Summary
The antigen receptor for natural killer T cells (NKT TCR) bind CD1d-restricted microbial and self lipid antigens, although the molecular basis of self-CD1d recognition is unclear. Here, we have characterized NKT TCR recognition of CD1d molecules loaded with natural self-antigens (Ags), and report the 2.3 Å resolution structure of an autoreactive NKT TCR-phosphatidylinositol-CD1d complex. NKT TCR recognition of self and foreign antigens was underpinned by a similar mode of germline-encoded recognition of CD1d. However, NKT TCR autoreactivity is mediated by unique sequences within the non-germline encoded CDR3β loop encoding for a hydrophobic motif that promotes self-association with CD1d. Accordingly, NKT cell autoreactivity may arise from the inherent affinity of the interaction between CD1d and the NKT TCR, resulting in the recognition of a broad range of CD1d restricted self-antigens. This demonstrates that multiple self-antigens can be recognized in a similar manner by autoreactive NKT TCRs.
Introduction
Natural Killer T (NKT) cells are innate-like T cells that recognize lipid antigens presented by the non-polymorphic major histocompatibility complex (MHC) class I-like molecule CD1d. NKT cells express a highly restricted semi-invariant T-cell receptor (NKT TCR) repertoire. Upon stimulation, NKT cells rapidly produce large amounts of cytokines and chemokines. In this manner, NKT cells can modulate the response of other innate and adaptive immune cells, such as T and B cells, NK cells and dendritic cells (DC). Consistent with this, NKT cells have been shown to influence the outcome of immune responses in a broad range of diseases, including cancer, autoimmunity, inflammation and infection (Bendelac et al., 2007; Kronenberg, 2005; Matsuda et al., 2008).
NKT TCRs are typically composed of an invariant TCRα chain formed by a canonical rearrangement between the Vα14 and Jα18 gene segments in mice or their orthologs Vα24 and Jα18 in humans (Dellabona et al., 1994; Lantz and Bendelac, 1994). This TCRα chain associates with TCRβ chains limited in their Vβ usage (Vβ11 in humans and Vβ8s, Vβ7 and Vβ2 in mice) but with extensive junctional diversity in their complementary determining region 3 (CDR3β) (Matsuda et al., 2001).
NKT cells can recognize CD1d molecules bound to a number of different glycolipids. As some of these glycolipids are microbial in origin it is thought that NKT cells contribute to responses to invading bacteria. However, NKT cells also react with CD1d-expressing APCs in the absence of infectious organisms, suggesting that NKT TCRs can recognize CD1d plus endogenous lipids and thus play a role in autoimmunity or indirectly play a role in innate response to infections. Several studies have investigated the nature of the endogenous lipid antigen(s) presented by CD1d molecules in various immunological contexts. Over 170 lipid species, comprising glycerophospholipids and sphingolipids, have been identified from human CD1d monomers produced by the human B cell line 721.221 (Cox et al., 2009). Another study revealed that phosphatidylcholine, lysophospholipids and sphingomyelin represent the main lipids associated with human CD1d molecules expressed in this cell line (Yuan et al., 2009). Several glycosphingolipids, including GM1a, GD1a and GM2, are also found bound by mouse CD1d expressed by myeloid APCs (Muindi et al., 2010). Finally, various molecular species of phosphatidylcholine with different fatty acid compositions are found loaded into mouse CD1d molecules produced by insect cells (Giabbai et al., 2005). These studies reveal the great diversity of self-lipids that can be found associated with CD1d molecules when produced in different systems.
The structures of several mouse and human NKT TCRs in complex with CD1d bound to the marine sponge-derived antigen α-Galactosylceramide (αGC) (Borg et al., 2007; Pellicci et al., 2009) or related microbial glycolipids (Li et al., 2010) have recently been solved. These structures show that NKT TCRs, regardless of their precise TCRβ sequence, dock similarly on CD1d. The location and details of the docking are quite different from those seen in the engagement of conventional TCRs to their MHC plus peptide ligands (Godfrey et al., 2008). A “hot spot” on the NKT TCR, composed only of germline-encoded residues, is essential for recognition of CD1d plus several structurally diverse antigens, including isoglobotrihexosylceramide (iGb3), which has been proposed to be one of the self antigens responsible for NKT cell autoreactivity (Zhou et al., 2004). These studies also suggest that the diverse CDR3β loop can modulate and fine tune the interaction of NKT TCRs with CD1d-glycolipid antigen complexes (Mallevaey et al., 2009). Presently, whilst the structural basis of NKT TCR-CD1d-αGC recognition is established, it is unclear how the NKT TCR can recognize more diverse CD1d-restricted ligands including CD1d presenting self-antigens during autoreactivity, which underpins NKT cell activation in autoimmunity and cancer.
Self recognition by NKT cells is an important issue (Gapin, 2010). It is involved in positive selection of NKT cell precursors in the thymus (Bendelac et al., 2007) but is also implicated in many diseases including cancer (Bellone et al., 2010; Swann et al., 2007; Swann et al., 2009), ischemia-reperfusion injury, sickle cell disease (Lappas et al., 2006; Wallace et al., 2009) and innate responses to infection (Brigl and Brenner, 2009; Brigl et al., 2003; Mattner et al., 2005; Paget et al., 2007). Although the mechanisms underlying this “self-reactivity” are clearly an important issue, so far they remain poorly defined. The problem is difficult to investigate with the normal range of NKT TCRs because they have, on the whole, relatively low affinity for their endogenous ligands. To improve our ability to study NKT TCRs reaction with self, we isolated NKT TCRs that have high affinity for self ligands. With this approach, via functional and structural analyses, we showed that NKT TCR reactivity against a panel of CD1d-self lipid complexes is caused via the CDR3β loop directly mediating contacts with the monomorphic CD1d molecule, while the self antigen exclusively contacted the invariant α-chain. These results provide a molecular basis for understanding NKT cell autoreactivity, and suggest that a range of different CD1d-restricted self-lipids can be recognized in a similar manner by the NKT TCR.
Results
NKT TCR CDR3β loop modulates αGC-CD1d tetramer binding
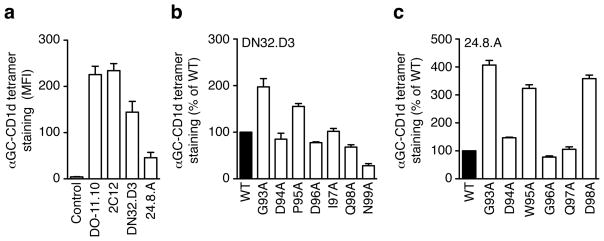
Previous mutational data suggests that the hypervariable CDR3β loop of NKT TCRs modulates the NKT TCR affinity for CD1d-antigen complexes (Mallevaey et al., 2009; Scott Browne et al., 2007), although the basis for this is unclear. To further assess the role of the CDR3β loop, we chose to clone 4 different Vβ8.2 chains with various CDR3β-Jβ rearrangements derived from NKT hybridomas (2C12) (Brossay et al., 1998a), DN32.D3 (Lantz and Bendelac, 1994) and 24.8.A (Gumperz et al., 2000) and the Vβ8.2 DO-11.10 TCRβ chain that we used previously (Scott Browne et al., 2007). We expressed each TCRβ chain separately in the TCR-negative 5KC hybridoma together with the invariant Vα14-Jα18 chain. After gating for identical amounts of TCR expression, all four hybridomas stained with αGC-CD1d tetramer with various intensities, with DO-11.10 and 2C12 being the highest, DN32.D3 moderate and 24.8.A the weakest (Fig 1a). Only minor variations on αGC-CD1d tetramer binding were detected in a previous alanine scanning of the DO-11.10 CDR3β loop (Scott Browne et al., 2007). Therefore, we also substituted individual residues in the CDR3β of DN32.D3 and 24.8.A with alanine and repeated the analysis with each mutant chain expressed together with the wild-type Vα14 chain. For DN32.D3, the βD94A, βD96A, βI97A and βQ98A substitutions had a minor or no effect on tetramer binding when compared to the wild-type TCR, while the βG93A and βP95A mutants enhanced interaction with the tetramer and the βN99A almost completely abolished staining (Fig 1b). For 24.8.A, the βD94A, βG96A and βQ97A substitutions had little effect on tetramer binding (Figure 1c), while the βG93A, βW95A and βD98A residue substitutions greatly enhanced tetramer staining up to 3 to 4-fold, demonstrating that the wild-type 24.8.A CDR3β is less than optimal for αGC-CD1d recognition. Together, these data demonstrate that amino acid composition of the hypervariable CDR3β loop can positively or negatively modulate NKT TCR recognition of the αGC-CD1d complex.
Figure 1.
TCR CDR3β loop modulates αGC-CD1d tetramer binding. (a) Staining of hybridomas expressing the Vα14 TCRα chain paired with the Vβ8.2 chain in the context of the indicated CDR3β-Jβ rearrangement. The MFI of αGC-CD1d tetramer staining was determined for a narrow TCR gate. Data represent the mean + s.e.m. of two independent experiments. (b, c) Staining of hybridomas expressing the Vα14i TCRα chain paired with wild-type or CDR3β alanine substitutions of the DN32.D3 (b) or 24.8.A (c) TCRβ chains. The MFI of αGC-CD1d tetramer staining was determined for a narrow TCR gate and is normalized to wild-type MFI (set as 100%). Data represents the mean + s.e.m. of two independent experiments.
CDR3β loops mediate self-CD1d complexes recognition
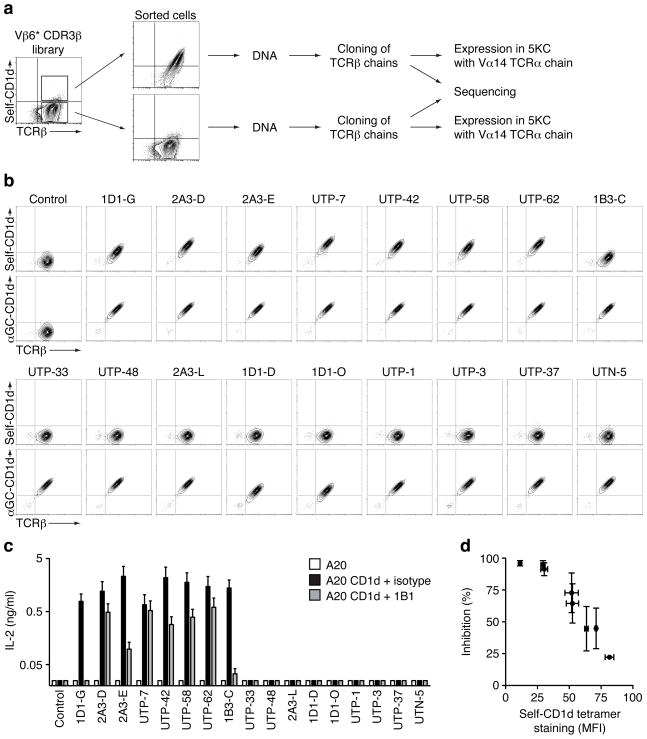
Next, we tested if the CDR3β loop could also modulate the affinity of the TCR for CD1d-self complexes. We have previously identified a population of cells, out of a library of hybridomas expressing NKT TCRs with randomized CDR3β loops, which could interact with CD1d tetramers prepared with recombinant mouse CD1d protein monomers produced in HEK 293 human cell lines (Mallevaey et al., 2009). These CD1d tetramers were presumably loaded with naturally expressed self-antigen(s). To identify the nature of the CDR3β responsible for this reactivity, we sorted self-CD1d tetramer-positive and -negative cells and PCR amplified the TCRβ chain (Fig 2a). Each TCRβ chain was sequenced and cloned into a retroviral vector and expressed separately with the invariant Vα14 chain into TCRα−β− hybridomas. The cells were then stained with the self- and αGC-CD1d tetramers. All hybridomas bound the αGC-CD1d tetramer and seven out of seventeen hybridomas also bound the CD1d tetramers loaded with natural self-antigen(s) (Fig 2b). These results demonstrate that particular CDR3β loop sequences can confer self-reactivity while retaining αGC reactivity. Sequence analysis of the CDR3β loops expressed by the highly self-reactive TCRs (mean fluorescent intensity (MFI) > 30) revealed the preferential usage of the hydrophobic residues leucine and/or isoleucine at the first and second position of the CDR3β loop, whereas the sequences found within the self-CD1d tetramer-negative cells appeared diverse (Supplementary Table 1).
Figure 2.
CDR3β loops and self-CD1d complex recognition. (a) Strategy used to identify CDR3β sequences that allow or not for self-CD1d complexes recognition. (b) Staining of hybridomas expressing the Vα14 TCRα chain paired with the indicated TCRβ chain. Data are representative of three independent experiments. (c) Enzyme-linked immunosorbent assay of interleukin-2 (IL-2) production by the indicated hybridomas in response to A20 cells transfected or not with mCD1d, in the presence of 10μg/ml 1B1 antibody or isotype control. Data represent the mean + s.e.m. of two independent experiments. (d) Staining by self-CD1d tetramers and inhibition of IL-2 production by 1B1 antibody for the hybridomas expressing self-CD1d-reactive NKT TCRs. Control, hybridoma expressing the Vα14i TCRα chain paired with the Vβ6 DO-11.10 TCRβ chain.
Hybridomas expressing TCRs that bound CD1d tetramers naturally loaded with self-lipid(s) responded well to A20 cells transfected with CD1d. This reactivity was inhibited by the addition of anti-CD1d (Fig 2c). Interestingly, the percentage of inhibition obtained by the addition of anti-CD1d inversely correlated to the relative avidity of each TCR expressed by the hybridoma, as measured by self-CD1d tetramer binding (Fig 2d). By contrast, hybridomas with TCRs that did not bind CD1d tetramers naturally loaded with self-lipid(s) were not autoreactive. Altogether, these data demonstrate that a hydrophobic stretch of amino acids within the CDR3β loop of αGC-CD1d tetramer-positive NKT TCRs can determine self-reactivity.
Crystal structure of an NKT TCR-self antigen-CD1d complex
To gain further insight as to how the CDR3β loop controls the reactivity with CD1d molecules naturally loaded with self-antigen(s), we expressed and refolded the autoreactive NKT TCR 2A3-D (Fig 2 and Supplementary Table 1), then formed and crystallized the complex with mouse CD1d molecules complexed to the self-antigen phosphatidylinositol (PI) (Supplementary Table 2). The electron density at the NKT TCR-CD1d interface was unambiguous (Supplementary Figure 1), thereby permitting detailed analysis into the structural basis of autoreactivity by the 2A3-D NKT TCR. NKT TCR-CD1d contacts are described below, while NKT TCR-PI contacts are detailed later in “Self-antigen permissibility of the NKT TCR”.
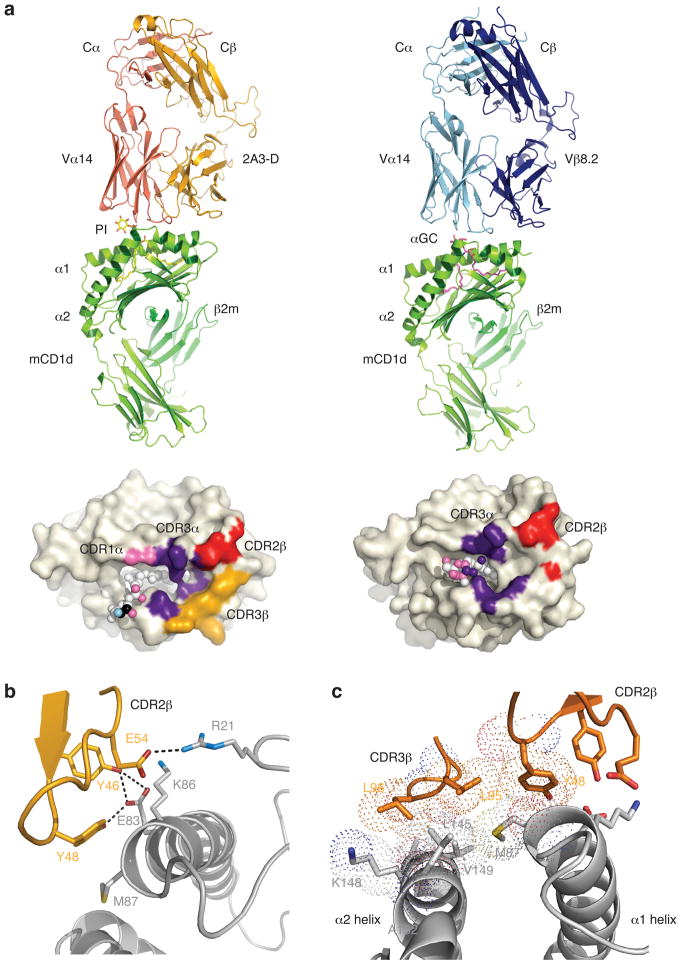
The NKT TCR docked in a parallel, tilted mode, directly above the F′-pocket of CD1d, as observed previously in NKT TCR-αGC-CD1d complexes, thereby providing immediate insight that the recognition of self-CD1d closely approximates the recognition of non-self CD1d-ligands (Fig 3a). Further, the conformation of the CD1d Ag-binding clefts when bound to the self-Ag and αGC were very similar (not shown), indicating that the auto-reactivity was attributable to an inherent property within the 2A3-D NKT TCR. Within this docking framework, the invariant CDR3α loop played a predominant role, interacting with both CD1d α-helices, whereas the Vα14 chain made a small contribution by contacting V72 and S76 of CD1d (Figure 3a, Supplementary Table 3).
Figure 3.
NKT TCR recognition of CD1d-PI. (a) Overview of the autoreactive 2A3-D NKT TCR-CD1d-PI complex (left hand side) compared to the Vβ8.2 NKT TCR-CD1d-αGC complex. Associated footprints on CD1d are shown underneath, and highlight the greater role of the CDR3β loop (gold) in the autoreactive complex. (b) Interactions mediated via the CDR2β loop in the 2A3-D NKT TCR-CD1d-PI complex (c) The two hydrophobic residues (Leu 95, Leu 96) in CDR3β act as a hydrophobic cap by shielding from solvent a surface exposed hydrophobic patch on CD1d. vdw radii shown as dots.
The 2A3-D NKT TCR was isolated from a library expressing modified Vβ6 chains in which the CDR2β loop had been replaced with the corresponding sequence found in the Vβ8.2 CDR2β loop, such as to provide the appropriate contact residues important for CD1d recognition (Mallevaey et al., 2009). The two tyrosine residues (βY46 and βY48) of the modified CDR2β loop converged on the same region of CD1d, with βY46 forming H-bonds with E83 and a salt bridge with K86, and βY48 sitting against M87 and H-bonding to E83. Furthermore, βE54 H-bonded to the main chain of CD1d and formed a salt bridge with K86 (Fig 3b, Supplementary Table 3); collectively these interactions were comparable to that previously observed in the Vβ8.2 NKT TCR-αGC-CD1d complex. The buried surface area of the autoreactive NKT TCR CD1d-complex was 940 Å2, compared to that of 760 Å2 of the Vβ8.2 NKT TCR-CD1d complex. Notably, while the CDR3β loop of the NKT TCR has previously been shown to play at most a minor role in contacting CD1d, for the 2A3-D NKT TCR, it played a more prominent role, exclusively interacting with CD1d, positioned away from the PI head group (Fig 3a). Specifically, the di-leucine motif at the tip of the CDR3β loop made direct contacts with L145 and K148 of CD1d. Further, these two leucine residues (βL95 and βL96) appeared to act as a hydrophobic cap, sitting above, and subsequently shielding from solvent, a hydrophobic region on CD1d, that comprised A152, V149, the aliphatic moiety of K148, L145 and M87 (Fig 3c). The hydrophobicity of this region was also extended by βL95 directly contacting βY48, the latter of which contacted M87 (Fig 3c).
Collectively, the structure showed that the hydrophobic nature of the tip of the CDR3β loop played an important role in driving NKT TCR autoreactivity, by sequestering a surface exposed, hydrophobic region of the CD1d-Ag binding cleft.
Mutational analysis of 2A3-D TCR
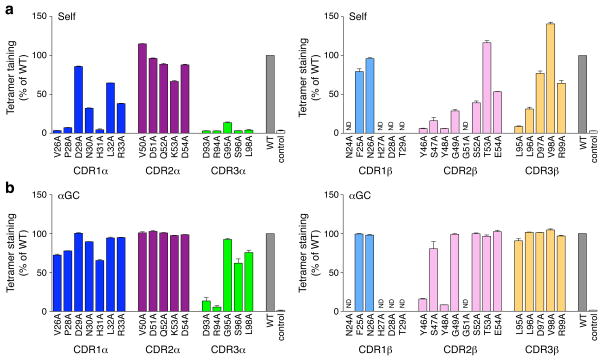
Next alanine substitution was applied to each residue in the CDR loops of the 2A3-D NKT TCR and each mutant was analyzed using a tetramer-binding assay. As shown in Figure 4, the effect of mutations in the three CDRs of the 2A3-D TCRα chain to the binding of self-CD1d tetramers was very similar to the pattern of reactivity of the NKT TCR with the DO11.10 TCRβ chain interacting with the αGC-CD1d complex (Scott Browne et al., 2007). The “hot spot”, composed of the CDR1α, CDR3α and CDR2β loops, was also important for the recognition of the natural self-antigen(s)-CD1d complex suggesting the mode of recognition of natural self antigen(s) by the NKT TCR is likely to be very similar to that of αGC. Interestingly, the βL95A and βL96A substitutions in the CDR3β loop largely abrogated self-CD1d tetramer binding, indicating that the hydrophobic interactions mediated via these residues are critically important for the interaction. We also tested the effect of these mutations on the binding to αGC-CD1d tetramers. Addition of αGC in tetramer preparations greatly enhanced staining intensities for all hybridomas. Only four residues, CDR3α D93 and R94 and CDR2β Y46 and Y48, were essential to the recognition of the αGC-CD1d complex, thereby defining the “core” residues within the hot spot necessary for interacting with the antigenic-CD1d complex. Interestingly, while the two leucine residues (95 and 96) in the CDR3β loop were important for recognition of the self-CD1d tetramers, we only detected a 10% reduction in αGC-CD1d tetramer binding as compared to the wild-type counterpart for the βL95A substitution and the βL96A substitution had no effect. Thus, while the CDR3β loop is not critically important for αGC-CD1d recognition, the two hydrophobic leucine residues at position β95 and β96 of the 2A3-D CDR3β were critical to confer reactivity to CD1d molecules loaded with natural self-lipids. The importance of this hydrophobic motif for the recognition of self-CD1d was further supported by the βL95I and βL95V mutations, which had either no effect or mild effects on self-CD1d tetramer binding, respectively (see Supplementary Figure 2).
Figure 4.
Mutational analysis of the 2A3-D NKT TCR. Staining of hybridomas expressing mutant versions (horizontal axes) of the Vα14i-2A3-Dβ TCRα (left) or TCRβ (right) with mouse CD1d tetramers loaded with naturally expressed self-antigens(s) (a) or αGC (b). Dark blue, CDR1α, magenta, CDR2α, green, CDR3α, light blue, CDR1β, light pink, CDR2β, yellow, CDR3β. WT, unsubstituted Vα14i-2A3-Dβ TCR (wild-type controls); control, Vα14i-Vβ6-DOβ TCR. ND, not done. The MFI of tetramer staining for each mutant was determined for a narrow TCR gate and is normalized to wild-type MFI (set as 100%). Data represent the mean normalized MFI + s.e.m. of at least two independent experiments.
Grafting of autoreactive CDR3β loop transfers autoreactivity
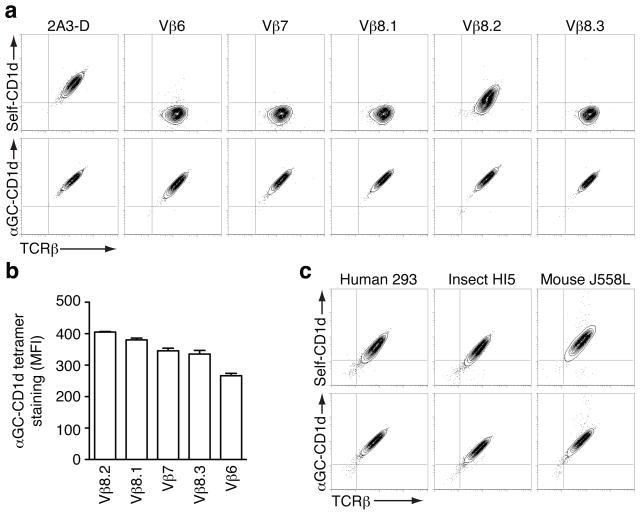
We cloned the Vβ6, Vβ7, Vβ8.1, Vβ8.2 and Vβ8.3 chains separately with the 2A3-D CDR3β loop to test whether it would be sufficient to provide reactivity to self-CD1d molecules. 5KC hybridoma containing the Vα14 invariant chain were transduced with each TCRβ construct and tested for their interaction with the CD1d tetramers loaded with natural self-lipid(s) and αGC-CD1d tetramers. Importantly, in a similar analysis performed with all mouse Vβ chains containing the DO-11.10 CDR3β loop, no reactivity to CD1d loaded with natural self-lipid(s) was detected but NKT TCRs demonstrated a wide range of avidity for CD1d-antigen complexes as a function of Vβ usage, with Vβ8.2 displaying the highest relative avidity while others, such as Vβ6, showed no interaction with CD1d-antigen complexes (Mallevaey et al., 2009). With this new CDR3β loop however, all NKT TCRs stained intensely with αGC-CD1d tetramers, and the Vβ8.2-containing NKT TCR now stained with self-CD1d tetramers (Fig 5a). Interestingly, the hierarchy of relative avidity conferred by different Vβ usage within the TCR (Vβ8.2 > Vβ8.1 ≥ Vβ7 > Vβ8.3 > Vβ6) was conserved irrespective of the CDR3β loop used (Fig 5b). Altogether, these results show that the 2A3-D CDR3β loop similarly increases the overall affinity of the NKT TCR for the CD1d-antigen complexes, including those comprised of self-antigens. We next tested whether CD1d tetramers derived from either human 293 cells, insect cells, or mouse B cell lymphoma J558L (Schumann et al., 2003), differed in their ability to bind the 2A3-D TCR. 2A3-D hybridomas stained similarly in each case (Fig 5c) and furthermore, similar staining was observed when αGC was added to the reagents. This important result suggests that the 2A3-D TCR can interact with mouse CD1d molecules loaded, perhaps, with a variety of different self lipid(s) and certainly with self lipids from a variety of cell sources.
Figure 5.
Grafting of autoreactive CDR3β loop transfers autoreactivity. Staining of hybridomas expressing the Vα14 TCRα chain paired with the indicated Vβ chains in the context of the 2A3-D CDR3β. (a) Representative dot plots of three independent experiments. (b) The MFI of αGC-CD1d tetramer staining was determined for a narrow TCR gate. Data represent the mean + s.e.m. of three independent experiments. (c) Reactivity of the 2A3-D NKT TCR with CD1d molecules. Staining of the 2A3-D hybridoma with tetramers made of mouse CD1d monomers, or mouse CD1d:Ig complexes, produced in the indicated cell lines. Data are representative of three independent experiments.
Conserved recognition of self and exogenous antigens
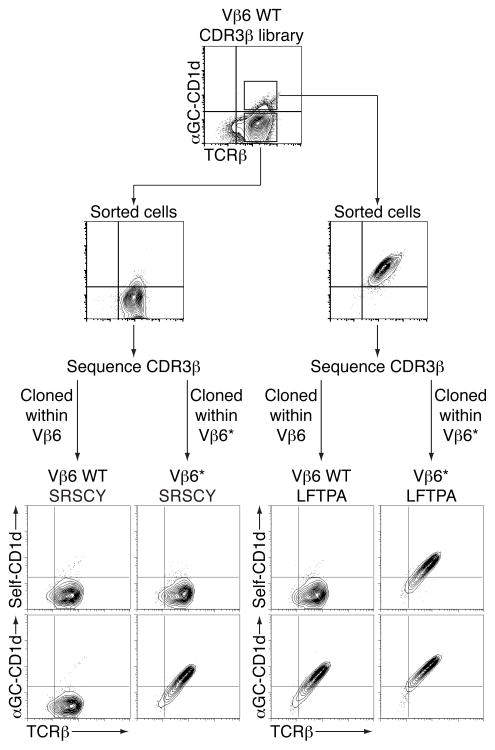
To extend the idea that the 2A3-D CDR3β loop can confer enhanced CD1d recognition independently of the antigen, we first isolated αGC-CD1d reactive and non-reactive hybridomas from a CDR3β library of Vβ6-expressing NKT TCR and sequenced their TCRβ chains (Mallevaey et al., 2009). The isolated CDR3β sequences were then cloned into wild-type or CDR2β-modified Vβ6 chains (denoted as Vβ6*) and transduced into 5KC together with the Vα14i chain (Fig 6). Staining intensities for self-CD1d tetramers and αGC-CD1d tetramers were compared. As expected, Vβ6-containing NKT TCRs bearing a CDR3β sequence with hydrophobic residues at position 1 and 2 (LFTPA) stained with αGC-CD1d tetramer (Fig 6). By simply transferring this CDR3β loops into the Vβ6* chains, the resulting TCR could now interact with the self-CD1d tetramers. An NKT TCR with a CDR3β sequence without the hydrophobic residues at the beginning of the loop (SRSCY) showed no interaction with self- or αGC-CD1d tetramers when this loop was expressed within wild-type Vβ6 chain, but stained with αGC-CD1d tetramer when the loop was expressed in the context of the Vβ6* chain. The experiment was repeated with other CDR3β loops derived from the two groups with similar results (data not shown). These results suggest that particular CDR3β loops can modulate the recognition by the NKT TCR of CD1d molecules presenting either self or exogenous lipid antigens similarly.
Figure 6.
Conserved recognition of self and exogenous antigens. Staining of hybridomas expressing the Vα14 TCRα chain paired with the indicated TCRβ chain. Data are representative of three independent experiments.
Self-antigen permissibility of the NKT TCR
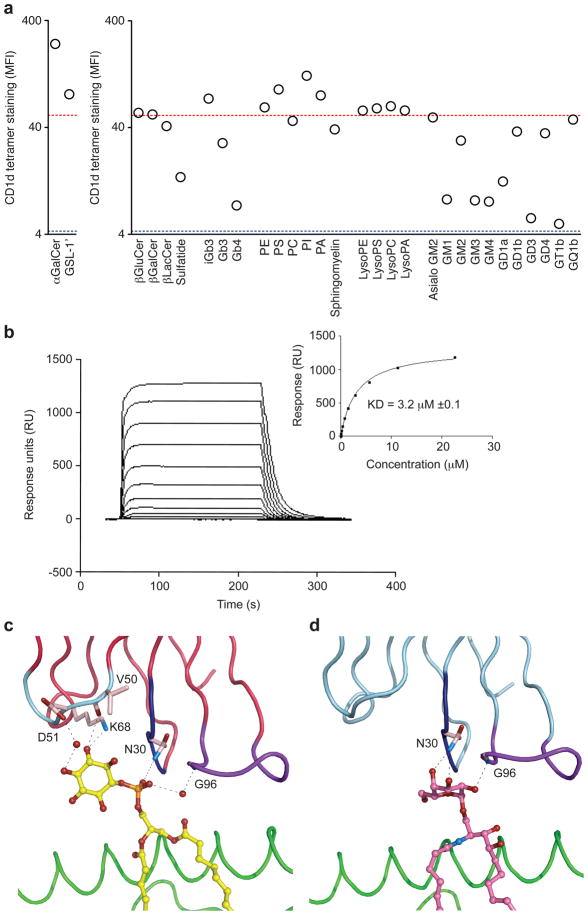
What is the nature of the self-lipid(s) or glycolipid(s) that are being recognized by the autoreactive NKT TCRs? To start addressing this question, we generated 28 different CD1d tetramers independently loaded with a panel of self-lipids that have either been found associated with CD1d molecules (Cox et al., 2009; Yuan et al., 2009) or have been identified in the mouse thymus by mass spectrometry (Li et al., 2009). We also produced four control tetramers using the same volume of solvent used to solubilize the various lipids. These various tetramers were then used to independently stain the hybridoma expressing the NKT TCR composed of the Vβ8.2 chain with the 2A3-D CDR3β loop (Fig 5). The choice of this particular TCR was dictated by its slightly lower apparent avidity for the CD1d tetramer loaded with natural self-lipids compared to the 2A3-D TCR (Fig 5a). Addition of solvent alone to the mCD1d monomers before tetramerization did not hamper the capacity of the CD1d tetramers loaded with natural self-lipid(s) to interact with the Vβ8.2-2A3-D NKT TCR (data not shown). Tetramers made with β-glucosylceramide (β-GluCer), β-galactosylceramide (β-GalCer), phosphatidylethanolamine (PE), phosphatidylcholine (PC), gangliotriaosylceramide (asialo-GM2), tetrasialoganglioside GQ1b and all lysophospholipids also stained similarly to the tetramer loaded with natural self-lipids (Fig 7a). However, addition of iGb3, phosphatidylserine (PS), phosphatidylinositol (PI) or phosphatidic acid (PA) to the tetramers led to an increase of tetramer binding for a similar level of TCR expression (Fig 7a). In contrast, addition of β-lactosylceramide (β-LacCer), sulfatide, globotrihexosylceramide (Gb3), globoside (Gb4), monosialogangliosides GM1, GM2, GM3, GM4, disialogangliosides GD1a, GD1b, GD3 or trisialoganglioside GT1b reduced, sometimes back to background values, the binding to the NKT TCR (Fig 7a). These results show that not all potential self-antigens are recognized by these high affinity NKT TCRs and further demonstrate that several, structurally distinct, self-lipids can be recognized by the NKT TCR.
Figure 7.
NKT TCR Recognition of CD1d-self Ag. (a) Antigen permissiveness by the NKT TCR. Staining of the hybridoma expressing the Vα14-Vβ8.2 TCR containing the CDR3β of 2A3-D with CD1d tetramers loaded with the indicated glycolipids or phospholipids (x axes). The MFI of CD1d tetramer staining was determined for a narrow TCR gate. Red dotted line, MFI with CD1d tetramer loaded with vehicle only (self-CD1d tetramer), blue dotted line, MFI with no addition of CD1d tetramer. Data represent the mean of two independent experiments. (b) Binding of the 2A3-D NKT TCR to mCD1d-PI as assessed by SPR. NKT TCR were injected over streptavidin immobilized mCD1d-PI and over an empty flow cell. Sensorgrams show the binding (response units, RU) of decreasing concentrations of TCR (22.5, 11.25, 5.625, 2.8125, 1.406 μM, 703, 351, 176, 87.9 43.9 nM) to the mCD1d-PI following subtraction of the control flow cell. Insets show saturation plots demonstrating equilibrium binding of NKT TCR to immobilized CD1d-PI. (c) Interactions between 2A3-D NKT TCR and PI (d) Interactions between Vβ8.2 NKT TCR and α-GC.
To gain insight as to how some self-antigens might prevent recognition by the autoreactive TCR, we analyzed the structure of CD1d-sulfatide (Zajonc et al., 2005) and that of the 2.5 Å resolution structure of the bulky disialogangliosides GD3 in binary complex with CD1d (Supplementary Table 4). The autoreactive NKT TCR complex was docked onto these non-permissive CD1d-self Ag complexes, and both the sulfatide and GD3 self-antigens appear incompatible with the conserved mode of NKT TCR recognition (Supplementary Figure 4). In both cases, the head groups obstructed the incoming NKT TCR due to steric hindrance with the invariant α-chain, thereby providing a plausible explanation to the lack of tetramer binding to the autoreactive TCR (Fig 7a).
Given that the 2A3-D TCR recognized the PI self-antigen with the highest avidity (Fig 7a), we determined, via surface plasmon resonance (SPR), its affinity for the PI-CD1d and the αGC-CD1d complexes. These affinity measurements were compared to that of another non-autoreactive Vβ8.2 NKT TCR (Pellicci et al., 2009). Interestingly, the 2A3-D TCR bound to αGC-CD1d with such high affinity that it would not readily dissociate (Supplementary Fig 3), indicating very low nM binding affinity, while the affinity for the non-autoreactive Vβ8.2 NKT TCR-αGC-CD1d interaction was ≈ 94nM (Pellicci et al., 2009). In contrast, the 2A3-D TCR bound to PI-CD1d with an affinity of 3 μM (Fig 7b), whereas no appreciable binding was observed for the Vβ8.2 NKT TCR-PI-CD1d interaction (not shown), indicating that the 2A3-D TCR had a higher propensity to bind CD1d when compared to that of the Vβ8.2 NKT TCR.
We subsequently analyzed the contacts between the 2A3-D TCR and the PI self-antigen. As expected, the polar head group of PI is exposed, and thus able to be directly contacted by the NKT TCR. Comparison of the structures of the 2A3-D TCR-PI-CD1d complex with the Vα14-Vβ8.2 NKT TCR-αGC-CD1d complex showed that the conformation of the CD1d or the CDR loops of the respective NKT TCRs were very similar (Supplementary Figure 5), indicating that there was no requirement for increased plasticity of the NKT TCR to accommodate the bulkier head group of PI. The PI head group is oriented such that the phosphate moiety abuts the CDR1α loop, while the inositol ring sits underneath, and contacts, the CDR2α loop and extends into bulk solvent, with the 3′ OH and 4′ OH groups being fully solvent-exposed (Fig 7c). Surprisingly, the PI head group does not make any direct interactions with the Jα18-encoded CDR3α loop, with its interactions being limited to a water mediated contact between the phosphate moiety and the CDR3α loop. Instead, the phosphate moiety packs against P28 and forms an H-bond with N30 from the CDR1α loop. Only the 6′ and 5′ OH groups of the inositol ring are sequestered by the NKT TCR, with the 6′ OH group H-bonding to N30α, while the 5′ OH H-bonds to the main chain of the CDR2α loop, and further, forms an H-bond to K68, a framework residue from the invariant α-chain (Fig 7c). Further, the conformation of the inositol ring is stabilized by vdw interactions between the CD1d-α2-helix and the CDR2α loop of the NKT TCR.
The plane of the inositol ring sits approximately perpendicular to the CD1d-Ag binding cleft in stark contrast to the more parallel-packed conformation of the galactosyl-head group of αGC (Fig 7d), revealing that the mode of interactions between this self and foreign Ag differ markedly. The αGC head group sits directly underneath the CDR1α loop and abuts the CDR3α loop, with the 2′ -, 3′ - and 4′-OH groups being sequestered closely by the NKT TCR, which contrasts that of the inositol ring-mediated interactions. The only site of commonality between the PI and αGC mediated interactions is where 4′ OH of αGC approximates the position of the phosphate moiety of PI, and thereby mimics the interactions with Asn30α from the CDR1α loop.
Accordingly, there is a marked difference in the way in which the invariant NKT TCRα chain directly contacts the PI self-Ag when compared to the foreign αGC agonist. Nevertheless, by conserving the same docking mode onto CD1d, the autoreactive NKT TCR can accommodate the recognition of various self and non-self antigens.
DISCUSSION
The recognition of self-antigens presented by CD1d is an intrinsic characteristic of NKT cells and relates both to the normal functioning and aberrant activity of NKT cells. Our findings provide a molecular basis for understanding NKT autoreactivity.
Here we show that a hydrophobic motif within the CDR3β loop of the NKT TCR can modulate the affinity of the NKT TCR for self antigen-CD1d complexes. Notably, hybridomas engineered to express NKT TCRs incorporating this CDR3β loop were highly autoreactive to a variety of CD1d-expressing APCs (data not shown). We have previously shown that, in general, changes in the NKT TCR β chain affect the affinity of the TCR for a ligand, but do not affect the order of the hierarchy with which this TCR reacts with different ligands (Mallevaey et al., 2009). Therefore the results presented here with the high affinity 2A3-D TCR are representative of the relative abilities of other NKT TCRs to react with different ligands. The results are thus relevant to our understanding of the specificities of NKT TCRs in general. The crystal structure of the autoreactive NKT TCR-CD1d-PI complex demonstrates that this motif directly interacts with CD1d, indicating that self-reactivity is not mediated via Ag-specific contacts, as the CDR3β loop is positioned distant from the self-antigen, in this case PI. This CDR3β sequence acted as a hydrophobic cap sequestering an otherwise surface exposed hydrophobic patch on the α2-helix on CD1d, thereby leading to an energetically-favored association. We suggest that many CDR3β sequences containing hydrophobic residues at these positions would favor NKT cell autoreactivity. However, the presence of such a hydrophobic cap is probably not the only means by which the CDR3β loop can increase the overall affinity of the NKT TCR for CD1d, as no obvious correlation between CD1d binding affinity and CDR3β loop sequences was observed in a recent report using autoreactive human NKT TCRs (Matulis et al., 2010).
Previous data had suggested that differences of reactivity between NKT cell hybridomas generated from different tissues were the result of the presentation of different self-antigens by CD1d expressed by different APCs (Brossay et al., 1998b; Park et al., 1998). Our current data indicate that autoreactive NKT cells might in fact be directed towards a set of self-antigens common to many APC types and that antigen-independent mechanisms might be responsible for the difference in observed responses.
Parts of our study were conducted using NKT TCRs composed of Vβ6 chains in which the CDR2β loop had been replaced with that of Vβ8.2 (Mallevaey et al., 2009), and therefore it could be argued that these chimeric TCRs might not be reflective of “native” NKT TCRs. Our mutational and structural analyses, however, demonstrated that these NKT TCRs dock onto CD1d in a manner essentially identical to what has been previously reported for another NKT TCR using a Vβ8.2 chain (Pellicci et al., 2009). Thus, the gain of affinity of these TCRs for self-antigen-CD1d molecules was mediated only by the increased energy provided by the unique CDR3β loops. Furthermore, transfer of these unique CDR3β loops into native Vβ8.2 chains was sufficient to generate NKT TCRs capable of interacting with self-CD1d tetramer. Altogether, these results demonstrate that recognition by these autoreactive TCRs reflects normal NKT TCR recognition and reinforce the idea that the NKT cell repertoire is on a sliding scale of avidity, which is modulated by the CDR2β and CDR3β loops (Mallevaey et al., 2009). Our crystallographic data also reveal that the mode in which NKT TCRs recognize foreign CD1d-restricted Ags mirrors that of NKT TCR recognition of self, thereby further highlighting the innate-style properties of the NKT TCR. Namely, the energetic basis of NKT TCR recognition of both CD1d-αGC and CD1d-self-Ags was underpinned by the germline-encoded CDR1α, CDR3α and CDR2β loops. Recognition of “self” also required the involvement of the CDR3β loop, while this loop was dispensable for the recognition of αGC, which can be attributed to differences of energy provided to the interaction by the different antigens, with the “self” antigens being of low affinity. These results suggest that the natural TCR repertoire of NKT cells is inherently reactive to, but has a sub-optimal affinity for self-antigens. Notably, the region of the α2 helix of CD1d that was contacted by the CDR3β loop of the NKT TCR has been speculated to move depending upon the lipid tail lengths of CD1d-restricted glycolipids (McCarthy et al., 2007). Therefore, it is possible that different self-antigens might be able to modulate the overall affinity of CD1d recognition through such a mechanism.
While the autoreactive NKT TCR recognized a broad panel of self-lipids, it did not recognize certain lipids well. These findings do not parallel a previous report regarding the isolation of rare NKT cells specific for phospholipids (Gumperz et al., 2000). Instead, the autoreactive NKT TCRs we described here were not antigen specific and could accommodate the recognition of several, but not all, self and foreign glycolipids. It is possible that certain lipids have to change conformation to enable NKT TCR-CD1d ligation, as has been recently observed for a microbial lipid (Li et al., 2010), whereas the more “bulky” or rigid antigens hinder recognition. As CD1d molecules at the surface of APCs are most likely loaded with a variety of different self antigens (Cox et al., 2009; Muindi et al., 2010; Yuan et al., 2009), our results suggest that autoreactivity of NKT cells might derive from a shift in the balance between self antigens that can be recognized by the NKT TCR and self antigens that cannot. Different immune conditions might be able to alter this balance, through changes in APC type and activation status, TLR signaling (Brigl et al., 2003; Mattner et al., 2005; Paget et al., 2007; Salio et al., 2007), CD1d expression (Sköld et al., 2005) or cytokine expression (Wesley et al., 2008).
The determination of the autoreactive NKT TCR-PI-CD1d self-complex provides compelling insight into how the NKT TCR can accommodate ligands that are markedly structurally dissimilar to that of αGC or closely related Ags. In contrast to the close sequestration of the galactosyl-headgroup by the NKT TCR, the bulkier polar head group of PI is orientated towards the bulk solvent. It is anticipated that the autoreactive NKT TCR will recognize other self-ligands that are similar to PI (eg PC, PE, PS, PA) in an analogous manner. Further, it is suggestive that the β-linked self glycosphingolipid Ag, iGb3, may adopt a similar binding mode, with the 3 sugar moieties of iGb3 orientating towards the α-chain.
NKT cells expressing TCRs capable of interacting with CD1d tetramer loaded with natural self antigen(s) are not detected directly ex vivo. This might be the result of several non-exclusive possibilities. First, the affinity of the NKT TCR for “self” is much lower than the affinity for αGC (Cantu et al., 2003; Kjer-Nielsen et al., 2006; Pellicci et al., 2009; Sidobre et al., 2004), making it difficult to detect by direct tetramer staining. Second, unique CDR3β sequences encoding for hydrophobic residues in the middle of the loop were necessary for this reactivity. The random generation of such CDR3β sequences by the recombination machinery is only achieved by nucleotide addition, making the generation of such NKT TCRs an infrequent event. Finally, it is possible that NKT precursor cells in the thymus expressing TCRs with too high affinity for self-antigen(s) might undergo negative selection that limits the generation of NKT cells with high self-CD1d tetramer reactivity.
Altogether, we have demonstrated that NKT cell autoreactivity can be the result of a hydrophobic motif within the CDR3β loop of the NKT TCR that mediates direct recognition with CD1d in an antigen-independent manner. We also provide structural basis of how an NKT TCR can recognize structurally distinct ligands. Conceptually, our data represent a large step in our understanding of NKT-cell autoreactivity in general, including the identification of a set of candidate self-antigens that can be recognized by the autoreactive TCR. The next challenge will be to determine which, and how, any of these self-antigen(s) plays an active role in NKT cell biology.
METHODS
Reagents
Mouse CD1d monomers, produced in HEK 293 cells, were obtained from the National Institutes of Health Tetramer Core Facility. Mouse CD1d monomers produced in insect Hi5 cells were a generous gift from Dr. M. Kronenberg (La Jolla Institute for Allergy and Immunology). Mouse CD1d:Ig protein (Dimer X), produced in the mouse B cell lymphoma J558L was purchased from BD Biosciences. α-galactosylceramide (αGC), isoglobotrihexosylceramide (iGb3), globotrihexosylceramide (Gb3) were purchased from Alexis Biochemicals. Phosphatidylinositol (PI), phosphatitic acid (PA) and lysophospholipids were purchased from Avanti Polar Lipids. All other glycolipids and phospholipids were purchased from Matreya, LLC.
TCRαβ constructs and retroviral plasmids
Wild-type and mutant TCRα chains were generated as previously described (Scott Browne et al., 2007). TCRβ constructs were cloned into mouse stem cell virus-based plasmids with an internal ribosome entry site (IRES) plus sequence encoding for the human nerve growth factor receptor as a reporter. In some instances, DNA was purified from sorted hybridomas and TCRβ chains were cloned as described above. All TCR V-gene segments are named and their amino acids are numbered according to the International Union of Immunological Societies–Arden compilation (Arden et al., 1995). This system of numbering is based on the known amino termini of several V-gene segments. This numbering follows a system developed for immunoglobulin V elements in which the V-framework residues are numbered consistently. In this way, the cysteine residues that form the intrachain disulfide bond are numbered consistently. The varying lengths of the V loops are dealt with by consistent numbering of the most common length with omission of numbers for shorter loops and the addition of lower-case letters to accommodate longer loops.
Cell lines and retroviral packaging
TCRα and β constructs were expressed by retroviral transduction of 5KC C3 or 5KC D8, two clones derived from the 5KC-78.3.20 TCRα- and TCRβ-negative hybridoma (White et al., 1993) that were selected for the loss of CD1d expression (not shown). Retroviral plasmids were transfected into Phoenix cells together with the pCLEco accessory plasmid with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s specifications. Retrovirus-containing supernatants were harvested 48 hr after transfection, centrifuged, and filtered for removal of debris. Hybridomas were “spin-infected” at 3300 × g for 90 min at 37°C in retrovirus-containing supernatants supplemented with polybrene (8 μg/ml). Hybridomas were then sorted on a MoFlo cell sorter (Dakocytomation) on the basis of retroviral reporter and TCRβ expression.
Tetramer staining
Glycolipids and phospholipids were dissolved in various solvents as indicated in supplementary Table 5 and diluted to working concentration in a vehicle solution (0.9% NaCl 0.05% [vol/vol] Tween 20). Biotinylated recombinant mouse CD1d proteins were incubated overnight with the indicated glycolipid or phospholipid in vehicle solution and then tetramerized by the addition of phycoerythrin-conjugated streptavidin. Alternatively, the “self” tetramer was prepared identically with the addition of vehicle only. Control tetramers were prepared by the addition of the same volume of solvent in vehicle solution. Antigen loading of mCD1d:Ig protein and preparation of mCD1d:Ig staining reagents were performed according to the manufacturer’s specifications. Hybridomas were stained for 45 min at 25°C with the indicated tetramer plus antibody to TCRβ (anti-TCRβ; H57-597; eBioscience). Cells were analyzed by flow cytometry on FACScalibur (BD Biosciences) or LSR II (BD Biosciences) instruments and data were analyzed in FlowJo (Treestar).
Hybridoma stimulation
A total of 5 × 104 hybridomas were cultured for 20 hr with 5 × 104 A20 cells transfected or not with mouse CD1d, in complete RPMI medium containing 10% (vol/vol) FCS, in the presence of 10 μg/ml anti-mCD1d (1B1; eBioscience) or isotype control. Hybridoma responses were measured by an IL-2 enzyme-linked immunosorbent assay in accordance with standard protocols.
Protein production and purification
Mouse CD1d (mCD1d) was produced as described previously (Matsuda et al., 2000). In brief, mCD1d was made using the baculovirus pBacp10pH (provided by Dr. Mitchell Kronenberg, La Jolla Institute for Allergy and Immunology, CA, USA). Recombinant mCD1d was produced with a BirA tag followed by a 6 histidine tag, using the High Five insect cell line (Invitrogen). Soluble mCD1d protein was purified using Ni-Agarose affinity purification followed by size exclusion through a Superdex 200 16/60 gel filtration column. Soluble NKT TCR Vα14 and 2A3-D chains were expressed in E. coli strain BL21 and produced in inclusion bodies. The inclusion body protein was harvested, refolded and purified as described by Garboczi et al. (Garboczi et al., 1996), except the addition of 5 M urea in the refolding buffer.
Phosphotidylinositol (PI) lipid loading of mCD1d
mCD1d lipid loading was carried out by incubating mCD1d with a 3 molar excess of GD3 overnight at room temperature. The loaded species was subject to reloading with PI (Avanti Polar Lipids) under the same conditions. After each loading stage, anion exchange purifications were performed to separate loaded and unloaded species.
Complex of 2A3-D NKT TCR with mCD1d
Purified NKT TCR and mCD1d-PI were mixed and incubated at room temperature with gentle agitation for 45 minutes. The ternary complex (TCR, mCD1d-PI) was separated from any TCR and mCD1d monomer by size exclusion through a Superdex 200 16/60 column. Purified protein was then spin concentrated to 10 mg/ml.
Crystallization, structure determination and refinement
Hanging drop vapour diffusion crystallisation experiments were performed by mixing 1 μl of protein, 1 μl of mother liquor (13% polyethylene glycol 6000 and 0.1 M sodium citrate pH 6.1). Crystals were cryoprotected by a slowly passing the crystal through mother liquor drops, sequentially increasing the glycerol concentration to 15%. Diffraction data from the ternary complex crystals were collected with synchrotron radiation and processed using the Mosflm (Supplementary Table 2). Using the NKT TCR Vα14-Vβ7-CD1d-αGC structure (PDB 3HE7), with CDR loops, lipid and waters removed, Phaser was able to yield an interpretable 2.9 Å experimental map, revealing a single molecule in the asymmetric unit as predicted from the Matthew’s coefficient. The model was then rigid-body refined into the experimental map before careful model building with COOT (Emsley and Cowtan, 2004). This was then used as the initial model for refinement of a higher 2.3 Å resolution data set. PI could be easily placed in the unbiased difference density (Supplementary Figure 1), which adopted a single conformation. Iterative model building and refinement with PHENIX (Zwart et al., 2008) yielded a final model with final statistics shown in Supplementary Table 2. The quality of the structure was assessed with programs within the CCP4 suite. CD1d-GD3 was crystallized using crystallization conditions similar to that used for other CD1d-binary structures (Zajonc et al., 2008) and refined to 2.5 Å resolution (see Supplementary Table 4).
Surface plasmon resonance measurements and analysis
All surface plasmon resonance (SPR) experiments were carried out at 25°C on a Biacore 3000 instrument using HBS buffer (10 mM HEPES-HCl (pH 7.4), 150 mM NaCl and 0.005% surfactant P20 supplied by manufacturer). Equilibrium affinity measurements of the NKT TCR-PI-CD1d and NKT TCR-αGC-CD1d interaction were determined by SPR and have been described previously (Kjer-Nielsen et al., 2006), with the CD1d-PI being specifically coupled to a SA sensor chip (GE Healthcare). The NKT TCR was passed over the sensor chip at 5 μL/min for 150 sec at 25°C and the final response was subtracted from that of an empty flow cell. All measurements were minimally in duplicate, and the BIAevaluation Version 3.1 (Biacore AB) was used for all data analysis.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Health (AI076463 and AI078246 to L.G. and AI187785 to P.M.); NIH training grant T32 AI07405 to J. P. S. -B. and M. H. Y.; Howard Hughes Medical Institute (P. M.); the Cancer Council of Victoria, and the National Health and Medical Research Council of Australia. D. I. G. is supported by an NHMRC fellowship; J. R. is supported by an Australian Research Council Federation Fellowship. T. M., A. J. C., J. R. and L. G. designed, did the experimental work, analyzed the experiments and prepared the manuscript. D. G. P., O. P., J. L. M., M. H. Y., D. I. G., J. M., P. M. provided intellectual insight and crystallographic data. T. M., J. P. S. -B., D. I. G., P. M., J. R. and L. G. devised the project and wrote the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arden B, Clark SP, Kabelitz D, Mak TW. Mouse T-cell receptor variable gene segment families. Immunogenetics. 1995;42:501–530. doi: 10.1007/BF00172177. [DOI] [PubMed] [Google Scholar]
- Bellone M, Ceccon M, Grioni M, Jachetti E, Calcinoto A, Napolitano A, Freschi M, Casorati G, Dellabona P. iNKT cells control mouse spontaneous carcinoma independently of tumor-specific cytotoxic T cells. PLos One. 2010;5:e8646. doi: 10.1371/journal.pone.0008646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A, Savage PB, Teyton L. The Biology of NKT Cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, Rossjohn J. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- Brigl M, Brenner MB. How invariant natural killer T cells respond to infection by recognizing microbial or endogenous lipid antigens. Sem Immunol. 2009;22:79–86. doi: 10.1016/j.smim.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Brigl M, Bry L, Kent SC, Gumperz JE, Brenner MB. Mechanism of CD1d-restricted natural killer T cell activation during microbial infection. Nat Immunol. 2003;4:1230–1237. doi: 10.1038/ni1002. [DOI] [PubMed] [Google Scholar]
- Brossay L, Naidenko O, Burdin N, Matsuda J, Sakai T, Kronenberg M. Structural requirements for galactosylceramide recognition by CD1- restricted NK T cells. J Immunol. 1998a;161:5124–5128. [PubMed] [Google Scholar]
- Brossay L, Tangri S, Bix M, Cardell S, Locksley R, Kronenberg M. Mouse CD1-autoreactive T cells have diverse patterns of reactivity to CD1+ targets. J Immunol. 1998b;160:3681–3688. [PubMed] [Google Scholar]
- Cantu C, 3rd, Benlagha K, Savage PB, Bendelac A, Teyton L. The Paradox of Immune Molecular Recognition of α-Galactosylceramide: Low Affinity, Low Specificity for CD1d, High Affinity for αβ TCRs. J Immunol. 2003;170:4673–4682. doi: 10.4049/jimmunol.170.9.4673. [DOI] [PubMed] [Google Scholar]
- Cox D, Fox L, Tian R, Bardet W, Skaley M, Mojsilovic D, Gumperz J, Hildebrand W. Determination of cellular lipids bound to human CD1d molecules. PLoS ONE. 2009;4(5):e5325. doi: 10.1371/journal.pone.0005325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellabona P, Padovan E, Casorati G, Brockhaus M, Lanzavecchia A. An invariant Vα24-JαQ/Vβ11 T cell receptor is expressed in all individuals by clonally expanded CD4−8− T cells. J Exp Med. 1994;180:1171–1176. doi: 10.1084/jem.180.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gapin L. iNKT cell autoreactivity; what is ‘self’ and how is it recognized? Nat Rev Immunol. 2010;10:272–277. doi: 10.1038/nri2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- Giabbai B, Sidobre S, Crispin MD, Sanchez-Ruiz Y, Bachi A, Kronenberg M, Wilson IA, Degano M. Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: a molecular basis for NKT cell activation. J Immunol. 2005;175:977–984. doi: 10.4049/jimmunol.175.2.977. [DOI] [PubMed] [Google Scholar]
- Godfrey DI, Rossjohn J, McCluskey J. The fidelity, occasional promiscuity, and versatility of T cell receptor recognition. Immunity. 2008;28:304–314. doi: 10.1016/j.immuni.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Gumperz JE, Roy C, Makowska A, Lum D, Sugita M, Podrebarac T, Koezuka Y, Porcelli SA, Cardell S, Brenner MB, Behar SM. Murine CD1d-restricted T cell recognition of cellular lipids. Immunity. 2000;12:211–221. doi: 10.1016/s1074-7613(00)80174-0. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Borg NA, Pellicci DG, Beddoe T, Kostenko L, Clements CS, Williamson NA, Smyth MJ, Besra GS, Reid HH, et al. A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition. J Exp Med. 2006;203:661–673. doi: 10.1084/jem.20051777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg M. Toward an Understanding of NKT Cell Biology: Progress and Paradoxes. Annu Rev Immunol. 2005;23:877–900. doi: 10.1146/annurev.immunol.23.021704.115742. [DOI] [PubMed] [Google Scholar]
- Lantz O, Bendelac A. An invariant T cell receptor α chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4−8− T cells in mice and humans. J Exp Med. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappas CM, Day YJ, Marshall MA, Engelhard VH, Linden J. Adenosine A2A receptor activation reduces hepatic ischemia reperfusion injury by inhibiting CD1d-dependent NKT cells. J Exp Med. 2006;203:2639–2648. doi: 10.1084/jem.20061097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Girardi E, Wang J, Dawen Yu E, Painter GF, Kronenberg M, Zajonc DM. The Vα14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode. J Exp Med. 2010 doi: 10.1084/jem.20101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Thapa P, Hawke D, Kondo Y, Furukawa K, Furukawa K, Hsu FF, Adlercreutz D, Weadge J, Palcic M, et al. Immunologic glycosphingolipidomics and NKT cell development in mouse thymus. J Proteome Res. 2009;8:2740–2751. doi: 10.1021/pr801040h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallevaey T, Scott Browne J, Matsuda JL, Young MH, Pellicci DG, Patel O, Thakur M, Kjer-Nielsen L, Richardson SK, Cerundolo V, et al. T cell receptor CDR2β and CDR3β loops collaborate functionally to shape the iNKT cell repertoire. Immunity. 2009;31:60–71. doi: 10.1016/j.immuni.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda JL, Gapin L, Fazilleau N, Warren K, Naidenko OV, Kronenberg M. Natural killer T cells reactive to a single glycolipid exhibit a highly diverse T cell receptor β repertoire and small clone size. Proc Natl Acad Sci U S A. 2001;98:12636–12641. doi: 10.1073/pnas.221445298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda JL, Mallevaey T, Scott-Browne J, Gapin L. CD1d-restricted iNKT cells, the ‘Swiss-Army knife’ of the immune system. Curr Opin Immunol. 2008;20:358–368. doi: 10.1016/j.coi.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang CR, Koezuka Y, Kronenberg M. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattner J, Debord KL, Ismail N, Goff RD, Cantu C, 3rd, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- Matulis G, Sanderson JP, Lissin NM, Asparuhova MB, Bommineni GR, Schümperli D, Schmidt RR, Villiger PM, Jakobsen BK, Gadola SD. Innate-like control of human iNKT cell autoreactivity via the hypervariable CDR3b loop. PLoS Biol. 2010;8:e1000402. doi: 10.1371/journal.pbio.1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy C, Shepherd D, Fleire S, Stronge VS, Koch M, Illarionov PA, Bossi G, Salio M, Denkberg G, Reddington F, et al. The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation. J Exp Med. 2007;204:1131–1144. doi: 10.1084/jem.20062342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muindi K, Cernadas M, Watts GFM, Royle L, Neville DCA, Dwek RA, Besra GS, Rudd PM, Butters TD, Brenner MB. Activation state and intracellular trafficking contribute to the repertoire of endogenous glycosphingolipids presented by CD1d. Proc Natl Acad Sci U S A. 2010;107:3052–3057. doi: 10.1073/pnas.0915056107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget C, Mallevaey T, Speak AO, Torres D, Fontaine J, Sheehan KC, Capron M, Ryffel B, Faveeuw C, Leite de Moraes M, et al. Activation of invariant NKT cells by toll-like receptor 9-stimulated dendritic cells requires type I interferon and charged glycosphingolipids. Immunity. 2007;27:597–609. doi: 10.1016/j.immuni.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Park SH, Roark JH, Bendelac A. Tissue-specific recognition of mouse CD1 molecules. J Immunol. 1998;160:3128–3134. [PubMed] [Google Scholar]
- Pellicci DG, Patel O, Kjer-Nielsen L, Pang SS, Kyparissoudis K, Sullivan LC, Brooks AG, Reid HH, Smyth MJ, Mallevaey T, et al. Differential Vβ8.2 and Vβ7-mediated NKT T-cell receptor recognition of CD1d-α-galactosylceramide. Immunity. 2009;31:47–59. doi: 10.1016/j.immuni.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salio M, Speak AO, Shepherd D, Polzella P, Illarionov PA, Veerapen N, Besra GS, Platt FM, Cerundolo V. Modulation of human natural killer T cell ligands on TLR-mediated antigen-presenting cell activation. Proc Natl Acad Sci U S A. 2007;104:20490–20495. doi: 10.1073/pnas.0710145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann J, Voyle RB, Wei BY, MacDonald HR. Cutting Edge: Influence of the TCR Vβ Domain on the Avidity of CD1d:α-Galactosylceramide Binding by Invariant Vα14 NKT Cells. J Immunol. 2003;170:5815–5819. doi: 10.4049/jimmunol.170.12.5815. [DOI] [PubMed] [Google Scholar]
- Scott Browne J, Matsuda JL, Mallevaey T, Borg NA, White J, McCluskey J, Rossjohn J, Kappler J, Marrack P, Gapin L. Germline-encoded recognition of diverse glycolipids by NKT cells. Nat Immunol. 2007;8:1105–1113. doi: 10.1038/ni1510. [DOI] [PubMed] [Google Scholar]
- Sidobre S, Hammond KJ, Benazet-Sidobre L, Maltsev SD, Richardson SK, Ndonye RM, Howell AR, Sakai T, Besra GS, Porcelli SA, Kronenberg M. The T cell antigen receptor expressed by Vα14i NKT cells has a unique mode of glycosphingolipid antigen recognition. Proc Natl Acad Sci U S A. 2004;101:12254–12259. doi: 10.1073/pnas.0404632101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sköld M, Xiong X, Illarionov PA, Besra GS, Behar SM. Interplay of cytokines and microbial signals in regulation of CD1d expression and NKT cell activation. J Immunol. 2005;175:3584–3593. doi: 10.4049/jimmunol.175.6.3584. [DOI] [PubMed] [Google Scholar]
- Swann JB, Coquet JM, Smyth MJ, Godfrey DI. CD1-restricted T cells and tumor immunity. Curr Top Microbiol Immunol. 2007;314:293–323. doi: 10.1007/978-3-540-69511-0_12. [DOI] [PubMed] [Google Scholar]
- Swann JB, Uldrich AP, van Dommelen S, Sharkey J, Murray WK, Godfrey DI, Smyth MJ. Type I natural killer T cells suppress tumors caused by p53 loss in mice. Blood. 2009;113:6382–6385. doi: 10.1182/blood-2009-01-198564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace KL, Marshall MA, Ramos SI, Lannigan JA, Field JJ, Strieter RM, Linden J. NKT cells mediate pulmonary inflammation and dysfunction in murine sickle cell disease through production of IFNγ and CXCR3 chemokines. Blood. 2009;114:667–676. doi: 10.1182/blood-2009-02-205492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley JD, Tessmer MS, Chaukos D, Brossay L. NK cell-like behavior of Vα14i NK T cells during MCMV infection. PLoS Pathogens. 2008;4:e1000106. doi: 10.1371/journal.ppat.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Pullen A, Choi K, Marrack P, Kappler JW. Antigen recognition properties of mutant Vβ3+ T cell receptors are consistent with an immunoglobulin-like structure for the receptor. J Exp Med. 1993;177:119–125. doi: 10.1084/jem.177.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Kang SJ, Evans JE, Cresswell P. Natural lipid ligands associated with human CD1d targeted to different subcellular compartments. J Immunol. 2009;182:4784–4791. doi: 10.4049/jimmunol.0803981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc DM, Maricic I, Wu D, Halder R, Roy K, Wong CH, Kumar V, Wilson IA. Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity. J Exp Med. 2005;202:1517–1526. doi: 10.1084/jem.20051625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc DM, Savage PB, Bendelac A, Wilson IA, Teyton L. Crystal structures of mouse CD1d-iGb3 complex and its cognate Vα14 T cell receptor suggest a model for dual recognition of foreign and self glycolipids. J Mol Biol. 2008;377:1104–1116. doi: 10.1016/j.jmb.2008.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Mattner J, Cantu C, 3rd, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu YP, Yamashita T, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, et al. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.