Summary
Adherens junctions, which play a central role in intercellular adhesion, comprise clusters of type I classical cadherins that bind via extracellular domains extended from opposing cell surfaces. We show that a molecular layer seen in crystal structures of E- and N-cadherin ectodomains reported here and in the C-cadherin structure corresponds to the extracellular architecture of adherens junctions. In all three ectodomain crystals, cadherins dimerize through a trans adhesive interface and are connected by a second, cis, interface. Assemblies formed by E-cadherin ectodomains coated on liposomes also appear to adopt this structure. Fluorescent imaging of junctions formed from wild-type and mutant E-cadherins in cultured cells confirm conclusions derived from structural evidence. Mutations that interfere with the trans interface ablate adhesion, whereas cis interface mutations disrupt stable junction formation. Our observations are consistent with a model for junction assembly involving strong trans and weak cis interactions localized in the ectodomain.
Introduction
The clustering of cell-surface proteins into multi-protein assemblies, or junctions, is a hallmark of many cell and membrane adhesion processes (Dean et al., 2003; Grakoui et al., 1999). Adhesion is initiated through the trans (cell-to-cell) dimerization of adhesion proteins embedded in opposing membranes which then undergo lateral, cis, clustering through still poorly-understood mechanisms (Hong et al., 2010; Yap et al., 1997). The assemblies that are formed in this way are likely to provide templates for the recruitment of cytoplasmic proteins which may then initiate downstream signaling events. Although the structures of individual trans dimers of numerous adhesion proteins have now been revealed (Arac et al., 2007; Boggon et al., 2002; Somers et al., 2000), atomic level structures of larger assemblies have not been determined. Here we present crystal structures of the complete ectodomains of E- and N-cadherins, and show that their crystals share a common molecular layer that is formed by a similar array of cis and trans interactions. Functional assays suggest that this molecular layer represents the three-dimensional extracellular structure of adherens junctions.
Adherens junctions are intercellular structures that are formed by clusters of trans dimers of classical cadherins. They are characterized by defined intermembrane spacing such that apposed cell membranes appear parallel and cytoplasmic plaques that link to F-actin (Farquhar and Palade, 1963; McNutt and Weinstein, 1973). Type I classical cadherins and the related type II cadherins mediate Ca2+-dependent adhesion between the cells of vertebrates (Takeichi, 1988). Type I and II cadherin domain structures are similar, each with an ectodomain composed of five tandem extracellular cadherin (EC) repeats preceded by a signal sequence and a pro-domain that must be removed by proteolysis for adhesive function, a single membrane-spanning region and cytoplasmic regions with binding sites for β-, γ- and p120-catenins. The latter are thought to mediate connections to the actin cytoskeleton and to regulate cadherin turnover and recycling (Huber and Weis, 2001; Ishiyama et al., 2010; Ozawa et al., 1989).
The crystal structure of the whole (EC1-EC5) ectodomain from C-cadherin, a type I classical cadherin from Xenopus laevis, represents the only complete cadherin ectodomain structure published to date (Boggon et al., 2002). This structure, which is consistent with numerous other structures of adhesive type I and type II ectodomain fragments (Harrison et al., 2010; Haussinger et al., 2004; Parisini et al., 2007; Shapiro et al., 1995), reveals a “strand swap” trans interface in which the N-terminal β-strand from the EC1 domain of each paired cadherin exchanges with that of the partner molecule. A second functionally important trans interface, involving the linker region between the EC1 and EC2 domains, has also been identified and constitutes a kinetic intermediate on the path to the formation of strand swapped dimers (Harrison et al., 2010; Nagar et al., 1996; Pertz et al., 1999). However, despite a detailed understanding of trans dimerization, little is known about the mode of cadherin assembly in adherens junctions. Cryo-electron tomography (cryo-ET) structures of related desmosome junctions have been reported (Al-Amoudi et al., 2007; He et al., 2003). Desmosomes, which are anchored by the intermediate filament system, are also mediated by interactions between the ectodomains of specialized cadherins, desmocollins and desmogleins (Delva et al., 2009). The known determinants of strand swapping in type I cadherins (Posy et al., 2008) are conserved in desmosomal cadherins, suggesting a similar mode for trans binding between their membrane distal EC1 domains.
Here we show that the crystal structures of the complete ectodomains of N- and E-cadherin, along with the previously determined structure of C-cadherin, reveal the extracellular organization of adherens junctions. Although the three proteins form crystals in unrelated lattices, all crystals contain a molecular layer assembled via two interfaces: the well-characterized trans strand swap adhesive interface between EC1 domains, and a lateral cis interface in which a different EC1 domain surface interacts with a region of the EC2 domain of a neighboring molecule. Whereas the adhesive trans interface forms between cadherins oriented as if presented from apposed cell surfaces, the cis interface is formed between cadherins positioned as if emanating from the same cell surface.
Cryo-EM imaging of artificial junctions between cadherin coated liposomes and fluorescent imaging of adherens junctions in transfected cells reveal an essential role for the cis interface in the assembly of cadherin junctions. We suggest a mechanism for junction assembly involving cooperative cis and trans interactions that is likely to be relevant to other systems that mediate membrane apposition and transmembrane signaling.
Results
Ectodomain structures of E-cadherin and N-cadherin
We determined crystal structures of the mature ectodomains from mouse E-cadherin and mouse N-cadherin to 3.4Å and 3.2Å resolution, respectively. Data and refinement statistics are listed in Table 1.
Table 1. Data collection and refinement statistics.
| E-cadherin EC1-5 | N-cadherin EC1-5 | E-cadherin EC1-2 V81D |
E-cadherin EC1-2 L175D |
|
|---|---|---|---|---|
| Data Collection | ||||
| Space group | C2 | C2221 | C222 | C222 |
| a, b, c (Å) | 119.1, 79.7, 176.0 | 91.4, 111.6, 262.1 | 142.7, 168.9, 131.1 | 140.9, 169.5, 131.4 |
| α, β, γ (°) | 90, 98.5, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Molecules per asu | 2 | 1 | 2 | 2 |
| Resolution (Å) | 3.4 (3.52-3.4) | 3.2 (3.31-3.2) | 2.7 (2.8-2.7) | 2.75 (2.85-2.75) |
| Rsym | 0.123 (0.381) | 0.113 (0.348) | 0.135 (0.539) | 0.116 (0.505) |
| I / σI | 6.5 (2.0) | 12.6 (4.0) | 10.9 (3.1) | 11.2 (4.3) |
| Completeness (%) | 99.5 (99.2) | 88.1 (69.9) | 99.8 (99.8) | 100 (100) |
| Redundancy | 3.2 (3.0) | 5.6 (5.1) | 5.6 (5.4) | 6.9 (6.7) |
| Refinement | ||||
| Resolution (Å) | 20-3.4 | 20-3.2 | 20–2.7 | 20-2.75 |
| Number of reflections | 22536 | 20206 | 41130 | 39137 |
| Rwork / Rfree | 0.230/0.293 | 0.229/0.267 | 0.214/0.245 | 0.201/0.224 |
| Average B-factors (Å2) | 65.9 | 61.6 | 52.6 | 38.9 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.009 | 0.007 | 0.019 | 0.026 |
| Bond angles (°) | 1.42 | 1.18 | 1.77 | 2.16 |
| Ramachandran Statistics | ||||
| Favored (%) | 89.8 | 93.1 | 96.7 | 96.4 |
| Allowed (%) | 10.2 | 6.7 | 3.3 | 3.6 |
| Disallowed (%) | 0 | 0.2 | 0 | 0 |
One crystal was used per dataset. Values in parentheses are for the highest resolution shell.
Rfree = Rwork calculated using 5% of the reflection data chosen randomly and omitted from the start of refinement.
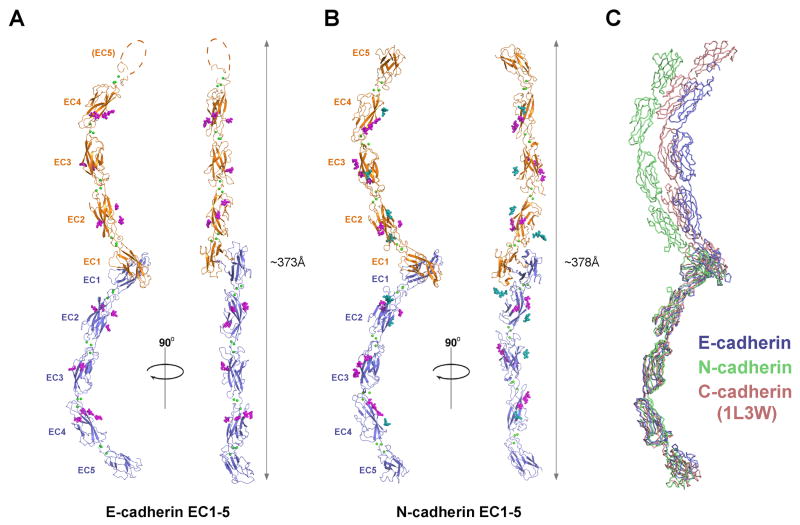
The overall structures of the E- and N-cadherin ectodomains are very similar to that observed in the previously published structure of C-cadherin (Boggon et al., 2002). In the E-cadherin structure, two molecules are present in the crystallographic asymmetric unit: one in which all five EC domains are resolved (residues 1 to 538 of the mature protein) and another in which the membrane proximal EC5 domain is disordered (Fig. 1A). In the N-cadherin ectodomain structure, the crystallographic asymmetric unit contains a single molecule in which EC1 to EC5 (residues 1 to 542 of the mature protein) are well ordered (Fig. 1B). In both ectodomain structures, five EC domains, each adopting a characteristic seven-stranded β-barrel fold, are arranged in tandem to form an elongated, curved structure that is stabilized by binding of three Ca2+ ions between each set of successive EC domains, with twelve bound Ca2+ ions in total (Fig. 1).
Figure 1. Ectodomain structures of mouse E- and N-cadherin.
(A) E-cadherin crystal structure showing a strand swapped trans dimer (ribbon view). Protomers are colored blue and orange; calcium ions bound in the interdomain linker regions are shown as green spheres. Strand swapped dimerization occurs between EC1 domains and is anchored by exchange of Trp2 residues (sticks). O-linked glycosylation observed in the structure is shown as magenta spheres. The EC5 domain of one protomer was not resolved and is represented by a dotted line. (B) N-cadherin strand swapped dimer crystal structure, shown in the same representation as for E-cadherin in panel A. N-linked glycosylation is shown as blue spheres. (C) Strand swapped trans dimer structures of E-, N- and C-cadherin (1L3W) superposed over a single protomer, to compare dimer angles. A closer comparison of the curvature of single ectodomains of E-, N- and C-cadherin is shown in Supp. Fig. 1B. See also Supp. Fig. 1 and Table 1.
Both E- and N-cadherin form strand swapped dimers in the crystal lattice that previous structural and mutation studies have identified as the adhesive trans binding interface of classical cadherins (Boggon et al., 2002; Harrison et al., 2010; Haussinger et al., 2004; Kitagawa et al., 2000; Parisini et al., 2007; Shapiro et al., 1995; Troyanovsky et al., 2003) (Fig. 1). In the dimer, EC1 domains of each protomer closely interact and symmetrically exchange their N-terminal β-strands, which contain a conserved tryptophan residue, Trp2. The Trp2 side chains docks into a conserved hydrophobic pocket in the partner EC1 domain and docking is stabilized by an intermolecular salt bridge and hydrogen bonds, as described for previous structures of adhesive fragments of type I cadherins (Supp. Fig. 1A) (Harrison et al., 2010; Haussinger et al., 2004; Parisini et al., 2007). The strand swap interface orients the partner ectodomains in a trans configuration, as if extending from opposing cell membranes, with a distance between the C-termini of approximately 373 Å for E-cadherin, 378Å for N-cadherin and 384 Å for C-cadherin (Boggon et al., 2002) (Fig. 1).
Comparison of the ectodomain structures of E-, N- and C-cadherin (Boggon et al., 2002) reveals that the individual protomers superpose over all five EC domains, with pairwise rmsd values of 3.7Å or less for 485-510 aligned Cα atoms (Fig. 1C). Thus, the pronounced overall curvature of the ectodomain appears to be a stable feature that is conserved in type I classical cadherins. However, ectodomain curvature is not identical in the three cadherins and moderate differences are evident when only the three EC1 domains are superimposed and differences in the entire ectodomain structures are considered (Supp. Fig. 1B). These differences arise mainly from variations in interdomain angles in the linkers between EC2-3 (angular difference between E-, N- and C-cadherin of up to ∼15°), EC3-4 (up to ∼16°) and EC4-5 (up to ∼19°). The relative orientations of ectodomains in trans dimers of E-, N- and C-cadherin also vary (Fig. 1C). Angles between EC1 domains in the trans dimers range from ∼52° (N-cadherin) to ∼88° (E-cadherin), in accord with previous structural studies with smaller fragments of E- and N-cadherin, suggesting that the angle of the trans dimer can vary substantially (Harrison et al., 2010; Haussinger et al., 2004; Parisini et al., 2007; Shapiro et al., 1995). Other notable structural features of the E- and N-cadherin ectodomains are shown in Supp. Fig 1.
A structurally conserved cis interface
In the published structure of C-cadherin EC1-EC5 (Boggon et al., 2002), a potential cis interface was identified in the crystal lattice that could in principle promote lateral association of cadherins on the cell surface. Remarkably, we observe the same cis interface in the crystal lattices of E- and N-cadherin. The cis interface and the strand swapped trans dimer interface described above are the only interactions observed in all three (E-, N- and C-) crystal lattices, which are maintained by a variety of other non-conserved crystal contacts (Supp. Table 1). The presence of a common contact in unrelated lattices suggests the possibility of a functional role.
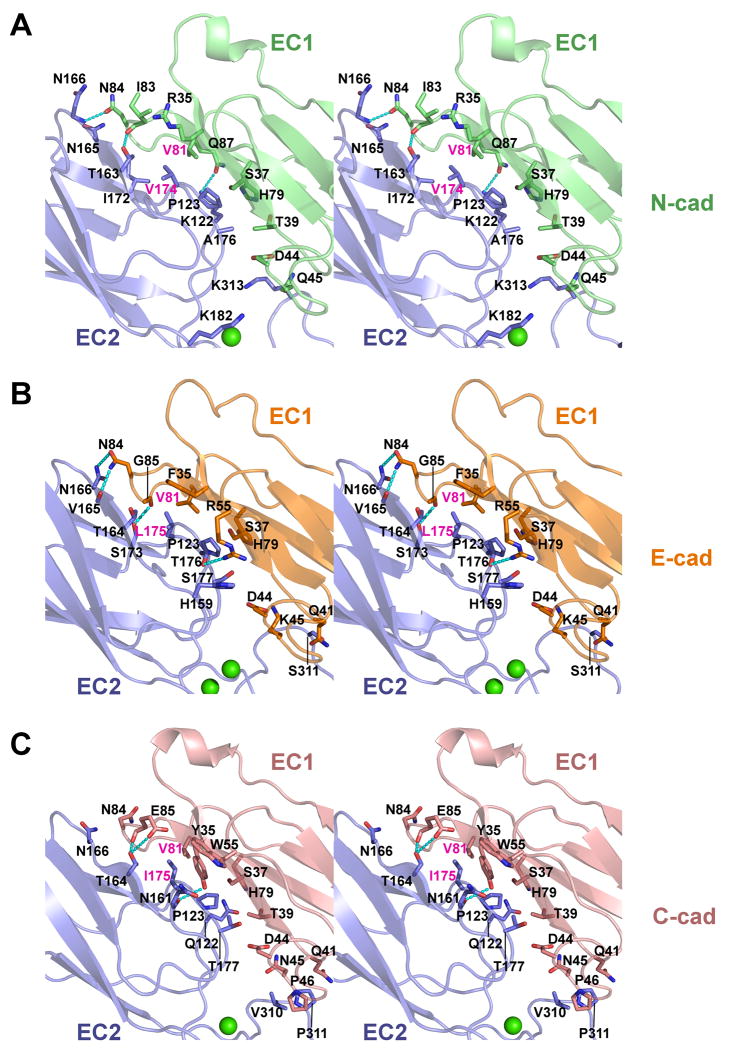
The cis interface in E-, N- and C-cadherin comprises a non-symmetrical interaction between the EC1 domain of one protomer and the EC2 domain of a partner cadherin, with additional minor contributions from the EC2-3 linker and the apex of EC3 (Fig. 2). In EC1, the concave face formed by the C, F and G strands and the FG and CD loops, opposite the trans dimer interface, binds to the convex face near the base of EC2 formed by strands B, D and E and loops AB and EF (Fig. 2A-C). The quasi β-helix between the C and D strands of EC1 contributes part of the interface and is positioned close to the EC2-3 calcium binding linker and to the FG loop at the apex of EC3. The interface buries a total surface area of 1120-1356Å2 in E-, N- and C-cadherin (Supp. Table 1) and involves essentially identical regions of the protein in each, though not all buried residues are conserved between sub-types (Fig. 2A-C, Supp. Fig. 2A). A small hydrophobic core is formed by the side chains of Val81 in EC1 and Pro123 and Leu175 (Val174 in N-cadherin, Ile175 in C-cadherin) in EC2. The side chains of Phe35 (E-cadherin), Tyr35 (C-cadherin) and the aliphatic part of Arg35 (N-cadherin) are also positioned close to the hydrophobic region. Several intermolecular hydrogen bonds are observed in each of the interfaces, particularly between the FG loop of EC1 and the D/E strands in EC2, however, specific hydrogen bonding patterns are not conserved between the three cadherins.
Figure 2. A conserved cis interface in E-, N- and C-cadherin.
Stereo views of cis interfaces observed in crystal structures of (A) mouse N-cadherin, (B) mouse E-cadherin and (C) Xenopus C-cadherin (1L3W) are shown in ribbon representation. Interfaces are formed between a concave surface of EC1 (colored green, orange and salmon for E-, N- and C-cadherin) and a convex surface of EC2 of a partner molecule oriented in parallel (blue). Regions of EC3 involved in contacts are also shown. Side chains of residues contributing at least 10Å2 buried surface area to the interface are displayed as sticks. Hydrogen bonds are shown as dashed lines, calcium ions are shown as green spheres. Residues selected for mutation (see text) are labeled in magenta. See also Supp. Fig. 2 and Supp. Table 1.
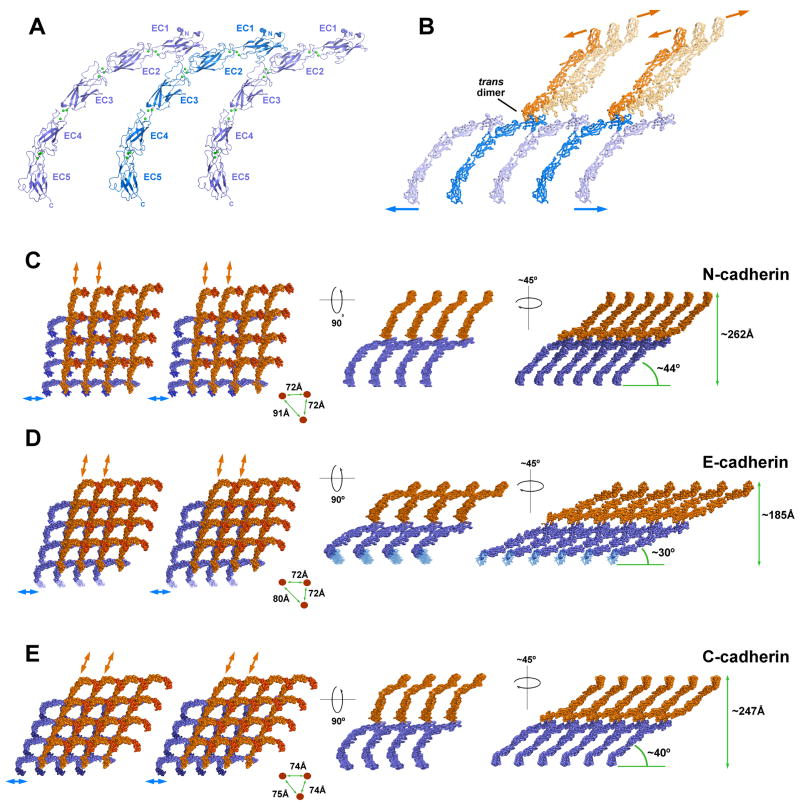
The cis interface orients partner cadherin ectodomains in parallel, as if extending from the same cell surface (Fig. 3A). Each cadherin ectodomain can simultaneously engage in two identical cis interactions: one via its EC1 domain, providing the concave side of the interface and a second via its EC2/3 region, providing the convex side. Thus, the cis interface arranges cadherin ectodomains into linear arrays (Fig. 3A). Additionally, each cadherin can engage in a single strand swapped trans interaction with a molecule oriented as if on the apposing cell membrane, itself able to participate in linear arrays (Fig. 3B, orange molecules). Thus, the trans dimer orients opposing linear arrays of cadherins at an almost perpendicular angle so that each line of molecules can bind in trans with multiple parallel lines on the opposing side. In this way, two interfaces (cis and trans) are sufficient to elaborate a molecular layer in which opposing cis oriented arrays intersect via trans dimer interactions (Fig. 3C-E). The layer extends in two dimensions and positions oppositely oriented cadherin C-termini in parallel planes equivalent to apposed membranes. This molecular layer suggests a possible structure for the extracellular arrangement of cadherins in adherens junctions.
Figure 3. A molecular layer formed by cis and trans interactions in crystal lattices of E-, N- and C-cadherin.
(A) Linear array formed by cis interactions between parallel ectodomains of N-cadherin. Identical interactions are observed in the crystal lattices of E-, and C-cadherin (1L3W). (B) One cis array (blue) engaged in strand swapped trans interactions with two cis arrays (orange) that are oriented as if emanating from an opposing cell. Arrows indicate directions in which linear rows of cis dimers would extend. Note the almost perpendicular angle between the opposing linear arrays. For clarity, only two trans interactions are shown (bolder shading). N-cadherin is depicted; similar interactions are observed in E-cadherin and C-cadherin. (C, D, E) Left panels show stereo views of segments of the molecular layer formed by cis and trans interactions in the crystal lattices of N-, E-and C-cadherin (1L3W), viewed perpendicular to the plane of the layer and oriented with blue cis arrays horizontal. Lattice segments comprising 4×4 trans dimers are shown; EC5 domains are shaded to aid orientation. Right panels show two projections of the molecular layer viewed along the proposed plane of the membranes. Distances between C-termini in right panels determined from crystal lattice dimensions (N-, C-cadherin) or from measurement in Pymol (E-cadherin).
The arrangement of the molecular layer differs in some respects between E-, N- and C-cadherin (Fig. 3C-E). In particular, the minimum angle between trans dimers and the presumptive plane of the membrane varies within a range of approximately 14°, giving a range of predicted intermembrane distances of 185-262Å (Fig. 3C-E, right panels). These sub-type differences are due primarily to variations in the curvature of individual ectodomains (See Supp. Fig. 1B) and, to a lesser extent, to variations in the angle of the trans dimer interface (See Fig. 1C), both of which may be flexible. Different packing forces in the respective crystal lattices and possible small differences between the low energy states of the different proteins could underlie the observed variations. Interestingly, the predicted intermembrane distances fall within the range reported for adherens junctions which covers a significant range between 150 and 300Å (Farquhar and Palade, 1963; Fawcett and McNutt, 1969; McNutt and Weinstein, 1973; Miyaguchi, 2000).
Sequence conservation suggests a functional cis interface
The observation of similar cis interfaces for three type I classical cadherins in unrelated crystal lattices is suggestive of potential biological relevance. In addition, analysis of sequence conservation indicates that residues in the cis interface, like those of the known trans interfaces, are significantly more conserved than other surface residues, providing independent data suggesting a biological role for the cis interface. Cis interface residues in EC1-2 show 23% identity (7/31 residues) between E-, N-, P-, M- and R-cadherins (mouse and human) and C-cadherin (frog), compared with 9% identity (6/66 residues) for non-interface surface residues (see Supp. Fig. 2 for full analysis).
Site-directed mutants that disrupt the cis interface
To investigate the relevance of the cis interface in cadherin adhesive function we designed mutations to disrupt this interaction in E-cadherin. The mutations V81D and L175D were targeted to the concave (EC1) and convex (EC2) sides of the interface, respectively. Both were intended to disrupt the small hydrophobic core of the cis interface (see Fig. 2A-C) by introducing a negatively charged aspartate side chain. The mutated residues are distal from the trans dimer face so that they should specifically target cis binding.
Crystal structures of E-cadherin EC1-EC2 V81D and L175D mutants were determined at 2.7Å and 2.8Å resolution (Table 1). Folding of the EC1 and EC2 domains in both mutants was identical to that observed in previously published wild-type structures (Supp. Fig. 3A) (Harrison et al., 2010; Haussinger et al., 2004; Parisini et al., 2007). Furthermore, two molecules comprising the crystallographic asymmetric unit formed strand swapped trans dimers in both mutant structures (Supp. Fig. 3B).
In all ten structures of EC1-2 fragments of E-cadherin published to date, the putative cis interface has been observed in the crystal lattice, despite the absence of the EC3 domain (Supp. Fig. 3C) (Harrison et al., 2010; Haussinger et al., 2004; Nagar et al., 1996; Parisini et al., 2007; Pertz et al., 1999). Notably, the cis interface is not present in the V81D and L175D structures reported here; instead the EC2 side of the interface is exposed to solvent and the EC1 face engages in an unrelated crystal contact (Supp. Fig 3D, E). The absence of the cis interface in our structures suggests that the V81D and L175D mutations disrupt the interaction as intended.
In agreement with the structural data, equilibrium analytical ultracentrifugation (AUC) experiments showed that V81D and L175D mutations introduced into E-cadherin EC1-2 or EC1-5 ectodomain constructs did not substantially alter dimerization affinities, suggesting normal trans dimer formation (Supp. Table 2). Trans dimer mutant E-cadherin EC1-5 (W2A K14E) was monomeric in the analysis, indicating that only trans dimer interactions are detectable in solution and suggesting the affinity of the cis interaction to be weaker than the detection limit of the assay (KD ∼1mM).
Assembly of junction-like structures between cadherin-coated liposomes
To determine if the junction-like molecular layer found in the three type I cadherin ectodomain crystal structures can also form in a membrane-associated context, we employed a liposome-based assay system. Liposomes were prepared using phospholipid mixtures that included Ni2+-chelated head groups to tether C-terminally 6xHis-tagged E-cadherin ectodomains on the surface. Time course measurements of light scattering at 650nm revealed that liposomes coated with wild-type E-cadherin ectodomains showed an increase in light scattering that typically reached a maximal plateau over ∼30 minutes (Supp. Fig. 4). By contrast, liposomes that were either uncoated or were coated with the non-adhesive E-cadherin trans-dimerization mutant W2A K14E (Harrison et al., 2010) showed no increase in light scattering. Liposome aggregation for the cis interface mutant V81D L175D was slightly diminished from wild-type protein, but nevertheless indicative of adhesive binding (Supp. Fig. 4).
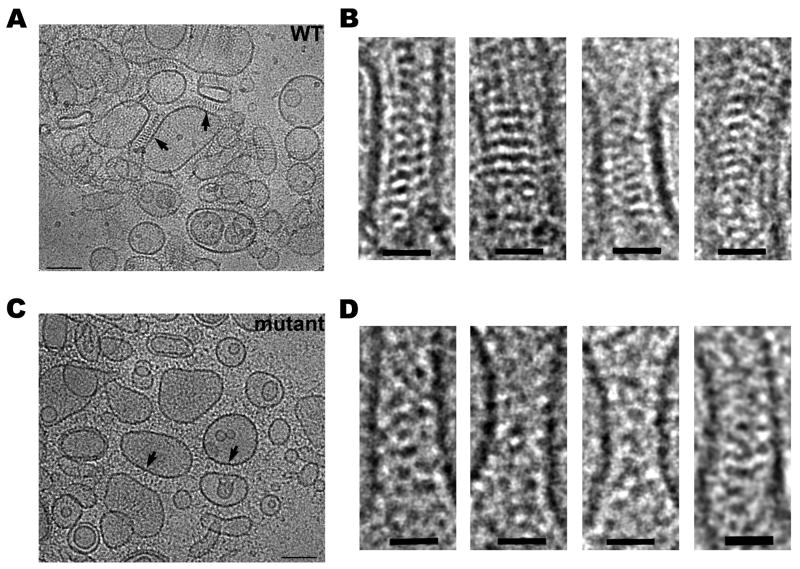
Micrographs of frozen-hydrated samples of aggregated liposomes were examined using cryo-EM. Liposomes coated with wild-type E-cadherin ectodomains reveal junction-like structures in the space between apposed liposomes (Fig. 4A). The mechanical strength resulting from the assembly of ordered junction-like structures by cadherin molecules is apparently sufficient to induce the flattening of the membrane surface of liposomes (Fig. 4A). In these junction-like structures, cadherin molecules are arranged in a periodic manner revealing a characteristic saw tooth pattern of high densities (Fig. 4B). Cadherin molecules are periodically positioned at ∼70Å intervals, consistent with the arrangement of cadherin ectodomains in the crystal structures, and protrude from the membrane surface at angles that can deviate from the membrane perpendicular by up to ∼40°. Intermembrane distance at the junctions varies between ∼300 and ∼340Å. These values are slightly larger than the measurements from the molecular layer identified in the crystal lattices (see Fig. 3C-E). The discrepancy likely arises from the presence of a disordered segment near the membrane attachment point. The E-cadherin ectodomain ends at residue D533, but five additional residues are found in a segment without secondary structure at the C-terminus; our ectodomain construct also includes six additional disordered residues preceding the 6-histidine tag at the extreme C-terminus. In a fully extended conformation this flexible linker could add up to 40Å to each cadherin monomer, or up to 80Å for a trans-bonded pair. Consistent with this, electron density is weak directly proximal to the membrane surface. Nevertheless, intermembrane distances measured here are broadly consistent with the reported dimensions of adherens junctions (Farquhar and Palade, 1963; Fawcett and McNutt, 1969; McNutt and Weinstein, 1973; Miyaguchi, 2000).
Figure 4. Electron microscopy of artificial junctions between E-cadherin coated liposomes.
(A) Cryo-electron microscopy of liposomes coated with wild-type mouse E-cadherin EC1-5 after 40 minutes of aggregation. Arrows indicate junction-like structures formed between apposed membranes. (B) Close-up views of selected junctions between wild-type E-cadherin coated liposomes. Note the ordered arrangement of electron-dense material in the intermembrane space. (C) Aggregated liposomes coated with the cis interface mutant V81D L175D of mouse E-cadherin EC1-5. Junction-like regions are indicated by arrows as in A. (D) Close up views of the mutant junctions. Note the absence of highly ordered intermembrane density compared to wild-type. Scale bars 100nm (A, C) and 30nm (B, D). See also Supp. Fig. 4 and Supp. Table 2.
We performed similar experiments with the cis interface mutant E-cadherin ectodomain V81D L175D, which fails to form cis interactions in crystals (see Supp. Fig. 3). Images of samples prepared with this mutant show electron dense material in the space between adherent liposomes, but this density fails to show identifiable features suggesting that the cadherin ectodomains are no longer arranged in ordered layers which can provide a clear projection image (Fig. 4C, D). This is in clear contrast to the ordered junction-like structures observed with the wild-type protein. The intermembrane distance also varies more substantially for the cis mutant-mediated adherent liposomes, indicating that the mutant cadherins in this contact region fail to form ordered structures.
The cis interface controls stability of cadherin junctions between cells
To determine the role of cadherin cis interactions in a cellular context, we introduced the cis interface mutation V81D V175D into cell surface expressed human E-cadherin tagged at the C-terminus with the photoconvertible fluorescent protein Dendra2. Transfection experiments were first performed with the human epidermoid carcinoma cell line A-431, previously shown to efficiently incorporate Dendra2-tagged E-cadherin into junctions along with endogenously-expressed E-cadherin (Hong et al., 2010).
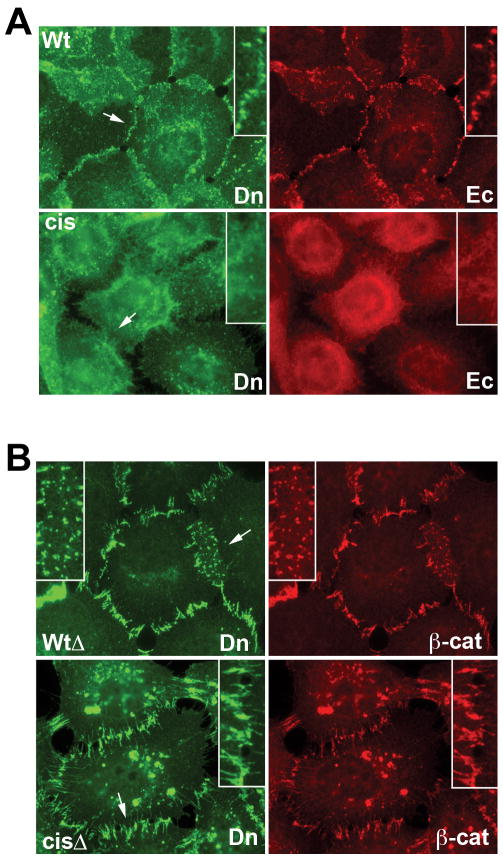
In A431 cells transfected with full-length cis interface mutant Ecadcis-Dendra, immunofluorescence microscopy revealed that in contrast to the parental protein Ecadwt-Dendra, the transfected mutant failed to assemble into cell-cell junctions (Fig. 5A). Furthermore, Ecadcis-Dendra acted in a dominant negative manner, ablating the endogenous adherens junctions in the transfected cells (Fig. 5A, Ec). The dominant-negative properties of Ecadcis-Dendra strongly suggest that the cis interface is essential for adherens junction formation, however, its exact role is not defined in these experiments since all cadherin function is ablated.
Figure 5. Effects of cis-mutations on subcellular distribution of full-length and catenin-uncoupled E-cadherin in A-431 cells.
(A) A-431 cells expressing full-length Dendra2-tagged human E-cadherin (Wt) or its V81D V175D cis mutant (Cis) were double-stained using rabbit anti-Dendra2 antibody against the recombinant cadherins (Dn, green) and mAB C20820 against endogenous cadherin (Ec, red). Magnifications of selected regions (arrows) are inset. Note the dominant negative phenotype of the cis mutant. (B) A-431 cells expressing catenin-uncoupled E-cadherin-Dendra (WtΔ) or its V81D V175D cis-mutant (CisΔ) stained with anti-Dendra2 (Dn, green) and anti-β-catenin to mark endogenous cadherins (β-cat, red). Recombinant and endogenous cadherins co-cluster at junctions in both cell lines. See also Supp. Fig. 3.
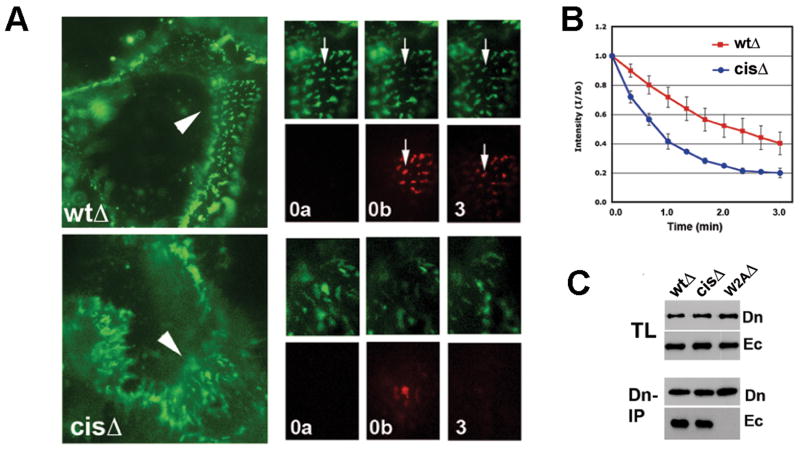
Dominant negative properties of mutant cadherins can sometimes be overcome by uncoupling them from cytoplasmic binding partners (Nieman et al., 1999). Therefore, in order to examine the cis mutants in cells with intact endogenous adherens junctions, we used a cadherin construct lacking most of the cadherin cytoplasmic domain, designated EcadΔ-Dendra (Hong et al., 2010). This protein is additionally uncoupled from clathrin-mediated endocytosis, allowing it to be stably present on the cell surface where it co-clusters with endogenous cadherin in the junctions of A-431 cells (Fig. 5B, WtΔ) (Hong et al., 2010). When cis interface mutations were introduced into these constructs, the mutant protein, EcadcisΔ-Dendra was also efficiently recruited to the cell-cell contacts of transfected A-431 cells, where it colocalized precisely with endogenous cadherins in the same junctions (Fig. 5B, CisΔ). Live imaging, however, revealed a dramatic effect of the cis interface mutation on the dynamics of these junctions: instead of the relatively stationary cadherin-containing clusters observed in the EcadwtΔ-Dendra control cells, clusters incorporating the cis mutant were extremely mobile, continuously and rapidly changing their shape and distribution (Movies S1-2). Moreover, Dendra activation assays revealed that the residence time of the mutant protein in these clusters was much shorter than that of the parental cadherin (Fig. 6A, B). Co-culture experiments showed that mixed junctions between wild-type and mutant cadherins were similarly unstable (Supp. Fig. 5), indicating, as predicted from our structures, that cis interactions on both sides are required to stabilize junctions. In each case, junctional instability must be due to changes in the cis interaction since immunoprecipitation experiments with the transfected A431 cells showed that EcadcisΔ-Dendra formed trans strand swap dimers equivalent to wild-type protein (Fig. 6C).
Figure 6. Cis mutations destabilize cadherin junctions in A-431 cells.
(A) Time-lapse analysis of photoactivated junctions in A-431 cells expressing fluorescent Ecadwt Δ-Dendra (WtΔ) or its V81D V175D cis interface mutant EcadcisΔ -Dendra (cisΔ). Left are low magnification images of initial frames; sequences are shown on the right (see movies S3 and S4). A 5 μm region of cell-cell contact (arrowhead) was photoactivated and cells were imaged in green (normal Dendra2 fluorescence) and red (photoconverted Dendra2) channels. ‘0a’: before photoactivation; ‘0b’ immediately after activation; ‘3’: after 3 minutes. Arrows in each sequence indicate the same cadherin cluster. (B) Changes in intensity of red fluorescence in individual junctions after Dendra2 activation, averaged from four independent experiments (n = 30). Initial red fluorescence is considered to be 1.0. Error bars represent SD (n = 20). (C) Co-immunoprecipitation assay with A-431 cells expressing EcadwtΔ-Dendra (WtΔ), cis mutant EcadcisΔ-Dendra (CisΔ) or trans dimer mutant (W2AΔ). Total lysates (TL) and anti-Dendra immunoprecipitates (Dn-IP) were probed with anti-Dendra (Dn) and for co-immunoprecipitated endogenous cadherin by anti-E-cadherin C20820 (Ec). See also Supp. Fig. 5 and Supp. Movies S1-S4.
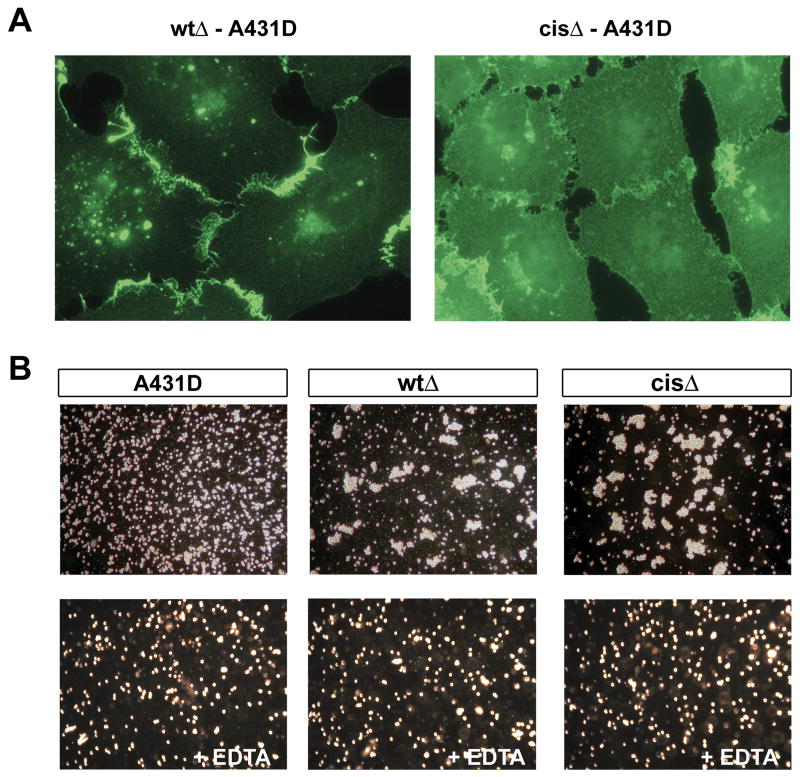
We carried out complementary transfection experiments using A-431D cells, which lack endogenous cadherins (Lewis et al., 1997, Supp. Fig. 6). In these transfected cells, both Ecadwt-Dendra and EcadwtΔ-Dendra localize efficiently to cell-cell junctions with minimal staining in other regions of the plasma membrane (Fig. 7A and Supp. Fig. 6). In contrast, the cis interface mutants Ecadcis-Dendra and EcadcisΔ-Dendra are distributed more diffusely over the cell surface with only slightly increased staining in the contact zone (Fig. 7A and Supp. Fig. 6). These results clearly show that E-cadherin cis interface mutants are unable to assemble junctions in the absence of endogenous wild-type cadherins. We also assessed the aggregation properties of the EcadwtΔ and EcadcisΔ A431D cells in “short term” aggregation assays (Fig. 7B). Remarkably, both wild-type and cis mutant transfectants could mediate cell aggregation with apparently equal facility. This observation suggests that, as for experiments with cadherin-coated liposomes (see Fig. 4 and Supp. Fig. 4), trans interactions are sufficient to mediate aggregation, while cis interactions are further required for assembly of junctions.
Figure 7. Subcellular distribution of E-cadherin cis mutants in A-431D cells.
(A) Dendra fluorescence of A-431D cells expressing equal wild-type EcadwtΔ-Dendra or cis mutant V81D V175D (EcadcisΔ-Dendra). (B) Short term aggregation assays with the two cell lines. Images show cells after 30 minutes of shaking aggregation in 3mM calcium (upper panels) or 3mM EDTA (lower panels). Parental A-431D cells show no aggregation (A-431D). See also Supp. Fig. 6 for results with full-length versions of the above constructs.
Discussion
We have presented data from several independent sources which, taken together, strongly suggest that the molecular layer observed in the crystal structures of E-, N- and C-cadherin corresponds to the arrangement of cadherin ectodomains within adherens junctions. Here we briefly review this evidence and discuss the implications of these results for the mechanism of junction assembly.
Adherens junction structure
Evidence supporting the idea that the molecular layer common to the type I cadherin structures represents the extracellular arrangement of adherens junction includes: (1) All three whole cadherin ectodomain structures, despite their presentation in independent lattices, form crystals containing similar molecular layers defined by two interactions – the strand swap trans interface, and the lateral cis interface. The geometry of this molecular layer is consistent with requirement that adhesive cadherins emanate from opposing membranes, and suggests an inter-membrane spacing of ∼185-262Å. This is likely to correspond to a continuous range of possible intermembrane distances, since molecular modeling shows that most of the difference in spacing between the three cadherins is due to ectodomain curvature, which is likely to be somewhat variable despite rigidification by interdomain calcium binding (Y.W. and B.H., unpublished results). The range of estimated intermembrane spacing from the crystal lattices is consistent with the known dimensions of adherens junctions (Farquhar and Palade, 1963; Fawcett and McNutt, 1969; McNutt and Weinstein, 1973; Miyaguchi, 2000), though artifacts of sample preparation, coupled with undefined projection angle in these published studies contribute uncertainty to the reported measurements. (2) Both sets of interfacial residues exhibit a higher level of sequence conservation than do other surface residues. (3) Liposome experiments show that junction-like structures can be formed by the ectodomain alone, in agreement with cell studies showing that E-cadherin lacking the cytoplasmic region can form adherens junctions (Hong et al., 2010; Ozaki et al., 2010). Thus, all necessary molecular interactions required for initial junction formation are localized in the ectodomain and would be expected to form in crystals, where protein concentrations are high. (4) Adhesion between liposomes coated with E-cadherin ectodomains is mediated by an ordered structure seen in electron micrographs that is consistent with the lattice structure observed in the cadherin crystals. Adhesion also occurs when the cis interface is ablated (Supp. Fig. 4), but an ordered junction-like structure is no longer observed (Fig. 4). (5) In a cellular context, ablation of the cis interface interferes with localization of cadherins to intercellular junctions. Consistent with the results for cadherin-coated liposomes, cell adhesion occurs in the absence of cis interactions but the extent of cadherin accumulation at cell-cell contact regions is diminished and the resulting junctions are highly unstable.
A prominent feature of the junction structure revealed by the type I cadherin crystals is that the two monomers in each trans strand swapped dimer orient their cis interface surfaces in nearly perpendicular directions. Each trans dimer thereby participates in two linear arrays, each formed by cis interactions involving one of its monomers (Fig. 3). The net effect is the formation of a 2D lattice which, we argue, represents the extracellular structure of adherens junctions.
Although type I, type II, and desmosomal cadherins share the same strand swap mode of trans interaction (Boggon et al., 2002; Patel et al., 2006; Shapiro and Weis, 2009), differences in sequence and structure suggest that the cis interface identified here is likely to be specific to type I cadherins. Type II cadherins notably lack the pseudo-β helix region which plays a role in formation of the type I cadherin cis interface. Furthermore, whereas virtually all type I cadherin multi-domain crystal structures include cis interface interactions (Boggon et al., 2002; Harrison et al., 2010; Haussinger et al., 2004; Nagar et al., 1996; Parisini et al., 2007; Pertz et al., 1999), the two published multi-domain type II cadherin structures (Patel et al., 2006) and two additional unpublished structures from our lab (JB, OJH, BH, and LS, unpublished) all lack a similar interface. Nevertheless, there is significant evidence for adherens junction formation by type II cadherins (Kiener et al., 2006; Uehara, 2006), and we thus infer that alternative lateral interfaces must be utilized.
Desmosomal cadherins also appear to lack the sequence determinants found for the cis interface of type I cadherins. This is consistent with the very different structure determined for desmosomes by cryo-ET (Al-Amoudi et al., 2007), which depicts the same strand swap adhesive interface, but the arrangement of protomers in cis appears to be entirely different. Inter-membrane spacings reported for desmosomes (∼250-350Å) (Al-Amoudi et al., 2007; McNutt and Weinstein, 1973) are consistently larger than for adherens junctions (∼150-300Å) (Farquhar and Palade, 1963; Fawcett and McNutt, 1969; McNutt and Weinstein, 1973; Miyaguchi, 2000), despite the fact that both are composed of cadherin ectodomains with lengths expected to be nearly identical. The difference in spacing may be related to the angle at which the ectodomains protrude from the membrane, since fitting of the C-cadherin ectodomain crystal structure to cryo-EM maps of desmosomes reveal cadherin ectodomains oriented almost perpendicular to the cell membrane (Al-Amoudi et al., 2007).
Adhesion receptor clustering in other systems
The idea that specific cis and trans ectodomain interactions are responsible for the assembly of cell adhesion proteins into ordered junctions and signaling clusters has emerged from structural studies in a number of systems (Aricescu and Jones, 2007; Freigang et al., 2000; He et al., 2009; Kostrewa et al., 2001; Soroka et al., 2003). However, the clustering mechanisms proposed for these adhesion proteins each involve the formation of linear “zippers”. For L1 and JAM-A the zipper is formed through alternation of cis and trans interactions (He et al., 2009; Kostrewa et al., 2001), while for axonin it results from trans interactions alone (Freigang et al., 2000). However, structures such as junctions are two dimensional entities and indeed a one dimensional array is unlikely to be stable in the absence of additional interactions. Thus, it is likely that if these one-dimensional structures reflect the arrangement of natural junctions and signaling clusters then additional, yet-undefined, interactions assemble these linear arrays into higher order structures. A number of one dimensional models have been proposed for NCAM clustering (Kiselyov et al., 2005) but a two-dimensional model derived from a three domain crystal structure has also been suggested (Soroka et al., 2003). The complex formed between EphA2 and ephrin-A5 provides an example of a crystallographically identified two-dimensional array that has been characterized and functionally validated (Seiradake et al., 2010). Crystals of EphA2/ephrin-A5 reveal a two-dimensional lattice that contains multiple distinct cis and trans interfaces which together define an ordered structure consistent with requirements of cell surface geometry. Site-directed mutagenesis verifies the importance of some of these interfaces in cluster formation and in signal transduction initiated by Eph/ephrin interactions (Himanen et al., 2010; Seiradake et al., 2010).
The cadherin system is far simpler. Two biologically validated interfaces alone, one that functions exclusively in trans and the other exclusively in cis, together define a two-dimensional molecular lattice consistent with the geometry of adherens junctions. Here we report on the strength of these interactions which, aided by the relative simplicity of the system, enable us to describe the junction formation process in more detailed structural and energetic terms than has been possible previously. Our liposome experiments show that junction structures can spontaneously assemble and that the trans and cis interactions we describe are required for or this process to occur. Moreover, we show both for liposomes and for transfected cells, that trans interactions alone can drive adhesion, albeit weaker than when cis interactions are present, and that cis interactions are required for the formation of an ordered structure.
Forces that drive junction formation
The results of this study demonstrate that adherens junctions are assembled through a combination of trans and cis interactions. Trans interactions are clearly the stronger of the two and, in the case of E-cadherin, correspond to a KD, as measured in solution, of about 100 μM (Supp. Table 2). This, by most definitions, corresponds to a “weak” interaction but given the fact that cell adhesion involves multiple proteins, the combined cell-cell interaction free energy can be quite large (Chen et al., 2005; Katsamba et al., 2009). Indeed trans interactions are sufficient on their own to drive cell and liposome aggregation (Fig. 7, Supp. Fig. 4). Even in the absence of cis interactions cadherins can still accumulate in contact regions, but to a much lesser extent (Figs. 5, 7), by a “diffusion trap” mechanism whereby monomers that diffuse into this region form trans dimers, and are unable to leave until the dimers dissociate. The role of this mechanism has been described in a recent study of the interactions of cadherin coated beads with epithelial cells (Perez et al., 2008). The effect of the diffusion trap mechanism is evident from our results using the E-cadherin cis mutant V81D L175D. Liposomes coated with this mutant still adhere, although to a slightly lesser extent than wild type (Supp. Fig. 4). Moreover, in transfected A-431D cell lines, which do not express endogenous wild type proteins, there is some accumulation of cis mutant proteins in inter-cellular contact regions, although much less than in wild type (Fig. 7A). The difference reflects the contribution of cis interactions to cadherin accumulation.
The role of cis interactions is also evident in our experiments with transfected A-431 cells, which also express endogenous E-cadherin. In this case cis mutants interfere with junction formation in a dominant negative fashion (Fig. 5). Cis mutants that are uncoupled from cytoplasmic interactions (EcadcisΔ-Dendra) accumulate into junctions together with wild-type proteins, presumably via the diffusion trap mechanism, but, as evidenced by the reduced junctional residence time of the mutant protein, the resulting junctions are less stable and more dynamic than those formed by wild-type protein alone (Fig. 6). These results clearly indicate the importance of forming a junction structure stabilized by both trans and cis interactions.
The central role of cis interactions in junction formation is remarkable since the interactions are too weak to be detected in solution. Indeed, cis interactions are not on their own able to produce observable clusters on an isolated cell surface, yet combined with trans interactions they can produce stable and ordered structures. How is this possible? We recently investigated the physical principles that underlie junction formation based in part on simulations that mimic the organization of cadherins in the 2D lattice seen in crystal structures (Wu et al., 2010). We find that junction formation can be viewed as a transition from a diffuse two-dimensional “gas” phase composed of cadherin monomers freely diffusing in the plasma membrane to a structured “solid” phase consisting of trans cadherin dimers that make lateral cis interactions. The transition is cooperative so that trans and cis interactions enforce one another and can result in junction formation even when cis interactions are weak. In addition there is strong evidence for molecular cooperativity between cis and trans interactions; that is, the formation of trans dimers increases the strength of cis interactions (Zhang et al., 2009). The relative simplicity of the cadherin system has made it possible for us to describe junction formation in quantitative terms. Nevertheless, the underlying principles should apply to the formation of ordered assemblies in more complex multi-component systems.
Biological roles of ordered junctions
Our results show that cadherins can drive cell aggregation even in the absence of the cis interactions required to form junctions. Thus, the role of ordered junctions is likely to extend beyond adhesion alone. One attractive possibility is that the ordered type I cadherin junction structure, which can spontaneously assemble only upon cell-cell contact, may provide a mechanism for transducing outside-in signals across the plasma membrane. Engagement of cadherins at sites of cell-cell contact is known to initiate signaling through several different pathways (Li et al., 2001; Watanabe et al., 2009). In addition to effects on diverse cellular behaviors such as motility and survival (McCrea et al., 2009), these signaling mechanisms are thought to modulate cytoskeletal activity leading to stabilization and expansion of the nascent cell-cell junctions (Kovacs et al., 2002; Vasioukhin et al., 2000). Currently, the mechanism by which engagement of cadherin ectodomains between apposed cells leads to signaling across the plasma membrane is not well understood. It appears likely that cadherin-mediated signaling could be triggered by assembly of a defined supermolecular complex, as is characteristic of many transmembrane signaling events (Cochran et al., 2001; Hubbard and Till, 2000). The extracellular adherens junction structures suggested here for C-, E-, and N-cadherins all have lateral distances that would place adjacent cadherin transmembrane segments ∼72-74Å apart in a characteristic array (See Fig. 3). This ordered structure should, in principle, impose an arrangement on the cytoplasmic domains that could act as a scaffold for assembly of a defined molecular complex to initiate signaling responses and trigger further expansion and organization of the junction by the cytoskeleton.
Materials and Methods
Protein expression and structure determination
Mouse E-cadherin (Asp1 to Ala544 of the mature protein) and mouse N-cadherin (Asp1 to Val553) ectodomains were expressed in human embryonic kidney 293 cells; EC1-2 fragments of mouse E-cadherin (Asp1 to Asp213) were expressed in E.coli. Cloning, expression, purification and crystallization of the wild-type and mutant proteins are detailed in Extended Experimental Procedures. X-ray data was collected from single crystals at 100K using a wavelength of 0.979Å at the X4A and X4C beamlines of the National Synchrotron Light Source, Brookhaven National Laboratory. Data processing and structure determination by molecular replacement are described in Extended Experimental Procedures.
Liposome aggregation assays and cryo-EM
Liposomes composed of a 9:1 molar ratio of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and the nickel salt of 1,2-dioleoyl-sn-glycero-3-([N(5-amino-1-carboxypentyl)iminodiaceticacid]-succinyl) (DOGS-NTA) were prepared and incubated with 6xHis tagged E-cadherin ectodomain proteins at room temperature while aggregation was monitored by OD650 measurement in a spectrophotometer. For cryo-EM, aggregated liposomes were applied to 300 mesh copper TEM grids with R 2/1 Quantifoil carbon film and were vitrified by blotting and plunge-freezing in liquid ethane. Frozen grids were transferred to a Tecnai Polara F30 TEM (FEI) and imaged at 300 kV under low-dose conditions at ∼10 μm underfocus. Microscope magnification was 39,000-59,000×. See Extended Experimental Procedures for detailed methods.
Cell transfection experiments
Transfection, growth, and immunofluorescence microscopy of human A-431, and A-431D cells were performed as described (Troyanovsky et al., 2007). The plasmid pRc-EcDendra-Δ748-KL encoding human E-cadherin truncated mutant EcadΔ-Dendra was described (Hong et al., 2010). For co-culture and immunoprecipitation experiments, Dendra tag was replaced with mCherry and 6xMyc, respectively. Wild-type and mutant cadherin transfectants expressing equal, moderate levels of transgenes were sorted by flow cytometry and used for experiments.
Live cell imaging was performed as described (Hong et al., 2010). To analyze cadherin junctional turnover, we used a Dendra photoactivation assay (Hong et al., 2010). A circular region of interest (φ=5μm) was photoactivated by 3s of 402 nm light and time-lapse images were taken in both FITC and TRITC filter sets. For Dendra- and mCherry cocultures, a FRAP assay was used: Dendra green fluorescence was fully photoconverted by 4s of 402 nm light in a region of interest and recovery of green fluorescence was then monitored over time. Further details and methods for image analysis, immunoprecipitation and cell aggregation assays in Extended Experimental Procedures.
Supplementary Material
Acknowledgments
We are thankful to Dr. J.K. Wahl (University of Nebraska) for providing A-431D cells. The work has been supported by Grants AR44016-04 (S.M.T), R01 GM062270 (L.S) and T32 GM082797 (B.H) from the National Institutes of Health and MCB-0918535 from the National Science Foundation (B.H). The financial support of the US-Israel Binational Science Foundation (Grant No. 2006-401, to A.B-S., B.H., and L.S.) and the Israel Science Foundation (Grant No. 659/06 to ABS) is gratefully acknowledged. X-ray data were acquired at the X4A and X4C beamlines of the National Synchrotron Light Source, Brookhaven National Laboratory; the beamlines are operated by the New York Structural Biology Center.
Footnotes
Accession numbers: The structures of mouse E-cadherin EC1-5, mouse N-cadherin EC1-5, mouse E-cadherin EC1-2 V81D mutant and mouse E-cadherin EC1-2 L175D mutant have been deposited in the PDB with accession codes 3Q2V, 3Q2W, 3Q2L and 3Q2N, respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Amoudi A, Diez DC, Betts MJ, Frangakis AS. The molecular architecture of cadherins in native epidermal desmosomes. Nature. 2007;450:832–837. doi: 10.1038/nature05994. [DOI] [PubMed] [Google Scholar]
- Arac D, Boucard AA, Ozkan E, Strop P, Newell E, Sudhof TC, Brunger AT. Structures of neuroligin-1 and the neuroligin-1/neurexin-1 beta complex reveal specific protein-protein and protein-Ca2+ interactions. Neuron. 2007;56:992–1003. doi: 10.1016/j.neuron.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Aricescu AR, Jones EY. Immunoglobulin superfamily cell adhesion molecules: zippers and signals. Curr Opin Cell Biol. 2007;19:543–550. doi: 10.1016/j.ceb.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- Chen C, Posy S, Ben-Shaul A, Shapiro L, Honig B. Specificity of cell-cell adhesion by classical cadherins: Critical role for low-affinity dimerization through beta-strand swapping. Proc Natl Acad Sci U S A. 2005;102:8531–8536. doi: 10.1073/pnas.0503319102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran JR, Aivazian D, Cameron TO, Stern LJ. Receptor clustering and transmembrane signaling in T cells. Trends Biochem Sci. 2001;26:304–310. doi: 10.1016/s0968-0004(01)01815-1. [DOI] [PubMed] [Google Scholar]
- Dean C, Scholl FG, Choih J, DeMaria S, Berger J, Isacoff E, Scheiffele P. Neurexin mediates the assembly of presynaptic terminals. Nat Neurosci. 2003;6:708–716. doi: 10.1038/nn1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delva E, Tucker DK, Kowalczyk AP. The desmosome. Cold Spring Harb Perspect Biol. 2009;1:a002543. doi: 10.1101/cshperspect.a002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar MG, Palade GE. Junctional complexes in various epithelia. J Cell Biol. 1963;17:375–412. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett DW, McNutt NS. The ultrastructure of the cat myocardium. I. Ventricular papillary muscle. J Cell Biol. 1969;42:1–45. doi: 10.1083/jcb.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freigang J, Proba K, Leder L, Diederichs K, Sonderegger P, Welte W. The crystal structure of the ligand binding module of axonin-1/TAG-1 suggests a zipper mechanism for neural cell adhesion. Cell. 2000;101:425–433. doi: 10.1016/s0092-8674(00)80852-1. [DOI] [PubMed] [Google Scholar]
- Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- Harrison OJ, Bahna F, Katsamba PS, Jin X, Brasch J, Vendome J, Ahlsen G, Carroll KJ, Price SR, Honig B, et al. Two-step adhesive binding by classical cadherins. Nat Struct Mol Biol. 2010;17:348–357. doi: 10.1038/nsmb.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussinger D, Ahrens T, Aberle T, Engel J, Stetefeld J, Grzesiek S. Proteolytic E-cadherin activation followed by solution NMR and X-ray crystallography. EMBO J. 2004;23:1699–1708. doi: 10.1038/sj.emboj.7600192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Cowin P, Stokes D. Untangling desmosomal knots with electron tomography. Science. 2003;302:109–113. doi: 10.1126/science.1086957. [DOI] [PubMed] [Google Scholar]
- He Y, Jensen GJ, Bjorkman PJ. Cryo-electron tomography of homophilic adhesion mediated by the neural cell adhesion molecule L1. Structure. 2009;17:460–471. doi: 10.1016/j.str.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen JP, Yermekbayeva L, Janes PW, Walker JR, Xu K, Atapattu L, Rajashankar KR, Mensinga A, Lackmann M, Nikolov DB, et al. Architecture of Eph receptor clusters. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10860–10865. doi: 10.1073/pnas.1004148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Troyanovsky RB, Troyanovsky SM. Spontaneous assembly and active disassembly balance adherens junction homeostasis. Proc Natl Acad Sci U S A. 2010;107:3528–3533. doi: 10.1073/pnas.0911027107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR, Till JH. Protein tyrosine kinase structure and function. Annu Rev Biochem. 2000;69:373–398. doi: 10.1146/annurev.biochem.69.1.373. [DOI] [PubMed] [Google Scholar]
- Huber AH, Weis WI. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- Ishiyama N, Lee SH, Liu S, Li GY, Smith MJ, Reichardt LF, Ikura M. Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion. Cell. 2010;141:117–128. doi: 10.1016/j.cell.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Katsamba P, Carroll K, Ahlsen G, Bahna F, Vendome J, Posy S, Rajebhosale M, Price S, Jessell TM, Ben-Shaul A, et al. Linking molecular affinity and cellular specificity in cadherin-mediated adhesion. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0905349106. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiener HP, Stipp CS, Allen PG, Higgins JM, Brenner MB. The cadherin-11 cytoplasmic juxtamembrane domain promotes alpha-catenin turnover at adherens junctions and intercellular motility. Mol Biol Cell. 2006;17:2366–2376. doi: 10.1091/mbc.E05-08-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselyov VV, Soroka V, Berezin V, Bock E. Structural biology of NCAM homophilic binding and activation of FGFR. J Neurochem. 2005;94:1169–1179. doi: 10.1111/j.1471-4159.2005.03284.x. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Natori M, Murase S, Hirano S, Taketani S, Suzuki S. Mutation analysis of cadherin-4 reveals amino acid residues of EC1 important for the structure and function. Biochem Biophys Res Commun. 2000;271:358–363. doi: 10.1006/bbrc.2000.2636. [DOI] [PubMed] [Google Scholar]
- Kostrewa D, Brockhaus M, D'Arcy A, Dale GE, Nelboeck P, Schmid G, Mueller F, Bazzoni G, Dejana E, Bartfai T, et al. X-ray structure of junctional adhesion molecule: structural basis for homophilic adhesion via a novel dimerization motif. EMBO J. 2001;20:4391–4398. doi: 10.1093/emboj/20.16.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs EM, Goodwin M, Ali RG, Paterson AD, Yap AS. Cadherin-directed actin assembly: E-cadherin physically associates with the Arp2/3 complex to direct actin assembly in nascent adhesive contacts. Curr Biol. 2002;12:379–382. doi: 10.1016/s0960-9822(02)00661-9. [DOI] [PubMed] [Google Scholar]
- Lewis JE, Wahl JK, 3rd, Sass KM, Jensen PJ, Johnson KR, Wheelock MJ. Cross-talk between adherens junctions and desmosomes depends on plakoglobin. J Cell Biol. 1997;136:919–934. doi: 10.1083/jcb.136.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Satyamoorthy K, Herlyn M. N-cadherin-mediated intercellular interactions promote survival and migration of melanoma cells. Cancer Res. 2001;61:3819–3825. [PubMed] [Google Scholar]
- McCrea PD, Gu D, Balda MS. Junctional music that the nucleus hears: cell-cell contact signaling and the modulation of gene activity. Cold Spring Harb Perspect Biol. 2009;1:a002923. doi: 10.1101/cshperspect.a002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNutt NS, Weinstein RS. Membrane ultrastructure at mammalian intercellular junctions. Prog Biophys Mol Biol. 1973;26:45–101. doi: 10.1016/0079-6107(73)90017-5. [DOI] [PubMed] [Google Scholar]
- Miyaguchi K. Ultrastructure of the zonula adherens revealed by rapid-freeze deep-etching. J Struct Biol. 2000;132:169–178. doi: 10.1006/jsbi.2000.4244. [DOI] [PubMed] [Google Scholar]
- Nagar B, Overduin M, Ikura M, Rini JM. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature. 1996;380:360–364. doi: 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- Nieman MT, Kim JB, Johnson KR, Wheelock MJ. Mechanism of extracellular domain-deleted dominant negative cadherins. J Cell Sci. 1999;112(Pt 10):1621–1632. doi: 10.1242/jcs.112.10.1621. [DOI] [PubMed] [Google Scholar]
- Ozaki C, Obata S, Yamanaka H, Tominaga S, Suzuki ST. The extracellular domains of E- and N-cadherin determine the scattered punctate localization in epithelial cells and the cytoplasmic domains modulate the localization. J Biochem. 2010;147:415–425. doi: 10.1093/jb/mvp192. [DOI] [PubMed] [Google Scholar]
- Ozawa M, Baribault H, Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisini E, Higgins JM, Liu JH, Brenner MB, Wang JH. The Crystal Structure of Human E-cadherin Domains 1 and 2, and Comparison with other Cadherins in the Context of Adhesion Mechanism. J Mol Biol. 2007;373:401–411. doi: 10.1016/j.jmb.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale M, Arkus N, Schieren I, Jessell TM, Honig B, Price SR, et al. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell. 2006;124:1255–1268. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Perez TD, Tamada M, Sheetz MP, Nelson WJ. Immediate-early signaling induced by E-cadherin engagement and adhesion. J Biol Chem. 2008;283:5014–5022. doi: 10.1074/jbc.M705209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertz O, Bozic D, Koch AW, Fauser C, Brancaccio A, Engel J. A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation. EMBO J. 1999;18:1738–1747. doi: 10.1093/emboj/18.7.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posy S, Shapiro L, Honig B. Sequence and structural determinants of strand swapping in cadherin domains: do all cadherins bind through the same adhesive interface? J Mol Biol. 2008;378:952–966. doi: 10.1016/j.jmb.2008.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiradake E, Harlos K, Sutton G, Aricescu AR, Jones EY. An extracellular steric seeding mechanism for Eph-ephrin signaling platform assembly. Nature Structural & Molecular Biology. 2010;17:398–U327. doi: 10.1038/nsmb.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L, Fannon A, Kwong P, Thompson A, Lehmann M, Grubel G, Legrand J, Als-Nielsen J, Colman D, Hendrickson W. Structural basis of cell-cell adhesion by cadherins. Nature. 1995;374:327–337. doi: 10.1038/374327a0. [DOI] [PubMed] [Google Scholar]
- Shapiro L, Weis WI. Structure and biochemistry of cadherins and catenins. Cold Spring Harb Perspect Biol. 2009;1:a003053. doi: 10.1101/cshperspect.a003053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers WS, Tang J, Shaw GD, Camphausen RT. Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1. Cell. 2000;103:467–479. doi: 10.1016/s0092-8674(00)00138-0. [DOI] [PubMed] [Google Scholar]
- Soroka V, Kolkova K, Kastrup JS, Diederichs K, Breed J, Kiselyov VV, Poulsen FM, Larsen IK, Welte W, Berezin V, et al. Structure and interactions of NCAM Ig1-2-3 suggest a novel zipper mechanism for homophilic adhesion. Structure. 2003;11:1291–1301. doi: 10.1016/j.str.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Takeichi M. The cadherins: cell-cell adhesion molecules controlling animal morphogenesis. Development. 1988;102:639–655. doi: 10.1242/dev.102.4.639. [DOI] [PubMed] [Google Scholar]
- Troyanovsky R, Sokolov E, Troyanovsky S. Adhesive and lateral E-cadherin dimers are mediated by the same interface. Mol Cell Biol. 2003;23:7965–7972. doi: 10.1128/MCB.23.22.7965-7972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanovsky RB, Laur O, Troyanovsky SM. Stable and unstable cadherin dimers: mechanisms of formation and roles in cell adhesion. Mol Biol Cell. 2007;18:4343–4352. doi: 10.1091/mbc.E07-01-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara K. Distribution of adherens junction mediated by VE-cadherin complex in rat spleen sinus endothelial cells. Cell Tissue Res. 2006;323:417–424. doi: 10.1007/s00441-005-0064-5. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V, Bauer C, Yin M, Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 2000;100:209–219. doi: 10.1016/s0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Sato K, Kaibuchi K. Cadherin-mediated intercellular adhesion and signaling cascades involving small GTPases. Cold Spring Harb Perspect Biol. 2009;1:a003020. doi: 10.1101/cshperspect.a003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Jin X, Harrison OJ, Shapiro L, Honig B, Ben-Shaul A. Cooperativity between trans and cis interactions in cadherin-mediated junction formation. Proc Natl Acad Sci U S A. 2010 Oct 12;107(41):17592–7. doi: 10.1073/pnas.1011247107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap AS, Brieher WM, Pruschy M, Gumbiner BM. Lateral clustering of the adhesive ectodomain: a fundamental determinant of cadherin function. Curr Biol. 1997;7:308–315. doi: 10.1016/s0960-9822(06)00154-0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sivasankar S, Nelson WJ, Chu S. Resolving cadherin interactions and binding cooperativity at the single-molecule level. Proc Natl Acad Sci U S A. 2009;106:109–114. doi: 10.1073/pnas.0811350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.