis. In total, 199,788 (wild type) and 265,234 (pat1.114) spectra were identified and searched against both the S. pombe protein data set (Wood et al. 2002; Hertz-Fowler et al. 2004) and the six-frame translation database. Overall, 167 novel peptides were identified at an FDR < 2% (Table S1). These peptides were mapped onto the existing genome annotation (Wood et al. 2002) and subjected to manual curation; canonical gene structures not contained within an alternative ORF of a known gene were defined. We also set the criterion that gene structures should include a methionine initiation codon either within the predicted ORF or, by extension, to include a canonical splice site. Using these criteria, we predicted nine novel genes (Table 1 and Table S2) and corrected the annotation at another 14 loci, including one status change from pseudogene to coding (Table 2 and Table S3). Five additional peptides provided experimental confirmation for novel protein predictions independently submitted to GeneDB over the course of this study. The remaining 133 peptides, which lacked the necessary up- and downstream signals of gene structure, were considered to be false positives (in correspondence with the estimated FDR), since they cannot physically be extended to create valid gene structures. This means that these peptides have an immediate upstream stop codon that has no intervening methionine or no acceptor that could provide the 3′ of an intron to negotiate the stop codon, or they are in a different/reverse frame of a known coding gene. Thus, the rejected fragments must be true false positives on the basis of our current knowledge of the biological mechanisms that make a functional protein.
TABLE 1.
Novel genes detected by the pipeline
| Method | GeneDB accession | Assigned gene name | Description/product | Transcriptional evidence (PCR/RNASeq) | Deletion phenotype |
|---|---|---|---|---|---|
| CG | SPAC222.19 | tam1 | Sequence orphan | Yes/Yes | NA |
| CG | SPAC17G8.15 | new1 | Histone-like transcription factor | Yes/Yes | Slow growth |
| CG | SPAC1805.18 | new2 | Sequence orphan | Yes/Yes | NA |
| CG | SPAC1486.11 | new3, fmc1 | Mitochondrial matrix protein | Yes/Yes | NA |
| CG and PG | SPAC3H5.13 | new4 | Conserved eukaryotic protein | Yes/Yes | NA |
| CG and PG | SPAC15A10.17 | new5, coa2 | Cytochrome C oxidase assembly factor | Yes/Yes | Elongated cellular morphology |
| CG and PG | SPAC4F10.22 | tam2, cmc4 | Mitochondrial intermembrane space protein | Yes/Yes | Viable |
| CG | SPAC1B3.21 | tam3 | Mitochondrial conserved protein | Yes/Yes | NA |
| CG | SPRRNA.56 | NA | rRNA | Yes/Yes | NA |
| CG | SPAC9G1.15c | tam4, mzt1 | Mitotic-spindle organizing protein | Yes/Yes | NA |
| CG | SPAC13F5.08 | new6, vts1 | Vps20-associated protein | Yes/Yes | NA |
| CG | SPAC6B12.19 | new7, rsa3 | Ribosome assembly protein Rsa3 | Yes/Yes | NA |
| CG | SPAC3G9.17 | new8 | Holo-[acyl-carrier-protein] synthase | Yes/Yes | Slow growth |
| PG | SPAC16E8.18c | tam5 | Sequence orphan | Yes/Yes | NA |
| CG | SPAC926.10 | new9 | Sequence orphan | Yes/Yes | NA |
| CG | SPAC19G12.17 | new10 | Enhancer of rudimentary homolog | Yes/Yes | NA |
| PG | SPAC23A1.20 | new11 | Pseudogene, truncated protein, no N-term methionine | Yes/Yes | NA |
| CG | SPAC1F7.14 | tam6 | Mitochondrial conserved protein | Yes/Yes | NA |
| CG | SPAC4D7.15 | new12 | Sequence orphan | Yes/Yes | NA |
| CG and PG | SPAC4D7.14 | new13 | Conserved fungal protein | Yes/Yes | NA |
| PG | SPBC839.20 | new14 | Dubious | No/No | NA |
| CG | SPBC409.23 | tam7 | Conserved fungal protein | Yes/Yes | Slow growth |
| CG & PF | SPBC8D2.23 | new15 | Mitochondrial protein, ribosomal subunit L35 related | Yes/Yes | NA |
| CG | SPBC3H7.18 | tam8 | Sequence orphan | Yes/Yes | NA |
| CG | SPBC1711.18 | tam9 | Mitochondrial ribosomal protein subunit L36, MrpL36/YmL36 | Yes/Yes | NA |
| CG | SPBC24C6.13 | new16 | Conserved eukaryotic protein | Yes/Yes | NA |
| CG | SPBC32F12.16 | new17 | Sequence orphan | Yes/Yes | NA |
| CG | SPBC30D10.21 | new18 | Mitochondrial protein | Yes/Yes | NA |
| CG | SPBC887.22 | new19 | Signal peptidase complex subunit Spc1 | No/Yes | NA |
| CG | SPBC839.19 | new20 | Conserved eukaryotic protein, broadly conserved (to Metazoa) | Yes/Yes | NA |
| CG and PF | SPBC530.16 | new21, ksh1 | DUF1242 family protein | Yes/Yes | Inviable |
| CG and PG | SPBC14C8.19 | tam10 | Sequence orphan | Yes/Yes | NA |
| PG | SPBC17D1.17 | tam11 | Sequence orphan | Yes/Yes | Elongated cellular morphology |
| CG | SPBC1105.19 | tam12 | Sequence orphan | Yes/Yes | NA |
| CG | SPBC56F2.15 | tam13 | Sequence orphan | Yes/Yes | NA |
| PF | SPCP20C8.04 | new22 | DUF1773 (S. pombe specific family) | No/No | NA |
| CG | SPCC330.20 | tam14 | Sequence orphan—conserved outside Schizosaccharomyces | Yes/Yes | NA |
| CG | SPCC63.09 | new23 | Dubious | Yes/No | NA |
| CG, PG, PF | SPCC4B3.20 | new24, cmc2 | Copper-binding protein of the mitochondrial inner membrane Cmc1 | Yes/Yes | NA |
| CG | SPCC330.21 | new25 | Dubious | Yes/Yes | NA |
CG, comparative genomics; PG, proteogenomics; PF, Pfam scan.
TABLE 2.
Novel extensions and revised gene structures
| Identification method | GeneDB accession/name | Status | Peptide sequence | Domain |
|---|---|---|---|---|
| PG | SPAC823.04 | Revised gene structure | KKHAPQELSSK | NA |
| PG | SPAC688.11, end4 | Revised gene structure | LQSDHMQSDASLMTSVR | NA |
| PG | SPAC29A4.03c, mrps9 | Revised gene structure | IIAAEEDVPK | NA |
| PG | SPAC688.04c, gst3 | Revised gene structure | IVWMLEELKVPYEIK | NA |
| PG | SPAPB17E12.14c | Revised gene structure | LAIIMVGLPAR | NA |
| PG | SPBC29A10.17 | Revised gene structure | TPPSAEADMSLR | NA |
| PG | SPBP8B7.31 | Revised gene structure | NSLNILSFLK | NA |
| PG | SPBC651.06, mug166 | Revised gene structure | TEIASAQAVMHDLLR | NA |
| PG | SPBC25H2.11c, spt7 | Revised gene structure | FGSLTSSFK | NA |
| PG | SPAC29A4.03c, mrps9 | Revised gene structure | LLNELNEIVPEYR | NA |
| PG | SPBC14C8.09c | Part of known protein | SLEILDMLLQSR | NA |
| PG | SPBC14C8.09c | Part of known protein | LLNGLPLNLVK | NA |
| PG | SPCC4G3.12c, not2 | Revised gene structure | QYMLESLLPIIR | NA |
| PG | SPCC4G3.12c, not2 | Revised gene structure | TGSTEELTETPADENAK | NA |
| PG | SPCC4G3.12c, not2 | Revised gene structure | TGSTEELTETPADENAKQYMLESLLPIIR | NA |
| PG | SPCC1322.16, phb2 | Revised gene structure | QRPFQQMNDLMKR | NA |
| PG | SPCC1442.04c | Part of known protein | NSVNKTETIREELDQAASK | NA |
| PG | SPAC11D3.11c | Status change from pseudogene to coding | MSVDPNIAALAK | NA |
| PG | SPBC13E7.01, cwf22 | Part of known protein | ALQAQLTDVNTPEYQR | NA |
| PG | SPBC13E7.01, cwf22 | Part of known protein | SINGLINKVNK | NA |
| PG | SPBC4F6.10, vps901 | Revised gene structure | LSSSIDSLLEMS | NA |
| PG | SPBC16H5.12c | Revised gene structure | SDIQAEPSTGTLENK | NA |
| PG | SPBC16H5.12c | Revised gene structure | TYSDSENNSTLDVSAQVLTLDNKSR | NA |
| PF | SPAC186.06 | Status change from coding to pseudogene | NA | PhzC-PhzF |
| PF | SPAC186.06 | Revised gene structure | NA | PhzC-PhzF |
| PF | SPCP20C8.01c | Part of known protein | NA | DUF1773 |
| CG | SPAC3A11.03 | Revised gene structure | NA | NA |
| CG | SPBC31F10.08, mde2 | Part of known protein | NA | NA |
| CG | SPBC1711.10c, npl4 | Revised gene structure | NA | NA |
| CG | SPBC1709.12, rid1 | Revised gene structure | NA | NA |
| CG | SPAC13F5.07c | Revised gene structure | NA | NA |
| CG | SPAC18B11.05, gpi18 | Revised gene structure | NA | NA |
| CG | SPCP20C8.01c | Part of known protein | NA | NA |
| CG | SPCC1235.16, vma21 | Part of known protein | NA | NA |
| CG | SPBC18H10.08c, ubp4 | Part of known protein | NA | NA |
| CG | SPACUNK4.14, mdb1 | Revised gene structure | NA | NA |
| CG | SPAC13F5.07c | Revised gene structure | NA | NA |
CG, comparative genomics; PG, proteogenomics; PF, Pfam scan.
All other peptides that could be extended to make a protein structure (regardless of other criteria) were included. Those that were unsupported by homology, or other methods, were designated as “dubious,” pending further investigation.
To ensure that the 133 rejected loci were indeed false predictions, we also sought transcriptional evidence at these loci, using an RNA sequencing data set (AB SOLiD v3+) produced from asynchronous wild type (972 h−) and pat1.114 cultures immediately before temperature shift to 32° (0 hr) and at 3, 5, and 10 hr after temperature shift, as before (two biological replicates were processed; D. Bitton, A. Grallert, J. Bradford, Y. Li, T. Yates, P. Scutt, Y. Hey, S. Pepper, I. Hagan, and C. Miller, unpublished results). As expected, at the majority of these loci (89), no 50mer sequence reads aligned perfectly to the genome (in any sample), while at the remaining 44 loci some transcriptional activity was observed in at least one of the 10 samples tested. Nevertheless, in most cases, the predicted peptide was in an alternate reading frame to that of a known protein (29 loci), making it impossible to distinguish between transcription that would result in the novel peptide and transcription that would result in the known protein being expressed. An additional set of loci was on the complementary strand to a known protein (8 loci) or positioned within untranslated regions (UTRs) (2 loci), in line with their initial classification as false positives (Table S4). The remaining 5 loci were located in intergenic regions and displayed low transcriptional activity, but the lack of conventional signals of gene structure precluded prediction of the complete architecture of these genes.
The relationship between S. pombe and other fungi was then assessed using comparative genomics. We extended the InParanoid algorithm (Remm et al. 2001) to make use of a complete, unbiased, six-frame translation of the S. pombe genome. We compared this data set to six other fungi (S. japonicus, S. octosporus, S. cryophilus, S. cerevisiae, A. fumigatus, and N. crassa) via their six-frame protein translations. To estimate the FDR, searches were also performed against databases produced by random permutation of the original data. The resultant ortholog sets were compared (Table S5) to produce a list of all conserved loci in S. pombe (44,900; FDR 0%; no matches in the permuted data set). These were then mapped onto an existing annotation. The vast majority of conserved sequences (99.9%) were found to be within existing genes. However, an additional 59 hits to six frame translations of the genomes of the most recently diverged species (S. japonicus, S. cryophilus, and S. octosporus; Figure 1) were identified in the fission yeast intergenic regions. These were then subjected to the same manual curation criteria used to filter the MS/MS data above. A further 35 novel genes, six gene extensions, and one protein-coding gene, previously annotated as a pseudogene, were identified. Five of these 35 genes overlapped with those found by proteogenomics.
A major challenge with any prediction algorithm is to find the right balance between true and false positives. Typically, parameter settings are chosen to err on the side of caution; underprediction is generally preferred instead of populating the databases with incorrectly identified genes. Since we were able to predict homology to S. cerevisiae for 16 of these novel loci using PSI-Blast (Altschul et al. 1997), we believe that the settings used here were similarly stringent. Importantly, since no matches were found to be significant with the permuted data, it remains likely that more protein-coding genes may remain.
We then scanned gene-free regions of the fission yeast six-frame translation for matches to Pfam protein domains (Finn et al. 2010). Six additional protein-coding domains were identified outside the known protein-coding regions (P-value < 3.9 × 10−07). Three of these genes had been identified within the comparative genomics work flow, and one also matched the peptide data. One domain led to the discovery of a novel protein-coding gene, not identified by the other methods, and one a gene that had been previously annotated as a pseudogene. Two domains were found to extend a known human MAWBP homolog (SPAC186.06). However, this domain extension introduces two frameshifts, making it unlikely that this gene is correctly translated or that there are errors in the genomic sequence.
We next sought to confirm transcription at these loci by RT-PCR and RNA sequencing. Transcription was confirmed at 38/40 of the loci in either the asynchronous wild-type or pat1.114-induced meiotic cultures (Table 1, Table S2; Figure S1, Figure S2). The inherent synchrony of sexual differentiation that is induced by a shift of a pat1.114 diploid strain to 32° enabled us to qualitatively assess whether the level of transcription for any of the new genes fluctuated with progression through the meiotic differentiation program. Fourteen transcripts were differentially expressed during meiosis. Using 5′ and 3′ RACE assays, we established the complete gene architecture of 33 predicted genes and 2 gene extensions, including the 5′ and/or 3′ UTRs. For 9 loci, data sets generated in the mass spectrometric analysis supported the existence of a bona fide protein-coding gene. Epitope “tags” were fused, in frame, to the 3′ end of the ORFs of two genes using standard PCR cassette-mediated tagging (Bahler et al. 1998). Western blot analysis confirmed that both genes were translated into proteins (Figure S3).
We refer to the novel protein-coding genes with no apparent induction in meiosis as new1–new25, and the 14 genes with transcripts altered in meiosis as tam1–tam14. A recent study conducted by Hutchins et al. (2010) identified tam4 as encoding the S. pombe counterpart of a novel component of the microtubule-nucleating γ-tubulin ring complex called Mozart (although the Hutchins et al. study prediction uses an initiator methionine that is 5′ of the ORF that we propose).
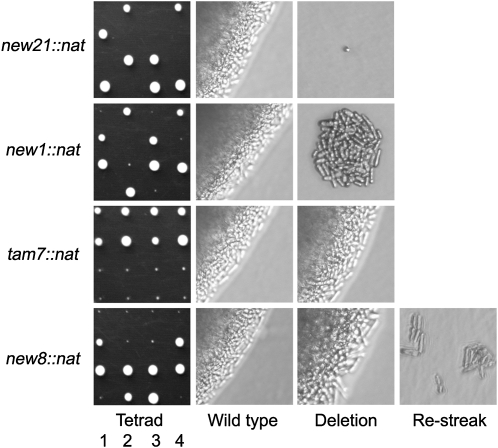
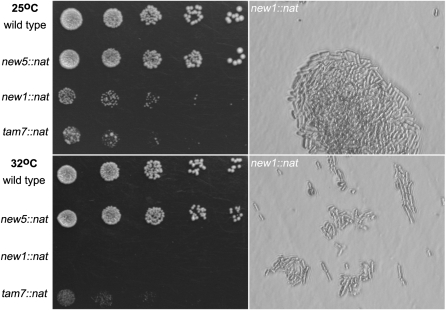
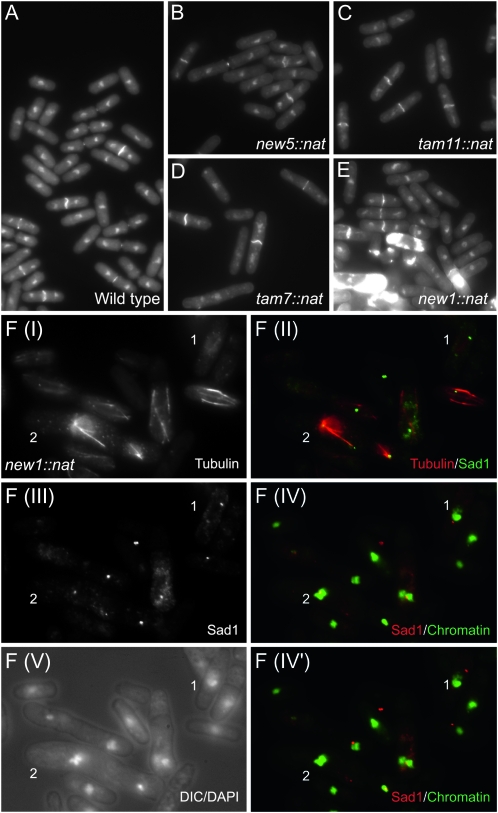
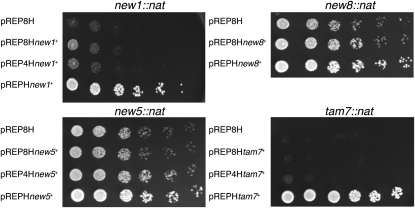
Next, three criteria were used to select seven genes (tam2, new1, new5, tam7, new8, tam11, and new21) for functional assessment by one-step PCR-mediated destruction (Bahler et al. 1998): evidence of transcription, the absence of close neighbors, and the prediction of a phenotype based on predictions arising from deletion of the S. cerevisiae homolog. While tam2.Δ was viable, new21 was not (Figure 2); it has human (TMEM167A) and S. cerevisiae (YNL024C-A) orthologs whose functions are suggested to lie within the secretory pathway (Wendler et al. 2009). new5.Δ and tam11.Δ displayed an elongated cellular morphology at division, typical of cells in which commitment to mitosis is delayed (Figure 3; Figure 4, B and C; Figure 5). Both new8.Δ and tam7.Δ cells grew extremely slowly at all temperatures tested (25°, 30°, 32°, and 36°; Figure 2 and Figure 3). tam7.Δ cells were also delayed in mitotic commitment (Figure 4D). The new1.Δ strain exhibited slow growth at 25° but was inviable at 32° (Figure 2 and Figure 3). Cell division was severely perturbed by the loss of this putative histone-like transcription factor (Table 1 and Table S2). Aberrant cell-wall deposits accumulated during septation (Figure 4E) alongside a range of mitotic defects including errors in chromosome segregation (cell type 1; Figure 4F; Figure S4; Figure S5) and spindle formation (cell type 2; Figure 4F; Figure S4; Figure S5).
Figure 2.—
Tetrad analysis of knockout strains. Asci from the appropriate new∷nat/new+ ade6.M210/ade6.M216 h+/h− diploid strains were dissected on rich yeast extract supplemented (YES) medium at 25°. Images of colony formation on the left demonstrate the essentiality of new21+ and the slow-growth phenotypes of new1∷nat, tam7∷nat, and new8∷nat cells. The bright-field microscopy images show increased magnification of new/tam+ (middle) and newx∷nat/tamx∷nat (right) colonies (denoted as “Wild type” and “Deletion”). The far right panel of the new8∷nat cells shows the inability of the microcolonies formed at 25° to grow 4 days after restreaking onto fresh YES at 25°.
Figure 3.—
Spot-test analysis reveals the temperature-dependent lethality of new1∷nat and tam7∷nat. Cells from cultures of the indicated strains grown to mid-log phase in rich YES liquid medium were plated in fivefold serial dilutions so that the final dilution plated around five cells on YES plates that were then incubated at the indicated temperatures. The limited ability of the plated new1∷nat cells to grow and divide at 32° is clear from a comparison of the magnified images, highlighting growth of the same strain on a YES plate at 25° (top right) and 32° (bottom right).
Figure 4.—
Phenotypic characterization of knockout strains. (A–E) Calcofluor DAPI staining of the indicated strains. S. pombe cells grow by linear extension until they reach a critical size threshold. Nuclear division is then initiated, followed by the contraction of the cytokinetic F-actin ring, resulting in separation of the two cytoplasms of the incipient daughter cells. F-actin ring contraction is coupled with the deposition of the cell-wall material of the primary septum that stains strongly as a white bar across the cell equator between the separated nuclei (Marks and Hyams 1985). This primary septum is subsequently degraded to separate the two daughter cells. Thus, the length of cells with the transecting bright calcofluor-positive stain is a direct indication of the timing at which cells commit to mitosis (Nurse 1975). It is clear from B–D that the cells with this bright bar in strains new5.Δ, tam11.Δ, and tam7.Δ are longer than the wild-type controls in A and so are delayed in division. (E) The accumulation of excessive regions of calcofluor staining in new1.Δ cells is indicative of severe defects in septation in some cells in the culture. (F) Anti-tubulin, anti-Sad1 immunofluorescence of new1.Δ cells. DAPI staining of chromatin either alone or in combination with differential interference contrast imaging visualizes the position of the chromatin relative to the cell periphery, as indicated on each panel in the figure. A range of mitotic defects was apparent, including, most notably, a failure in chromosome segregation along elongating spindles (cell 1) and the formation of monopolar spindles with expansive arrays of red microtubules extending from single green foci of Sad1 staining [cell 2: F(II)]. Highly condensed, unsegregated chromosomes cluster around these Sad1 foci [cell 2: F(IV)]. F(IV) and F(IV′) show the same DAPI images of chromatin merged with two different focal planes in the Sad1 channel because the different spindle pole bodies (SPBs) in cells 1 and 2 reside in different focal planes.
Figure 5.—
Spot-test analysis demonstrates complementation of the compromised growth phenotype of new1∷nat, new5∷nat, new8∷nat, and tam7∷nat following exogenous provision of mRNA encoding the appropriate ORF. Cells from cultures of the indicated strains were grown to mid-log phase in rich YES liquid medium containing 100 μg·ml−1 hygromycin to select for the presence of the plasmid and plated in fivefold serial dilutions, so that the final dilution plated around five cells on YES plates that were then incubated at 25°. The provision of the appropriate reading frame rescued the slow-growth phenotype in each case. The subtlety of the growth defect in new5∷nat meant that the differential colony size relative to wild-type controls is only transiently visible in spot-test analyses (compare the clear complementation here with the apparent wild-type growth in new5∷nat cells in Figure 3); however, it is clear that, even in this case, the exogenous provision of the appropriate mRNA complements the slow growth arising from deletion of this gene.
Complementation analysis was used to address whether the phenotypes observed in the “deletion” strains genuinely arose from the loss of the protein corresponding to the ORF from the proteome or from some other consequence of the alteration to the genomic sequence. The new1+, new5+, new8+, new21+, and tam7+ ORFs were cloned into versions of the pREP1, pREP41, and pREP81 vectors in which the LEU2+ gene had been replaced with hygR (pREPH pREP4H and pREP8H, respectively; A. Grallert and I. M. Hagan, unpublished results) and transformed into the appropriate heterozygous deletion/wild-type ORF diploids. Haploid deletion strains harboring the plasmids were subsequently isolated and subjected to spot-test analysis at 25°. In each case, a strain harboring the empty pREP8H vector served as a control to define the basal level of growth of the deletion strain. The provision of the corresponding ORF complemented the slow-growth phenotype of new1∷nat, new5∷nat, new8∷nat, and tam7∷nat (Figure 5). Transformation of the new21∷nat/new21+ heterozygous diploid strain with any pREP or its derivative failed repeatedly. We assume that this is due to the poor growth of this diploid, which would be consistent with a requirement for two copies of new21+ for healthy growth of diploid S. pombe cells.
DISCUSSION
The systematic pipeline developed here (Figure 6) has expanded the gene repertoire of the fission yeast S. pombe with the identification of 39 new genes and one rRNA gene (equivalent to an ∼0.8% increase to the proteome) (Table 1 and Table S2). We have also refined the sequence coordinates that define 21 more loci (Table 2; Table S3; Figure 7A). The verification of the majority of the genes by RT-PCR, RNA sequencing, and RACE, the supporting evidence at the protein level for 10 genes, and the association of a deletion phenotype for six of seven candidates tested strongly suggest that all are bona fide genes. However, our appraisal of the new11 locus illustrates the challenge in setting a threshold for the definition of a gene. Despite substantial evidence, including MS/MS data, transcriptional support, and most of the signals of conventional gene structure, our inability to locate a suitable 5′ “ATG” leads us to annotate this “gene” as a “pseudogene.” In other words, considerable potential remains for extending the catalog of protein-coding genes in S. pombe still further as new technologies emerge and our understanding of gene structure advances.
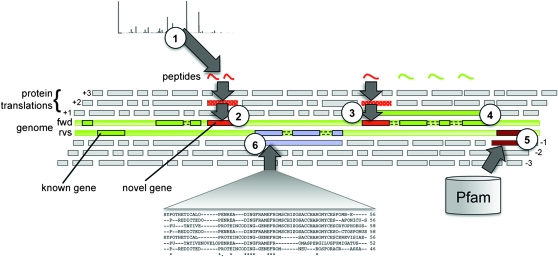
Figure 6.—
A cartoon representing the gene detection pipeline. The entire genome is translated in all three forward and reverse reading frames and segmented at putative stop codons. Data from different sources are compared to these translations and mapped back to the genome. Matching features that do not overlap with existing genes are used to infer novel genes and/or to refine gene structures. (1) MS/MS data searched against all putative protein sequences that could be expressed from the genome identify novel protein-coding genes (2) and refine 5′ and 3′ ends of others (3). MS/MS data matching pseudogenes, and genes annotated as noncoding, confirm translation and lead to altered annotation (4). Pfam searches against six-frame translations identify additional protein domains (5). Comparative genomic searches between whole-genome protein translations of six fungal species to S. pombe are used to infer novel protein coding genes (6).
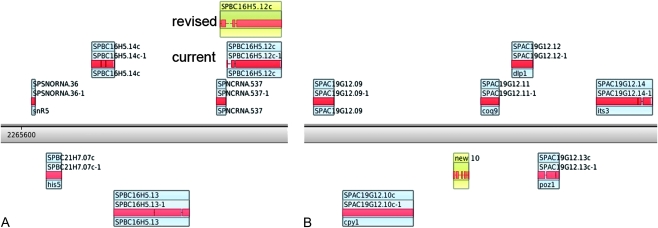
Figure 7.—
Examples of loci identified by the pipeline and confirmed by 5′ and 3′ RACE. Part of the S. pombe genome represented by the X:Map genome browser (http://xmap.picr.man.ac.uk) is shown. The chromosome runs across the middle of the figure with genes on the forward strand shown above the center line and those on the reverse strand, below. Boxes represent genes, with the transcript structure and exons indicated by the smaller boxes within. (A) The 5′ extension to SPBC16H512c identified by two novel peptides. The original and revised gene structures are shown; the revised gene structure incorporates a locus previously annotated as a ncRNA (noncoding RNA). (B) Location of new10, enhancer of rudimentary homolog-like protein, identified by comparative genomics.
Comparisons with S. cerevisiae databases predict localization for 18 genes and an inferred function for 13. For example, new10 (Figure 7B), which was identified solely by comparative genomics, is broadly conserved from humans to protozoa. The gene encodes enhancer of rudimentary homolog, a putative transcriptional repressor implicated in cell cycle regulation and significantly upregulated in malignant breast and ovarian cancer cells (Zafrakas et al. 2008).
The length of the majority of the novel protein-coding genes was below the 100-amino-acid cutoff set for the original annotation pipeline (Wood et al. 2002). Since similar size criteria have been used for other organisms, it is reasonable to expect that the application of similar direct pipelines to interrogate genome annotation in other model organisms should provide similar refinements to their gene complement and bring us closer to the desired catalog of protein-coding genes.
Acknowledgments
This work was supported by Cancer Research UK grant no. C147/A6058. V.W. is supported by Cancer Research UK, and work on bioinformatics resources for S. pombe at Cambridge University is supported by Wellcome Trust grant WT090548MA to S.G. Oliver and V.W. Genome sequences of S. japonicus and S. octosporus were generated by the Broad Institute and that of S. cryophilus was generated by the Stowers Institute.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.123497/DC1.
References
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie, III et al., 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14 943–951. [DOI] [PubMed] [Google Scholar]
- Basi, G., E. Schmid and K. Maundrell, 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123 131–136. [DOI] [PubMed] [Google Scholar]
- Bitton, D. A., D. L. Smith, Y. Connolly, P. J. Scutt and C. J. Miller, 2010. An integrated mass-spectrometry pipeline identifies novel protein coding-regions in the human genome. PLoS One 5 e8949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellana, N. E., S. H. Payne, Z. Shen, M. Stanke, V. Bafna et al., 2008. Discovery and revision of Arabidopsis genes by proteogenomics. Proc. Natl. Acad. Sci. USA 105 21034–21038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutrow, N., D. A. Nix, D. Holt, B. Milash, B. Dalley et al., 2008. Dynamic transcriptome of Schizosaccharomyces pombe shown by RNA-DNA hybrid mapping. Nat. Genet. 40 977–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fermin, D., B. B. Allen, T. W. Blackwell, R. Menon, M. Adamski et al., 2006. Novel gene and gene model detection using a whole genome open reading frame analysis in proteomics. Genome Biol. 7 R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, R. D., J. Mistry, J. Tate, P. Coggill, A. Heger et al., 2010. The Pfam protein families database. Nucleic Acids Res. 38 D211–D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk, D. G., C. A. Ball, K. Dolinski, S. R. Engel, E. L. Hong et al., 2006. Saccharomyces cerevisiae S288C genome annotation: a working hypothesis. Yeast 23 857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling et al., 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5 R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, N., S. Tanner, N. Jaitly, J. N. Adkins, M. Lipton et al., 2007. Whole proteome analysis of post-translational modifications: applications of mass-spectrometry for proteogenomic annotation. Genome Res. 17 1362–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan, I., and M. Yanagida, 1995. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 129 1033–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentges, P., B. Van Driessche, L. Tafforeau, J. Vandenhaute and A. M. Carr, 2005. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast 22 1013–1019. [DOI] [PubMed] [Google Scholar]
- Hertz-Fowler, C., C. S. Peacock, V. Wood, M. Aslett, A. Kerhornou et al., 2004. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 32 D339–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins, J. R., Y. Toyoda, B. Hegemann, I. Poser, J.-K. Heriche et al., 2010. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino, Y., and M. Yamamoto, 1985. Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol. Gen. Genet. MGG 198 416–421. [DOI] [PubMed] [Google Scholar]
- Jones, A. R., J. A. Siepen, S. J. Hubbard and N. W. Paton, 2009. Improving sensitivity in proteome studies by analysis of false discovery rates for multiple search engines. Proteomics 9 1220–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall, L., J. D. Storey, M. J. MacCoss and W. S. Noble, 2008. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J. Proteome Res. 7 29–34. [DOI] [PubMed] [Google Scholar]
- Li, P., and M. McLeod, 1996. Molecular mimicry in development: identification of ste11+ as a substrate and mei3+ as a pseudosubstrate inhibitor of ran1+ kinase. Cell 87 869–880. [DOI] [PubMed] [Google Scholar]
- Lyne, R., G. Burns, J. Mata, C. Penkett, G. Rustici et al., 2003. Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics 4 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks, J., and J. S. Hyams, 1985. Localization of F-actin through the cell-division cycle of Schizosaccharomyces pombe. Eur. J. Cell Biol. 39 27–32. [Google Scholar]
- Mata, J., R. Lyne, G. Burns and J. Bahler, 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32 143–147. [DOI] [PubMed] [Google Scholar]
- Maundrell, K., 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123 127–130. [DOI] [PubMed] [Google Scholar]
- McLeod, M., and D. Beach, 1988. A specific inhibitor of the ran1+ protein kinase regulates entry into meiosis in Schizosaccharomyces pombe. Nature 332 509–514. [DOI] [PubMed] [Google Scholar]
- Moreno, S., A. Klar and P. Nurse, 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194 795–823. [DOI] [PubMed] [Google Scholar]
- Nurse, P., 1975. Genetic control of cell size at cell division in yeast. Nature 256 547–551. [DOI] [PubMed] [Google Scholar]
- Nurse, P., 1985. Mutants of the fission yeast Schizosaccharomyces pombe which alter the shift between cell proliferation and sporulation. Mol. Gen. Genet. MGG 198 497–502. [Google Scholar]
- Quintales, L., M. Sanchez and F. Antequera, 2010. Analysis of DNA strand-specific differential expression with high density tiling microarrays. BMC Bioinformatics 11 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm, M., C. E. Storm and E. L. Sonnhammer, 2001. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 314 1041–1052. [DOI] [PubMed] [Google Scholar]
- Schrimpf, S. P., M. Weiss, L. Reiter, C. H. Ahrens, M. Jovanovic et al., 2009. Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol. 7 e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilov, I. V., S. L. Seymour, A. A. Patel, A. Loboda, W. H. Tang et al., 2007. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6 1638–1655. [DOI] [PubMed] [Google Scholar]
- Tang, W. H., I. V. Shilov and S. L. Seymour, 2008. Nonlinear fitting method for determining local false discovery rates from decoy database searches. J. Proteome Res. 7 3661–3667. [DOI] [PubMed] [Google Scholar]
- Tanner, S., Z. Shen, J. Ng, L. Florea, R. Guigo et al., 2007. Improving gene annotation using peptide mass spectrometry. Genome Res. 17 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y., S. Shinozaki-Yabana, Y. Chikashige, Y. Hiraoka and M. Yamamoto, 1997. Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature 386 187–190. [DOI] [PubMed] [Google Scholar]
- Wendler, F., A. K. Gillingham, R. Sinka, C. Rosa-Ferreira, D. E. Gordon et al., 2009. A genome-wide RNA interference screen identifies two novel components of the metazoan secretory pathway. Embo J. 29 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm, B. T., S. Marguerat, S. Watt, F. Schubert, V. Wood et al., 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453 1239–1243. [DOI] [PubMed] [Google Scholar]
- Wood, V., R. Gwilliam, M. A. Rajandream, M. Lyne, R. Lyne et al., 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415 871–880. [DOI] [PubMed] [Google Scholar]
- Zafrakas, M., I. Losen, R. Knuchel and E. Dahl, 2008. Enhancer of the rudimentary gene homologue (ERH) expression pattern in sporadic human breast cancer and normal breast tissue. BMC Cancer 8 145. [DOI] [PMC free article] [PubMed] [Google Scholar]