Abstract
Alkali production by oral streptococci is considered important for dental plaque ecology and caries moderation. Recently, malolactic fermentation (MLF) was identified as a major system for alkali production by oral streptococci, including Streptococcus mutans. Our major objectives in the work described in this paper were to further define the physiology and genetics of MLF of oral streptococci and its roles in protection against metabolic stress damage. l-Malic acid was rapidly fermented to l-lactic acid and CO2 by induced cells of wild-type S. mutans, but not by deletion mutants for mleS (malolactic enzyme) or mleP (malate permease). Mutants for mleR (the contiguous regulator gene) had intermediate capacities for MLF. Loss of capacity to catalyze MLF resulted in loss of capacity for protection against lethal acidification. MLF was also found to be protective against oxidative and starvation damage. The capacity of S. mutans to produce alkali from malate was greater than its capacity to produce acid from glycolysis at low pH values of 4 or 5. MLF acted additively with the arginine deiminase system for alkali production by Streptococcus sanguinis, but not with urease of Streptococcus salivarius. Malolactic fermentation is clearly a major process for alkali generation by oral streptococci and for protection against environmental stresses.
Keywords: oral streptococci, malolactic fermentation, stress protection
Introduction
Dental caries are the result of acidification of dental plaque, mainly by oral lactic acid bacteria, most notably Streptococcus mutans, but also other acid-producing plaque bacteria. Plaque acidification is not only damaging to teeth but also stressful for the acid-producing organisms, which have developed a variety of means to moderate acid stress. Other environmental stresses, such as oxidative or starvation stress, may act in conjunction with acidification in causing damage. The major means for resisting the damaging effects of plaque acidification involves membrane F-ATPases, specifically F1F0(H+)-ATPases in oral streptococci (Bender et al. 1986; Bender and Marquis 1987; Quivey et al. 2000). These proton-translocating ATPases act to expel protons from the cytoplasm and to maintain δ-pH across the cell membrane, with the cytoplasm less acidified than the environment. A variety of other adaptations, especially changes in membrane fatty acids, can also enhance acid tolerance (Fozo et al. 2004; Fozo and Quivey 2004).
Another common way for oral bacteria to protect themselves against acid damage is through the production of alkali (Burne and Marquis 2000). In fact, the acid–base cycles in supragingival plaque and saliva are significantly affected by microbial alkali production, especially during the pH-rise phase of the cycle (Wijeyeweera and Kleinberg 1989). For example, alkali production by the arginine deiminase system (ADS) has been found (Casiano-Colón and Marquis 1988) to be protective against acid damage partly because of increases in cellular and environmental pH, with resultant relief of acid stress. Alkali production is protective also under pH-stat conditions, presumably because it can be coupled to substrate-level ATP synthesis through carbamate kinase. ATP can then fuel excretion of protons across the cell membrane through F-ATPase.
Another major source of microbially produced alkali in the mouth involves cytoplasmic ureases, such as those of Streptococcus salivarius (Chen and Burne 1996) or Actinomyces naeslundii (Morou-Bermudez and Burne 2000), which can catalyze hydrolysis of urea normally present in saliva to CO2 and NH3, but without concomitant ATP production. Urease production appears to be regulated mainly in response to nitrogen starvation rather than acidification, so protection against acidification can be a secondary function for the enzyme.
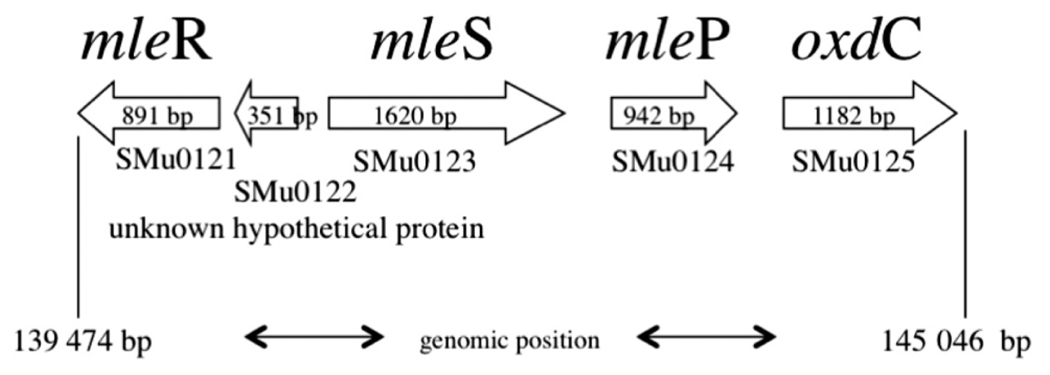
Despite its high level of acid tolerance, S. mutans is both ADS negative and urease negative. It does have an agmatine deiminase system (Griswold et al. 2006), but the system appears to function mainly for detoxification of agmatine, although it is active at low pH values and so may contribute to acid tolerance. Thus, highly acid-tolerant, cariogenic S. mutans appeared not to have a highly active metabolism for base production. However, we recently identified a different type of system for alkali production by oral lactic acid bacteria, including S. mutans. The system does not produce ammonia, but instead catalyzes decarboxylation of l-malic acid (Sheng and Marquis 2007). We found that most oral lactic acid bacteria carry out malolactic fermentation (MLF), in which l-malate is catabolized to l-lactate and CO2, leading to cytoplasmic alkalinization and also to ATP synthesis by means of F-ATPase acting in the synthase mode. The genome of S. mutans was found (Oralgen database; http://www.oralgen.lanl.gov) to contain the contiguous genes mleR (SMu0121) for a regulator of MLF, mleS (SMu0123) for malolactic enzyme, and mleP (SMu0124) for a malate permease (Fig. 1). The MLF system was found to be inducible with l-, but not d-, malate for suspension cultures or biofilms of S. mutans UA159. Many lactic acid bacteria can also decarboxylate citrate to yield CO2 and enhance acid tolerance, but not S. mutans (Korithoski et al. 2005).
Fig. 1.
Organization of the malolactic gene cluster in the Streptococcus mutans UA159 genome (Ajdić et al. 2002). The genes are numbered using the Oralgen nomenclature (http://www.oralgen.lanl.gov).
The results of previous work (Sheng and Marquis 2007) with intact cells and biofilms of S. mutans UA159 indicated that the optimal pH for MLF was approximately 4, and the minimum pH for fermentation was below 3. Alkali production from malate was protective against acid damage to S. mutans UA159 at low plaque pH values. Our objectives for this paper were to further characterize the physiology of MLF in oral streptococci and its interactions with other alkali-generating systems. In this paper, additional data are presented to indicate that MLF is a major factor in protecting S. mutans not only against acid damage but also oxidative and starvation damage.
Materials and methods
Bacteria
Streptococcus mutans UA159, Streptococcus sanguinis ATCC10556, and S. salivarius ATCC13914 are maintained routinely in our laboratory with weekly subculture on tryptic soy agar plates (Difco, Detroit, Mich.) to avoid selection of rapidly growing variants. Long-term storage is at −70 °C in 50% (v/v) glycerol–water solution. Suspension cultures induced for MLF were routinely grown at 37 °C in tryptone yeast extract (TY) medium (Belli and Marquis 1991) supplemented with 25 mmol/L glucose and 50 mmol/L l-malate until early stationary phase, when maximal malolactic fermentation activity was reached. The ADS of S. sanguinis was induced by growing cells in suspension cultures with a low level of glucose (5.6 mmol/L) and a high level of arginine (57.4 mmol/L). MLF could be independently induced in the cells by adding 50 mmol/L l-malate to the culture medium. Induction of MLF occurred in cultures of urease-positive S. salivarius grown in TY medium with 55 mmol/L glucose and 50 mmol/L l-malate.
Biofilms of S. mutans UA159 were grown on standard microscopic glass slides in fed-batch cultures, as previously described (Phan et al. 2000). The slides were placed initially in TY medium plus 29.2 mmol/L sucrose for the first 4 days, with daily transfer to fresh medium. Then, the biofilms were transferred to TY medium supplemented with 25 mmol/L glucose and 50 mmol/L l-malate, pH 6, for induction of MLF activity. Biofilms of S. sanguinis induced for MLF and ADS were grown following this same procedure but with addition of 57.4 mmol/L arginine to the medium.
Permeabilized cells were prepared by treating intact cells with toluene and then freezing and thawing, as described previously (Belli and Marquis 1991).
The UA159 strain of S. mutans was induced for decarboxylation of oxalate by static growth in TY medium supplemented with 25 mmol/L glucose, 20 mmol/L sodium oxalate, and 10 µmol/L MnCl2. Cells were harvested at the early stationary phase of culture growth.
Construction of mle deletion mutant strains of S. mutans
The annotated genome of S. mutans UA159 contains a region with genes highly orthologous to established malolactic operons (mle) in other bacteria (Ansanay et al. 1993; Denayrolles et al. 1994; Labarre et al. 1996a, 1996b). The gene arrangement and location in the S. mutans genome are shown in Fig. 1. Strains deleted for each mle gene were created as part of a larger project, and details of the constructions will be provided elsewhere. Briefly, the entire open reading frame of each mle gene was removed and replaced with a promoterless erythromycin resistance gene (ErmR). The ErmR marker was a promoterless, nonpolar, 738 base DNA fragment previously reported (Macrina et al. 1983). Mutant strains, transformed by established procedures (Perry and Kuramitsu 1981), were selected on agar containing brain–heart infusion medium (BHI) supplemented with erythromycin (10 g/mL). The location of the ErmR gene, in each strain, was confirmed by PCR amplification using 1 primer homologous to ErmR and 1 primer homologous to S. mutans UA159, lying outside the recombination region (Table 1). PCR-based characterization showed conclusively that the mleR, mleS, and mleP genes were all completely deleted in their respective strains (data not shown). Positive transformants were stored in BHI and glycerol.
Table 1.
PCR primers used in this study.
| Primer name | Oligodeoxynucleotide primer sequence (5′ → 3′)a | Genomic location of primer |
|---|---|---|
| MU0121P7UPFGA | GGTATCATCGGCAAGCACAAGG | 140 840 – 140 861 |
| MU0121P8DNRHA | CATGCTCAAATTGAGATGTTAGCC | 138 928 – 138 951 |
| MU0123P7UPFGA | TGACTATAAGGCCATTTGTTTCATAC | 140 462 – 140 487 |
| MU0123P8DNRHA | GTAAATTAGGAAGTAAGGCAGAGC | 143 178 – 143 201 |
| MU0124P7UPFGA | GGCAGAAGCATCGGCTCAAGACTTG | 142 242 – 142 266 |
| MU0124P8DNRHA | CTAACAGAAACTCACAGCCCTTATCC | 144 302 – 144 327 |
| SerB Erm Left | TCTGACGATAAGTTGAATAGATGAC | |
| SerB Erm Right | TTGTTTCAAAATGGGTCAATC |
Underlined primer sequences are homologous to the Streptococcus mutans UA159 chromosome.
Metabolite assays
Malate utilization was monitored with use of enzyme kits obtained from R-Biopharm AG (Darmstadt, Germany) containing l-malate dehydrogenase, as described previously (Sheng and Marquis 2007). The assay is specific for l-malate and is based on changes in absorption of light of 340 nm wavelength associated with NADH production from NAD+. Malolactic activity was expressed as micromoles of l-malate decarboxylated per hour per milligram of cell or biofilm dry mass.
Degradation of oxalate was assayed enzymatically with the use of kits (R-Biopharm AG) containing oxalate decarboxylase and formate dehydrogenase. NAD+ reduction was assayed spectrophotometrically.
Assays of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) activity were carried out as described previously (Baldeck and Marquis 2008).
Production of titratable acid and base was assayed by titration to maintain constant pH through addition of known volumes of standard KOH or HCl solutions, as described previously (Phan et al. 2004).
Protection against killing by acid, peroxide, and starvation
Protection against killing by acid, peroxide, and starvation was assayed by plate counting of samples of cell suspensions subjected to one of the stresses. Samples for plating were diluted immediately after being taken to eliminate the damaging stress. After hydrogen peroxide was used as the stressor, addition of catalase did not enhance recovery of viable cells or increase levels of specific enzymes in the cells.
Protection of cytoplasmic GAPDH was assayed by subjecting permeabilized cells to peroxide stress and then diluting the suspensions prior to assessing remaining levels of enzyme activity per unit of cell dry mass, as described previously (Baldeck and Marquis 2008).
Results
Effects of deletion mutations on MLF and protection against acid killing
Figure 1 shows the sequestered arrangement of genes for malolactic fermentation in the S. mutans UA159 chromosome based on information in the Oralgen database (Ajdić et al. 2002). The first gene SMu0121 (mleR) encodes a regulatory protein of the LysR family. SMu0122 is currently undefined. Deletion of the SMu0122 locus appeared, in our hands, to be a lethal event for the organism. We do not speculate on the mechanism of this lethality, but do point out that loss of any of the other open reading frames associated with mle XXXXX. The second annotated gene SMu0123 (mleS) codes for the malolactic enzyme, which catalyzes the decarboxylation of l-malic acid to l-lactic acid and CO2. The third gene, SMu0124 (mleP), codes for a malate permease, which catalyzes transport of l-malate across the cell membrane, possibly via electrogenic antiport in exchange for lactate (Sobczak and Lolkema 2005). The next downstream gene, SMu0125 (oxdC), encodes an oxalate decarboxylase, which may be responsible for the relatively low level of oxalate decarboxylase activity found for the organism, 0.6 µmol oxalate decarboxylated·mg cell dry mass−1·h−1 to yield CO2 and formate at an optimal pH of 5.5. However, oxalate decarboxylation is not part of MLF. It is of interest as another decarboxylation reaction resulting in alkalization and possibly ATP production through F(H+)-ATPase. Thus, there may be a functional connection between MLF and oxalate metabolism in S. mutans.
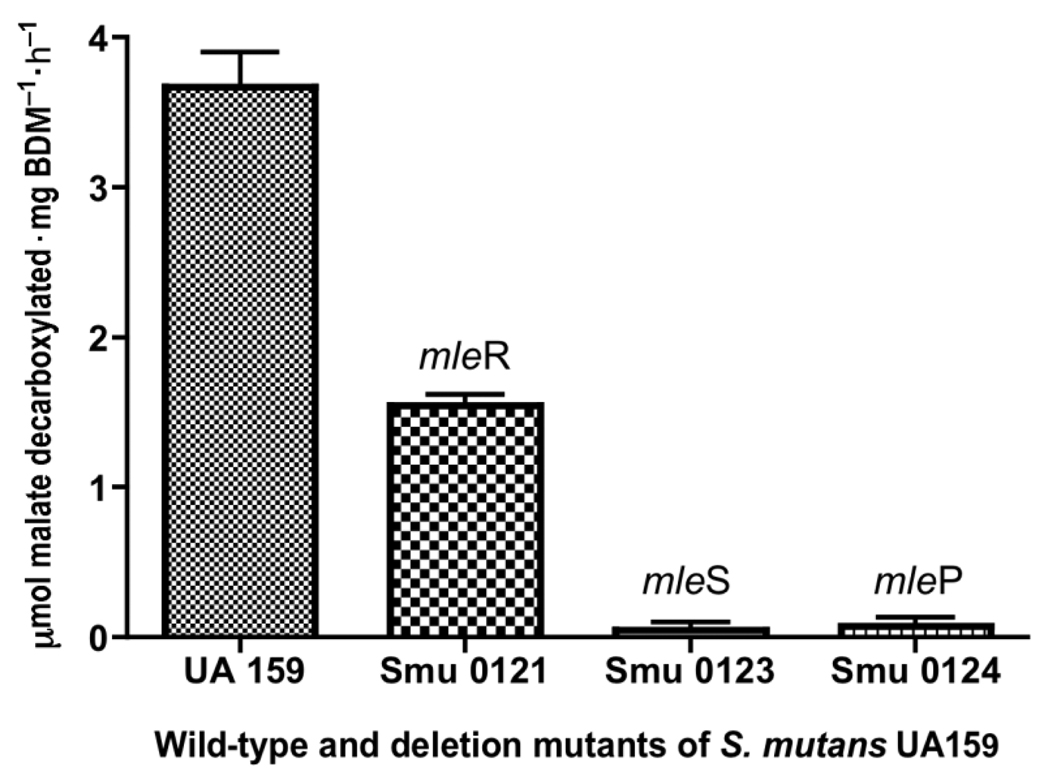
The effects of deletion mutagenesis on MLF by UA159 are indicated in Fig. 2. Under the experimental conditions, the malate-induced parent organism could catabolize an average of 3.7 µmol malate·mg cell dry mass−1·h−1 at the optimal pH of 4 and 37 °C in suspensions with saturating substrate levels of l-malate. The mleR-negative mutant had lower activity, only about 1.7 µmol·mg−1·h−1, or about 20% that of the parent activity. This result suggests that the MelR regulator is of the positive type, enhancing transcription of mleS and (or) mleP. The mleS-negative mutant was essentially devoid of MLF activity, while the mleP mutant had barely detectable MLF activity. This later finding is in line with our findings indicating that the MLF system in S. mutans UA159 is mainly transport limited, because it was difficult to demonstrate malate pooling within cells except at high malate concentrations of 50 mmol/L (data not shown).
Fig. 2.
Malolactic fermentation of Streptococcus mutans UA159 and deletion mutants in biofilms at constant pH of 4.0 and excess l-malate (50 mmol/L). n = 3; error bars indicate SD. BDM, biofilm dry mass.
Our previous findings (Sheng and Marquis 2007) indicated that MLF protects against acid killing under pH-stat conditions as well as under conditions in which the pH is allowed to increase to nonlethal values for the organism. In the current study, in pH-rise experiments with UA159 cells at a starting pH value of 3, overnight incubation resulted in a pH rise to about 7 but not above, presumably because MLF is optimal at pH 4, but nearly completely stopped at pH 7.
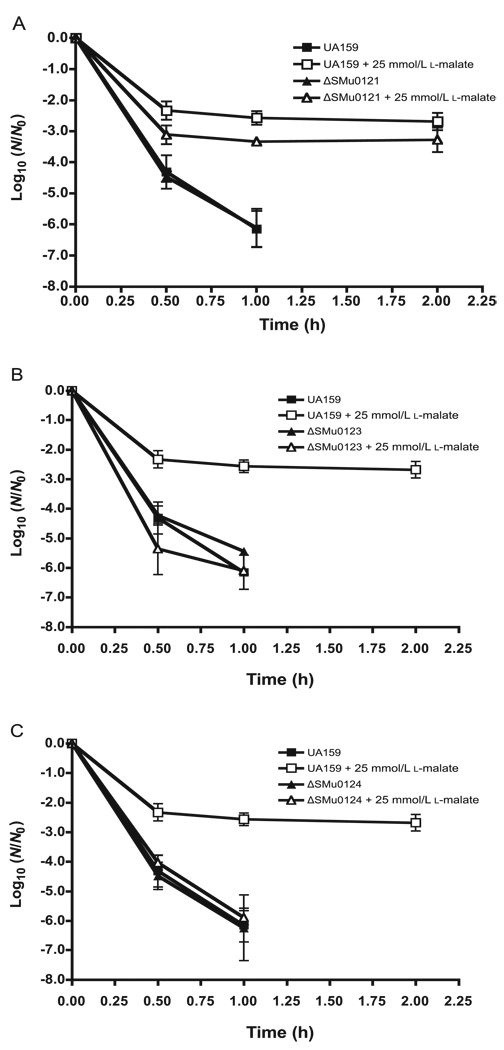
As shown by the data in Fig. 3, reduced MLF capacities of the mutants were reflected in reduced protection against acid killing of S. mutans UA159 cells in suspensions held at a constant pH value of 3, 1 pH unit below the optimal pH of 4 for MLF. As shown for the parent organism, exposure to pH 3 was lethal, but acid killing could be markedly reduced by addition of 25 mmol/L l-malate, a saturating level of malate for catabolism and protection against acid killing. In contrast, there was essentially no protection of the mleS or mleP mutants. Protection was only somewhat reduced for the mleR-deficient mutant. Overall, the data indicate that malate protection against acid killing of S. mutans UA159 requires MLF decarboxylase and transmembrane malate transport activities.
Fig. 3.
Effects of deletion mutations of mleR (A), mleS (B), or mleP (C) on MLF protection against acid killing of Streptococcus mutans UA159. Cells were exposed to a constant pH of 3.0 with or without 25 mmol/L l-malate. N0, the initial number of colony-forming units per millilitre; N, the number of colony-forming units at time t of acid exposure. Error bars indicate SD, n = 3.
MLF protection against oxidative damage
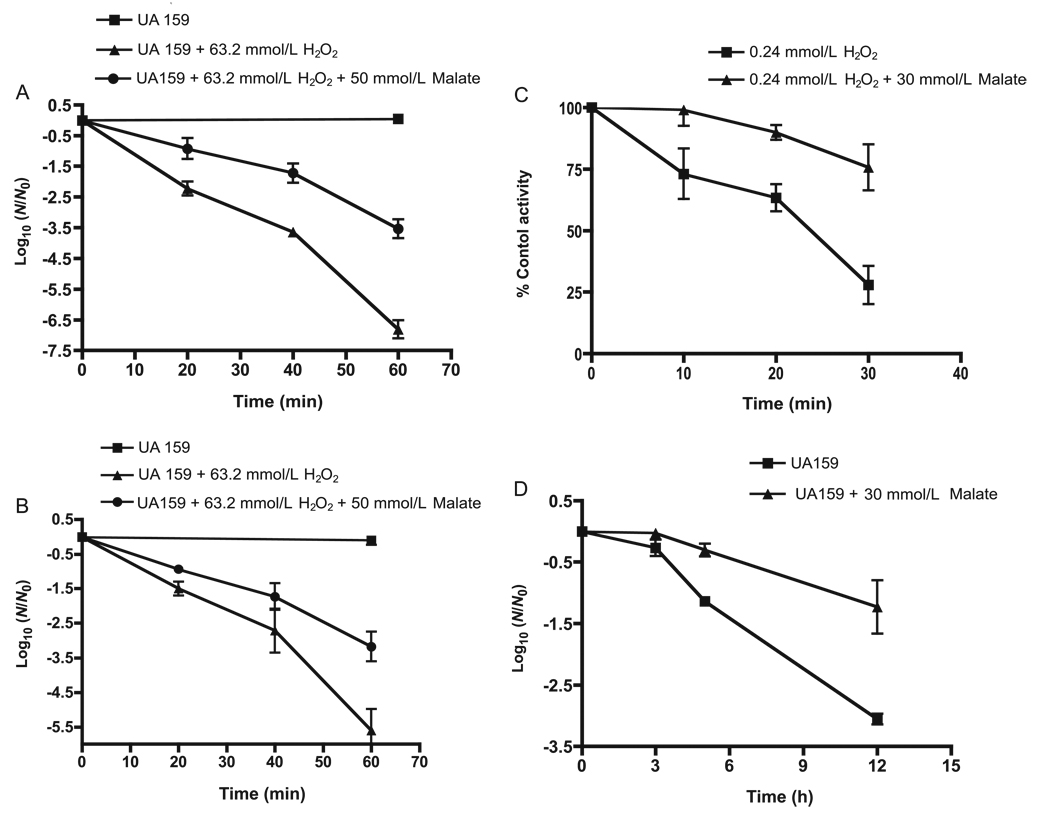
MLF could protect intact cells of S. mutans UA159 from oxidative damage, as shown by the sample data of Fig. 4. In this case, the damage assayed was lethal damage caused by exposure of suspension cells to a high level of hydrogen peroxide, 0.2% m/v, or approximately 63.2 mmol/L, at pH 5.0 (Fig. 4A). Again, the concentration of malate used, 50 mmol/L, was saturating for catabolism, but graded protection could be demonstrated with lower concentrations of malate. Similar data for protection of cells in suspensions exposed to hydrogen peroxide at a pH of 4.0 are shown (Fig. 4B). Protection against peroxide killing was much reduced at higher pH values of 6 or 7, at which MLF activity was reduced. The mleS deletion mutant was not protected against peroxide killing by malate. In fact, killing of the deletion mutant was actually enhanced by 50 mmol/L malate (data not shown). At pH 5, glucose also was marginally protective against peroxide killing but certainly not as protective as malate (data not shown). The difference between glucose and malate is most likely related at least partly to differences in the pH optima, about pH 6 for glycolysis compared to 4 for MLF.
Fig. 4.
Malate protection of Streptococcus mutans UA159 against oxidative and starvation damage. (A) Protection against killing by 0.2% (63.2 mmol/L) hydrogen peroxide at pH 5; (B) protection against peroxide killing at pH 4.0; (C) protection against inactivation at pH 5.0 by 0.00075% (0.24 mmol/L) hydrogen peroxide of glyceraldehyde-3-phosphate dehydrogenase in cells permeabilized for the enzyme assay; (D) protection against starvation killing of cells harvested from stationary-phase suspension cultures, ,washed, and then starved at 37 °C in standard salt solution (50 mmol/L KCl plus 1 mmol/L MgCl2) at pH 5.0. Error bars indicate SD; n = 3.
Malate protection could be demonstrated even with permeabilized cells against milder, bacteriostatic damage by hydrogen peroxide, which involves inhibition of glycolysis but not killing (Baldeck and Marquis 2008). As shown by the data presented in Fig. 4C, 30 mmol/L malate at pH 5 protected the enzyme GAPDH in permeabilized cells against inactivation by 0.00075% (approximately 0.24 mmol/L) hydrogen peroxide. This low level of peroxide has little effect on GAPDH in intact cells exposed to the peroxide, washed, and then permeabilized. A 50% inactivation of the enzyme in intact cells did occur in 15 min at peroxide levels of ca. 2 mmol/L (Baldeck and Marquis 2008). GAPDH is one of the most sensitive targets for bacteriostatic peroxide damage to cells of S. mutans. The damage to GAPDH mainly involves oxidation of enzyme sulfhydryl groups to sulfenic–sulfonic acids, which cannot be repaired (Weber et al. 2004). However, the damage is only bacteriostatic for intact cells, because the cells retain the capacity to synthesize new enzyme when transferred to fresh media. Intact cells treated with 2.3 mmol/L peroxide grew if plated on tryptic soy agar to yield colonies, which appeared only after 48 h of incubation compared with 24 h for untreated cells. Even at 48 h, the colonies were smaller than those from control cells. However, on subculture, they yielded normal-sized colonies.
MLF protection against death by starvation
As expected, malate was protective also against death by starvation, as indicated by the data in Fig. 4D. In this experiment, static cultures were grown until the early stationary phase. Cells were harvested by centrifugation, washed once with standard salt solution (50 mmol/L KCl plus 1 mmol/L MgCl2), and suspended in salt solution before incubation at 37 °C for 12 h at pH 5.0. Malate protection against starvation death was readily demonstrated. Glucose also was effective for protection.
Base production from MLF compared with acid production from glycolysis
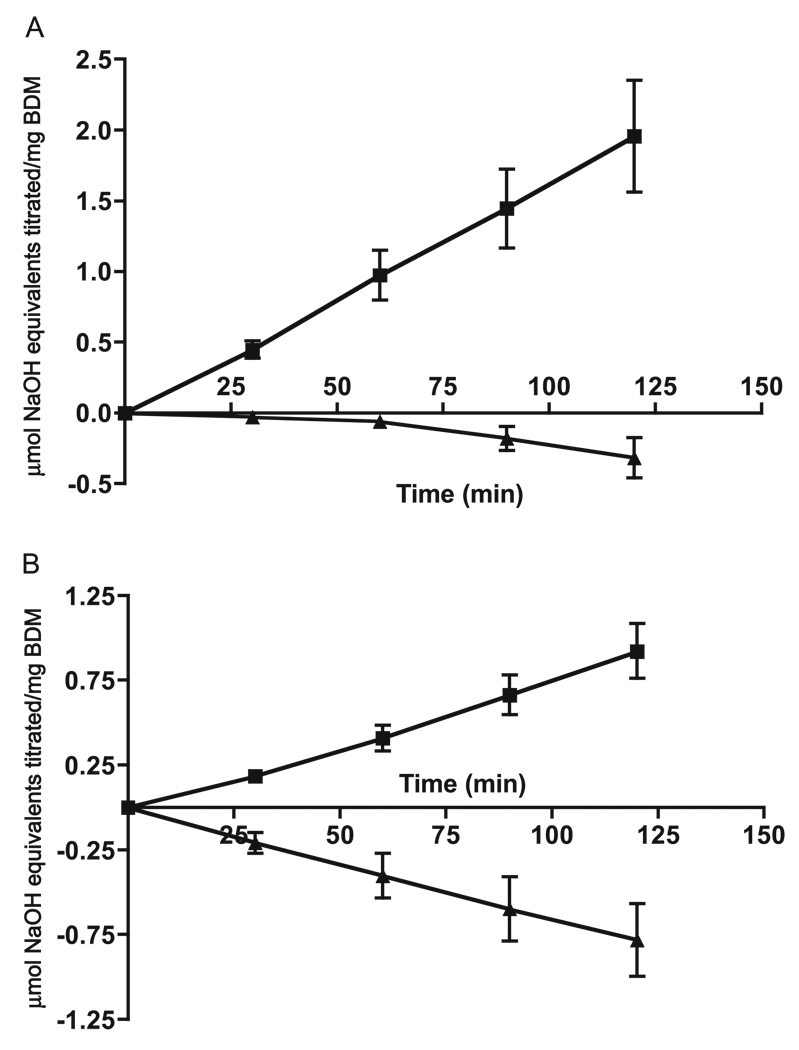
The capacity of malate-induced cells of S. mutans UA159 to produce alkali from malate can be more than sufficient to counter their capacity to produce acid from glycolysis, particularly at acid pH values characteristic of cariogenic plaque. Examples are presented in Fig. 5 for biofilms grown on glass slides in fed-batch cultures, as described previously (Baldeck and Marquis 2008). Initial biofilm growth was in TY–sucrose medium changed daily for 3 days to yield mature films with high levels of matrix polysaccharides on glass microscope slides. The biofilms were then induced for MLF in TY–malate medium (Fig. 6) and harvested after full induction of MLF. Induced biofilms exposed to 25 mmol/L glucose at pH 5 produced acid rapidly (Fig. 5A), assessed in terms of amounts of standardized KOH solution required to maintain constant pH. However, if the cells were given a mix of 25 mmol/L glucose and 25 mmol/L l-malate, there was no net acid production. In fact, after about 1 h, there was net production of alkali, and addition of HCl rather than KOH solution was required to maintain near-constant pH in the biofilms. The dominance of alkali production by biofilms exposed to 25 mmol/L glucose and 25 mmol/L l-malate was even more pronounced at pH 4 (Fig. 5B), presumably because glycolysis was slower at this lower pH value, while MLF was more rapid at pH 4, the optimal pH value for MLF.
Fig. 5.
Acid production from glucose (25 mmol/L) by Streptococcus mutans UA159 in suspensions with (filled triangles) or without (filled squares) neutralization through base production from MLF (25 mmol/L malate) at a constant pH value of 5 (A) or 4.0 (B). Values above 0.00 indicate acid production from glycolysis, while values below 0.00 indicate net base production from MLF plus glycolysis. Error bars indicate SD; n = 3. BDM, biofilm dry mass.
Fig. 6.
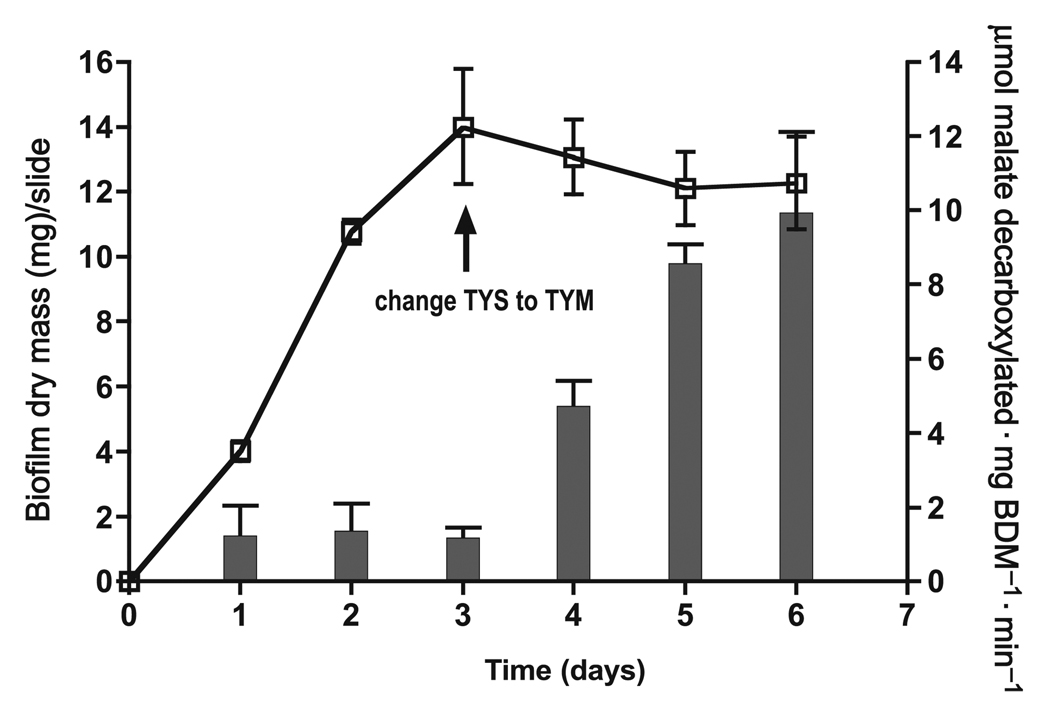
Induction of malolactic fermentation by 25 mmol/L l-malate in biofilms of Streptococcus mutans UA159. Induction involved a switch from TY–sucrose to TY–malate feeding of the biofilms, as indicated in the figure. Error bars indicate SD; n = 3. Open squares indicate biofilm growth, while filled bars indicate MLF. BDM, biofilm dry mass.
Similar data indicating dominance of alkali production over acid production at pH values below 5 could be demonstrated readily with suspension cells as well as with bioflms. The data presented in Fig. 6 indicate that MLF could be induced readily in biofilms even in the absence of net growth from the third to the sixth day. The biofilms clearly became induced for MLF, and capacities for catabolism of l-malate increased from about 1 to as high as about 10 nmol malate catabolized·mg biofilm dry mass (mg BDM)−1·min−1 (0.6 µmol·mg BDM·h−1, or about one-tenth the rate found for cell suspensions). The slow decarboxylation rate of biofilms compared with that of suspension cells was possibly due to diffusion limitations into biofilms. Still, induction and activity of MLF would be expected for oral biofilms in the human mouth and have also been demonstrated for supragingival plaque from humans (J. Sheng and R.E. Marquis, unpublished data).
Interactions of MLF with other alkali-generating systems of oral streptococci
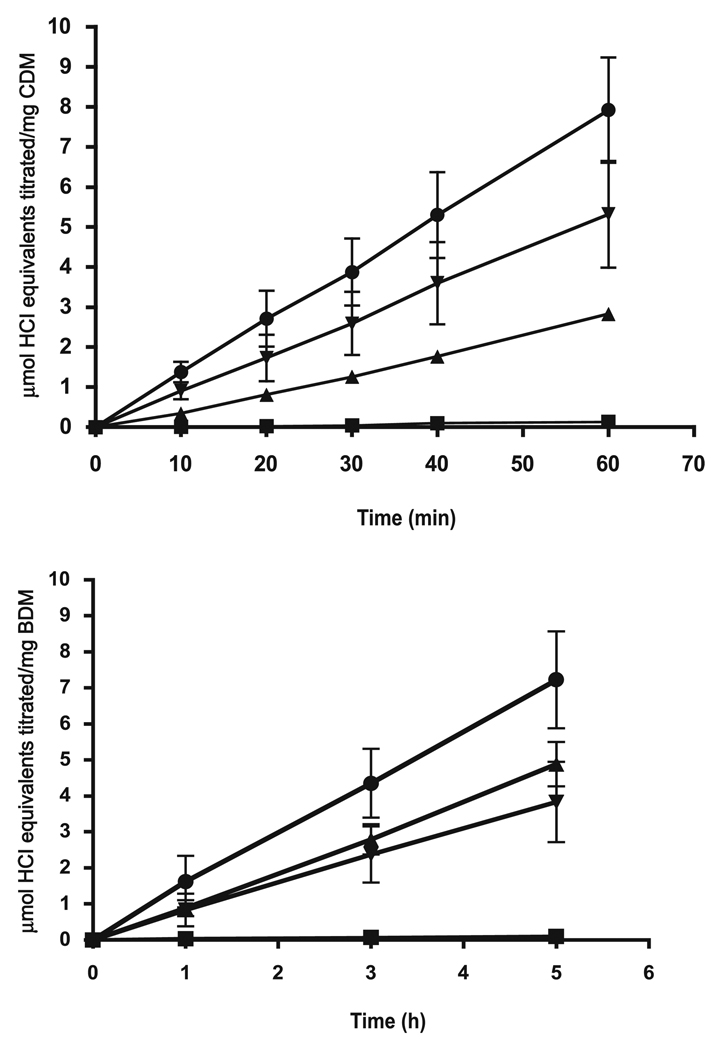
Many oral streptococci have more than one inducible system for alkali production. For example, S. mutans has both the agmatine deiminase system and MLF. Results of initial experiments with combinations of agmatine and malate added to induced S. mutans UA159 were not very conclusive because of the relatively high capacity of the MLF compared with that of the agmatine deiminase pathway, especially at low pH values. Thus, alkali production at pH 4 or 5 was mainly by MLF. A better system to explore interactions was that involving arginine deiminase and MLF, which can be coinduced in S. sanguinis ATCC 10556 with mixtures of arginine and l-malate. ADS is inducible with arginine but is subject to catabolite repression. MLF is inducible with l-malate and not very sensitive to catabolite repression, at least not that involving glucose. Both systems are relatively acid tolerant and function well at pH values below 5 (Casiano-Colón and Marquis 1988; Sheng and Marquis 2007). As shown by the data presented in Fig. 7, both systems were active for suspension cells (Fig. 7, upper panel) or biofilms (Fig. 7, lower panel) at pH 5.0 under pH-stat conditions. The actions of the 2 enzymes, when saturating levels of both arginine and malate were supplied, was approximately additive for alkali production. This same sort of additive action could be demonstrated also for Streptococcus ratti FA1, which is ADS positive and MLF positive. Our attempts to study positive interactions of urease and MLF with cells of S. salivarious ATCC 13914 were not very successful because of differences in pH optima for the systems. Urease is not very acid tolerant, while MLF requires acid pH values for full activity.
Fig. 7.
Alkali production at pH 5.0 by Streptococcus sanguinis ATCC10556 cells in suspensions (upper panel) or biofilms (lower panel) with no catabolite addition (filled squares), with addition of 25 mmol/L l-arginine (filled triangles), with 25 mmol/L l-malate (filled, inverted triangles), or with both 25 mmol/L l-arginine and 25 mmol/L l-malate (filled circles). Error bars indicate SD; n = 3. BDM, biofilm dry mass; CDM, cell dry mass.
Discussion
The results of previous studies (Sheng and Marquis 2007; Sheng et al. 2008) indicated that MLF protects cells of S. mutans, and presumably those of other MLF-positive oral lactic acid bacteria, against acid damage under pH-stat or pH-rise conditions. In this paper we show that deletion mutations in any one of the 3 annotated genes for MLF of S. mutans UA159 reduced the capacity of the organism to produce alkali from malate and to protect against lethal acid damage. Mutation of mleR resulted in only partial loss of function, while mutation of mleS led to essentially complete loss of function. Mutation of mleP also resulted in nearly complete loss of function, and this finding suggests that MLF is mainly transport limited, as also indicated by previous unsuccessful attempts to show significant intracellular pooling of malate by S. mutans UA159 cells, except at very high concentrations of malate.
The finding that MLF protects cells against lethal oxidative damage caused by hydrogen peroxide was somewhat surprising but in line with our view that catabolism in oral streptococci is generally protective against damage caused by a variety of environmental stresses, including acid, oxidative, and starvation stresses. Previously, Jang and Imlay (2007) reported that malate protected fumarase A of Escherichia coli against peroxide damage and could even reactivate damaged enzyme through repair of Fe–S clusters. The protection was interpreted in terms of substrate binding of malate and resultant shielding of sensitive sites of Fe–S clusters against reaction with peroxide. Our results indicate malate protection of GAPDH in permeabilized cells at low levels of peroxide, suggestive of similar enzyme protective effects to those shown for fumarase A.
It seems that the protection against peroxide damage that we could demonstrate at constant pH for intact cells may have a different mechanism, mainly related to generation of δ-pH and ATP by MLF. For protection in the intact-cell system, the malate had to be metabolized, and mleS or mleP deletion mutants showed essentially no protection. Peroxide damage to S. mutans cells was found previously to be greater at higher pH values (Baldeck and Marquis 2008). Therefore, any production of alkali in the cytoplasm should actually increase peroxide damage rather than protect against it. ATP or δ–ψ produced from MLF are likely to be protective elements, as they are for acid damage. However, an exact mechanism was not defined.
Alkali-producing systems other than MLF, notably ADS and urease, have been previously found to protect cells of oral streptococci against acid damage. Again, there appear to be at least 2 mechanisms for protection, one related simply to pH rise and the other to pH-stat conditions. The first is easily understood, in that pH rise would directly reduce acid stress. This stress reduction would likely be more important for the less acid-tolerant organisms in the microbiota (Horiuchi et al. 2009) and thus serve to reduce the selection for acid tolerance in mixed populations, such as those of dental plaque. The pH-independent protection could arise from a number of factors, but is probably associated with ATP synthesis and with changes in δ-pH across the cell membrane. Results of our previous studies with oral bacteria (Sheng and Marquis 2006) indicated that catabolites such as glucose and other metabolized sugars protect against acid killing under pH-stat conditions when no net acidification occurred from their metabolism. The main possibility is that ATP is able to function for protection, probably by fueling proton expulsion from the cytoplasm through the F-ATPase, with the resultant sparing of acid damage to sensitive targets. Energy-dependent repair of cell damage is another possibility.
Major questions arising from our work concern the effects of MLF on the physiology and pathogenicity of the plaque microbiota in the human mouth. Alkali production in dental plaque is generally considered to be protective against caries (Burne and Marquis 2000). In fact, there is now a set of clinically tested oral-care products containing arginine for inhibiting development of dental caries in humans (Acevedo et al. 2008).
Acknowledgements
This research was supported by grants R01 DE13683, R01 DE017425, and R01 DE06127 from the US National Institute of Dental and Craniofacial Research. We thank Elizabeth Grayhack and James Payne for help in constructing mutants used in this study.
Footnotes
Addendum
While our manuscript was under review, a paper on malolactic fermentation by S. mutans (Lemme et al. 2010) was published on the Internet. The published paper dealt primarily with regulation of the fermentation by MleR and environmental influences but overlapped with our study. The pictures developed by the 2 papers are generally in accord, except that our finding of protection against oxidative damage by malolactic fermentation was not found by Lemme et al. We do not feel that extensive comparative analysis of the 2 papers is desirable in this paper, since both will be available in the literature.
References
- Acevedo AM, Montero M, Rojas-Sanchez F, Machado C, Rivera LE, Wolff M, Kleinberg I. Clinical evaluation of the ability of CaviStat in a mint confection to inhibit the development of dental caries in children. J. Clin. Dent. 2008;19(1):1–8. PMID:18500152. [PubMed] [Google Scholar]
- Ajdić D, McShan WM, McLaughlin RE, Savić G, Chang J, Carson MB, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U.S.A. 2002;99(22):14434–14439. doi: 10.1073/pnas.172501299. doi:10.1073/pnas.172501299. PMID:12397186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansanay V, Dequin S, Blondin B, Barre P. Cloning, sequence and expression of the gene encoding the malolactic enzyme from Lactococcus lactis. FEBS Lett. 1993;332(1–2):74–80. doi: 10.1016/0014-5793(93)80488-g. doi:10.1016/0014-5793(93)80488-G. PMID:8405453. [DOI] [PubMed] [Google Scholar]
- Baldeck JD, Marquis RE. Targets for hydrogen-peroxide-induced damage to suspension and biofilm cells of Streptococcus mutans. Can. J. Microbiol. 2008;54(10):868–875. doi: 10.1139/w08-078. doi:10.1139/W08-078. PMID:18923556. [DOI] [PubMed] [Google Scholar]
- Belli WA, Marquis RE. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl. Environ. Microbiol. 1991;57(4):1134–1138. doi: 10.1128/aem.57.4.1134-1138.1991. PMID:1829347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender GR, Marquis RE. Membrane ATPases and acid tolerance of Actinomyces viscosus and Lactobacillus casei. Appl. Environ. Microbiol. 1987;53(9):2124–2128. doi: 10.1128/aem.53.9.2124-2128.1987. PMID:2445289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender GR, Sutton SVW, Marquis RE. Acid tolerance, proton permeabilities, and membrane ATPases of oral streptococci. Infect. Immun. 1986;53(2):331–338. doi: 10.1128/iai.53.2.331-338.1986. PMID:3015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne RA, Marquis RE. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol. Lett. 2000;193(1):1–6. doi: 10.1111/j.1574-6968.2000.tb09393.x. doi:10.1111/j.1574-6968.2000.tb09393.x. PMID:11094270. [DOI] [PubMed] [Google Scholar]
- Casiano-Coló;n A, Marquis RE. Role of the arginine deiminase system in protecting oral bacteria and an enzymatic basis for acid tolerance. Appl. Environ. Microbiol. 1988;54(6):1318–1324. doi: 10.1128/aem.54.6.1318-1324.1988. PMID:2843090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YY, Burne RA. Analysis of Streptococcus salivarius urease expression using continuous chemostat culture. FEMS Microbiol. Lett. 1996;135(2–3):223–229. doi: 10.1111/j.1574-6968.1996.tb07993.x. doi:10.1111/j.1574-6968.1996.tb07993.x. PMID:8595861. [DOI] [PubMed] [Google Scholar]
- Denayrolles M, Aigle M, Lonvaud-Funel A. Cloning and sequence analysis of the gene encoding Lactococcus lactis malolactic enzyme: relationships with malic enzymes. FEMS Microbiol. Lett. 1994;116(1):79–86. doi: 10.1111/j.1574-6968.1994.tb06679.x. doi:10.1111/j.1574-6968.1994.tb06679.x. PMID:8132158. [DOI] [PubMed] [Google Scholar]
- Fozo EM, Kajfasz JK, Quivey RG., Jr Low pH-induced membrane fatty acid alterations in oral bacteria. FEMS Microbiol. Lett. 2004;238(2):291–295. doi: 10.1016/j.femsle.2004.07.047. doi:10.1111/j.1574-6968.2004.tb09769.x. PMID:15358413. [DOI] [PubMed] [Google Scholar]
- Fozo EM, Quivey RG., Jr Shifts in the membrane fatty acid profile of Streptococcus mutans enhance survival in acidic environments. Appl. Environ. Microbiol. 2004;70(2):929–936. doi: 10.1128/AEM.70.2.929-936.2004. doi:10.1128/AEM.70.2.929-936.2004. PMID:14766573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold AR, Jameson-Lee M, Burne RA. Regulation and physiologic significance of the agmatine deiminase system of Streptococcus mutans UA159. J. Bacteriol. 2006;188(3):834–841. doi: 10.1128/JB.188.3.834-841.2006. doi:10.1128/JB.188.3.834-841.2006. PMID:16428386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi M, Washio J, Mayanagi H, Takahashi N. Transient acid-impairment of growth ability of oral Streptococcus, Actinomyces, and Lactobacillus: a possible ecological determinant in dental plaque. Oral Microbiol. Immunol. 2009;24(4):319–324. doi: 10.1111/j.1399-302X.2009.00517.x. doi:10.1111/j.1399-302X.2009.00517.x. PMID:19572895. [DOI] [PubMed] [Google Scholar]
- Jang S, Imlay JA. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron–sulfur enzymes. J. Biol. Chem. 2007;282(2):929–937. doi: 10.1074/jbc.M607646200. doi:10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korithoski B, Krastel K, Cvitkovitch DG. Transport and metabolism of citrate by Streptococcus mutans. J. Bacteriol. 2005;187(13):4451–4456. doi: 10.1128/JB.187.13.4451-4456.2005. doi:10.1128/JB.187.13.4451-4456.2005. PMID:15968054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarre C, Diviés C, Guzzo J. Genetic organization of the mle locus and identification of a mleR-like gene from Leuconostoc oenos. Appl. Environ. Microbiol. 1996a;62(12):4493–4498. doi: 10.1128/aem.62.12.4493-4498.1996. PMID:8953720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarre C, Guzzo J, Cavin JF, Diviés C. Cloning and characterization of the genes encoding the malolactic enzyme and the malate permease of Leuconostoc oenos. Appl. Environ. Microbiol. 1996b;62(4):1274–1282. doi: 10.1128/aem.62.4.1274-1282.1996. PMID:8919788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods. 2002;49(2):193–205. doi: 10.1016/s0167-7012(01)00369-4. doi:10.1016/S0167-7012(01)00369-4. PMID:11830305. [DOI] [PubMed] [Google Scholar]
- Lemme A, Sztajer H, Wagner-Döbler I. Characterization of mleR, a positive regulator of malolactic fermentation and part of the acid tolerance response in Streptococcus mutans. BMC Microbiol. 2010;10(1):58. doi: 10.1186/1471-2180-10-58. doi:10.1186/1471-2180-10-58. PMID:20178568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrina FL, Evans RP, Tobian JA, Hartley DL, Clewell DB, Jones KR. Novel shuttle plasmid vehicles for Escherichia–Streptococcus transgeneric cloning. Gene. 1983;25(1):145–150. doi: 10.1016/0378-1119(83)90176-2. doi:10.1016/0378-1119(83)90176-2. PMID:6319229. [DOI] [PubMed] [Google Scholar]
- Morou-Bermudez E, Burne RA. Analysis of urease expression in Actinomyces naeslundii WVU45. Infect. Immun. 2000;68(12):6670–6676. doi: 10.1128/iai.68.12.6670-6676.2000. doi:10.1128/IAI.68.12.6670-6676.2000. PMID:11083780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry D, Kuramitsu HK. Genetic transformation of Streptococcus mutans. Infect. Immun. 1981;32(3):1295–1297. doi: 10.1128/iai.32.3.1295-1297.1981. PMID:7251168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T-N, Reidmiller JS, Marquis RE. Sensitization of Actinomyces naeslundii and Streptococcus sanguis in biofilms and suspensions to acid damage by fluoride and other weak acids. Arch. Microbiol. 2000;174(4):248–255. doi: 10.1007/s002030000202. doi:10.1007/s002030000202. PMID:11081793. [DOI] [PubMed] [Google Scholar]
- Phan T-N, Buckner T, Sheng J, Baldeck JD, Marquis RE. Physiologic actions of zinc related to inhibition of acid and alkali production by oral streptococci in suspensions and biofilms. Oral Microbiol. Immunol. 2004;19(1):31–38. doi: 10.1046/j.0902-0055.2003.00109.x. doi:10.1046/j.0902-0055.2003.00109.x. PMID:14678472. [DOI] [PubMed] [Google Scholar]
- Quivey RG, Jr, Kuhnert WL, Hahn K. Adaptation of oral streptococci to low pH. Adv. Microb. Physiol. 2000;42:239–274. doi: 10.1016/s0065-2911(00)42004-7. doi:10.1016/S0065-2911(00)42004-7. PMID:10907552. [DOI] [PubMed] [Google Scholar]
- Sheng J, Marquis RE. Enhanced acid resistance of oral streptococci at lethal pH values associated with acid-tolerant catabolism and with ATP synthase activity. FEMS Microbiol. Lett. 2006;262(1):93–98. doi: 10.1111/j.1574-6968.2006.00374.x. doi:10.1111/j.1574-6968.2006.00374.x. PMID:16907744. [DOI] [PubMed] [Google Scholar]
- Sheng J, Marquis RE. Malolactic fermentation by Streptococcus mutans. FEMS Microbiol. Lett. 2007;272(2):196–201. doi: 10.1111/j.1574-6968.2007.00744.x. doi:10.1111/j.1574-6968.2007.00744.x. PMID:17490430. [DOI] [PubMed] [Google Scholar]
- Sheng J, Baldeck JD, Marquis RE. Physiology of malolactic fermentation by Streptococcus mutans. 86th General Session and Exhibition of the International Association for Dental Research; 2–5 July 2008; Toronto, Ont.. 2008. p. 119. Abstr. 1635. [Google Scholar]
- Sobczak I, Lolkema JS. The 2-hydroxycarboxylate transporter family: physiology, structure, and mechanism. Microbiol. Mol. Biol. Rev. 2005;69(4):665–695. doi: 10.1128/MMBR.69.4.665-695.2005. doi:10.1128/MMBR.69.4.665-695.2005. PMID:16339740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Engelmann S, Becher D, Hecker M. Oxidative stress triggers thiol oxidation in the glyceraldehyde-3-phosphate dehydrogenase of Staphylococcus aureus. Mol. Microbiol. 2004;52(1):133–140. doi: 10.1111/j.1365-2958.2004.03971.x. doi:10.1111/j.1365-2958.2004.03971.x. PMID:15049816. [DOI] [PubMed] [Google Scholar]
- Wijeyeweera RL, Kleinberg I. Acid–base pH curves in vitro with mixtures of pure cultures of human oral microorganisms. Arch. Oral Biol. 1989;34(1):55–64. doi: 10.1016/0003-9969(89)90046-0. doi:10.1016/0003-9969(89) 90046-0. PMID:2675801. [DOI] [PubMed] [Google Scholar]