Abstract
To identify genes potentially playing an important role in the progression of colorectal carcinoma (CRC), we screened global gene expression using cDNA expression array on 41 CRC tissue samples and 25 noncancerous colorectal tissue samples. Among the up-regulated genes, forkhead box M1 (FOXM1) has been shown to play a critical role in pathogenesis of various malignancies. Using immunohistochemistry on 448 Saudi CRC samples in tissue microarray format, FoxM1 protein overexpression was seen in 66% of CRC tissues and was significantly associated with poorly differentiated and highly proliferative tumors (P = 0.0200 and 0.0018, respectively). FoxM1 expression was also significantly associated with MMP-9 protein expression (P = 0.0002). In vitro data using CRC cell lines showed that inhibition of FoxM1 by thiostrepton resulted in inhibition of proliferation and induction of apoptosis in a dose-dependent manner. Overexpression of FoxM1 potentiated cell proliferation, cell transformation, and migration/invasion of CRC cells via up-regulation of FoxM1 target genes MMP2 and MMP9 and protected these cells from thiostrepton-mediated antiproliferative effects. Finally, in vivo, overexpression of FoxM1 promoted growth of CRC-cell line xenograft tumors in nude mice. Altogether, our data indicate that FoxM1 signaling contributes to aggressiveness in a subset of CRC and that the FOXM1 gene may serve as a useful molecular biomarker and potential therapeutic target.
Colorectal carcinoma (CRC) is still a cause of high morbidity and mortality rates. Significant improvements have been made in management of this disease, mainly through the introduction of adjuvant chemotherapy agents.1 Recently, advances in understanding of tumor biology have led to the development of targeted therapies, allowing progress in the treatment of CRC.2,3
Forkhead box protein M1 (FoxM1) is a member of the FoxM family and its deregulation has been implicated in pathogenesis of many cancers because of its ability to drive cell cycle progression and evasion of growth arrest.4 FoxM1 is known to be a key regulator of transition from G1 to S phase, and loss of FoxM1 expression has been reported to generate mitotic spindle defects leading to mitotic catastrophe.5–7 FoxM1 has been implicated in the carcinogenesis of tumor development in various cancers, including hepatocellular, prostate, lung, glioma, cervical, and gastric cancers.8–14 Recent studies showed that down-regulation of FoxM1 leads to inhibition of cell growth, migration, and invasion in a number of cancer types.14–17
In the present study, we first investigated expression levels of transcripts using cDNA microarray techniques in a series of CRC samples. FOXM1 was identified as one of the dysregulated genes in CRC. Overexpression of FOXM1 was further analyzed on a large collection of Middle Eastern CRC samples using tissue microarray (TMA) analysis. We then determine the potential role of FoxM1 expression in CRC development and progression using a well-established FoxM1 overexpression system, both in vitro and in vivo.
Materials and Methods
Expression Profiling
GeneChip technology (Affymetrix, Santa Clara, CA) was used to study gene expression in CRC samples. Affymetrix HG U133 plus 2.0 GeneChips containing approximately 47,000 human transcripts and variants were used, representing 39,000 genes. Microarray experiments were performed as described previously18 and according to the manufacturer's protocols. Samples passing Affymetrix quality control criteria, as specified in the manufacturer's documentation, were used for data analysis (35 CRC and 24 noncancerous matched samples).
Data Analysis
Expression profiles for 35 CRC and 24 noncancerous colorectal tissue samples were used. Principal component analysis was performed across the samples (Figure 1A) using Partek Genomics Suite 6.4 software (Partek, St. Louis, MO). The Strand Genomics Avadis software package (Strand Life Sciences, Bangalore, Karnataka, India) was used for hierarchical clustering. Affymetrix CEL files are available online at Gene Expression Omnibus (accession number GSE23878; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi).
Figure 1.
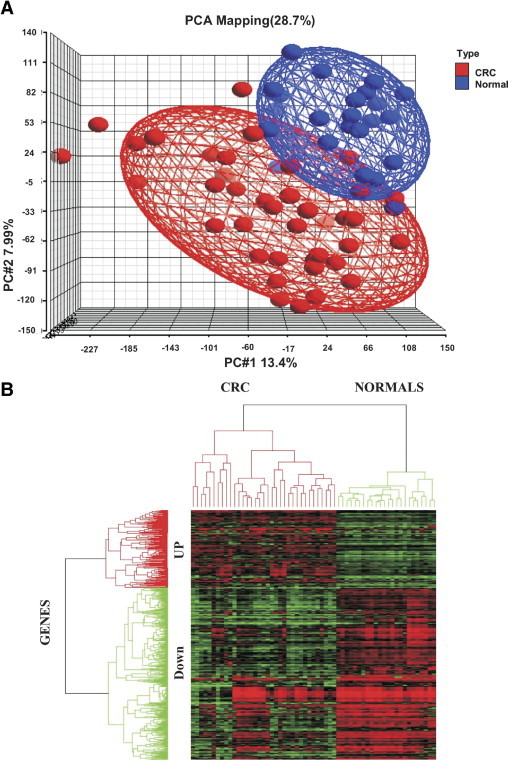
A: Principal component analysis showing distribution of colorectal cancer cases and normal controls separately. B: Hierarchical cluster analysis of the 479 differentially expressed genes (148 up-regulated and 331 down-regulated) in CRC and normal samples. Green indicates down-regulated expression and red indicates up-regulated expression, with intensity of color reflecting intensity of up- or down-regulation.
Patient Selection and Tissue Microarray Construction
A TMA containing 448 CRC, 307 normal matched colorectal mucosa, and 24 benign colorectal adenoma samples was constructed as described previously.19 The Colorectal Unit of the Department of Surgery at the King Faisal Specialist Hospital and Research Center provided long-term follow-up data. One pathologist (P.B.) reviewed all tumors for grade and histological subtype. The Institutional Review Board of the King Faisal Specialist Hospital and Research Center approved this study.
Immunohistochemistry
Tissue microarray slides were processed and stained manually, as described previously.19 Immunohistochemical detection of the transcription factor FoxM1 was performed using antibody from Santa Cruz Biotechnology (clone K-19; Santa Cruz, CA), as described previously.12 We also assessed expression of matrix metalloproteinase-9 (MMP-9). Primary antibodies used, dilutions, and incidence of positive cases are listed in Supplemental Table S1 (available at http://ajp.amjpathol.org). Quantification of TMA spots was performed as described previously.20 Each TMA spot was assigned an intensity score from 0 to 3 (I0, I1–I3) and proportion of the tumor staining for that intensity was recorded in 5% increments from 0 to 100 (P0, P1–P3). A final H score (range, 0–300) was obtained by adding the sum of scores obtained for each intensity I and proportion of area stained P, so that H score = I1 × P1 + I2 × P2 + I3 × P3. The CRCs were stratified into two groups, based on X-tile plots: one with complete absence or reduced staining (H score = 0–25) and the other with overexpression (H score > 25). X-tile plots were similarly used to stratify the CRC cases into two groups for MMP-9. X-tile plots were constructed for assessment of biomarker and optimization of cutoff points based on outcome, as described previously.20,21
Statistical Analysis
Contingency table analysis and χ2 tests were used to study relationship between clinicopathological variables and gene amplification. Survival curves were generated using the Kaplan-Meier method, with significance evaluated using the Mantel-Cox log-rank test. The limit of significance for all analyses was defined as a P value of 0.05; two-sided tests were used in all calculations. The JMP 7.0 software package (SAS Institute, Cary, NC) was used for data analyses.
Cell Culture
Colo-320, HCT-15, CX-1, DLD-1, and LOVO human colon adenocarcinoma and CL-11 human colon carcinoma cells were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany). All cell lines were tested for immunological markers and cytogenetics. The cell lines were also fingerprinted, and species was confirmed by isoelectric focusing of aspartate transaminase, malate dehydrogenase, and nucleoside phosphorylase. Cells were cultured in RPMI 1640 medium supplemented with 10% (v/v) fetal bovine serum, 100 U/ml penicillin, and 100 U/ml streptomycin at 37°C in humidified atmosphere containing 5% CO2. All of the experiments were performed in RPMI-1640 containing 5% serum.
Reagents and Antibodies
FoxM1 inhibitor (thiostrepton) was purchased from Tocris Cookson (Ellisville, MO). Antibodies against cleaved caspase-3, p-Akt, and BID (BH3 interacting domain death agonist) were purchased from Cell Signaling Technologies (Beverly, MA). FoxM1, cytochrome c, β-actin, caspase-3, and poly(ADP-ribose) polymerase (PARP) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). XIAP (X-chromosome linked inhibitor of apoptosis) and caspase-8 antibodies were purchased from R&D Systems (Minneapolis, MN). MMP-9 and MMP-2 antibodies were purchased from AnaSpec (San Jose, CA). Annexin V was purchased from Molecular Probes–Invitrogen (Eugene, OR). Human cDNA FoxM1 clone (catalog no. SC128214) vector alone (pCMV6-XL5) was purchased from OriGene Technologies (Rockville, MD).
Stable Transfection of CRC Cells
HCT-15 and LOVO cells were transfected with pCMV-XL5 and pCMV-XL5-FoxM1 plasmids using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Cells were incubated at 37°C for 6 hours, after which the lipid and plasmid complex was removed and fresh medium was added. Cells were grown for 48 hours after transfection. Clonal selection was performed according to the manufacturer's protocol, using the recommended amount of neomycin (G418). A single clone each was selected from vector-transfected HCT-15 and LOVO CRC cells alone, designated as HCT-15 vector and LOVO vector cells. Similarly, two HCT-15 and two LOVO FoxM1-overexpressing clones were selected and designated as HCT-15 clone 1, HCT-15 clone 3, LOVO clone 1, and LOVO clone 3, respectively.
Cell Growth Studies by MTT Assays
Cells (104) were incubated in triplicate in a 96-well plate in a final volume of 0.2 ml for 48 hour at 37°C. Cell viability assay using MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] was performed as described previously.22
Soft Agar Colony Assays
Soft agar colony experiments were performed according to the manufacturer's protocol (Chemicon International, Temecula, CA). Briefly, 2500 cells were plated in 0.5 ml culture medium containing 0.4% (v/v) top agar and 20% fetal bovine serum layered over a basal layer of 0.8% (v/v) agar and 20% fetal bovine serum with culture medium and were allowed to grow for 4 weeks, as described previously.17 After 4 weeks incubation, cells were stained at a final concentration of 1 mg/ml cell stain solution (solution supplied with the kit).
Cell Invasion and Migration Assays
Cell invasion and migration assay were performed using 24-well Transwell permeable supports with 8-μm pores (Corning Life Sciences, Lowell, MA). Cells were suspended in serum-free medium and were seeded into Transwell inserts either uncoated (for migration assay) or coated (for invasion assay) with growth factor-reduced Matrigel (BD Biosciences, Bedford, MA). Bottom wells were filled with complete medium; after 24 hours, cells were stained with Andwin Scientific Diff-Quik stain set (Fisher Scientific, Pittsburg, PA), and photographed under a fluorescent microscope and manual cell counts were obtained.
Enzyme-Linked Immunosorbent Assay for VEGF, MMP-2, and MMP-9
Cells were seeded in six-well plates. After 24 hours of seeding, cells were appropriately treated with and without 5 and 10 μmol/L thiostrepton for 48 hours, after which the culture media were collected and centrifuged to remove cellular debris. The assay was performed using vascular endothelial growth factor (VEGF) and MMP-2 enzyme-linked immunosorbent assay kits (R&D System, Minneapolis, MN), according to the manufacturer's instructions.
Gene Silencing Using siRNA
FoxM1 siRNA and scrambled control siRNA were purchased from Qiagen (Valencia, CA). Cells were transfected using Lipofectamine 2000 (Invitrogen) for 6 hours, after which the lipid and siRNA complex was removed and fresh growth medium was added. Cells were lysed 48 hours after transfection, and specific protein levels were determined by Western blot analysis with specific antibodies.
Cell Lysis and Immunoblotting
Cells were lysed as previously described.22 Proteins were immunoblotted with different antibodies and visualized by the enhanced chemiluminescence method (Amersham, Piscataway, NJ).
Animals and Xenograft Study
Six-week-old NU/J nude mice were obtained from Jackson Laboratories (Bar Harbor, ME) and were maintained in a pathogen-free animal facility for at least 1 week before use. All animal studies were done in accordance with institutional guidelines. For xenograft study, mice were inoculated subcutaneously into the right abdominal quadrant with 5 × 106 cells of HCT-15 vector, HCT-15 clone 1, and HCT-15 clone 3 in 200 μL PBS. The body weight and tumor volume of each mouse were monitored weekly. The tumor volume was measured as described previously.19 After 5 weeks treatment, mice were sacrificed and individual tumors were weighed, then snap-frozen in liquid nitrogen for storage.
Results
Affymetrix Data Analysis
Raw cell file data acquired by Affymetrix GCOS v1.2 software were imported into Partek Genomic suite 6.4 to generate principal component analysis across the samples (Figure 1A) for further analysis. The raw hybridization data were processed as described previously.18 In all, 1377 genes were identified as differentially expressed, with a cutoff P value of <0.05 and twofold expression changes between CRC and the noncancerous control (279 up-regulated and 1098 down-regulated). Further filtration applying a fold change >2.25 resolved the up-regulated genes list to 148. Similarly, 331 down-regulated genes were identified, applying a fold change >3 for down-regulated genes only (see Supplemental Tables S2 and S3 at http://ajp.amjpathol.org). Supervised analysis using complete linkage hierarchical cluster analysis with a distance matrix was performed using Avadis software (Strand Life Sciences). The resulting color-coded cluster image map is presented in Figure 1B, revealing the clustering of CRC and noncancerous tissues in separate groups. Among significantly overexpressed genes in CRC, compared with noncancerous control, were TACSTD2, BIRC5, CLDN1, IL8, FOXM1, AURKA, and ECT2 (see Supplemental Tables S1 and S2 at http://ajp.amjpathol.org). Real-time reverse-transcriptase quantitative PCRs were performed on 30 CRC samples for FOXM1, IL8, CLDN1, SPINK2, DES, and ADAMTS1 for validation of Affymetrix expression data (see Supplemental Figure S1 at http://ajp.amjpathol.org).
FoxM1 Expression in CRC Patients by Immunohistochemistry
Immunohistochemical analysis of FoxM1 expression was interpretable in 370 CRC spots and the incidence of FoxM1 expression (Figure 2A) was found to be 66.2% (245/370) (Table 1). FoxM1 expression was seen predominantly in the nuclear compartment and was more frequently detected in moderately or poorly differentiated CRCs than in well-differentiated CRCs (P = 0.0200). FoxM1 expression also correlated significantly with proliferative marker Ki-67 (P = 0.0018) and MMP-9 (P = 0.0002) (Table 1). FoxM1 expression was not associated with age, sex, histology subtype, or American Joint Committee on Cancer (AJCC) stage. The expression of FoxM1 in colorectal adenomas (49.25 ± 37.91) and CRCs (53.26 ± 39.51) was more than in normal colon (1.5 ± 5.0; P < 0.0001 and P < 0.0001, respectively). However, there was no difference in expression between CRC and adenomas (P = 0.6507; Figure 2B). These findings suggest that dysregulation in FoxM1 expression is an early event in colorectal carcinogenesis. CRC patients with overexpression of FoxM1 had an overall survival of 62.8% at 5 years, compared with 69.0% with low FoxM1 expression. Thus, there was no difference in overall survival (P = 0.4543).
Figure 2.
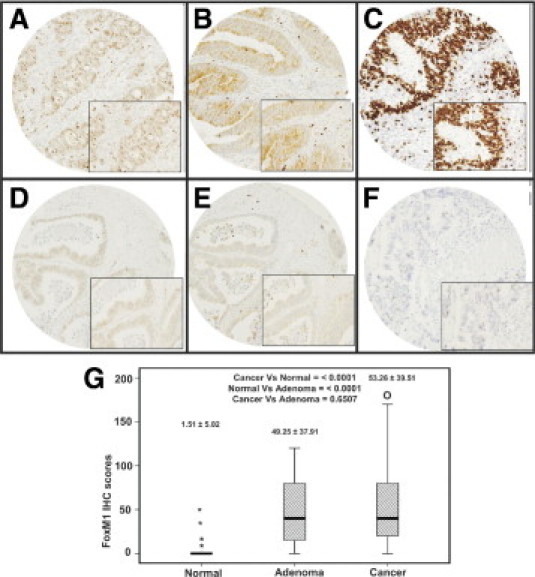
Tissue microarray based immunohistochemical analysis of FoxM1, MMP-9, and Ki-67 in CRC patients. A CRC array spot showing overexpression of FoxM1 (A), MMP-9 (C), and Ki-67 (E). CRC tissue array spots showing low expression of FoxM1 (B), MMP-9 (D), and Ki-67 (F). All images were taken on an Olympus BX 51 microscope, 20 × 0.70 objective (inset: magnified view, 40 × 0.85). G: Box-plots indicate the mean and SD of FoxM1 expression in three groups: adjacent normal colorectal mucosa, adenoma, and CRC. FoxM1 expression in colorectal adenomas (49.25 ± 37.91) and CRCs (mean 53.26 ± 39.51) was greater than in normal colon (1.51 ± 5.02) (P < 0.001 and P < 0.001, respectively); however, there was no difference in expression between CRC and adenomas (P = 0.6507). The asterisks indicate the wide range of FoxM1 expression in the normal colorectal mucosa group. FoxM1 expression in normal colorectal mucosa was in the range of 0 to 5 in >95% of the samples; however, a few samples (4%) showed expression score of 10, and 2 samples had extreme scores of 20 and 70. In the cancer group, the letter O indicates one CRC spot with an IHC score of 190, and it indicates extreme deviation from the mean.
Table 1.
Correlation Between FoxM1 IHC Status and Clinicopathological Parameters in Colorectal Carcinoma
| Total |
High expression |
Low expression |
|||||
|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | P value | |
| Sample size | n = 370 | n = 245 | 66.2 | n = 125 | 33.8 | ||
| Age | |||||||
| ≤0 years | 122 | 33.0 | 85 | 69.7 | 37 | 30.3 | 0.3219 |
| >50 years | 248 | 67.0 | 160 | 64.5 | 88 | 33.8 | |
| Sex | |||||||
| Male | 175 | 47.3 | 121 | 69.1 | 54 | 30.9 | 0.2590 |
| Female | 195 | 52.7 | 124 | 63.6 | 71 | 36.4 | |
| Tumor site | |||||||
| Left colon | 306 | 82.7 | 197 | 64.4 | 109 | 35.6 | 0.0954 |
| Right colon | 64 | 17.3 | 48 | 75.0 | 16 | 25.0 | |
| Histological type | |||||||
| Adenocarcinoma | 323 | 87.3 | 210 | 65.0 | 113 | 35.0 | 0.1914 |
| Mucinous carcinoma | 47 | 12.7 | 35 | 74.5 | 12 | 25.5 | |
| Tumor stage | |||||||
| I | 50 | 14.2 | 33 | 66.0 | 17 | 34.0 | 0.4073 |
| II | 120 | 34.1 | 74 | 61.7 | 46 | 38.3 | |
| III | 138 | 39.2 | 94 | 68.1 | 44 | 31.9 | |
| IV | 44 | 12.5 | 33 | 75.0 | 11 | 25.0 | |
| Differentiation | |||||||
| Well | 29 | 7.8 | 22 | 75.9 | 7 | 24.1 | 0.0200 |
| Moderate | 276 | 74.6 | 172 | 62.3 | 104 | 37.7 | |
| Poor | 65 | 17.6 | 51 | 78.5 | 14 | 21.5 | |
| Ki67⁎ | |||||||
| ≥50 | 304 | 86.4 | 211 | 69.4 | 93 | 30.6 | 0.0018 |
| <50 | 48 | 13.6 | 22 | 45.8 | 26 | 54.2 | |
| MMP-9 | |||||||
| >100 | 241 | 67.7 | 176 | 73.0 | 65 | 27.0 | 0.0002 |
| ≤100 | 115 | 32.3 | 61 | 53.0 | 54 | 47.0 | |
| Overall survival | |||||||
| 5 Years | 62.8 | 69.0 | 0.4543 | ||||
Analysis failure of some spots for these IHC markers was attributed to missing or nonrepresentative spots.
FoxM1 Expression in CRC Cell Lines and Effect on Cell Growth
The baseline expression of FoxM1 was determined in a panel of human CRC cancer cell lines in our initial in vitro experiments (Figure 3A), and a total of six cell lines. LOVO, CL-11, Colo-320, CX-1, DLD-1, and HCT-15 were selected for this study. Colo-320 showed the highest expression of FoxM1, whereas CL-11 and CX-1 expressed moderate levels of FoxM1. The other three cell lines (LOVO, DLD-1, and HCT-15) showed lower expression of FoxM1 (Figure 3A). In subsequent experiments, the effect of thiostrepton, an inhibitor of FoxM1,23 was assessed in CRC cells. Treatment of CRC cells with thiostrepton caused inhibition of viability in a dose-dependent manner that reached significance for most of the doses tested. (Figure 3B). Interestingly, of the four cell lines tested (ie, LOVO, Colo-320, DLD-1, and HCT-15), Colo-320 showed less response to thiostrepton-induced inhibition of viability, compared with other CRC cells.
Figure 3.
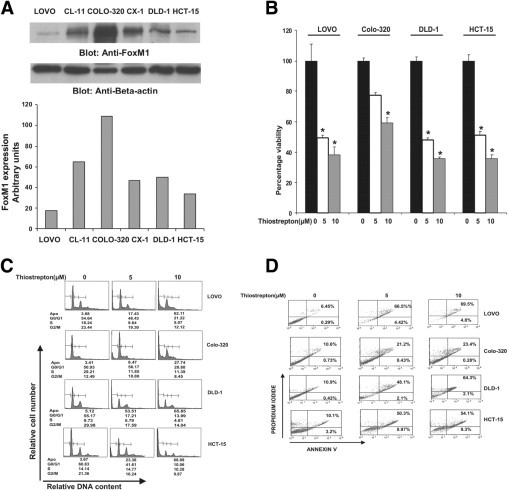
FoxM1 expression and its effect on cell growth in colorectal cancer (CRC) cell lines. A: Upper panel: LOVO, Cl-11, COLO-320, CX1, DLD-1, and HCT-15 cells were lysed and proteins were immunoblotted with FoxM1 and β-actin. Lower panel: The data obtained from the immunoblot analyses of FoxM1 were used to evaluate relative expression by spot densitometry. B: CRC cells were incubated with 5 and 10 μmol/L thiostrepton for 48 hours. Cell proliferation assays were performed using MTT as described in Materials and Methods. Data are reported as means ± SD of three independent experiments, with replicates of six wells for all of the doses and vehicle control for each experiment. *P < 0.05, Student's t-test. C: Thiostrepton-induced increase in subG1/Apo fraction of CRC cells. LOVO, Colo-320, DLD-1, and HCT-15 CRC cells were treated with 5 and 10 μmol/L thiostrepton for 48 hours. Thereafter, the cells were washed, fixed, and stained with propidium iodide and were analyzed for DNA content by flow cytometry as described in Materials and Methods. At least three independent experiments were performed for all of the cell lines. *P < 0.05, Student's t-test. D: Thiostrepton-mediated apoptosis in CRC cell lines.LOVO, COLO-320, DLD-1, and HCT-15 CRC cells were treated with 5 and 10 μmol/L thiostrepton for 48 hours and cells were subsequently stained with fluorescein-conjugated annexin-V and propidium iodide and analyzed by flow cytometry. Right lower (RL) quadrant denotes annexin v postive cells depicting alive apoptotic cells while right upper (RU) quadrant denotes annexin V and propidium iodide positive cells depicting dead apoptotic cells. Apoptosis is measured as the sum of RL and RU. X-axis denotes number of annexin V positive cells and y-axis denotes number of propidium iodide positive cells.
We determined whether thiostrepton-suppressed CRC cell growth was due to cell cycle arrest or apoptosis. CRC cells were treated with thiostrepton and cell-cycle fractions were determined by flow cytometry. Thiostrepton treatment caused an increase in sub-G1 population, accompanied by loss of G0/G1, S, and G2/M cell-cycle fractions (Figure 3C), suggesting that thiostrepton-mediated growth inhibition is due to apoptosis. Thiostrepton treatment had a more pronounced effect on LOVO, DLD-1, and HCT-15 cells than on Colo-320 cells. In addition, an increase in expression of p27 in CRC cell lines was detected in HCT-15 cells, relative to that in Colo-320 cells (data not shown). To further confirm that thiostrepton treatment caused apoptosis, CRC cells were treated with 5 and 10 μmol/L thiostrepton, and apoptotic cells were assayed by annexin V/propidium iodide dual staining. In the LOVO cell line, 70.9% and 75.3% apoptosis was seen in LOVO cell line at 5 and 10 μmol/L thiostrepton treatment, respectively (Figure 3D). Similar results were obtained in DLD1 and HCT-15 cell lines, showing a dose-dependent response. Colo-320 cells showed fewer apoptotic cells than the other three CRC cells, confirming our notion that FoxM1 expression can protect cells from thiostrepton-mediated apoptosis.
Overexpression of FoxM1 by cDNA Transfection Promotes CRC Cell Growth
To confirm whether overexpression of FoxM1 is associated with increased growth in CRC cells, HCT-15 and LOVO cells were transfected with either vector alone or FoxM1 cDNA, and stable clones were selected as described in Materials and Methods. The FoxM1 level was increased in both cell lines that were transfected with FoxM1 constructs, compared with vector alone (Figure 4A). We further determined that FoxM1-overexpressing clones were more proliferating, compared with vector-transfected clones (Figure 4B). We next determined the clonogenicity of FoxM1-overexpressing CRC cells. Formation of colonies was significantly higher in FoxM1-overexpressing HCT-15 clones 1 and 3, compared with HCT-15-vector cells (Figure 4C). These results from clonogenic assays were found to be consistent with the MTT data (Figure 4B), suggesting that FoxM1 expression influences the growth of CRC cells.
Figure 4.
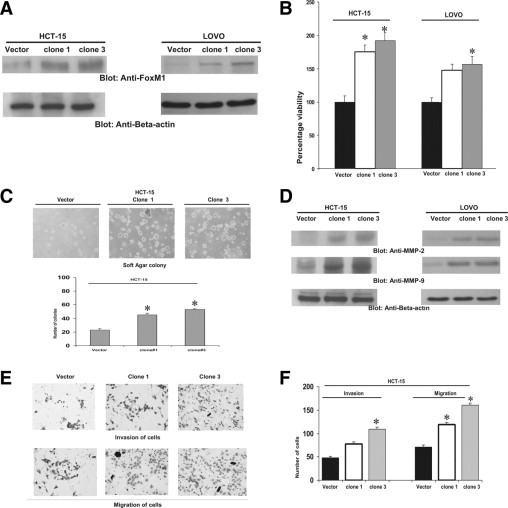
Overexpression of FoxM1 promoted growth and clonogenicity and induced invasion and migration in CRC cells. A: FoxM1 cDNA was transfected in HCT-15 and LOVO and stable clones were selected. HCT-15 and LOVO cells with vector alone or FoxM1-transfected clones were lysed and proteins were immunoblotted with FoxM1 and β-actin. B: Vector and FoxM1-overexpressing HCT-15 clones 1 and 3 and LOVO clones 1 and 2 were seeded in 96-well plates. After 48 hours, cell proliferation assays were performed using MTT as described in Materials and Methods. Data are reported as means ± SD of three independent experiments, with replicates of six wells for all of the doses and vehicle control for each experiment *P < 0.05, Student's t-test. C: Clonogenic assays were performed using vector and FoxM1-overexpressing HCT-15 clones 1 and 3 as described in Materials and Methods (upper panel). HCT-15 vector and HCT-15 clones 1 and 3 were plated in soft agar plates for 4 weeks, after which cells were stained and manually counted (lower panel). *P < 0.05, Student's t-test. D: HCT-15 and LOVO cells with vector alone or FoxM1-transfected clones were lysed and proteins were immunoblotted with MMP-2, MMP-9, and β-actin. E: Invasion-migration assays were performed using vector and FoxM1-overexpressing HCT-15 clones 1 and 3 as described in Materials and Methods. F: Manual counts of invasion and migration experiment of HCT-15 vector, clone 1, and clone 3 cells treated as just described for (E). *P < 0.005.
Overexpression of FoxM1 Increases MMP-2 and MMP-9 Protein Expression and Enhances Migration and Invasion of CRC Cells
Our clinical data showed a significant association between FoxM1 and MMP-9. We therefore investigated whether MMPs are regulated by FoxM1 in CRC cells. MMP-2 and MMP-9 protein levels were increased in FoxM1-overexpressing CRC clones, compared with vector (Figure 4D), suggesting that FoxM1 regulates the expression of MMPs in CRC. We further determined the effect of FoxM1 overexpression on CRC cell migration and invasion. FoxM1-overexpressing CRC clones showed a stronger penetration through the Matrigel-coated membrane, compared with the control vector-containing clones (Figure 4E). These results showed that FoxM1 regulated CRC cell migration and invasion via up-regulation of MMPs.
Overexpression of FoxM1 Protects Cells from Thiostrepton-Mediated Cell Growth Inhibition and Suppression of Migration/Invasion of CRC Cells
In the next series of experiments, we sought to determine whether overexpression of FoxM1 protects CRC cells from thiostrepton-mediated cell growth inhibition and suppression of migration and invasion. Vector and FoxM1-overexpressing CRC clones were treated with thiostrepton and cell proliferation was determined. The 10 μmol/L thiostrepton treatment caused a 51.2% inhibition in HCT-15 vector cells, but only 38% and 20% inhibition in HCT-15 clones 1 and 3, respectively (Figure 5A). Interestingly, on cell cycle analysis, HCT-15 vector showed 63% cells in G1 phase and 19.7% in S phase before treatment, compared with 38.43% in G1 phase and 48.8% cells in S phase for HCT-15 clone 3 (Figure 5B), a statistically significant difference (P < 0.05). These data suggest that FoxM1 overexpression led to increased proliferation of cells. Thiostrepton caused an increase in the apoptotic fraction of cells to 42.01% in HCT-15 vector, with subsequent decrease in G1 and S phase. In HCT-15 clone 3, however, there was an increase in apoptotic fraction to 22% along with an increase in G1 phase and decreased percentage of cells in S phase. These data suggest that, after treatment with thiostrepton, there was shift of cells toward the apoptotic fraction, a shift that was more pronounced in HCT-15 vector cells than in the FoxM1-overexpressing HCT-15 clone 3. Similarly, treatment of HCT-15 vector cells with 5 and 10 μmol/L thiostrepton caused 47% and 70% apoptosis, respectively, but only 26% and 41% apoptosis in FoxM1-overexpressing HCT-15 cells (see Supplemental Figure S2 at http://ajp.amjpathol.org). In addition, activation of caspase-3 and PARP was more pronounced in vector-containing clones, compared with FoxM1-overexpressing clones, after treatment with thiostrepton (Figure 5C).
Figure 5.
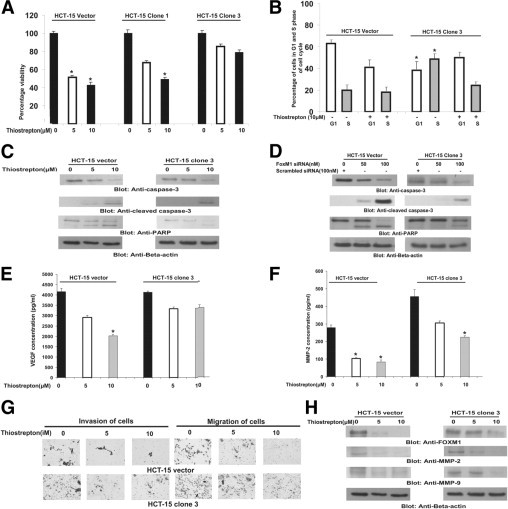
Overexpression of FoxM1 protects HCT-15 cells from thiostrepton-mediated inhibition, apoptosis, and inhibition of migration and invasion. A: Vector and FoxM1-transfected HCT-15 clones 1 and 3 were treated with 5 and 10 μmol/L thiostrepton for 48 hours. Cell proliferation assays were performed using MTT as described in Materials and Methods. Data are reported as means ± SD of three independent experiments, with replicates of six wells for all of the doses and vehicle control for each experiment. *P < 0.05, Student's t-test. B: Vector and FoxM1-transfected HCT-15 cells were treated with 5 and 10 μmol/L thiostrepton for 48 hours. Thereafter, the cells were washed, fixed, and stained with propidium iodide, and analyzed for DNA content by flow cytometry. At least three independent experiments were performed for all of the cell lines. *P < 0.05, Student's t-test. C: After treatment with 5 and 10 μmol/L thiostrepton for 48 hours, cells were lysed and proteins were separated on SDS-polyacrylamide gel electrophoresis and were immunoblotted with caspase-3, cleaved caspase-3, PARP, and β-actin. D: HCT-15 vector and HCT-15 clone 3 cells were transfected with scrambled siRNA and FoxM1 siRNA (100 nmol/L). After 48 hours, cells were lysed and proteins were immunoblotted with antibodies against caspase-3, cleaved caspase-3, PARP, and β-actin. HCT-15 vector and FoxM1-transfected HCT-15 clone 3 were treated with 5 and 10 μmol/L thiostrepton for 48 hours and activity of VEGF (E) and of MMP-2 (F) was determined by enzyme-linked immunosorbent assay. *P < 0.05, Student's t-test. G: HCT-15 vector and FoxM1-transfected HCT-15 clone 3 cells were treated with 5 and 10 μmol/L thiostrepton and invasion and migration assays were performed. H: After treatment with 5 and 10 μmol/L thiostrepton for 48 hours, cells were lysed and proteins were separated on SDS-PAGE and immunoblotted with FoxM1, MMP-2, MMP-9, and β-actin.
To confirm the specific role of thiostrepton in CRC cells, we transfected HCT-15 vector and clone 3 cells with siRNA targeted against FoxM1 and found that activation and cleavage of caspase-3 and PARP was more pronounced in HCT-15 vector cells, compared with HCT-15 clone 3 cells, suggesting that FoxM1 overexpression partially protects HCT-15 cells from apoptosis (Figure 5D). Thiostrepton-mediated inhibition of cell transformation was less effective in FoxM1-overexpressing clones (see Supplemental Figure S3 at http://ajp.amjpathol.org). Enzyme-linked immunosorbent assay experiments also suggest that overexpression of FoxM1 in CRC cells partially protected cells from thiostrepton-induced decreased secretion of VEGF and MMP-2 (Figure 5, E and F). Similarly, thiostrepton treatment efficiently inhibited migration and invasiveness in vector clones, compared with FoxM1-overexpressing CRC cells (Figure 5G) (see also Supplemental Figure S4 at http://ajp.amjpathol.org). Finally, we studied the effect of thiostrepton treatment on expression of MMP-2 and MMP-9. Expression of MMP-2 and MMP-9 decreased significantly in HCT-15 vector cells at 5 μmol/L thiostrepton treatment, whereas a lesser effect was seen on MMP expression at this concentration in FoxM1-overexpressing HCT-15 cells (Figure 5H) (see also Supplemental Figure S3 at http://ajp.amjpathol.org). These data suggest that FoxM1 overexpression protects CRC cells from thiostrepton-induced inhibition of cell transformation, migration, and invasion.
Effect of FoxM1 in CRC Tumorigenesis in Nude Mice
Finally, we sought to determine the effect of FoxM1 expression on CRC cell xenograft tumor growth in vivo using nude mice. The mice were inoculated subcutaneously into the right abdominal quadrant with 5 × 106 cells of HCT-15 vector, HCT-15 clone1, or HCT-15 clone 3, in groups of five. After 5 weeks, mice were sacrificed and tumors were collected. HCT-15 clones 1 and 3 caused the formation of larger tumors in mice, compared with HCT-15 vector cells (Figure 6A). The greater tumor volume in mice with HCT-15 clones 1 and 3 reached significance (P < 0.05) at the end of the fourth week, compared with HCT-15 vector cells. A significant increase in tumor weight (Figure 6B) was observed in mice with HCT-15 clone 3, compared with HCT-15 vector-induced xenografts (P < 0.05). Additionally, images of tumor with HCT-15 clones 1 and 3 after necropsy showed larger tumor size, compared with vector-containing cells (Figure 6C). Because our in vitro data showed that FoxM1 expression up-regulated MMPs, we tested whether overexpression of FoxM1 in vivo altered the expression of these proteins. We analyzed MMP-2 and MMP-9 levels in primary tumors derived from HCT-15 vector-inoculated mice and in tumors with HCT-15 clones 1 and 3 by Western blot analysis. In these mice, the levels of MMP-2 and MMP-9 were markedly increased in tumors with overexpressing HCT-15 clones 1 and 3, compared with tumors with vector HCT-15 cells (Figure 6D).
Figure 6.
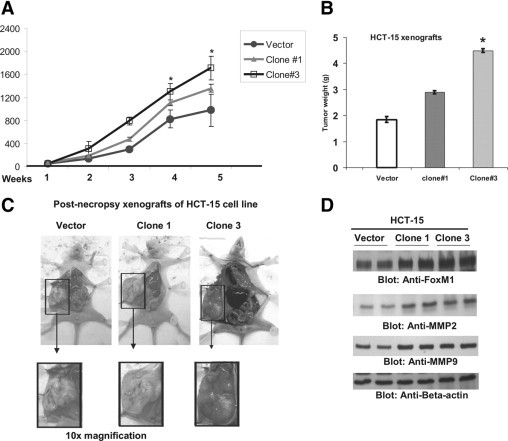
FoxM1 overexpression promotes colorectal cancer tumor growth. Nude mice were injected subcutaneously with HCT-15 vector and HCT-15 clone 1 and clone 3 cells. A: The volume of each tumor was measured every week. The average (n = 6) tumor volume in each group of mice was calculated, *P < 0.05. B: After 6 weeks of treatment, mice were sacrificed and the mean tumor weight (±SD) was calculated in each group. *P < 0.05, Student's t-test. C: Representative tumor images of each group of mice after necropsy. Insets show ×10 magnification. D: Whole-cell homogenates from mice injected with HCT-15 vector and HCT-15 clone 1 and clone 3 were immunoblotted with antibodies against FoxM1, MMP-2, MMP-9, and β-actin.
Discussion
In an attempt to identify genes that might be of importance in the malignant progression of CRC in Middle Eastern populations and genes that could be targeted in cancer therapy, we screened and validated the global gene expression in CRC using cDNA expression array and TMA. In all, 41 CRC patient samples were compared with 25 noncancerous colorectal tissue samples by use of cDNA. We identified a list of genes that are differentially expressed between primary tumors and normal samples through an analysis of microarray expression profiles. Quantitative real-time PCR was performed to confirm the differential expression of selected genes. Several genes involved in cell cycle regulation, proliferation, cell signaling, and carcinogenesis were identified. Interestingly, among these genes, FOXM1 was found to be significantly up-regulated in CRC, compared with noncancerous colorectal tissue. These data are in concordance with another study, by Pilarsky et al,24 showing by microarray analysis that FoxM1 was overexpressed in various epithelial cancers, including those of the colon, pancreas, and kidney.
To further identify the effect of the FOXM1 gene on the clinicopathological outcome of Middle Eastern CRC, protein expression of FoxM1 was examined in a large cohort of Saudi CRC using TMA technology. FoxM1 overexpression was seen in >65% of CRC, and it was significantly associated with a more aggressive phenotype (characterized by poorly differentiated tumors with a high proliferative index and high MMP-9 expression). These clinical data, and recent studies highlighting the critical role of FoxM1 in carcinogenesis,10–17 prompted us to study the effect of FoxM1 inhibition on CRC in vivo and in vitro. In the present study, we first confirmed the role of FoxM1 in cell proliferation of CRC cell lines. We found that down-regulation of FoxM1 by thiostrepton, an inhibitor of FoxM1, resulted in a significant decrease in the ability of CRC cell lines to proliferate, as well as reduction in oncogenicity of these cells. On the other hand, overexpression of FoxM1 resulted in elevated levels of proliferation and increased oncogenic potential of CRC cells. Previous reports have also suggested a role of FoxM1 in the proliferation and growth of CRC.25,26 Yoshida et al25 examined the role of FoxM1 in colon cancer development and proliferation by using FoxM1b transgenic mice and conditional FoxM1 knockout mice. Azoxymethane/dextran sulfate sodium-treated, Rosa26-FoxM1b transgenic mice showed an increase in the number and size of colorectal tumors, compared with wild-type mice, and a FoxM1-depleted colon cancer cell line showed reduced DNA replication and anchorage-independent growth.25 In addition, FoxM1 is, along with zinc finger protein GLI1, a key downstream signaling component of the sonic hedgehog signaling pathway in CRC, which when stimulated leads to up-regulation of FoxM1 and to cell proliferation in both human CRC patient samples and CRC cell lines.26 In agreement with these other studies, we observed a significantly higher FoxM1 expression in both CRC and adenoma, compared with the adjacent paired normal colorectal mucosa. Based on these observations, we can speculate that FoxM1 dysregulation is an early event in colorectal carcinogenesis and that FoxM1 up-regulation leads to increased proliferation and transformation, thereby providing a growth advantage.
FoxM1 accumulates mainly in the cytoplasm in late G1 and S phases, and nuclear translocation occurs before entry into G2-M phase.4 The importance of FoxM1 with respect to the cell cycle is well recognized,15,16,27–30 and we observed diminished G1-S progression as a consequence of down-regulation of FoxM1. We also observed an increased expression of cyclin-dependent kinase inhibitors such as p27, which are known to negatively regulate cell cycle progression.31–34 Our observations confirm previous reports supporting the role of FoxM1 in CRC progression.
We then found direct clinical evidence of a strong correlation among FoxM1-expressing tumors and MMPs in Middle Eastern CRC samples. MMPs are crucial in the process of tumor invasion and metastasis, and MMP-2 and MMP-9 are directly linked with angiogenesis and degradation of the basement membrane collagen leading to metastasis.35 We found that down-regulation of FoxM1 by thiostrepton treatment in CRC cell lines led to reduced expression of MMP-2 and MMP-9, as well as decreasing the invasive and migration capability of MMPs in CRC cells. Conversely, overexpression of FoxM1 in CRC cells contributed directly to overexpression of MMP-2 and MMP-9, and induced invasive and migratory potential of CRC cells. Interestingly, thiostrepton-induced inhibition had less effect on MMP-2 and MMP-9 expression, and on invasion and migration of CRC cells in FoxM1-overexpressing clone, suggesting that FoxM1 expression plays a critical role in the invasiveness and migratory activity of CRC cells by regulating MMP expression.
VEGF is another important factor in tumor cell invasion, angiogenesis, and metastasis.36 A strong correlation between expression of FoxM1 and VEGF has been reported in both breast and pancreatic cancers.16,17 In the present study, by using a well established FoxM1 overexpression system, we were able to show that altered FoxM1 expression significantly affected VEGF expression in CRC. For example, inhibition of FoxM1 expression significantly suppressed VEGF expression in CRC cells that expressed low amounts of FoxM1. In those clones that overexpressed FoxM1, however, thiostrepton-induced inhibition of VEGF release was less appreciable, thereby validating the oncogenic role of FoxM1 in CRC.
Our in vivo studies of the effect of FoxM1 overexpression on tumor growth in a murine xenograft model are consistent with results obtained in vitro using CRC cell lines. FoxM1 overexpression contributed significantly on the tumorigenicity via up-regulation of its target proteins, MMP-2 and -9.
In summary, our results show that FoxM1 expression was increased in Middle Eastern CRC, compared with normal colorectal mucosa. Our data also suggest that FoxM1 expression plays a pivotal role in colorectal carcinogenesis, in that it is associated with poorly differentiated tumors with a high proliferative index and high MMP-9 expression. We have also presented experimental evidence that down-regulation of FoxM1 caused inhibition of cell proliferation and invasion and migration of CRC cells. Overexpression of FoxM1 enhanced aggressiveness and tumorigenicity via up-regulation of MMP-2 and -9 of CRC cells. Based on these findings, we conclude that an aberrant FoxM1 signaling pathway plays a critical role in the pathogenesis of CRC, and we identify FOXM1 as a potential biomarker and a novel therapeutic target in distinct molecular subtypes of CRC.
Acknowledgments
We acknowledge Mehar Sultana, Valorie Balde, Saeeda O. Ahmed, Ruben D. Santos, Selah Fulgencio, and Aishah S. Olyan for their technical assistance and Zeeshan Quadri for data analysis.
Footnotes
Supported by King Abdulaziz City for Science and Technology and the National Comprehensive Plan for Science and Technology.
S.U., M.A., and A.H. contributed equally to this work.
Supplemental material for this article can be found at http://ajp.amjpathol.org and at 10.1016/j.ajpath.2010.10.020.
Supplementary data
Verification of 6 selected genes by quantitative real-time PCR analysis. Bar graph representing expression of these differentially expressed genes in CRC samples as measured by oligonucleotide microarray (dark bars) and quantitative real-time PCR (white bars) analyses. A fold change of 1 indicates equal expression in both groups. The error bars represent standard deviations of the mean expression values as measured by qRT-PCR.
HCT-15 vector and FoxM1-transfected HCT-15 clone 3 cells were treated with 5 and 10 μmol/L thiostrepton for 48 hours and apoptosis was analyzed by flow cytometry.
A, B: Clonogenic assays were performed using vector and HCT-15 clone 3 as described in Materials and Methods. HCT-15 vector and HCT-15 clone 3 were plated in Soft agar plates for 4 weeks after cells were stained and manually counted. The experiments were repeated at least three times, and representative data are presented.
HCT-15 vector and HCT-15 clone 3 cells were treated with 5 and 10 μmol/L thiostrepton and invasion and migration assays were performed. Bar graph denotes manual counts of invasion (A) and migration (B). The experiments were repeated at least three times, and representative data are presented.
References
- 1.Douillard J.Y., Cunningham D., Roth A.D., Navarro M., James R.D., Karasek P., Jandik P., Iveson T., Carmichael J., Alakl M., Gruia G., Awad L., Rougier P. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: a multicentre randomised trial [Erratum appeared in Lancet 2000;355:1372] Lancet. 2000;355:1041–1047. doi: 10.1016/s0140-6736(00)02034-1. [DOI] [PubMed] [Google Scholar]
- 2.Khamly K., Jefford M., Michael M., Zalcberg J. Beyond 5-fluorouracil: new horizons in systemic therapy for advanced colorectal cancer. Expert Opin Investig Drugs. 2005;14:607–628. doi: 10.1517/13543784.14.6.607. [DOI] [PubMed] [Google Scholar]
- 3.Cunningham D., Humblet Y., Siena S., Khayat D., Bleiberg H., Santoro A., Bets D., Mueser M., Harstrick A., Verslype C., Chau I., Van Cutsem E. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 4.Myatt S.S., Lam E.W. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 5.Ye H., Holterman A.X., Yoo K.W., Franks R.R., Costa R.H. Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S phase. Mol Cell Biol. 1999;19:8570–8580. doi: 10.1128/mcb.19.12.8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X., Kiyokawa H., Dennewitz M.B., Costa R.H. The forkhead box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc Natl Acad Sci USA. 2002;99:16881–16886. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wonsey D.R., Follettie M.T. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65:5181–5189. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]
- 8.Kalinichenko V.V., Major M.L., Wang X., Petrovic V., Kuechle J., Yoder H.M., Dennewitz M.B., Shin B., Datta A., Raychaudhuri P., Costa R.H. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalin T.V., Wang I.C., Ackerson T.J., Major M.L., Detrisac C.J., Kalinichenko V.V., Lyubimov A., Costa R.H. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006;66:1712–1720. doi: 10.1158/0008-5472.CAN-05-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim I.M., Ackerson T., Ramakrishna S., Tretiakova M., Wang I.C., Kalin T.V., Major M.L., Gusarova G.A., Yoder H.M., Costa R.H., Kalinichenko V.V. The forkhead box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66:2153–2161. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- 11.Liu M., Dai B., Kang S.H., Ban K., Huang F.J., Lang F.F., Aldape K.D., Xie T.X., Pelloski C.E., Xie K., Sawaya R., Huang S. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66:3593–3602. doi: 10.1158/0008-5472.CAN-05-2912. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y., Zhang N., Dai B., Liu M., Sawaya R., Xie K., Huang S. FoxM1B transcriptionally regulates vascular endothelial growth factor expression and promotes the angiogenesis and growth of glioma cells. Cancer Res. 2008;68:8733–8742. doi: 10.1158/0008-5472.CAN-08-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan D.W., Yu S.Y., Chiu P.M., Yao K.M., Liu V.W., Cheung A.N., Ngan H.Y. Over-expression of FOXM1 transcription factor is associated with cervical cancer progression and pathogenesis. J Pathol. 2008;215:245–252. doi: 10.1002/path.2355. [DOI] [PubMed] [Google Scholar]
- 14.Li Q., Zhang N., Jia Z., Le X., Dai B., Wei D., Huang S., Tan D., Xie K. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res. 2009;69:3501–3509. doi: 10.1158/0008-5472.CAN-08-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang I.C., Chen Y.J., Hughes D.E., Ackerson T., Major M.L., Kalinichenko V.V., Costa R.H., Raychaudhuri P., Tyner A.L., Lau L.F. FoxM1 regulates transcription of JNK1 to promote the G1/S transition and tumor cell invasiveness. J Biol Chem. 2008;283:20770–20778. doi: 10.1074/jbc.M709892200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z., Banerjee S., Kong D., Li Y., Sarkar F.H. Down-regulation of forkhead box M1 transcription factor leads to the inhibition of invasion and angiogenesis of pancreatic cancer cells. Cancer Res. 2007;67:8293–8300. doi: 10.1158/0008-5472.CAN-07-1265. [DOI] [PubMed] [Google Scholar]
- 17.Ahmad A., Wang Z., Kong D., Ali S., Li Y., Banerjee S., Ali R., Sarkar F.H. FoxM1 down-regulation leads to inhibition of proliferation, migration and invasion of breast cancer cells through the modulation of extra-cellular matrix degrading factors. Breast Cancer Res Treat. 2009;122:337–346. doi: 10.1007/s10549-009-0572-1. [DOI] [PubMed] [Google Scholar]
- 18.Siraj A.K., Bavi P., Abubaker J., Jehan Z., Sultana M., Al-Dayel F., Al-Nuaim A., Alzahrani A., Ahmed M., Al-Sanea O., Uddin S., Al-Kuraya K.S. Genome-wide expression analysis of Middle Eastern papillary thyroid cancer reveals c-MET as a novel target for cancer therapy. J Pathol. 2007;213:190–199. doi: 10.1002/path.2215. [DOI] [PubMed] [Google Scholar]
- 19.Uddin S., Hussain A.R., Ahmed M., Abubaker J., Al-Sanea N., Abduljabbar A., Ashari L.H., Alhomoud S., Al-Dayel F., Bavi P., Al-Kuraya K.S. High prevalence of fatty acid synthase expression in colorectal cancers in Middle Eastern patients and its potential role as a therapeutic target. Am J Gastroenterol. 2009;104:1790–1801. doi: 10.1038/ajg.2009.230. [DOI] [PubMed] [Google Scholar]
- 20.Uddin S., Siraj A.K., Al-Rasheed M., Ahmed M., Bu R., Myers J.N., Al-Nuaim A., Al-Sobhi S., Al-Dayel F., Bavi P., Hussain A.R., Al-Kuraya K.S. Fatty acid synthase and AKT pathway signaling in a subset of papillary thyroid cancers. J Clin Endocrinol Metab. 2008;93:4088–4097. doi: 10.1210/jc.2008-0503. [DOI] [PubMed] [Google Scholar]
- 21.Camp R.L., Dolled-Filhart M., Rimm D.L. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 22.Hussain A.R., Al-Jomah N.A., Siraj A.K., Manogaran P., Al-Hussein K., Abubaker J., Platanias L.C., Al-Kuraya K.S., Uddin S. Sanguinarine-dependent induction of apoptosis in primary effusion lymphoma cells. Cancer Res. 2007;67:3888–3897. doi: 10.1158/0008-5472.CAN-06-3764. [DOI] [PubMed] [Google Scholar]
- 23.Bhat U.G., Halasi M., Gartel A.L. Thiazole antibiotics target FoxM1 and induce apoptosis in human cancer cells. PLoS One. 2009;4:e5592. doi: 10.1371/journal.pone.0005592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pilarsky C., Wenzig M., Specht T., Saeger H.D., Grützmann R. Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data. Neoplasia. 2004;6:744–750. doi: 10.1593/neo.04277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida Y., Wang I.C., Yoder H.M., Davidson N.O., Costa R.H. The forkhead box M1 transcription factor contributes to the development and growth of mouse colorectal cancer. Gastroenterology. 2007;132:1420–1431. doi: 10.1053/j.gastro.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 26.Douard R., Moutereau S., Pernet P., Chimingqi M., Allory Y., Manivet P., Conti M., Vaubourdolle M., Cugnenc P.H., Loric S. Sonic Hedgehog-dependent proliferation in a series of patients with colorectal cancer. Surgery. 2006;139:665–670. doi: 10.1016/j.surg.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 27.Laoukili J., Kooistra M.R., Brás A., Kauw J., Kerkhoven R.M., Morrison A., Clevers H., Medema R.H. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 28.Laoukili J., Stahl M., Medema R.H. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102. doi: 10.1016/j.bbcan.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Major M.L., Lepe R., Costa R.H. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol Cell Biol. 2004;24:2649–2661. doi: 10.1128/MCB.24.7.2649-2661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan Y., Raychaudhuri P., Costa R.H. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol. 2007;27:1007–1016. doi: 10.1128/MCB.01068-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee M.H., Yang H.Y. Negative regulators of cyclin-dependent kinases and their roles in cancers. Cell Mol Life Sci. 2001;58:1907–1922. doi: 10.1007/PL00000826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coqueret O. New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol. 2003;13:65–70. doi: 10.1016/s0962-8924(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 33.Wang X., Krupczak-Hollis K., Tan Y., Dennewitz M.B., Adami G.R., Costa R.H. Increased hepatic Forkhead Box M1B (FoxM1B) levels in old-aged mice stimulated liver regeneration through diminished p27Kip1 protein levels and increased Cdc25B expression. J Biol Chem. 2002;277:44310–44316. doi: 10.1074/jbc.M207510200. [DOI] [PubMed] [Google Scholar]
- 34.Wang I.C., Chen Y.J., Hughes D., Petrovic V., Major M.L., Park H.J., Tan Y., Ackerson T., Costa R.H. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–10894. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curran S., Murray G.I. Matrix metalloproteinases: molecular aspects of their roles in tumour invasion and metastasis. Eur J Cancer. 2000;36:1621–1630. doi: 10.1016/s0959-8049(00)00156-8. [DOI] [PubMed] [Google Scholar]
- 36.Loges S., Mazzone M., Hohensinner P., Carmeliet P. Silencing or fueling metastasis with VEGF inhibitors: antiangiogenesis revisited. Cancer Cell. 2009;15:167–170. doi: 10.1016/j.ccr.2009.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Verification of 6 selected genes by quantitative real-time PCR analysis. Bar graph representing expression of these differentially expressed genes in CRC samples as measured by oligonucleotide microarray (dark bars) and quantitative real-time PCR (white bars) analyses. A fold change of 1 indicates equal expression in both groups. The error bars represent standard deviations of the mean expression values as measured by qRT-PCR.
HCT-15 vector and FoxM1-transfected HCT-15 clone 3 cells were treated with 5 and 10 μmol/L thiostrepton for 48 hours and apoptosis was analyzed by flow cytometry.
A, B: Clonogenic assays were performed using vector and HCT-15 clone 3 as described in Materials and Methods. HCT-15 vector and HCT-15 clone 3 were plated in Soft agar plates for 4 weeks after cells were stained and manually counted. The experiments were repeated at least three times, and representative data are presented.
HCT-15 vector and HCT-15 clone 3 cells were treated with 5 and 10 μmol/L thiostrepton and invasion and migration assays were performed. Bar graph denotes manual counts of invasion (A) and migration (B). The experiments were repeated at least three times, and representative data are presented.


