Abstract
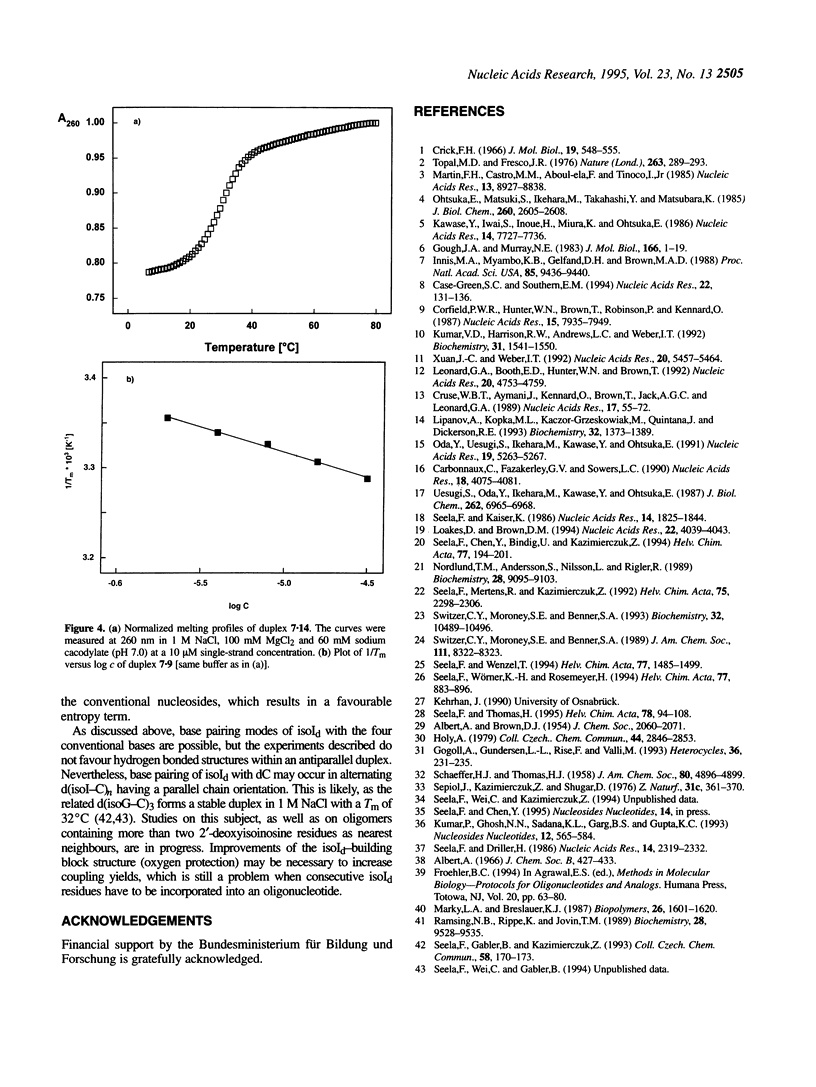
The fluorescent nucleoside 2'-deoxyisoinosine (2, isoId) has been incorporated into oligonucleotides. For this purpose the phosphonate 3a and the phosphoramidite 3b, as well as the polymer-linked 3d, have been synthesized and oligonucleotides were prepared by P(III) solid-phase chemistry. One or two isoId-residues were introduced into the oligomer d(T12), replacing dT either in the middle or at the 3'- and 5'-ends. The isoId-containing oligomers were hybridized with a modified d(A)12 containing the conventional nucleosides (dA, dT, dG and dC) opposite to isoId. The replacement of one dT by isoId in the centre of the duplex reduced the Tm value by approximately 15 degrees C and a decrease of approximately 25 degrees C was found when two isoId residues were incorporated. Thermodynamic data were determined from the melting curves. The destabilization was almost independent of the four naturally occurring nucleosides located opposite to isoId. The isoId (2) seems to be stacked in the duplex when dT-dA base pairs are the nearest neighbours; an internal loop is formed in the case of oligomers containing two consecutive isold residues.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carbonnaux C., Fazakerley G. V., Sowers L. C. An NMR structural study of deaminated base pairs in DNA. Nucleic Acids Res. 1990 Jul 25;18(14):4075–4081. doi: 10.1093/nar/18.14.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corfield P. W., Hunter W. N., Brown T., Robinson P., Kennard O. Inosine.adenine base pairs in a B-DNA duplex. Nucleic Acids Res. 1987 Oct 12;15(19):7935–7949. doi: 10.1093/nar/15.19.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Cruse W. B., Aymani J., Kennard O., Brown T., Jack A. G., Leonard G. A. Refined crystal structure of an octanucleotide duplex with I.T. mismatched base pairs. Nucleic Acids Res. 1989 Jan 11;17(1):55–72. doi: 10.1093/nar/17.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J. A., Murray N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983 May 5;166(1):1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- Innis M. A., Myambo K. B., Gelfand D. H., Brow M. A. DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9436–9440. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase Y., Iwai S., Inoue H., Miura K., Ohtsuka E. Studies on nucleic acid interactions. I. Stabilities of mini-duplexes (dG2A4XA4G2-dC2T4YT4C2) and self-complementary d(GGGAAXYTTCCC) containing deoxyinosine and other mismatched bases. Nucleic Acids Res. 1986 Oct 10;14(19):7727–7736. doi: 10.1093/nar/14.19.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V. D., Harrison R. W., Andrews L. C., Weber I. T. Crystal structure at 1.5-A resolution of d(CGCICICG), an octanucleotide containing inosine, and its comparison with d(CGCG) and d(CGCGCG) structures. Biochemistry. 1992 Feb 11;31(5):1541–1550. doi: 10.1021/bi00120a035. [DOI] [PubMed] [Google Scholar]
- Leonard G. A., Booth E. D., Hunter W. N., Brown T. The conformational variability of an adenosine.inosine base-pair in a synthetic DNA dodecamer. Nucleic Acids Res. 1992 Sep 25;20(18):4753–4759. doi: 10.1093/nar/20.18.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipanov A., Kopka M. L., Kaczor-Grzeskowiak M., Quintana J., Dickerson R. E. Structure of the B-DNA decamer C-C-A-A-C-I-T-T-G-G in two different space groups: conformational flexibility of B-DNA. Biochemistry. 1993 Feb 9;32(5):1373–1389. doi: 10.1021/bi00056a024. [DOI] [PubMed] [Google Scholar]
- Loakes D., Brown D. M. 5-Nitroindole as an universal base analogue. Nucleic Acids Res. 1994 Oct 11;22(20):4039–4043. doi: 10.1093/nar/22.20.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marky L. A., Breslauer K. J. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987 Sep;26(9):1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- Martin F. H., Castro M. M., Aboul-ela F., Tinoco I., Jr Base pairing involving deoxyinosine: implications for probe design. Nucleic Acids Res. 1985 Dec 20;13(24):8927–8938. doi: 10.1093/nar/13.24.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund T. M., Andersson S., Nilsson L., Rigler R., Gräslund A., McLaughlin L. W. Structure and dynamics of a fluorescent DNA oligomer containing the EcoRI recognition sequence: fluorescence, molecular dynamics, and NMR studies. Biochemistry. 1989 Nov 14;28(23):9095–9103. doi: 10.1021/bi00449a021. [DOI] [PubMed] [Google Scholar]
- Oda Y., Uesugi S., Ikehara M., Kawase Y., Ohtsuka E. NMR studies for identification of dI:dG mismatch base-pairing structure in DNA. Nucleic Acids Res. 1991 Oct 11;19(19):5263–5267. doi: 10.1093/nar/19.19.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka E., Matsuki S., Ikehara M., Takahashi Y., Matsubara K. An alternative approach to deoxyoligonucleotides as hybridization probes by insertion of deoxyinosine at ambiguous codon positions. J Biol Chem. 1985 Mar 10;260(5):2605–2608. [PubMed] [Google Scholar]
- Ramsing N. B., Rippe K., Jovin T. M. Helix-coil transition of parallel-stranded DNA. Thermodynamics of hairpin and linear duplex oligonucleotides. Biochemistry. 1989 Nov 28;28(24):9528–9535. doi: 10.1021/bi00450a042. [DOI] [PubMed] [Google Scholar]
- Seela F., Driller H. Palindromic oligonucleotides containing 7-deaza-2'-deoxyguanosine: solid-phase synthesis of d[(p)GG*AATTCC] octamers and recognition by the endodeoxyribonuclease EcoRI. Nucleic Acids Res. 1986 Mar 11;14(5):2319–2332. doi: 10.1093/nar/14.5.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seela F., Kaiser K. Phosphoramidites of base-modified 2'-deoxyinosine isosteres and solid-phase synthesis of d(GCI*CGC) oligomers containing an ambiguous base. Nucleic Acids Res. 1986 Feb 25;14(4):1825–1844. doi: 10.1093/nar/14.4.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepiol J., Kazimierczuk Z., Shugar D. Tautomerism of isoguanosine and solvent-induced keto-enol equilibrium. Z Naturforsch C. 1976 Jul-Aug;31(7-8):361–370. doi: 10.1515/znc-1976-7-803. [DOI] [PubMed] [Google Scholar]
- Switzer C. Y., Moroney S. E., Benner S. A. Enzymatic recognition of the base pair between isocytidine and isoguanosine. Biochemistry. 1993 Oct 5;32(39):10489–10496. doi: 10.1021/bi00090a027. [DOI] [PubMed] [Google Scholar]
- Topal M. D., Fresco J. R. Base pairing and fidelity in codon-anticodon interaction. Nature. 1976 Sep 23;263(5575):289–293. doi: 10.1038/263289a0. [DOI] [PubMed] [Google Scholar]
- Uesugi S., Oda Y., Ikehara M., Kawase Y., Ohtsuka E. Identification of I:A mismatch base-pairing structure in DNA. J Biol Chem. 1987 May 25;262(15):6965–6968. [PubMed] [Google Scholar]
- Xuan J. C., Weber I. T. Crystal structure of a B-DNA dodecamer containing inosine, d(CGCIAATTCGCG), at 2.4 A resolution and its comparison with other B-DNA dodecamers. Nucleic Acids Res. 1992 Oct 25;20(20):5457–5464. doi: 10.1093/nar/20.20.5457. [DOI] [PMC free article] [PubMed] [Google Scholar]



