Abstract
Changes in gene regulation are thought to play an important role in speciation and adaptation, especially in primates. However, we still know relatively little about the mechanisms underlying regulatory evolution. In particular, the extent to which epigenetic modifications underlie gene expression differences between primates is not yet known. Our study focuses on an epigenetic histone modification, H3K4me3, which is thought to promote transcription. To investigate the contribution of H3K4me3 to regulatory differences between species, we collected gene expression data and identified H3K4me3-associated genomic regions in lymphoblastoid cell lines (LCLs) from humans, chimpanzees, and rhesus macaques, using three cell lines from each species. We found strong evidence for conservation of H3K4me3 localization in primates. Moreover, regardless of species, H3K4me3 is consistently enriched near annotated transcription start sites (TSS), and highly expressed genes are more likely than lowly expressed genes to have the histone modification near their TSS. Interestingly, we observed an enrichment of interspecies differences in H3K4me3 at the TSS of genes that are differentially expressed between species. We estimate that as much as 7% of gene expression differences between the LCLs of humans, chimpanzees, and rhesus macaques may be explained, at least in part, by changes in the status of H3K4me3 histone modifications. Our results suggest a modest, yet important role for epigenetic changes in gene expression differences between primates.
COMPARATIVE studies of gene expression have identified a large number of differentially expressed genes among primate species (Enard et al. 2002; Cáceres et al. 2003; Karaman et al. 2003; Khaitovich et al. 2004, 2005; Gilad et al. 2006; Blekhman et al. 2008, 2009, 2010; Babbitt et al. 2010a). In a number of cases, these studies also pointed to possible connections between interspecies differences in gene regulation and differences in ultimate physiological or morphological phenotypes (Rockman et al. 2005; Loisel et al. 2006; Pollard et al. 2006; Prabhakar et al. 2008; Warner et al. 2009; Babbitt et al. 2010b). However, we still know little about the relative importance of different regulatory mechanisms to interspecies differences in gene expression levels (Chabot et al. 2007; Blekhman et al. 2009). In particular, little is known about the relative contribution of changes in epigenetic modifications to regulatory variation in primates.
Comparative studies of one class of epigenetic marker, DNA methylation, suggest that interprimate differences in epigenetic modifications may be abundant. For example, Gama-Sosa et al. (1983) found that relative global methylation levels across tissues generally differ between human and three other primate species (with the exception of hypermethylation in the brain and the thymus, which was observed in all studied species). Focusing on individual loci, Enard et al. (2004) compared methylation profiles of 36 genes in livers, brains, and lymphocytes from humans and chimpanzees and found significant interspecies methylation level differences in 22 of the 36 genes, in at least one tissue.
A somewhat different picture may be emerging from comparative studies of a different class of epigenetic markers, histone modifications; however, to our knowledge, no comparisons of histone modifications in primates have yet been published. An early comparative study of 15 genomic regions associated with histone acetylation in humans found evidence for conservation of the histone acetylation status of 10 of the orthologous regions in mouse (Roh et al. 2007). On a larger scale, characterization of several types of histone modifications on human chromosomes 21 and 22, and the syntenic chromosomes in mouse, indicated that the genomic locations of these epigenetic markers at orthologous loci are generally strongly conserved, even in the absence of sequence conservation (Bernstein et al. 2005; Wilson et al. 2008). Interestingly, the conservation of histone modification patterns was highest in genomic regions proximal to annotated orthologous genes.
With few exceptions, however (e.g., with respect to DNA methylation; Farcas et al. 2009), comparative studies in primates have not explored the extent to which epigenetic differences between species underlie interspecies differences in gene regulation. To take first steps toward this goal, we compared gene expression levels and histone modification data in samples from humans and two of our closest extant evolutionary relatives, chimpanzees (Pan troglodytes) and rhesus macaques (Macaca mulatta).
To facilitate inferences of causality in our data, we chose to focus on the trimethylation of lysine 4 of histone H3 (H3K4me3), because its role in gene regulation is relatively well understood. Specifically, the observation that RNA polymerase II (polII) colocalizes with genomic regions characterized by H3K4me3 led to the hypothesis that polII is one of the factors that help recruit the methyltransferases that establish H3K4me3 marks (Gerber and Shilatifard 2003; Hampsey and Reinberg 2003; Ng et al. 2003; Heintzman et al. 2007; Ruthenburg et al. 2007). Once established, H3K4me3 facilitates the recruitment of the BPTF subunit of the nucleosome-remodeling factor (NURF), which promotes chromatin accessibility (Li et al. 2006; Wysocka et al. 2006; Ruthenburg et al. 2007). Additionally, H3K4me3 blocks binding of the NuRD nucleosome-remodeling and deacetylase compressor complex, which decreases chromatin accessibility (Nishioka et al. 2002; Zegerman et al. 2002). The H3K4me3 modifications, therefore, are thought to result in more accessible chromatin and to facilitate active transcription. General support for this mechanism comes from the observation that H3K4me3 is enriched near the transcription start sites (TSS) of active genes. Moreover, highly expressed genes are more likely than lowly expressed genes to be associated with H3K4me3 modifications (Santos-Rosa et al. 2002; Schneider et al. 2004; Schübeler et al. 2004; Pokholok et al. 2005; Karlic et al. 2010).
In addition, several functional studies have found that disruption of the histone methyltransferases that catalyze H3K4me3 leads to a decrease in transcription levels. For example, Nislow et al. (1997) showed that knocking out the yeast SET1 gene, whose protein methylates H3K4, leads to lower gene expression levels of a subset of target genes. Similarly, Wang et al. (2009) demonstrated that disrupting the methyltransferase domain of Mll1 in mouse embryonic fibroblasts leads to decreased levels of H3K4me3 and concomitant lower expression levels of a large number of genes. Using a different system, Nishioka et al. (2002) showed that when the human methyltransferase SETD7 is recruited to reporter constructs, it specifically methylates H3K4 and promotes higher reporter gene expression levels.
Thus, a large body of work has established a causal link between H3K4me3 and gene regulation. In what follows, we use this link to query the proportion of genome-wide gene expression differences between closely related primate species that may be explained by corresponding differences in H3K4me3.
MATERIALS AND METHODS
Samples, study design, and cell culture:
Three lymphoblastoid cell lines (LCLs) were used from each primate species: human, chimpanzee, and rhesus macaque. LCLs were obtained from Coriell (http://www.coriell.org/), New Iberia Research Center (University of Louisiana at Lafayette), and New England Primate Research Center (NEPRC, Harvard Medical School) and were all derived from male individuals. Details on all samples can be found in Table S1. Regardless of species, cells were maintained at identical conditions of 37° with 5% CO2 in RPMI media with 15% FBS, supplemented with 2 mm l-glutamate, 100 IU/ml penicillin, and 100 μg/ml streptomycin.
Chromatin immunoprecipitation:
To determine localization of H3K4me3, we used ChIPseq (see supporting information, File S1 for details on the ChIP protocol). Each ChIP sample, and a pooled input control from each species, was sequenced in one lane of an Illumina GAII flow cell. Sequenced reads were mapped to the human (hg18, March 2006), chimpanzee (panTro2, March 2006), or rhesus macaque (rheMac2, January 2006) genomes, as appropriate, using MAQ (Li et al. 2008) version 0.6.8. After excluding reads with a MAQ quality score <10, we used MACS (Zhang et al. 2008) with a P-value cutoff of 0.0005 (Table S2) to identify peaks of H3K4me3-associated genomic regions and to estimate a false discovery rate (FDR) for each peak on the basis of the input sequence data from each species. In principle, at this point we could begin to compare H3K4me3 across individuals and species by investigating whether each modified site is observed in all, or in a subset, of the three species. However, a naïve comparison of the lists of genomic regions associated with H3K4me3 modifications across species ignores the fact that evidence for an H3K4me3 peak in one species provides information about the likelihood of an H3K4me3 peak in closely related species. To take this into account, we classified H3K4me3-associated genomic regions by using a two-step FDR cutoff.
Specifically, we first used a stringent FDR cutoff of 2% to classify H3K4me3+ regions in each individual. Subsequently, these regions were mapped to the genomes of the other two species with the BLAST-like alignment tool (BLAT) to determine orthology (see File S1), and the FDR threshold for observing peaks at the orthologous sites in all other samples was relaxed to 5%. The choice of specific thresholds was based on the overall distribution of FDR values (see Figure S2). A species was considered positive for H3K4me3 at a given genomic region if at least one individual from the species was positive for H3K4me3 at that region.
To analyze H3K4me3 marks in the context of TSS, a set of orthologous TSS positions in the three species was identified (see File S1 and Table S4). Ensembl ensGene TSS coordinates were downloaded for human (hg18) from Galaxy (http://main.g2.bx.psu.edu/) and were mapped to the other two genomes (panTro2 and rheMac2) with both LiftOver (http://hgdownload.cse.ucsc.edu/admin/exe/) and BLAT (http://genome.ucsc.edu/), resulting in 35,232 unique TSS from 25,160 genes. TSS were considered H3K4me3+ for a sample if a H3K4me3-associated region was present within 1 kb of the TSS, and a gene was considered H3K4me3+ in a species if at least one of its TSS in at least one individual was classified H3K4me3+.
RNA sequencing and analysis of differences in gene expression levels:
RNA was extracted from each LCL sample using a Qiagen RNeasy mini kit. We confirmed high quality of the RNA using Agilent's 2100 Bioanalyzer (RIN ≥ 9.3 for all samples). Samples for RNA sequencing were prepared as previously described (Marioni et al. 2008). Each sample was sequenced in one lane of an Illumina GAII flow cell, and reads were mapped to the human (hg18), chimpanzee (panTro2), or rhesus macaque (rheMac2) genomes, as appropriate, with MAQ (Li et al. 2008) version 0.6.8. We then mapped the reads to a set of previously annotated 150,107 orthologous exons from 20,689 genes in the three species (Blekhman et al. 2010), excluding reads with MAQ mapping quality scores <10. Estimates of gene expression levels were obtained by summing the number of reads mapping to all exons of a gene. Gene expression was analyzed with an approach similar to that used previously (Marioni et al. 2008; Blekhman et al. 2010) (see File S1 for more details).
Comparing expression profiles and H3K4me3-associated regions:
Of the 14,526 genes with expression data and the 25,160 genes with orthologous TSS data, 12,559 genes overlapped (File S2). We estimated the proportion of genes whose interspecies differences in gene expression levels might be explained by corresponding differences in H3K4me3 presence at TSS, using various thresholds of statistical significance to classify genes as differentially expressed. We performed this analysis for gene expression differences across all ranges and by restricting our analysis to gene expression levels within and across the range in which H3K4me3 also varies. We also performed this analysis using subsampled ChIPseq data in which equal numbers of sequencing reads across samples were used to classify H3K4me3-associated regions in each sample.
We examined enrichment of GO annotations (The Gene Ontology Consortium (Ashburner et al. 2000), via the Web tool GeneTrail (http://genetrail.bioinf.uni-sb.de/) (Backes et al. 2007)) for the genes whose interspecies differences in gene expression levels could potentially be explained by differences in H3K4me3, using differentially expressed genes between species with H3K4me3 presence in any species as a background. An FDR correction was applied to the resulting P-values (Table S5, Table S6, and Table S7).
RESULTS
To investigate the relationship between changes in H3K4me3 status and differences in gene expression levels between humans and nonhuman primates, we collected gene expression data and identified genomic regions associated with the H3K4me3 modification in lymphoblastoid cell lines (LCLs) from humans, chimpanzees, and rhesus macaques.
Genome-wide profiles of H3K4me3:
We used chromatin immunoprecipitation followed by massively parallel sequencing (ChIPseq) to identify genomic regions associated with the H3K4me3 modification in LCLs from three individuals from each of the three species. Specifically, following ChIP with an antibody against H3K4me3, we sequenced enriched chromatin from each LCL with an Illumina Genome Analyzer II (GAII), using one lane of a flow cell per sample. As a control, we sequenced three pools of input chromatin, one from each species (see materials and methods, File S1, and Figure S1 for a description of sample processing, the ChIPseq protocol, and examples of positive control regions).
We obtained, on average, 17.8 m (± 0.7 m) reads per sequenced lane (Table S3). We used MAQ (Li et al. 2008) to align the sequence reads to their respective reference genomes (human, hg18; chimpanzee, panTro2; or rhesus macaque, rheMac2), filtered reads on the basis of mapping quality (see materials and methods), and used MACS (Zhang et al. 2008) to identify, in each sample, genomic regions with peaks of aligned sequencing reads, which correspond to regions associated with the H3K4me3 modification. To facilitate a comparison of H3K4me3 across species, we identified the orthologous sequences of all H3K4me3-associated regions in the genomes of human, chimpanzee, and rhesus macaque (initially, without applying an FDR cutoff; see materials and methods and File S1).
To minimize the number of falsely identified interspecies differences, we applied two statistical cutoffs to classify genomic regions as associated with H3K4me3. Specifically, conditional on observing an H3K4me3-associated region with high confidence (namely, using a stringent cutoff) in one individual, we assumed that a H3K4me3 modification was more likely to occur in the same region in other individuals as well, and accordingly relaxed the statistical cutoff for the classification of such secondary observations (see materials and methods, File S1, and Figure S2 for more details). Essentially, we used information across samples to increase the power to detect H3K4me3 peaks in any sample. We then merged overlapping H3K4me3-associated genomic regions across individuals (regardless of species) to define boundaries for H3K4me3-associated metaregions. We performed this step to account for possible ambiguity in the classification of the exact boundaries of the peaks. For all subsequent analyses, we considered the H3K4me3-associated metaregions (which are referred to throughout as H3K4me3-associated regions). Using this approach, we classified 19,105 genomic regions as associated with H3K4me3 modification in at least one individual, with an average of 12,394 H3K4me3 peaks per individual, with relatively little variation across individuals or species (SEM = 188; Table S3).
Overall, we found a high level of overlap in the localization of H3K4me3 across individuals. This property of the data is robust with respect to the specific statistical cutoffs used to classify H3K4me3-associated regions (Figure S3). Of all the regions associated with H3K4me3 in any individual, 57.7% were associated with H3K4me3 in at least one individual from each of the three species (Figure S4). As expected, we observed a higher overlap between individuals from the same species (77.7 ± 0.4%) than between individuals from different species (65.5 ± 0.6%). Moreover, consistent with the known phylogeny of the three species, the overlap in H3K4me3-associated regions is higher between human and chimpanzee individuals (69.5 ± 0.3%) than between rhesus macaque and either human (63.9 ± 0.4%) or chimpanzee individuals (63.2 ± 0.3%). These properties of the data are robust with respect to a broad range of statistical cutoffs used to classify genomic regions as associated with H3K4me3 modification (Figure S3 and Figure S4).
H3K4me3 near transcription start sites:
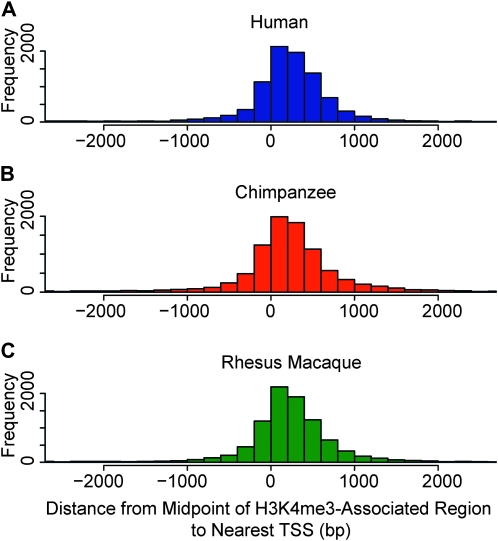
We next investigated whether the previously established (Santos-Rosa et al. 2002; Schneider et al. 2004; Schübeler et al. 2004; Pokholok et al. 2005) enrichment of H3K4me3 near TSS is observed in our data. To do so, we identified a set of TSS most likely to be orthologous across the three species (see materials and methods). Indeed, we found that H3K4me3-associated regions are enriched near annotated TSS in all three species (Figure 1). Our data also recapitulate the previously observed slight asymmetry in the distribution of H3K4me3 near TSS (e.g., Barski et al. 2007). On average, 61.2 ± 1.5% of all H3K4me3 peaks were found within 1 kb of a TSS, regardless of species. The enrichment of H3K4me3 near TSS is also robust with respect to a wide range of statistical cutoffs (Figure S5). The H3K4me3 modifications within 1 kb of TSS are more likely to be conserved across species than those that are farther from TSS (Figure S3). These observations are consistent with a conserved role for H3K4me3 in mediating transcription initiation.
Figure 1.—
H3K4me3 modifications are enriched near TSS in all three species. Plotted are histograms of the distance from the middle of H3K4me3-associated regions to the nearest TSS in (A) humans (B) chimpanzees, and (C) rhesus macaques. Only H3K4me3-associated regions within 2.5 kb of annotated TSS are shown (62.8 ± 1.3% of the total number of regions).
Our subsequent analyses focused on the subset of H3K4me3-associated regions that fall near TSS. Specifically, for each individual, we classified each TSS as either H3K4me3+ or H3K4me3−, on the basis of the presence of H3K4me3 within 1 kb of the TSS. To further minimize falsely identified interspecies differences, we classified a TSS as H3K4me3+ for the entire species whenever we detected H3K4me3 near the TSS in at least one individual.
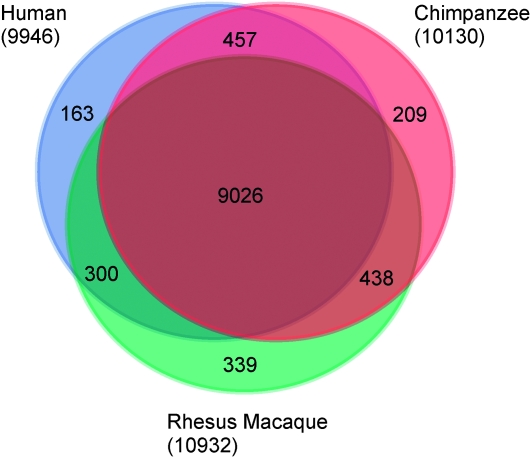
We then proceeded to classify genes as H3K4me3+ when at least one of their annotated TSS was H3K4me3+, on the basis of the criteria specified above. Using this approach, we classified 9026 of the 25,160 interrogated orthologous genes as H3K4me3+ in all three species and 163, 209, and 339 genes as H3K4me3+ only in humans, chimpanzees, or rhesus macaques, respectively (Figure 2).
Figure 2.—
Overlap of genes classified as H3K4me3+ across species. A gene was considered H3K4me3+ in a species when at least one individual had the histone modification. The number of total H3K4me3+ genes for each species is shown in parentheses; the number of total genes examined is 25,160. The same pattern was observed when equal (subsampled) numbers of ChIPseq reads from each individual were used for the analysis (Figure S6).
Relationship between variation in gene expression levels and H3K4me3:
To study interspecies differences in H3K4me3 in the context of gene expression differences between species, we extracted and sequenced RNA from the same nine cell lines, using one lane of a GAII flow cell for each sample (see materials and methods and File S1 for a detailed description of sample processing and quality control analysis). We focused on sequence reads that mapped to a previously annotated set of 150,107 orthologous exons (from 20,689 genes) in the three species (Blekhman et al. 2010). At these orthologous exons, overall patterns of interspecies variation in gene expression levels recapitulated the known phylogeny of the three species (Figure S7).
To identify genes whose expression levels differ between species, we analyzed the RNA sequencing data by using a Poisson mixed model including a fixed effect for each species and a random effect to account for variation between individuals from the same species (see File S1 for more details). Using this approach, we classified 2199, 5420, and 5702 genes as differentially expressed between humans and chimpanzees, humans and rhesus macaques, and chimpanzees and rhesus macaques, respectively, at an FDR of 5%.
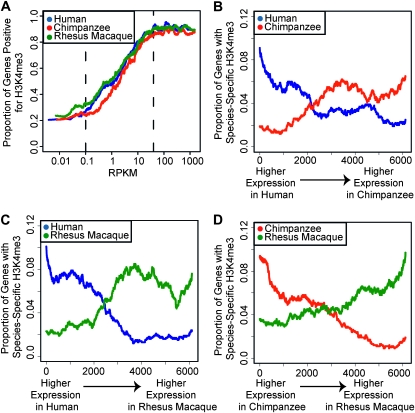
As observed previously (Santos-Rosa et al. 2002; Schneider et al. 2004; Schübeler et al. 2004; Pokholok et al. 2005), highly expressed genes are more likely to have the H3K4me3 modification at their TSS. This pattern was particularly striking across the distribution of moderately expressed genes (Figure 3A). For highly and lowly expressed genes outside of this range, the proportion of H3K4me3+ genes remains roughly constant.
Figure 3.—
H3K4me3 modifications are correlated with gene expression levels. (A) The proportion of H3K4me3+ genes is plotted for sliding windows (n = 500) of genes ordered by increasing expression levels in human (blue), chimpanzee (red), and rhesus macaque (green). Expression level (x-axis) is plotted as reads per kilobase of mappable exon, per million mapped reads (RPKM). Dotted lines bracket the range of expression values for which H3K4me3 levels also varied. (B–D) The proportion of genes that are H3K4me3+ in one species but H3K4me3− in the other is plotted for sliding windows (n = 1000) of genes ranked by the difference in expression levels between (B) human and chimpanzee, (C) human and rhesus macaque, or (D) chimpanzee and rhesus macaque. For example, in B, data points in blue correspond to the proportion of genes that are H3K4me3+ in human but H3K4me3− in chimpanzee, among the 1000 genes in a given window. Similar patterns were observed when equal (subsampled) numbers of ChIPseq reads from each individual were used for the analysis (Figure S8).
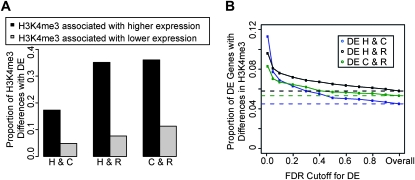
We therefore examined the relationship between H3K4me3 and differences in gene expression levels between species, focusing specifically on the range of expression levels that were correlated with H3K4me3 presence at the TSS. Within and across this range, genes detected as differentially expressed between species were more likely to be classified as H3K4me3+ in the species in which the gene was expressed at a higher level, and as H3K4me3− in the species in which the gene was expressed at a lower level (Figures 3, B–D; see Figure 4 for examples). This pattern provides strong evidence for a mechanism that relates interspecies differences in H3K4me3 status to differences in gene expression levels between species. Indeed, overall (across all expression ranges), genes whose expression levels, as well as their H3K4me3 status, differ across species were three to five times more likely to be classified as H3K4me3+ in the species in which the gene was expressed at a higher level (sign test P < 10−14; Figure 5A and Figure S9).
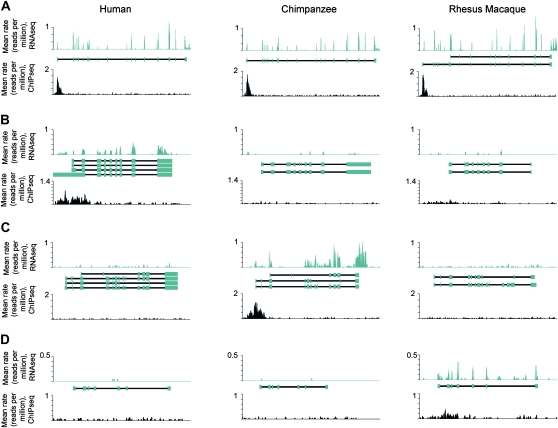
Figure 4.—
Examples of patterns of H3K4me3 peaks and gene expression levels across species. In each plot, the x-axis is distance along a genomic region containing a gene. The relative locations of Ensembl annotated exons are shown as blue boxes in the middle, with TSS to the left. The y-axes are the species-averaged rates at which each base was sequenced, scaled by a factor of 106, for H3K4me3 ChIPseq (top plots within each panel), and RNAseq (bottom plots within each panel) data from human (left), chimpanzee (middle), and rhesus macaque (right) samples. Shown are examples of (A) no difference in gene expression levels and no difference in H3K4me3 marks across species (RBM28), (B) species-specific higher gene expression level and lineage-specific H3K4me3 mark in human (ELF3), (C) chimpanzee (SSH3L), or (D) rhesus macaque (LCN15).
Figure 5.—
The association between interspecies differences in gene expression levels and H3K4me3 modifications. (A) We plotted the proportion of genes that are positive for H3K4me3 in one species and negative in another, which also were classified as differentially expressed in either the expected (dark bars) or unexpected (light bars) direction (at FDR ≤ 5%). (B) For each pairwise species comparison, we plotted the proportion of differentially expressed genes that show interspecies differences in H3K4me3 status in the expected direction (y-axis), over a range of FDR cutoffs used for classifying genes as differentially expressed between the species (x-axis). The dotted lines correspond to the overall (genome-wide) proportion of genes that show differences in H3K4me3 status in the expected direction, between human and chimpanzee (blue), between human and rhesus macaque (black), and between chimpanzee and rhesus macaque (green). In both panels, we refer to each species as “H” (human) “C” (chimpanzee), or “R” (rhesus macaque). Similar patterns were observed when equal (subsampled) numbers of ChIPseq reads from each individual were used for the analysis (Figure S9).
We explored the extent to which interspecies differences in gene expression levels might be explained, at least in part, by corresponding differences in H3K4me3 presence at the TSS. Because the results of such an analysis would differ on the basis of the choice of statistical cutoffs for the classification of genes as differentially expressed, we used a range of cutoffs. Importantly, more stringent statistical cutoffs resulted in higher estimates of the proportion of interspecies gene expression differences that can potentially be explained by differences in H3K4me3 status between the species (Figure 5B). Similarly, higher fold expression level differences across species were also associated with a higher proportion of differences in H3K4me3 status (Figure S9). If we conservatively interpret the overall genome-wide level of pairwise interspecies differences in H3K4me3 status as background (with no functional significance), we estimate that for a range of typically used FDR cutoffs (1–10%), interspecies differences in H3K4me3 presence at TSS could explain as little as 2.4% (205/2978 at FDR = 10%, with a background correction of 4.5%; Figure 5B) and as much as 6.8% (62/549 at FDR = 1%, with the same background correction) of gene expression differences between human and chimpanzee. Similarly, we estimate that interspecies differences in H3K4me3 could explain from 1.8% (411/5403 with a background correction of 5.8%) to 3.8% (251/2616) of gene expression differences between human and rhesus macaque, and from 1.4% (402/6004 with a background correction of 5.3%) to 3.0% (243/2933) of gene expression differences between chimpanzee and rhesus macaque (Figure 5B and see Figure S9 for a similar analysis using only genes whose expression levels fall within or across the range where H3K4me3 also varies).
We further classified differences in H3K4me3 status between humans and chimpanzees as ancestral or derived in each lineage, on the basis of the assumption that the H3K4me3 status of rhesus macaques represents the ancestral state. Interestingly, we found that when differences in gene expression levels between humans and chimpanzees could be explained by corresponding interspecies differences in H3K4me3 status, we could often predict, on the basis of the ancestral H3K4me3 status, the relative gene expression levels in rhesus macaques (for example, in 121/146 or 83% of cases, when interspecies gene expression differences were classified at FDR < 0.05; sign test P < 10−15).
Analysis of functional enrichments:
We examined the functional annotations (based on Gene Ontology; Ashburner et al. 2000) of the genes whose interspecies differences in gene expression levels could potentially be explained by differences in H3K4me3 status (compared to a background of differentially expressed genes between species, which are associated with H3K4me3 in any of the species). Among notable observations (see Table S5, Table S6, and Table S7 for complete lists of results), we found enrichments of genes involved in developmental processes (FDR < 10−4) and genes related to the plasma membrane (FDR < 10−6), among genes that were differentially expressed, possibly due to changes in H3K4me3 status, in all three pairwise species comparisons. Among genes whose regulation is inferred to be affected by interspecies changes in H3K4me3 we also found a significant enrichment of genes encoding for proteins with receptor activity (FDR < 0.002) and system development (FDR < 10−3). These results were consistent regardless of the species pairwise comparisons we made, indicating that common biological functions may be regulated by changes to epigenetic mechanism in primates. These observations are interesting in light of the established role of epigenetic markers in development in general (Kouzarides 2007) and specifically in B cell development (Busslinger 2004), and the importance of lymphocyte membrane-bound receptors in immune function (Alberts et al. 2002), a phenotype on which there has been strong selective pressures in primates (Barreiro and Quintana-Murci 2010).
DISCUSSION
Comparative studies of regulatory mechanisms in primates are challenging because of the practical and ethical constraints on genetic experimentation in apes and because of the limited availability of tissue samples (especially from chimpanzees). The difficulty in obtaining primate tissue samples often renders comparative studies somewhat difficult to interpret, for example because the cellular composition cannot be controlled across samples from different species (Blekhman et al. 2008, 2009). Here, we chose to work with LCLs, an abundant source of material, which allows us to work on the same cell type in all three primate species.
The usefulness of LCLs for studies of gene regulatory mechanisms has been strongly established. Indeed, nearly all genome-wide surveys of functional regulatory variation in humans have been conducted in LCLs. Cell lines offer convenience and replicability and are often the only resource available for functional studies in humans and nonhuman apes. However, while convenient, work with LCLs is often criticized because the Epstein–Barr virus transformation is associated with a number of specific artifacts. For example, cell lines often carry chromosomal abnormalities (Redon et al. 2006), have certain altered patterns of gene expression and DNA methylation (Hannula et al. 2001; Carter et al. 2002), and may have batch effects related to preparation and/or growth rates (Akey et al. 2007; Choy et al. 2008), which could be more pronounced for LCLs from different species.
Reassuringly, recent observations indicate that functional studies in cell lines uncover a substantial amount of genetic variation that affects gene expression levels in primary tissues. Studies in LCLs have resulted in numerous important insights into mechanisms of gene regulation (Monks et al. 2004; Morley et al. 2004; Cheung et al. 2005; Stranger et al. 2005, 2007; Dixon et al. 2007; Frazer et al. 2007; Moffatt et al. 2007; Veyrieras et al. 2008; Ge et al. 2009). In addition, regulatory architecture found in LCLs is often replicated in primary tissues (Bullaughey et al. 2009; Dimas et al. 2009; Verlaan et al. 2009; Zeller et al. 2010), with the most recent study estimating that ∼70% of cis expression quantitative trait loci (eQTL) identified in LCLs can be replicated in primary skin tissue (Ding et al. 2010).
With respect to comparative studies in primates, Khaitovich et al. (2006) have shown that patterns of interspecies gene expression differences across LCLs and primary tissues are similar. Specifically, they classified (at P < 0.001) similar numbers of transcripts as differentially expressed between human and chimpanzee in LCLs (1369), heart (1094), and brain (1403). Moreover, the overlap in interspecies differentially expressed transcripts between LCLs and each of the investigated primary tissues (6–15%) was similar to the observed overlap across primary tissues (5–19%). Thus, while LCLs are not a perfect model for comparative studies of gene regulatory mechanisms, on balance, cell lines are a useful system (and at times, the only available system) for such studies.
Interspecies differences in H3K4me3:
In our comparative study of primate LCLs, we found an enrichment of H3K4me3 near TSS in all three species, consistent with previous observations in more distantly related species, e.g., in mouse (Bernstein et al. 2005), chicken (Schneider et al. 2004), Drosophila (Schübeler et al. 2004), Arabidopsis (Zhang et al. 2009), and yeast (Santos-Rosa et al. 2002; Pokholok et al. 2005). This remarkably conserved pattern strongly implies that the functional mechanism by which H3K4me3 relates to gene regulation (Li et al. 2006; Wysocka et al. 2006) is shared across species. We further observed that the relationship between H3K4me3 and gene expression is strongest for moderate expression levels. This property of the data suggests that H3K4me3 may play an important role in promoting transcription initiation to moderate levels, while other mechanisms (for example, binding of enhancer or repressor elements) are likely required to achieve finer regulation. Interestingly, consistent with previous reports (Bernstein et al. 2005; Wilson et al. 2008), we found no correlation between interspecies differences in H3K4me3 status and sequence divergence at the corresponding genomic regions, either proximal or distal to TSS (Figure S10).
Overall, we estimated that up to 7% (10% if we restrict our analysis to the informative expression level range, Figure S9) of interspecies differences in gene expression levels might be explained, at least in part, by differences in H3K4me3 status across species. We note that the inference of causality relies exclusively on the previously proposed mechanism by which H3K4me3 promotes accessible chromatin and facilitates active transcription (Nishioka et al. 2002; Li et al. 2006; Wysocka et al. 2006; Ruthenburg et al. 2007).
That said, the notion that corresponding interspecies differences in H3K4me3 status and gene expression levels are causally related is supported by our data. Indeed, we observed that interspecies differences in H3K4me3 status can be used to predict gene expression differences between species more effectively than the converse (Figure 5A and Figure S9). In addition, we found that lineage-specific gains or loses of H3K4me3 are associated with lineage-specific changes in gene expression levels much more often than expected by chance alone.
Robustness of the results with respect to the classification approach:
We classified a gene as positive for H3K4me3 in each species when at least one individual of that species was positive for H3K4me3. This approach was motivated by the choice of a highly stringent criterion with which we classified genomic regions as associated with H3K4me3, as well as by the understanding that ChIPseq is inherently a low-power technique, such that false negatives (but not false positives) are expected to be frequent. Nevertheless, we are unable to exclude the possibility that the observed within-species variation in H3K4me3 modifications reflects polymorphisms rather than false negatives or positives. It is important, therefore, to ensure that our conclusions are robust with respect to the choice of how to classify a “species” trait.
To do so, we reanalyzed the data by (1) classifying a species' H3K4me3 status at each genomic region by a “majority rule,” and (2) by only considering H3K4me3-associated regions for which all three individuals within each species agree with respect to their H3K4me3 status.
The application of the first approach resulted in the exclusion of 3670 (19%) of the regions that were classified as H3K4me3+ in the original analysis, suggesting that typically, sites were classified as H3K4me3+ in more than one individual per species. Of the remaining 15,435 H3K4me3+ regions, 59% were shared across all three species (compared with 58% in the original analysis; Figure S4). The percentage of genes with significant interspecies differences in expression levels (at FDR of 1–10%) that also shows corresponding differences in H3K4me3 status (in the expected direction) is slightly higher than that observed in the original analysis (Figure S9; for comparisons between humans and chimpanzees, 3.1–7.3%; humans and rhesus macaques, 2.4–4.6%; and chimpanzees and rhesus macaques, 1.5–3.9%).
In turn, the application of the second approach resulted in the exclusions of more than half (56%) the regions that were classified as H3K4me3+ in the original analysis. This is a very strict classification approach, which reflects the unlikely possibility that the false positive rates associated with our ChIPseq experiment are very high and the false negative rates extremely low. Naturally, since we exclude any region in which all three individuals of a species do not agree, the remaining H3K4me3-associated regions showed a higher overlap between species compared with the original analysis. Specifically, 8442 regions were classified as positive for H3K4me3 in all three individuals of any species, of which 81% were shared across all three species (Figure S4). Importantly, however, the percentage of genes with significant interspecies differences in expression levels that also show corresponding differences in H3K4me3 status is similar to that observed in the original analysis (Figure S9; for comparisons between humans and chimpanzees, 1.6–4.6%; humans and rhesus macaques, 1.2–2.8%; and chimpanzees and rhesus macaques, 0.9–1.9%).
Our original analysis, therefore, is generally conservative with respect to the estimates of the proportion of interspecies gene expression differences that might be explained by corresponding differences in H3K4me3 between the species. More generally, our qualitative results are robust with respect to the specific choice on how to classify a gene as H3K4me3+ positive in a given species.
Summary
Our results suggest that changes in H3K4me3 likely contribute modestly to differences in gene expression levels between primates. We confirmed that these qualitative results of our comparative genomic study do not rely on specific arbitrary choices of statistical cutoffs (Figure S3, Figure S4, Figure S5, Figure S6, Figure S8, Figure S9, and Figure S10). That said, it is reasonable to assume that we are somewhat underestimating the number of interspecies differences in H3K4me3 status, because we consistently chose approaches that will minimize falsely identified differences between species. In addition, it is important to note that while we can identify large interspecies differences in H3K4me3 status using our data (effectively, focusing on qualitative differences), we would need higher quality data (higher than most ChIPseq datasets to date) to infer more subtle quantitative differences.
Our study joins similar efforts (Odom et al. 2007; Wilson et al. 2008; McManus et al. 2010) in taking first steps toward understanding the basis for gene expression differences between species. By collecting comparative data on other regulatory mechanisms, as well as by extending our analysis to consider quantitative interspecies differences in dynamic regulatory interactions, we hope to ultimately develop a better understanding of the relative contributions of the different mechanisms to regulatory differences across primates.
Acknowledgments
We thank L. Barreiro and all members of the Gilad lab for discussions and/or for comments on the manuscript. We thank the New England Primate Research Center and the New Iberia Research Center (University of Louisiana at Lafayette) for the primate LCLs. The University of Louisiana at Lafayette New Iberia Research Center is funded by National Institutes of Health/National Center for Research Resources (NIH/NCRR) grants RR015087, RR014491, and RR016483, and the Genetics Core of the New England Primate Research Center by NIH/NCRR grant RR00168. This work was supported by National Institute of General Medical Sciences grant GM084996 to Y.G.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.126177/DC1.
Available freely online through the author-supported open access option.
The RNAseq and ChIPseq data are available at the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under series accession no. GSE24111.
References
- Akey, J. M., S. Biswas, J. T. Leek and J. D. Storey, 2007. On the design and analysis of gene expression studies in human populations. Nat. Genet. 39 807–808, author reply 808–809. [DOI] [PubMed] [Google Scholar]
- Alberts, B., A. Johnson, J. Lewis, M. Raff, K. Roberts et al., 2002. Molecular Biology of the Cell. Garland Science, New York.
- Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler et al., 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbitt, C. C., O. Fedrigo, A. D. Pfefferle, A. P. Boyle, J. E. Horvath et al., 2010. a Both noncoding and protein-coding RNAs contribute to gene expression evolution in the primate brain. Genome Biol. Evol. 2 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbitt, C. C., J. S. Silverman, R. Haygood, J. M. Reininga, M. V. Rockman et al., 2010. b Multiple functional variants in cis modulate PDYN expression. Mol. Biol. Evol. 27 465–479. [DOI] [PubMed] [Google Scholar]
- Backes, C., A. Keller, J. Kuentzer, B. Kneissl, N. Comtesse et al., 2007. GeneTrail–advanced gene set enrichment analysis. Nucleic Acids Res. 35 W186–W192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro, L. B., and L. Quintana-Murci, 2010. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat. Rev. Genet. 11 17–30. [DOI] [PubMed] [Google Scholar]
- Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones et al., 2007. High-resolution profiling of histone methylations in the human genome. Cell 129 823–837. [DOI] [PubMed] [Google Scholar]
- Bernstein, B. E., M. Kamal, K. Lindblad-Toh, S. Bekiranov, D. K. Bailey et al., 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120 169–181. [DOI] [PubMed] [Google Scholar]
- Blekhman, R., A. Oshlack, A. E. Chabot, G. K. Smyth and Y. Gilad, 2008. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Genet. 4 e1000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blekhman, R., A. Oshlack and Y. Gilad, 2009. Segmental duplications contribute to gene expression differences between humans and chimpanzees. Genetics 182 627–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blekhman, R., J. Marioni, P. Zumbo, M. Stephens and Y. Gilad, 2010. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 20 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullaughey, K., C. I. Chavarria, G. Coop and Y. Gilad, 2009. Expression quantitative trait loci detected in cell lines are often present in primary tissues. Hum. Mol. Genet. 18 4296–4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger, M., 2004. Transcriptional control of early B cell development. Annu. Rev. Immunol. 22 55–79. [DOI] [PubMed] [Google Scholar]
- Cáceres, M., J. Lachuer, M. A. Zapala, J. C. Redmond, L. Kudo et al., 2003. Elevated gene expression levels distinguish human from non-human primate brains. Proc. Natl. Acad. Sci. USA 100 13030–13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, K. L., E. Cahir-McFarland and E. Kieff, 2002. Epstein-Barr virus-induced changes in B-lymphocyte gene expression. J. Virol. 76 10427–10436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot, A., R. A. Shrit, R. Blekhman and Y. Gilad, 2007. Using reporter gene assays to identify cis regulatory differences between humans and chimpanzees. Genetics 176 2069–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, V. G., R. S. Spielman, K. G. Ewens, T. M. Weber, M. Morley et al., 2005. Mapping determinants of human gene expression by regional and genome-wide association. Nature 437 1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy, E., R. Yelensky, S. Bonakdar, R. M. Plenge, R. Saxena et al., 2008. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 4 e1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimas, A. S., S. Deutsch, B. E. Stranger, S. B. Montgomery, C. Borel et al., 2009. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 325 1246–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, J., J. E. Gudjonsson, L. Liang, P. E. Stuart, Y. Li et al., 2010. Gene expression in skin and lymphoblastoid cells: refined statistical method reveals extensive overlap in cis-eQTL signals. Am. J. Hum. Genet. 87 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, A. L., L. Liang, M. F. Moffatt, W. Chen, S. Heath et al., 2007. A genome-wide association study of global gene expression. Nat. Genet. 39 1202–1207. [DOI] [PubMed] [Google Scholar]
- Enard, W., P. Khaitovich, J. Klose, S. Zollner, F. Heissig et al., 2002. Intra- and interspecific variation in primate gene expression patterns. Science 296 340–343. [DOI] [PubMed] [Google Scholar]
- Enard, W., A. Fassbender, F. Model, P. Adorjan, S. Paabo et al., 2004. Differences in DNA methylation patterns between humans and chimpanzees. Curr. Biol. 14 R148–R149. [PubMed] [Google Scholar]
- Farcas, R., E. Schneider, K. Frauenknecht, I. Kondova, R. Bontrop et al., 2009. Differences in DNA methylation patterns and expression of the CCRK gene in human and nonhuman primate cortices. Mol. Biol. Evol. 26 1379–1389. [DOI] [PubMed] [Google Scholar]
- Frazer, K. A., D. G. Ballinger, D. R. Cox, D. A. Hinds, L. L. Stuve et al., 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Sosa, M. A., R. M. Midgett, V. A. Slagel, S. Githens, K. C. Kuo et al., 1983. Tissue-specific differences in DNA methylation in various mammals. Biochim. Biophys. Acta 740 212–219. [DOI] [PubMed] [Google Scholar]
- Ge, B., D. K. Pokholok, T. Kwan, E. Grundberg, L. Morcos et al., 2009. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat. Genet. 41 1216–1222. [DOI] [PubMed] [Google Scholar]
- Gerber, M., and A. Shilatifard, 2003. Transcriptional elongation by RNA polymerase II and histone methylation. J. Biol. Chem. 278 26303–26306. [DOI] [PubMed] [Google Scholar]
- Gilad, Y., A. Oshlack, G. K. Smyth, T. P. Speed and K. P. White, 2006. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature 440 242–245. [DOI] [PubMed] [Google Scholar]
- Hampsey, M., and D. Reinberg, 2003. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell 113 429–432. [DOI] [PubMed] [Google Scholar]
- Hannula, K., M. Lipsanen-Nyman, S. W. Scherer, C. Holmberg, P. Hoglund et al., 2001. Maternal and paternal chromosomes 7 show differential methylation of many genes in lymphoblast DNA. Genomics 73 1–9. [DOI] [PubMed] [Google Scholar]
- Heintzman, N. D., R. K. Stuart, G. Hon, Y. Fu, C. W. Ching et al., 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39 311–318. [DOI] [PubMed] [Google Scholar]
- Karaman, M. W., M. L. Houck, L. G. Chemnick, S. Nagpal, D. Chawannakul et al., 2003. Comparative analysis of gene-expression patterns in human and African great ape cultured fibroblasts. Genome Res. 13 1619–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlic, R., H. R. Chung, J. Lasserre, K. Vlahovicek and M. Vingron, 2010. Histone modification levels are predictive for gene expression. Proc. Natl. Acad. Sci. USA 107 2926–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich, P., B. Muetzel, X. She, M. Lachmann, I. Hellmann et al., 2004. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 14 1462–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich, P., I. Hellmann, W. Enard, K. Nowick, M. Leinweber et al., 2005. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science 309 1850–1854. [DOI] [PubMed] [Google Scholar]
- Khaitovich, P., W. Enard, M. Lachmann and S. Paabo, 2006. Evolution of primate gene expression. Nat. Rev. Genet. 7 693–702. [DOI] [PubMed] [Google Scholar]
- Kouzarides, T., 2007. Chromatin modifications and their function. Cell 128 693–705. [DOI] [PubMed] [Google Scholar]
- Li, H., S. Ilin, W. Wang, E. M. Duncan, J. Wysocka et al., 2006. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., J. Ruan and R. Durbin, 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel, D. A., M. V. Rockman, G. A. Wray, J. Altmann and S. C. Alberts, 2006. Ancient polymorphism and functional variation in the primate MHC-DQA1 5′ cis-regulatory region. Proc. Natl. Acad. Sci. USA 103 16331–16336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni, J. C., C. E. Mason, S. M. Mane, M. Stephens and Y. Gilad, 2008. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18 1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus, C. J., J. D. Coolon, M. O. Duff, J. Eipper-Mains, B. R. Graveley et al., 2010. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 20 816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt, M. F., M. Kabesch, L. Liang, A. L. Dixon, D. Strachan et al., 2007. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448 470–473. [DOI] [PubMed] [Google Scholar]
- Monks, S. A., A. Leonardson, H. Zhu, P. Cundiff, P. Pietrusiak et al., 2004. Genetic inheritance of gene expression in human cell lines. Am. J. Hum. Genet. 75 1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley, M., C. M. Molony, T. M. Weber, J. L. Devlin, K. G. Ewens et al., 2004. Genetic analysis of genome-wide variation in human gene expression. Nature 430 743–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, H. H., F. Robert, R. A. Young and K. Struhl, 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11 709–719. [DOI] [PubMed] [Google Scholar]
- Nishioka, K., S. Chuikov, K. Sarma, H. Erdjument-Bromage, C. D. Allis et al., 2002. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 16 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nislow, C., E. Ray and L. Pillus, 1997. A yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol. Biol. Cell 8 2421–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom, D. T., R. D. Dowell, E. S. Jacobsen, W. Gordon, T. W. Danford et al., 2007. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat. Genet. 39 730–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett et al., 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122 517–527. [DOI] [PubMed] [Google Scholar]
- Pollard, K. S., S. R. Salama, N. Lambert, M. A. Lambot, S. Coppens et al., 2006. An RNA gene expressed during cortical development evolved rapidly in humans. Nature 443 167–172. [DOI] [PubMed] [Google Scholar]
- Prabhakar, S., A. Visel, J. A. Akiyama, M. Shoukry, K. D. Lewis et al., 2008. Human-specific gain of function in a developmental enhancer. Science 321 1346–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon, R., S. Ishikawa, K. R. Fitch, L. Feuk, G. H. Perry et al., 2006. Global variation in copy number in the human genome. Nature 444 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman, M. V., M. W. Hahn, N. Soranzo, F. Zimprich, D. B. Goldstein et al., 2005. Ancient and recent positive selection transformed opioid cis-regulation in humans. PLoS Biol. 3: e387. [DOI] [PMC free article] [PubMed]
- Roh, T. Y., G. Wei, C. M. Farrell and K. Zhao, 2007. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 17 74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg, A. J., C. D. Allis and J. Wysocka, 2007. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell 25 15–30. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein et al., 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419 407–411. [DOI] [PubMed] [Google Scholar]
- Schneider, R., A. J. Bannister, F. A. Myers, A. W. Thorne, C. Crane-Robinson et al., 2004. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 6 73–77. [DOI] [PubMed] [Google Scholar]
- Schübeler, D., D. M. MacAlpine, D. Scalzo, C. Wirbelauer, C. Kooperberg et al., 2004. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 18 1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger, B. E., M. S. Forrest, A. G. Clark, M. J. Minichiello, S. Deutsch et al., 2005. Genome-wide associations of gene expression variation in humans. PLoS Genet. 1 e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger, B. E., A. C. Nica, M. S. Forrest, A. Dimas, C. P. Bird et al., 2007. Population genomics of human gene expression. Nat. Genet. 39 1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlaan, D. J., B. Ge, E. Grundberg, R. Hoberman, K. C. Lam et al., 2009. Targeted screening of cis-regulatory variation in human haplotypes. Genome Res. 19 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrieras, J. B., S. Kudaravalli, S. Y. Kim, E. T. Dermitzakis, Y. Gilad et al., 2008. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 4 e1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P., C. Lin, E. R. Smith, H. Guo, B. W. Sanderson et al., 2009. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol. Cell. Biol. 29 6074–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, L. R., C. C. Babbitt, A. E. Primus, T. F. Severson, R. Haygood et al., 2009. Functional consequences of genetic variation in primates on tyrosine hydroxylase (TH) expression in vitro. Brain Res. 1288 1–8. [DOI] [PubMed] [Google Scholar]
- Wilson, M. D., N. L. Barbosa-Morais, D. Schmidt, C. M. Conboy, L. Vanes et al., 2008. Species-specific transcription in mice carrying human chromosome 21. Science 322 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka, J., T. Swigut, H. Xiao, T. A. Milne, S. Y. Kwon et al., 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442 86–90. [DOI] [PubMed] [Google Scholar]
- Zegerman, P., B. Canas, D. Pappin and T. Kouzarides, 2002. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J. Biol. Chem. 277 11621–11624. [DOI] [PubMed] [Google Scholar]
- Zeller, T., P. Wild, S. Szymczak, M. Rotival, A. Schillert et al., 2010. Genetics and beyond–the transcriptome of human monocytes and disease susceptibility. PLoS One 5 e10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Y. V. Bernatavichute, S. Cokus, M. Pellegrini and S. E. Jacobsen, 2009. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10: R62. [DOI] [PMC free article] [PubMed]
- Zhang, Y., T. Liu, C. A. Meyer, J. Eeckhoute, D. S. Johnson et al., 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9 R137. [DOI] [PMC free article] [PubMed] [Google Scholar]