Abstract
Persistent tobacco use and excessive alcohol consumption are major public health concerns worldwide. Both alcohol and nicotine dependence (AD, ND) are genetically-influenced complex disorders that exhibit a high degree of comorbidity. To identify gene variants contributing to one or both of these addictions, we first conducted a pooling-based genome wide association study (GWAS) in an Australian population, using Illumina Infinium 1M arrays. Allele frequency differences were compared between pooled DNA from case and control groups for: (i) AD, 1224 cases and 1162 controls; (ii) ND, 1273 cases and 1113 controls; and (iii) comorbid AD and ND, 599 cases and 488 controls. Secondly, we carried out a GWAS in independent samples from the Netherlands for AD and for ND. Thirdly, we performed a meta-analysis of the 10,000 most significant AD- and ND-related SNPs from the Australian and Dutch samples. In the Australian GWAS, one SNP achieved genomewide significance (p < 5×10−8) for ND (rs964170 in ARHGAP10 on chromosome 4, p=4.43×10−8) and three others for comorbid AD/ND (rs7530302 near MARK1 on chromosome 1 (p=1.90×10−9), rs1784300 near DDX6 on chromosome 11 (p=2.60×10−9) and rs12882384 in KIAA1409 on chromosome 14 (p=4.86×10−8)). None of the SNPs achieved genomewide significance in the Australian/Dutch meta-analysis, but a gene network diagram based on the top-results revealed overrepresentation of genes coding for ion-channels and cell adhesion molecules. Further studies will be required before the detailed causes of comorbidity between AD and ND are understood.
Keywords: addiction, alcohol, comorbidity, genetics, smoking
Nicotine and alcohol dependence frequently occur together. Approximately 23% of individuals with nicotine dependence meet past-year criteria for alcohol abuse or dependence, and individuals with alcohol dependence are over 4 times more likely to be nicotine dependent (Grant et al., 2004). Individuals who consume both cigarettes and alcohol are at an increased risk for some forms of cancer (Kuper et al., 2000) and alcohol dependent individuals are actually at greater risk of dying from tobacco-related diseases than dying from an alcohol-related disease (Hurt et al., 1996).
Both nicotine dependence (ND) and alcohol dependence (AD) are highly heritable (Gelernter & Kranzler, 2009; Heath et al., 1997; Knopik et al., 2004; Lessov et al., 2004; Madden et al., 1999; Madden et al., 2004; Vink et al., 2005) and their co-occurrence appears, in part, to be accounted for by common genetic variation (Koopmans et al., 1997; True et al., 1999). A genetic correlation between a history of regular smoking and DSM-IIIR alcohol dependence has been observed even after controlling for personality and history of other psychopathology (Madden et al., 2000). Behavioral and genetic studies in both rodents and humans suggest cross-tolerance processes that contribute to the high phenotypic and genetic correlations of these two behaviors (Butt et al., 2003; Butt et al., 2004; Madden & Heath, 2002; Madden et al., 1997; Owens et al., 2003). Substance-specific genetic risk factors also play a role in the development of ND and AD (Hettema et al., 1999).
Genomewide association studies (GWAS) provide a means of identifying common polymorphisms contributing to genetic risk without restricting the scope of study to known or suspected candidate genes. Some GWAS results for ND (or related phenotypes) have been reported (Bierut et al., 2007; Caporaso et al., 2009; Drgon et al., 2009; Johnson et al., 2006; Liu et al., 2009; Rosenberger et al., 2005; Saccone et al., 2008; Thorgeirsson et al., 2008; Uhl et al., 2008a; Uhl et al., 2007; Uhl et al., 2008b; Vink et al., 2009). ND-related findings met genomewide significance criteria in only one case (Thorgeirsson et al., 2008) for 7 SNPs on chromosome 15 (mainly represented by rs1051730 in CHRNA3). Two previous GWAS have been reported on AD (Johnson et al., 2006; Treutlein et al., 2009); the latter reported genome-wide significant results, after combining samples, for two SNPs located near PECR.
Identification of predisposing genetic variants common to ND and AD, may inform and ultimately improve treatments tailored for dependence to these substances, but an appreciation of comorbid ND and AD when designing genetic studies of either disorder has been lacking. The aim of this study was to conduct a GWAS using DNA pools from Australian cases who met DSM-IV criteria for lifetime history of ND, AD or comorbid AD and ND. We tested for replication of the initial findings using GWA data available from Dutch subjects with ND and AD information. We performed a meta-analysis of AD and ND in the Australian sample and in two samples from the Netherlands. This is the first study to our knowledge that has systematically screened genetic variants to identify those that affect a person’s liability to developing both AD and ND.
METHODS
Australian Sample
Participants were twins initially recruited through the Australian Twin Registry, and other members of their families. Individuals were selected from (a) a large-sibship study (BIGSIB) designed to study families with 5 or more offspring sharing both biological parents and unselected for phenotype (Hansell et al., 2008; Saccone et al., 2007); (b) an alcohol extreme discordant and concordant (EDAC) study (Hansell et al., 2008), designed to focus on families extremely discordant or concordant for heavy drinking and alcohol dependence risk; and (c) the Nicotine Addition Genetics (NAG) study, which targeted families based on heavy smoking index cases with one or more full sibling smokers identified in previous interview and questionnaire surveys (Loukola et al., 2008; Pergadia et al., 2009; Saccone et al., 2007). Self-reported ancestry of the participants is predominantly Anglo-Celtic and Northern European (>90%) with information available on the birthplace and ethnicity of their four grandparents. Eighty percent (N=1906 unrelated individuals) of the pooled GWAS sample provided self-report on ancestry and birthplace of all four grandparents. Of these, 95.6% had all European grandparents (with 70% of these Northern European, N=1281), a further 1.7% indicated Australian Aboriginal ancestry and 1.5% reported some Asian or Middle Eastern ancestry. Three who reported ‘Australian’ ancestry are thought to have all grandparents born in Australia but with European ancestry. The remaining 1.1% had one or more grandparent from the Americas, Africa or the Pacific region. All individuals were included in the pooling experiment. All participants provided written informed consent under study protocols approved by the Queensland Institute of Medical Research (QIMR) Human Research Ethics Committee. In total, 9669 individuals from 4882 families, comprising 3115 twins, 5665 non-twin siblings, and 889 parents were interviewed.
Case and Control Definitions of Alcohol and Nicotine Dependence
The focus of this study was discovery of allelic associations for DSM-IV defined nicotine and alcohol dependence in unrelated individuals using a case-control pooling design (Risch & Merikangas, 1996; Sham et al., 2002). Information was collected using a computer-assisted telephone diagnostic interview (CATI). Self-reported symptoms of DSM-IIIR and DSM-IV alcohol dependence and quantity and frequency of alcohol consumption were measured using an adaptation of the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) (Bucholz et al., 1994) for telephone administration. The tobacco section for the CATI was derived from the Composite International Diagnostic Interview (CIDI) (Cottler et al., 1991), and incorporated the Fagerstrom Test for Nicotine Dependence (FTND) (Heatherton et al., 1991) and DSM-IIIR and DSM-IV assessments of nicotine dependence. For the purposes of this study, nicotine (ND) and alcohol dependent (AD) cases were defined using DSM-IV criteria. ND controls were defined as individuals exposed to smoking (all AD−/ND− subjects tried cigarettes at some point in their lifetime; see Table 1), but had never become dependent. Similarly, all AD controls were exposed to cigarettes and practically all to alcohol (99% of AD−/ND− subjects had a full drink of alcohol at some point in their lifetime) but did not meet criteria for DSM-IV AD (Table 1).
Table 1.
Characteristics of the Australian Case and Controls Pools.
| Pool | AD+/ND− | AD−/ND+ | AD+/ND+ | AD−/ND− |
|---|---|---|---|---|
| Pool description | AD Cases | ND Cases | Comorbid Cases | Controls |
| No. of subjects | 625 | 674 | 599 | 488 |
| Gender | ||||
| Male | 395 (63%) | 304 (45%) | 320 (53%) | 225 (46%) |
| Female | 230 (37%) | 370 (55%) | 279 (47%) | 263 (54%) |
| Age (years) | ||||
| Range | 21–73 | 25–78 | 25–73 | 30–79 |
| M ± SDa | 41.0 ± 8.0 | 45.5 ± 9.9 | 41.3 ± 8.1 | 46.7 ± 10.1 |
| Smoker status | ||||
| Ever smokersb | 100% | 100% | 100% | 100% |
| Regular smokersc | 51% | 100% | 100% | 36% |
| MaxCigsd | ||||
| Female (M ± SD) | 10.0 ± 13.6 | 33.4 ± 13.1 | 36.9 ± 16.2 | 6.3 ± 10.7 |
| Male (M ± SD) | 13.1 ± 16.5 | 42.3 ± 17.8 | 43.8 ± 16.2 | 9.8 ± 16.0 |
| DSM-IV ND symptom count | ||||
| 0–1 symptoms | 67% | 0% | 0% | 82% |
| ≥4 symptoms | 3% | 69% | 79% | 1% |
| Alcohol use status | ||||
| Ever drinkerse | 100% | 99.9% | 100% | 99% |
| Regular drinkersf | 100% | 95% | 100% | 91% |
| MaxDrinksg | ||||
| Female (M ± SD) | 14.5 ± 8.1 | 12.1 ± 10.1 | 19.0 ± 11.9 | 8.3 ± 6.1 |
| Male (M ± SD) | 29.9 ± 14.8 | 24.4 ± 12.3 | 33.6 ± 14.8 | 18.4 ± 11.9 |
| DSM-IV AD symptom count | ||||
| 0–1 symptom | 0% | 60% | 0% | 76% |
| ≥4 symptoms | 49% | 2% | 62% | 1% |
AD+, alcohol dependent; AD−, not alcohol dependent; ND+, nicotine dependent; ND−, not nicotine dependent.
Mean ± standard deviation.
Individuals who have at least tried cigarette smoking (“have you ever tried cigarette smoking, even a puff?”).
Individuals who have smoked 100 or more cigarettes in their lifetime.
The largest number of cigarettes an individual had ever smoked in a 24-hour period.
Individuals who have at least tried alcohol (“have you ever had a drink of alcohol?”).
Individuals who have consumed alcohol at least once a month for six months or more.
The largest number of alcoholic beverages an individual had ever consumed in a 24-hour period.
Three case pools were constructed: (i) AD subjects with no history of ND despite a history of cigarette experimentation (AD+/ND−; N=625); (ii) ND subjects with no history of AD (AD−/ND+; N=674), but in almost all cases a history of experimentation with alcohol; and (iii) comorbid AD and ND subjects (AD+/ND+; N=599); an AD−/ND− control pool (N=488) was also created (Table 1). Each pool was constructed separately for males and females to allow for the examination of possible sex-specific effects. No more than one individual per family was used in the pooling experiment.
DNA Pool Construction
Association analysis using pooled samples is based on the addition of equal amounts of DNA from each relevant individual to either Case or Control pools, followed by quantitative measurement of the signals from each allele at each typed SNP, as a measure of the case or control allele frequency (Macgregor et al., 2008). Genomic DNA was extracted from peripheral venous blood samples. DNA concentrations in each sample were measured using PicoGreen (Molecular Probes) for the quantitation of double-stranded DNA in solution on a Fluoroskan Ascent CF plate reader (Labsystems, Chicago, IL). DNA samples were initially diluted to 70 ng/μL using 1X TE before another quantification measurement using the PicoGreen method. Each sample was then diluted to 45 ng/μL, re-quantitified using Pico Green and diluted to a final concentration of 30 ng/μL. Pools were constructed by combining equal volumes (10 μL) of each DNA sample. Variance introduced by different pool sizes was taken into account during data analysis. All pipetting steps requiring volumes greater than 1 μL were performed on an Eppendorf EpMotion 5070 or EpMotion 5075 robot (Eppendorf AG, Hamburg, Germany).
Genotyping of Australian Sample
Array-based genotyping was performed on an Illumina BeadStation platform using Infinium 1M (singleton) v1 (1.05 million markers) arrays and Infinium 1M (duo) v3 (1.2 million markers) arrays (Illumina Inc. California, USA). The BeadStation software was set to output bead level intensity data (i.e., raw green/red beadscores). Each of the 8 pools was genotyped in triplicate twice; once on a BeadStation at QIMR (1M v1 arrays) and once on a Beadstation at deCODE Genetics (deCODE Genetics Inc., Reykjavik, Iceland) (1M v3 arrays). In each case, each pool was hybridized to three Illumina arrays. Thus, 6 replicates are available for each of the 8 pools.
Analysis of Pooled Genotype Data
A small number of SNPs had negative values for one or more beadscores. For some of these SNPs, the vast majority of beadscores had valid positive values with only a small number of slightly negative values. To allow such SNPs to be included in the analysis (hence maximizing genomic coverage), a small amount (+50) was added to negative beadscores. Each SNP was then assessed to see if >90% of SNPs had positive scores. If so, the small number (<10%) of negative scores were assumed to represent small positive values (+1). SNPs which still had one or more negative scores after this process were not analysed further. All SNPs designed to detect copy number variants (i.e., invariants) were discarded. SNPs were removed in the initial quality control based on their frequency in HapMap CEU reference samples (The International HapMap Consortium., 2003). SNPs with CEU minor allele frequency <1% were dropped from further analysis (this reduced the number of 1M v1 SNPs of interest to ~800k SNPs and the number of 1M v3 SNPs of interest to ~920k SNPs). Removing SNPs which are likely to be monomorphic in our Caucasian samples is desirable as the subsequent normalization and analysis steps are designed primarily for SNPs which are polymorphic. Furthermore, SNPs with very low (or zero) minor allele frequencies are unlikely to show significant results in genetic association studies.
The beadscores required normalization/calibration because green beadscores were generally larger than red beadscores. The Illumina 1M v1 arrays had 20 stripes per array, each with ~50000 SNPs. The Illumina 1M v3 arrays had 6 stripes per array, each with ~200000 SNPs. Note that these ‘stripes’ were previously denoted ‘strands’ but this is now changed to avoid confusion with top/bottom strand. Within each stripe, approximately half of the SNPs were from the Illumina ‘top’ strand (A/C and A/G SNPs) and half were from the Illumina ‘bottom’ strand (T/C and T/G SNPs). Normalization was done within stripe by rescaling the red beadscore to make the mean value of the pooling allele frequency (PAF) =0.5 (over all SNPs on that stripe); PAF was computed as the corrected red intensity divided by the total (corrected red plus green) intensity.
Due to an array reagent issue on the QIMR run arrays which affected ‘top’ strand SNPs more than ‘bottom’ strand SNPs, the degree of normalization (amount by which red beadscores were rescaled) on the arrays varied by strand. Whilst results are broadly similar on both strands, the array reagent issue meant that the results from “bottom” strand SNPs showed lower levels of pooling error. We hence focused on the set of “bottom” strand SNPs in QIMR run arrays. In the deCODE run arrays (where there were no array reagent issues), both strands performed similarly.
Each SNP had approximately 15 PAF estimates per array. For each pool, a small number of SNPs had fewer than 20 PAF estimates available and these SNPs were dropped. After normalization, SNPs were removed from the analysis if they had a -log10(p) value > 6 for the quality control score described in Macgregor et al. (2008). SNPs from the sex chromosomes were also dropped. Overall, on the deCODE run 1M v3 arrays, ~918000 SNPs passed quality control (the vast majority of omitted SNPs were dropped due to their having <1% frequency in the CEU HapMap samples). For the QIMR run 1M v1 arrays, ~375k SNPs from the “bottom” strand were included in the analysis with the deCODE generated data (i.e. ~40% of SNPs had data from 6 arrays per pool, with the remainder typed on 3 arrays per pool).
Statistical Analysis of Pool Genotype Data
Given our design, we ran stratified analyses with our Australian sample, i.e., testing for effects of ND after controlling for AD (AD+ND+ versus AD+ND− and AD−ND+ versus AD−ND−) and testing for the effects of AD (AD+ND+ versus AD−ND+ and AD+ND− versus AD−ND−), in addition to testing for comorbid effects (AD+ND+ versus AD−ND−). For each pair of pools (e.g., AD+/ND+ versus AD+/ND− QIMR arrays, AD+/ND+ versus AD+/ND− deCODE arrays, AD−/ND+ versus AD−/ND− QIMR arrays, etc.) run separately, a linear model based approach was used to test for allelic association in which the response variable is the set of pooling allele frequency (PAF) estimates for each SNP. The predictor variable is case/control status. The linear model was used to estimate the pooling error across all SNPs. A test statistic which corrects for the pooling error was used to rank SNPs, with p-values based on a Chi-Square distribution. The test statistic is:
T_simple is d2/V, where d is the mean case pool PAF minus the mean control pool PAF and V is binomial sampling variance. vare_pool_2 and varSNPspecific are the two components of the pooling error – see appendix 2 of Macgregor et al. (2006) for full details. Since this method is based upon contrasting case and control pools, the effect of unequal amplification of alleles is minimal as such effects cancel out.
Results from the different array sets were analyzed separately to allow for the possibility of different levels of pooling error in each experiment. The results for each SNP were then combined by calculating a composite version of T_2_X, which weighted the result by the inverse of the estimated variance for that SNP – the estimated variance comprised the binomial sampling variance, together with the terms 2vare_pool_2 and varSNPspecific. In cases where a particular SNP passed quality control for a subset of pools, only the relevant pools contributed to the test statistic. That is, failing quality control for some pools did not necessarily remove that SNP completely from the analysis (although the sample size for that SNP would be reduced)
Dutch Samples
To confirm the findings from the pool-based GWAS, we sought to replicate our findings in silico in two cohorts: the Netherlands Study of Depression and Anxiety (NESDA; Penninx et al., 2008) and the Netherlands Twin Registry (NTR; Boomsma et al., 2006). The NESDA and NTR studies were approved by the Central Ethics Committee on Research Involving Human Subjects of the VU University Medical Center Amsterdam. Subjects were selected for a GWA for major depressive disorder (Boomsma et al., 2008; Sullivan et al., 2009) and the sample included 1738 depressed cases and 1802 controls. Details on the recruitment, sample composition, screening procedure and phenotype assessment are described in detail elsewhere (Boomsma et al., 2008). The number of AD and ND cases and controls is summarized in Table 2.
Table 2.
Number of Dutch Alcohol and Nicotine Dependent Cases and Controls by Sex
| Sample | Female | Male | Total |
|---|---|---|---|
| AD cases (NESDA) | 178 | 140 | 318 |
| AD controls (NESDA) | 929 | 313 | 1242 |
| Total GWAS alcohol (NESDA) | 1107 | 453 | 1560 |
| AD cases (NTR) | 103 | 134 | 237 |
| AD controls (NTR) | 1015 | 511 | 1526 |
| Total GWAS alcohol (NTR) | 1118 | 645 | 1763 |
| ND cases (NESDA/NTR) | 368 | 243 | 611 |
| ND controls NESDA/NTR) | 555 | 280 | 835 |
| Total GWAS nicotine (NESDA/NTR) | 923 | 523 | 1446 |
Case and Control Definitions of Alcohol and Nicotine Dependence
In both the NESDA and NTR samples, ND cases (N=611) were defined as scoring 4 or more on the Fagerstrom Test for Nicotine Dependence (FTND) (Heatherton et al., 1991) whereas ND controls (N=835) had FTND scores <4. For NTR subjects longitudinal FTND data (3 timepoints: 20003 timepoints: 2002 and 2004) were available in smokers and ex-smokers whereas for NESDA subjects FTND data were available in current smokers only. For the ex-smokers from NESDA (N=523) it was not possible to determine their FTND score for the period they smoked so they were excluded from analyses. For both samples the ‘never smokers’ (N=1208) were excluded because their liability to ND is unknown. Alcohol dependence was assessed by the DSM-IV based lifetime CIDI interview (version 2.1) in the NESDA sample (N=1760), with 318 AD cases and 1242 controls identified. The 200 participants identified as alcohol abusers were excluded. In the NTR sample (N=1777), participants were defined as being alcohol dependent if they ever reported a score of 2 or higher (N=237) on the CAGE (Ewing, 1984) questionnaire and classified as a control if they never scored 2 or higher (N=1526). Subjects with missing data on the CAGE were excluded (N=14).
Genotyping of Dutch Samples
Genomewide association genotyping for 600,000 SNPs was conducted by Perlegen Sciences (Mountain View, CA, USA). After a series of quality control processes (Sullivan et al., 2009), individual genotype data for 427,037 autosomal SNPs were available. This set of SNPs was used as the basis for an imputation procedure whereby ~2.5M HapMap (The International HapMap Consortium., 2003) SNPs were imputed. Imputation was carried out in IMPUTE (Marchini et al., 2007) using the HAPMAP Phase II CEU data (Build 35) available on the IMPUTE website (http://www.stats.ox.ac.uk/~marchini/software/gwas/impute.html#). The threshold (for genotype uncertainty) to include SNPs in the analyses was 0.70. Genome wide association analyses were carried out in PLINK (Purcell et al., 2007) (option --assoc and --model, no covariates were included).
Meta-analysis of the Australian and Dutch Data
Meta-analysis was performed by extending the analysis of the different array sets run on the pools. First the difference between case and control allele frequencies was computed for each SNP in the Dutch data set. The results for each SNP were then combined by calculating a composite version of T_2_X which weighted the pooling (Australian data) and individual genotyping (Dutch data) frequency difference values by the inverse of their variance. For the pool data, the variance comprised the binomial sampling variance, together with the terms vare_pool_2 and varSNPspecific. For the individual genotyping data, the variance comprised just the binomial sampling variance. In a limited number of cases, the allele frequency estimates from pooling were different from those obtained from individual genotyping; this was due largely to the potential for unequal amplification of alleles in the pooling analysis. To minimize the effects of this on the meta-analysis results, SNPs with minor allele frequency less than 5% (as measured in the Dutch samples) were filtered out in the final analysis. Our study had very limited power to detect variants with small minor allele frequencies so this is unlikely to have adversely affected results.
Connectivity Diagram
SNPs with a p-value < 0.001 in the meta-analyses of AD (N=515 SNPs) and ND (N=386) were selected. Of these, 261 AD and 213 ND SNPs were located in or close to (<10,000 bp) 200 and 165 genes respectively. In total, 169 AD and 143 ND genes/gene-products were found in the IPA database (Ingenuity Systems, release IPA6.0). The Ingenuity database contains information about structure, biological function and subcellular localization of proteins as well as information about protein-protein interactions. The available information was summarized in a connectivity diagram and the gene-products were manually reorganized by grouping the proteins with related structure or function.
RESULTS
Genome-wide Association in the Australian Sample
The lowest p-values for ND, AD and comorbid AD/ND respectively were 4.43×10−8 (rs964170 in ARHGAP10 on chromosome 4), 3.91×10−7 (rs12761801 in CTBP2 on chromosome 10) and 1.91×10−9 (rs7530302 located in a region upstream of MARK1 on chromosome1), with two additional SNPs for comorbid AD/ND (rs1784300 near DDX6 on chromosome 11 and rs12882384 in KIAA1409 on chromosome 14) giving p-values less than 5×10−8. Tables 3, 4, and 5 list detailed results for the 30 SNPs with lowest p-values from the analysis of the ND, AD and comorbid AD/ND DNA pools in the Australian sample.
Table 3.
Thirty Most Significant Association Results From the Analysis of Nicotine Dependence in the Australian DNA-Pools
| Chr | Location (bp) | SNP | Gene | SNP Type | P-value |
|---|---|---|---|---|---|
| 4 | 148915406 | rs964170 | ARHGAP10 | Intron | 4.43 × 10−8 |
| 7 | 86179382 | rs2189816 | GRM3 | Intron | 5.48 × 10−7 |
| 8 | 121997550 | rs10505382 | 9.10 × 10−7 | ||
| 2 | 62611059 | rs1997247 | 9.70 × 10−7 | ||
| 2 | 157381074 | rs11683474 | 1.51 × 10−6 | ||
| 5 | 9981862 | rs1035961 | 1.80 × 10−6 | ||
| 20 | 41771289 | rs285171 | MYBL2 | Intron | 2.76 × 10−6 |
| 1 | 224621971 | rs3219123 | PARP1 | Intron | 6.23 × 10−6 |
| 20 | 3778462 | rs6084496 | VISA | Intron | 6.79 × 10−6 |
| 1 | 218731575 | rs7530302 | 6.87 × 10−6 | ||
| 6 | 132210055 | rs7768555 | ENPP1 | Intron | 8.24 × 10−6 |
| 4 | 62201416 | rs6551640 | LPHN3 | Intron | 9.59 × 10−6 |
| 10 | 74909983 | rs11000671 | PPP3CB | Intron | 1.02 × 10−5 |
| 10 | 115584715 | rs10567 | DCLRE1A | 3′ UTR | 1.12 × 10−5 |
| 8 | 6603162 | rs1046449 | AGPAT5 | 3′ UTR | 1.14 × 10−5 |
| 10 | 18677238 | rs12240995 | CACNB2 | Intron | 1.33 × 10−5 |
| 7 | 141412174 | rs4276595 | MGAM | Intron | 1.58 × 10−5 |
| 11 | 7466142 | rs12805648 | OLFML1 | Coding | 1.79 × 10−5 |
| 17 | 52767460 | rs9890811 | MSI2 | Intron | 1.83 × 10−5 |
| 1 | 219781759 | rs11118748 | 1.84 × 10−5 | ||
| 7 | 153598213 | rs10274025 | NULL | Intron | 1.85 × 10−5 |
| 1 | 244772762 | rs3120701 | TFB2M | Intron | 1.86 × 10−5 |
| 14 | 93055200 | rs12882384 | KIAA1409 | Intron | 1.94 × 10−5 |
| 13 | 34976427 | rs7332116 | 1.98 × 10−5 | ||
| 18 | 19916814 | rs2305024 | C18orf17 | Intron | 2.05 × 10−5 |
| 2 | 62925712 | rs1877026 | EHBP1 | Intron | 2.06 × 10−5 |
| 22 | 33497583 | rs137270 | 2.11 × 10−5 | ||
| 16 | 74809643 | rs2866619 | 2.38 × 10−5 | ||
| 14 | 77004340 | rs3825694 | AHSA1 | Intron | 2.42 × 10−5 |
| 9 | 367066 | rs12348944 | DOCK8 | Coding | 2.80 × 10−5 |
Table 4.
Thirty Most Significant Association Results From the Analysis of Alcohol Dependence in the Australian DNA-Pools
| Chr | Location (bp) | SNP | Gene | SNP Type | P-value |
|---|---|---|---|---|---|
| 10 | 126762971 | rs12761801 | CTBP2 | Intron | 3.91 × 10−7 |
| 12 | 92595143 | rs9668896 | 5.62 × 10−7 | ||
| 11 | 118184839 | rs1784300 | 6.28 × 10−7 | ||
| 12 | 51470992 | rs12312467 | KRT3 | Intron | 1.30 × 10−6 |
| 10 | 94192885 | rs2798253 | 1.93 × 10−6 | ||
| 18 | 67894444 | rs17085505 | 2.18 × 10−6 | ||
| 15 | 27805919 | rs7179270 | TJP1 | Coding | 2.35 × 10−6 |
| 18 | 26365184 | rs8085261 | 3.15 × 10−6 | ||
| 5 | 2544585 | rs462437 | 3.43 × 10−6 | ||
| 6 | 52171060 | rs2154225 | 3.90 × 10−6 | ||
| 9 | 4160457 | rs7851070 | 7.02 × 10−6 | ||
| 2 | 113537285 | rs2472188 | IL1F5 | 3′ UTR | 8.72 × 10−6 |
| 9 | 10851806 | rs2890930 | 9.23 × 10−6 | ||
| 9 | 1990684 | rs10512034 | 1.02 × 10−5 | ||
| 15 | 27788620 | rs17683205 | TJP1 | Intron | 1.07 × 10−5 |
| 4 | 54438003 | rs2102010 | 1.09 × 10−5 | ||
| 21 | 42351204 | rs17114247 | 1.13 × 10−5 | ||
| 4 | 185302289 | rs11132226 | ENPP6 | Intron | 1.16 × 10−5 |
| 9 | 107269811 | rs2149592 | FSD1L | Intron | 1.17 × 10−5 |
| 17 | 59931967 | rs16947824 | DDX5 | Intron | 1.43 × 10−5 |
| 10 | 100789822 | rs2487891 | HPSE2 | Intron | 1.48 × 10−5 |
| 10 | 122831260 | rs10886848 | 1.77 × 10−5 | ||
| 7 | 141339277 | rs6947481 | 1.88 × 10−5 | ||
| 8 | 1588552 | rs17064176 | DLGAP2 | Intron | 2.01 × 10−5 |
| 12 | 31686631 | rs11051507 | 2.07 × 10−5 | ||
| 8 | 103136803 | rs2155235 | 2.10 × 10−5 | ||
| 15 | 27766632 | rs10519663 | 2.25 × 10−5 | ||
| 2 | 199387355 | rs10189905 | 2.29 × 10−5 | ||
| 14 | 28523792 | rs12587874 | 2.30 × 10−5 | ||
| 9 | 132541165 | rs11244096 | PRDM12 | Intron | 2.38 × 10−5 |
Table 5.
Thirty Most Significant Association Results From the Analysis of Comorbid Alcohol and Nicotine Dependence in the Australian DNA-Pools
| Chr | Location (bp) | SNP | Gene | SNP Type | P-value |
|---|---|---|---|---|---|
| 1 | 218731575 | rs7530302 | 1.90 × 10−9 | ||
| 11 | 118184839 | rs1784300 | 2.60 × 10−9 | ||
| 14 | 93055200 | rs12882384 | KIAA1409 | Intron | 4.86 × 10−8 |
| 1 | 218668246 | rs7512221 | 7.44 × 10−7 | ||
| 10 | 126762971 | rs12761801 | CTBP2 | Intron | 7.89 × 10−7 |
| 18 | 3166377 | rs10153321 | MYOM1 | Intron | 8.73 × 10−7 |
| 9 | 124552591 | rs10985760 | OR1L6 | Coding | 8.79 × 10−7 |
| 2 | 45435823 | rs10167668 | 1.12 × 10−6 | ||
| 18 | 54567449 | rs2319974 | MALT1 | 3′ UTR | 1.17 × 10−6 |
| 21 | 42351204 | rs17114247 | 1.31 × 10−6 | ||
| 4 | 148915406 | rs964170 | ARHGAP10 | Intron | 1.49 × 10−6 |
| 2 | 157381074 | rs11683474 | 2.00 × 10−6 | ||
| 21 | 22071121 | rs8131512 | 2.10 × 10−6 | ||
| 12 | 92595143 | rs9668896 | 2.79 × 10−6 | ||
| 4 | 185302289 | rs11132226 | ENPP6 | Intron | 3.08 × 10−6 |
| 15 | 67031734 | rs4365239 | 3.26 × 10−6 | ||
| 8 | 8267771 | rs2921010 | PRAGMIN | Intron | 3.35 × 10−6 |
| 1 | 235038621 | rs7526063 | MTR | Intron | 4.42 × 10−6 |
| 1 | 218692709 | rs7553380 | 5.29 × 10−6 | ||
| 9 | 32855569 | rs6476383 | 5.68 × 10−6 | ||
| 7 | 8767093 | rs994247 | 5.68 × 10−6 | ||
| 1 | 224322577 | rs6664668 | H3F3A | Intron | 6.96 × 10−6 |
| 17 | 59931967 | rs16947824 | DDX5 | Intron | 7.93 × 10−6 |
| 3 | 162707232 | rs7644500 | 9.51 × 10−6 | ||
| 7 | 6118208 | rs12667392 | USP42 | Intron | 9.77 × 10−6 |
| 16 | 82112614 | rs4074375 | CDH13 | Intron | 1.14 × 10−5 |
| 9 | 10851806 | rs2890930 | 1.17 × 10−5 | ||
| 6 | 94996990 | rs9360055 | 1.26 × 10−5 | ||
| 7 | 86179382 | rs2189816 | GRM3 | Intron | 1.29 × 10−5 |
| 10 | 49120681 | rs169439 | FRMPD2 | Intron | 1.30 × 10−5 |
The SNP rs964170 within ARHGAP10, which ranked number one for ND, also ranked number 11 for the comorbid analyses. The SNP rs2189816 within GRM3, which ranked number two for ND (p=5.5×10−7), also ranked number 29 for the comorbid analyses. The SNP rs7530302 upstream of MARK1 which ranked first for comorbid AD/ND also showed suggestive association with ND (p=6.87×10−6) and nominal association with AD (p=3.70×10−4).
Genome-wide Association in Dutch Samples
The top results of the Australian sample (rs964170 for ND and rs12761801 for AD) were not replicated in the Dutch samples. In the Dutch analyses, the lowest p-values were 4.3×10−6 for ND (rs11631180 in TBC1D2B on chromosome 15), 2.3×10−7 for AD in NESDA sample (rs16882302 in BACH2 on chromosome 6) and 1.3×10−6 for AD in NTR sample (rs16953659 on chromosome 16) (see Tables S1-S3 in the Appendix). The results for ND show an association for rs2718771 and rs11771941 (p-values 4.87×10−5 and 4.99×10−5) which are both located in CNTNAP2 on chromosome 7. Interestingly, we also found a SNP (rs851712) in CNTNAP2 associated with AD in the NTR sample (p=2.61×10−6).
Meta-analysis and Connectivity Diagram
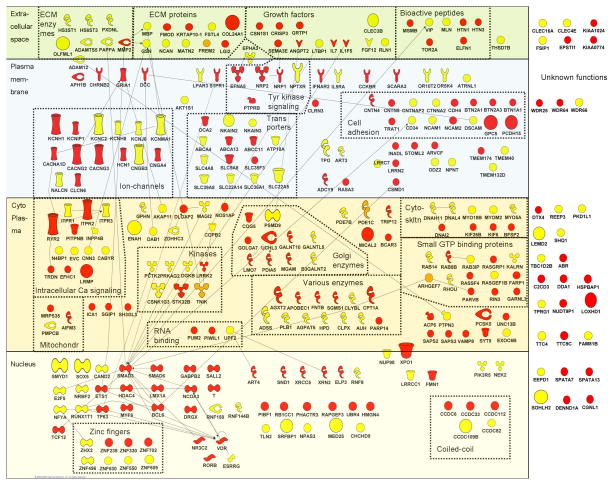
Tables 6 and 7 list detailed results for the 30 most significant SNPs from the meta-analysis of AD and ND, respectively. None of the SNPs reached genome-wide significance in the meta-analysis. To explore whether the marginally significant results contain SNPs from genes that form biological meaningful networks we constructed a connectivity diagram based on information in the Ingenuity database. The gene-products (N=308) were manually grouped based on the related structure or function of their encoded proteins. Figure 1 shows a group of 15 ion-channels, of which 6 contained one or more SNPs with a p-value smaller than 0.0001 in the meta-analyses, including the potassium voltage-gated channels KCNMA1 on chromosome 10 (p=8.91×10−6, rs592676) and KCNC2 on chromosome 12 (p=2.1×10−5, rs17803126) for ND. The Figure also shows a group of cell adhesion molecules. The most significant results are found for PCDH15 on chromosome 15 (p=1.58×10−5, rs1935468) and GPC5 (p=5.26×10−5, rs716623) on chromosome 13, both associated with AD. Other interesting results in this group are DSCAM on chromosome 21 (p=4.99×10−4, rs2837524) and CNTNAP2 (p=1.11×10−4, rs2707592), which are both associated with ND. CNTNAP2 is also ranked second for AD in the NTR sample (p=2.61×10−6, rs851712).
Table 6.
Thirty Most Significant Association Results From the Meta-Analysis of Nicotine Dependence in the Australian and Dutch Samples
| Chr | Location (bp) | SNP | Gene | SNP Type | P-value |
|---|---|---|---|---|---|
| 2 | 1529316 | rs6705087 | 4.80 × 10−6 | ||
| 11 | 7466142 | rs12805648 | OLFML1 | Coding | 1.35 × 10−5 |
| 12 | 120811195 | rs2230681 | PSMD9 | Coding | 1.36 × 10−5 |
| 5 | 39288779 | rs7700754 | 1.89 × 10−5 | ||
| 2 | 1524117 | rs4927632 | TPO | Intron | 2.04 × 10−5 |
| 12 | 73863133 | rs17803126 | KCNC2 | Intron | 2.08 × 10−5 |
| 22 | 37544720 | rs7288826 | NPTXR | 3′ UTR | 2.13 × 10−5 |
| 7 | 36044340 | rs10215376 | 2.53 × 10−5 | ||
| 4 | 10929509 | rs17383790 | 2.58 × 10−5 | ||
| 6 | 124483985 | rs594664 | NKAIN2 | Intron | 2.63 × 10−5 |
| 2 | 102845067 | rs12052617 | 2.92 × 10−5 | ||
| 1 | 223741626 | rs3795443 | ENAH | 3′ UTR | 4.56 × 10−5 |
| 11 | 73830888 | rs12800641 | 4.60 × 10−5 | ||
| 2 | 102927558 | rs11691730 | 4.65 × 10−5 | ||
| 5 | 122949002 | rs1579036 | CSNK1G3 | Intron | 4.88 × 10−5 |
| 2 | 1498071 | rs17732233 | TPO | Intron | 4.97 × 10−5 |
| 8 | 24918626 | rs2950347 | 5.28 × 10−5 | ||
| 7 | 31940499 | rs11770536 | PDE1C | Intron | 5.45 × 10−5 |
| 5 | 131755579 | rs274551 | SLC22A5 | Intron | 5.50 × 10−5 |
| 3 | 172663379 | rs12634193 | 6.05 × 10−5 | ||
| 5 | 121345819 | rs4496732 | SRFBP1 | Intron | 6.35 × 10−5 |
| 4 | 10742243 | rs4697674 | 6.85 × 10−5 | ||
| 12 | 24184569 | rs11047279 | SOX5 | Intron | 7.00 × 10−5 |
| 19 | 55030265 | rs752522 | MED25 | Intron | 7.84 × 10−5 |
| 3 | 45058932 | rs10514712 | 8.07 × 10−5 | ||
| 7 | 21413414 | rs12700284 | 8.44 × 10−5 | ||
| 8 | 87674247 | rs1992405 | CNGB3 | Intron | 8.47 × 10−5 |
| 4 | 110789544 | rs9996730 | CCDC109B | Intron | 8.82 × 10−5 |
| 10 | 78723963 | rs592676 | KCNMA1 | Intron | 8.91 × 10−5 |
| 6 | 33848360 | rs2296744 | LEMD2 | 3′ UTR | 8.99 × 10−5 |
Table 7.
Thirty Most Significant Association Results From the Meta-Analysis of Alcohol Dependence in the Australian and Dutch Samples
| Chr | Location (bp) | SNP | Gene | SNP Type | P-value |
|---|---|---|---|---|---|
| 7 | 82905406 | rs2247219 | SEMA3E | Intron | 6.23 × 10−6 |
| 11 | 105736354 | rs563816 | 7.17 × 10−6 | ||
| 1 | 29907191 | rs187954 | 8.65 × 10−6 | ||
| 10 | 55753731 | rs1935468 | PCDH15 | Intron | 1.58 × 10−5 |
| 1 | 11760686 | rs10864540 | C1orf167 | Intron | 1.72 × 10−5 |
| 11 | 68298116 | rs11228346 | CPT1A | Intron | 1.97 × 10−5 |
| 11 | 105738304 | rs618453 | 2.11 × 10−5 | ||
| 2 | 206348076 | rs3755233 | NRP2 | Intron | 2.13 × 10−5 |
| 13 | 37920191 | rs7328507 | 2.43 × 10−5 | ||
| 6 | 63922941 | rs6454050 | 2.57 × 10−5 | ||
| 8 | 54493239 | rs16919143 | 2.59 × 10−5 | ||
| 7 | 30843167 | rs10229281 | FLJ22374 | Intron | 2.85 × 10−5 |
| 22 | 35340813 | rs6000351 | CACNG2 | Intron | 2.95 × 10−5 |
| 13 | 96586447 | rs2282424 | 2.98 × 10−5 | ||
| 13 | 75069332 | rs4885322 | UCHL3 | Intron | 3.16 × 10−5 |
| 8 | 54529044 | rs10429416 | 3.17 × 10−5 | ||
| 5 | 106936959 | rs6881619 | EFNA5 | Intron | 3.24 × 10−5 |
| 4 | 5132896 | rs6811343 | STK32B | Intron | 3.43 × 10−5 |
| 5 | 35081502 | rs40200 | AGXT2 | Intron | 3.65 × 10−5 |
| 3 | 179187762 | rs6443523 | 3.66 × 10−5 | ||
| 1 | 101497539 | rs7532171 | 3.81 × 10−5 | ||
| 2 | 116787683 | rs10496515 | 3.92 × 10−5 | ||
| 9 | 77787045 | rs4416887 | PCSK5 | Intron | 3.96 × 10−5 |
| 6 | 83427171 | rs376096 | 4.04 × 10−5 | ||
| 17 | 2380774 | rs11655295 | 4.19 × 10−5 | ||
| 4 | 66266501 | rs4629506 | 4.28 × 10−5 | ||
| 1 | 235489612 | rs10495392 | RYR2 | Intron | 4.33 × 10−5 |
| 12 | 25135160 | rs1979522 | LRMP | Intron | 4.38 × 10−5 |
| 11 | 12177958 | rs10831759 | MICAL2 | Intron | 4.48 × 10−5 |
| 12 | 40033695 | rs10785246 | 4.67 × 10−5 |
Figure 1. Connectivity diagram of the proteins encoded by the top genes (SNP with p-value <.001 in or close to gene) from the meta-analyses for AD and ND.
SNPs with a p-value < 0.001 in the meta-analyses of AD (N=515 SNPs) and ND (N=386) were selected. Of these, 261 AD and 213 ND SNPs were located in or within 10,000 bp of 200 and 165 genes, respectively. Those gene names were entered in the IPA database (Ingenuity Systems, release IPA 6.0) and 169 AD and 143 ND genes/gene-products were found in the database (gene names that were not mapped included loci named LOCxxx). We applied a network-based approach that grouped these genes by the biological functions, cellular locations, and possible interactions of their encoded proteins.
The proteins are represented as nodes and are displayed in various shapes that represent the functional class of the gene product (see symbol legend). Nodes were manually reorganized with the ‘pathway designer’ and the location of the nodes is based on the subcellular location of the gene products: extracellular, plasma membrane, cytoplasm, nucleus or unknown localization. Small symbols indicate genes with lowest SNP 0.001 < p-value < 0.0001, large symbols indicate genes with p-value < 0.0001. Depicted in red are the genes for AD, in yellow are those for ND and in orange those that are associated with both AD and ND. The resulting connectivity diagram showed several noteworthy groups of genes belonging to the same functional class (see dotted boxes), of which some are described in more detail in the text.
DISCUSSION
We have used a GWAS strategy in a search for genes involved in AD, ND and comorbid AD/ND in samples from Australia and the Netherlands. Within the Australian data set, there were four genome-wide significant results (p<5×10−8): a SNP in ARHGAP10 for ND; and SNPs in KIAA1409 and near MARK1 and DDX6 for the comorbid analyses. There were no significant associations for AD alone.
The protein coded by ARHGAP10 is a member of the Rho GTPase activating protein (Rho-GAP) family which are negative regulators of Rho-GTPase signaling pathways related to actin cytoskeleton dynamics, cell proliferation, and differentiation (Basseres et al., 2002). The fact that this gene is highly expressed in muscle and brain further supports the hypothesis that ARHGAP10 is important for cell differentiation and might be implicated in neuronal plasticity (Basseres et al., 2002). KIAA1409 has not been extensively studied, but is believed to form part of a cation channel activated by neuropeptides substance P or neurotensin. MARK1 is a member of the MARK family of protein kinases and plays a key role in phosphorylation of microtubules involved in dendritic growth. Variants have been associated with autism spectrum disorders (ASDs) and over-expression of MARK1 in the prefrontal cortex of post-mortem brain tissue of patients with ASDs has been reported (Maussion et al., 2008). DDX6 lies at 121 cM on chromosome 11; this locus may harbor genes associated with smoking-related behavior (Li et al., 2008). We have previously found suggestive linkage for DSM-IV defined nicotine withdrawal at 123 cM (LOD=1.68; (Pergadia et al., 2009)), near this SNP. Others have reported linkage (LOD=1.97) for DSM-IV ND at 109 cM (Gelernter et al., 2007).
Another interesting result is GRM3 (metabotropic glutamate 3 receptor gene) on chromosome 7. In animal models of ND, chronic nicotine exposure was associated with increased activity of mGlu3 receptors (Kenny et al., 2003) and administration of an mGlu2/3 agonist was associated with decreased nicotine consumption, and nicotine self-administration was associated with a downregulation of mGlu2/3 receptors (Liechti et al., 2007). In humans, eight studies have found at least nominal associations with SNPs within GRM3 and schizophrenia (Chen et al., 2005; Egan et al., 2004; Fallin et al., 2005; Fujii et al., 2003; Marti et al., 2002; Mossner et al., 2008; Norton et al., 2005; Tochigi et al., 2006), and it is known that rates of smoking are very high (> 90%) in patients with schizophrenia (de Leon et al., 2002).
These associations in the Australian sample did not replicate in the Dutch samples. Furthermore, a meta-analysis on the Australian and Dutch samples, for AD and ND did not yield SNPs with genomewide significance. Pathway analysis has been proposed as a strategy to deal with highly polygenic traits in which effect sizes of single SNPs may be too low to be detected even in large studies (Wang et al., 2007). A number of open-access or proprietary systems are available to assess whether groups of genes in pathways are over-represented among the top genes in the GWA results. A feature of this approach is that prior knowledge in correctly assigning genes to particular pathways is crucial. Moreover, genes may have diverse functions which are not be reflected in pathway assignment. Vink et al. (2009) used a more liberal approach in which replicated genes were grouped by their biological functions, cellular locations, and possible interactions of their encoded proteins. The gene networks were visualised in a connectivity diagram. Using the same strategy, the top meta-analyses SNPs located in or close to genes were summarized in a connectivity diagram (Figure 1). Inspection of those top-ranked SNPs, in the light of potential gene networks involved in AD or ND, yielded a number of interesting findings. One group consisted of ion-channels, including several potassium voltage-gated ion channels, calcium ion-channels and several other types of ion-channels. KCNMA1 encodes a potassium large conductance calcium-activated channel. It has been postulated as a protein targeted by Lobeline which is a smoking cessation aid (Hu & Agarwal, 2009). In addition, chronic smoking has been shown to down-regulate KCNMA1 protein synthesis and mRNA expression in bronchial and bronchiolar smooth muscles in rats (Ye et al., 2004). KCNMA1 also affects the level of response to alcohol in humans (Schuckit et al., 2005) and C. elegans (Davies et al., 2003). Because KCNMA1 has been implicated in nicotine- and alcohol-related disorders in other studies (Davies et al., 2003; Hu & Agarwal, 2009; Schuckit et al., 2005; Ye et al., 2004) and several potassium voltage-gated ion channels were present in our top-30 results for ND it is likely that these ion channels play a role in the liability for ND and AD. A second group is formed by the genes coding for cell adhesion molecules. Several of these genes were also detected in previous GWAS to addiction phenotypes; PCDH15 (Uhl et al., 2008b), DSCAM (Liu et al., 2006; Uhl et al., 2008a), CNTN5 (Bice et al., 2009), CNTN4 (Bice et al., 2009). CNTNAP2 is a member of the neurexin family which act as cell adhesion molecules and receptors in synaptic signalling. Several neurexins are associated with addictive behaviors (e.g., NRXN3 (Hishimoto et al., 2007; Lachman et al., 2007; Novak et al., 2009)), and CNTNAP2 is previously associated with autism (Alarcon et al., 2008; Arking et al., 2008), schizophrenia (Friedman et al., 2008) and openness to experience (Terracciano et al., 2008).
Compared to the results of a GWA for smoking initiation and current smoking in the Dutch NESDA/NTR sample and 3 replication samples (Uhl et al., 2008a), some gene groups overlapped with the results of the present study (Figure 1), including the cell adhesion proteins DSCAM and CNTN4, transporters ABCA13 and ATP10A, and cytoskeleton proteins MYOM2 and DNAH11. In contrast, the study of Vink et al. (2009) showed a large group of glutamate signaling proteins which was not found in the present study (only GRIA1) whereas the group of genes involved in intracellular calcium signaling identified in the present study was not found by Vink et al. The overlapping groups (and genes) may reflect mechanisms involved in addictive behaviour in general, while the specific groups may reflect mechanisms specific for a phenotype, for example glutamate genes for smoking initiation and intracellular calcium signalling for nicotine dependence.
Our approach to discovery of genes affecting AD and ND has both strengths and limitations. The main strength is that we have taken a joint approach to alcohol and nicotine dependence, which are known to have a partially overlapping genetic basis. This approach should allow the identification of common genes and mechanisms for AD and ND. A main limitation is that, for the Australian sample, a DNA-pooling approach was used. Although DNA-pooling is theoretically sound (Macgregor et al., 2006; Macgregor et al., 2008) and can produce important results at a low genotyping cost (Brown et al., 2008; Melquist et al., 2007; Papassotiropoulos et al., 2006; Spinola et al., 2007; Steer et al., 2007), it was necessary to discount data from SNPs with low minor allele frequency and poor signal-to-noise ratios which might have been captured in an individual-genotyping design. A second limitation is the comparatively small sample size, although we estimate that power would be adequate to detect variants accounting for about 1–2% of variation in liability to AD or ND. Furthermore, we attempted to replicate the Australian results in another population with data available for both alcohol- and nicotine- dependence-related phenotypes. The lack of reproducible results on SNP level reinforces the view that genetic risk of AD or ND arises from multiple polymorphisms of individually small effect. Other published genomewide association studies of AD or ND have similarly found a paucity of large effects (e.g., Bierut et al., 2007; Treutlein et al., 2009). A third limitation of the current meta-analysis was the difference in dependence definitions in the three samples. Nicotine dependence was assessed using DSM-IV criteria for lifetime ND in the Australian sample and for FTND criteria in both Dutch samples. Alcohol dependence was defined as DSM-IV lifetime AD in the Australian and NESDA samples whereas the broader CAGE criteria were used in the NTR sample. A fourth limitation may be that specific genetic effects on AD may be harder to detect as it is much more likely to be comorbid with ND than vice versa. For instance in our general population Australian sample in ever smokers 32% of ND cases have a history of AD, whereas 56% of our AD cases have a history of ND. Also the small number of comorbid cases available within the Dutch samples precluded the comorbid AD+ND+ vs AD-ND- analysis in those samples. A final feature, which could be considered either a strength or a limitation, is recruitment of subjects from the general population rather than a clinical source. The severity of dependence may well be less among a population-based sample, but most alcohol-related problems occur in the large number of people who are only moderately affected; and smoking is also a problem affecting the general community. It should also be noted that this is a study of alcohol and nicotine dependency versus non-dependency. As such, we did not use a group of hypercontrols, but rather allowed AD and ND controls to represent the general population. This is similar to many other GWA studies in which controls are not explicitly sampled from the low tail of the risk distribution. For example, most controls in studies of cardiovascular disease are presumably carrying some atheromatous plaques but haven’t (yet) had the cardiovascular event which leads to the case diagnosis.
In conclusion, we have identified a number of gene networks (ion-channels, cell adhesion molecules) and genes (ARHGAP10, KIAA1409, GRM3) that may play a role in AD, ND or comorbid AD/ND although confirmation in a larger GWAS consortium is clearly needed.
Supplementary Material
Acknowledgments
This study was supported by NIH grants DA12854 (to P.A.F.M.), AA07728, AA07580, AA11998, and AA13321 (to A.C.H.), AA13320 (to R.D.T.), and DA019951 (to M.L.P.); and grants from the Australian National Health and Medical Research Council (496674 to S.M.) and Alcohol and Health Research Grant Scheme (to P.A.L.). We would like to thank the Australian families for their cooperation, and staff for their many contributions. We would also like to pay tribute to the memory of Dr. Richard Todd who was a senior investigator in this collaboration. We acknowledge support from NWO/ZonMW: Genetic determinants of risk behavior in relation to alcohol use and alcohol use disorder: a developmental perspective (311-60-008); Development of alcohol use disorders: the role of clinical, psychological, environmental, genetic and neurobiological factors (311-60-004); Genetic basis of anxiety and depression (904-61-090); Resolving cause and effect in the association between exercise and well-being (904-61-193); Twin-family database for behavior genomics studies (480-04-004); Twin research focusing on behavior (400-05-717), Center for Medical Systems Biology (NWO Genomics); Spinozapremie (SPI 56-464-14192). J.M.V. is financially supported by NWO (VENI 451-06-004). ABS was funded by the Center for Medical Systems Biology (CMSB). We also acknowledge support from: Geestkracht program (10-000-1002); matching funds from universities and mental health care institutes involved in NESDA (GGZ Buitenamstel-Geestgronden, Rivierduinen, University Medical Center Groningen, GGZ Lentis, GGZ Friesland, GGZ Drenthe); Centre for Neurogenomics and Cognitive Research (CNCR-VU). Genotyping was funded by the Genetic Association Information Network (GAIN) of the Foundation for the US National Institutes of Health, and analysis was supported by grants from GAIN and the NIMH (MH081802).
APPENDIX
Supplemental Tables S1–S3 may be found in the online Appendix of this article.
References
- Alarcon M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, Nelson SF, Cantor RM, Geschwind DH. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH, Chakravarti A. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basseres DS, Tizzei EV, Duarte AA, Costa FF, Saad ST. ARHGAP10, a novel human gene coding for a potentially cytoskeletal Rho-GTPase activating protein. Biochem Biophys Res Commun. 2002;294:579–585. doi: 10.1016/S0006-291X(02)00514-4. [DOI] [PubMed] [Google Scholar]
- Bice P, Valdar W, Zhang L, Liu L, Lai D, Grahame N, Flint J, Li TK, Lumeng L, Foroud T. Genomewide SNP screen to detect quantitative trait Loci for alcohol preference in the high alcohol preferring and low alcohol preferring mice. Alcohol Clin Exp Res. 2009;33:531–537. doi: 10.1111/j.1530-0277.2008.00866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, Swan GE, Rutter J, Bertelsen S, Fox L, Fugman D, Goate AM, Hinrichs AL, Konvicka K, Martin NG, Montgomery GW, Saccone NL, Saccone SF, Wang JC, Chase GA, Rice JP, Ballinger DG. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boomsma DI, de Geus EJ, Vink JM, Stubbe JH, Distel MA, Hottenga JJ, Posthuma D, van Beijsterveldt TC, Hudziak JJ, Bartels M, Willemsen G. Netherlands Twin Register: from twins to twin families. Twin Res Hum Genet. 2006;9:849–857. doi: 10.1375/183242706779462426. [DOI] [PubMed] [Google Scholar]
- Boomsma DI, Willemsen G, Sullivan PF, Heutink P, Meijer P, Sondervan D, Kluft C, Smit G, Nolen WA, Zitman FG, Smit JH, Hoogendijk WJ, van Dyck R, de Geus EJ, Penninx BW. Genome-wide association of major depression: description of samples for the GAIN Major Depressive Disorder Study: NTR and NESDA biobank projects. Eur J Hum Genet. 2008;16:335–342. doi: 10.1038/sj.ejhg.5201979. [DOI] [PubMed] [Google Scholar]
- Brown KM, Macgregor S, Montgomery GW, Craig DW, Zhao ZZ, Iyadurai K, Henders AK, Homer N, Campbell MJ, Stark M, Thomas S, Schmid H, Holland EA, Gillanders EM, Duffy DL, Maskiell JA, Jetann J, Ferguson M, Stephan DA, Cust AE, Whiteman D, Green A, Olsson H, Puig S, Ghiorzo P, Hansson J, Demenais F, Goldstein AM, Gruis NA, Elder DE, Bishop JN, Kefford RF, Giles GG, Armstrong BK, Aitken JF, Hopper JL, Martin NG, Trent JM, Mann GJ, Hayward NK. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat Genet. 2008;40:838–840. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, Reich T, Schmidt I, Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Butt CM, Hutton SR, Stitzel JA, Balogh SA, Owens JC, Collins AC. A polymorphism in the alpha4 nicotinic receptor gene (Chrna4) modulates enhancement of nicotinic receptor function by ethanol. Alcohol Clin Exp Res. 2003;27:733–742. doi: 10.1097/01.ALC.0000067973.41153.BC. [DOI] [PubMed] [Google Scholar]
- Butt CM, King NM, Stitzel JA, Collins AC. Interaction of the nicotinic cholinergic system with ethanol withdrawal. J Pharmacol Exp Ther. 2004;308:591–599. doi: 10.1124/jpet.103.059758. [DOI] [PubMed] [Google Scholar]
- Caporaso N, Gu F, Chatterjee N, Sheng-Chih J, Yu K, Yeager M, Chen C, Jacobs K, Wheeler W, Landi MT, Ziegler RG, Hunter DJ, Chanock S, Hankinson S, Kraft P, Bergen AW. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS ONE. 2009;4:e4653. doi: 10.1371/journal.pone.0004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, He G, Wu S, Xu Y, Feng G, Li Y, Wang L, He L. A case-control study of the relationship between the metabotropic glutamate receptor 3 gene and schizophrenia in the Chinese population. Schizophr Res. 2005;73:21–26. doi: 10.1016/j.schres.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Cottler LB, Robins LN, Grant BF, Blaine J, Towle LH, Wittchen HU, Sartorius N. The CIDI-core substance abuse and dependence questions: cross-cultural and nosological issues. The WHO/ADAMHA Field Trial. Br J Psychiatry. 1991;159:653–658. doi: 10.1192/bjp.159.5.653. [DOI] [PubMed] [Google Scholar]
- Davies AG, Pierce-Shimomura JT, Kim H, VanHoven MK, Thiele TR, Bonci A, Bargmann CI, McIntire SL. A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell. 2003;115:655–666. doi: 10.1016/s0092-8674(03)00979-6. [DOI] [PubMed] [Google Scholar]
- de Leon J, Diaz FJ, Rogers T, Browne D, Dinsmore L. Initiation of daily smoking and nicotine dependence in schizophrenia and mood disorders. Schizophr Res. 2002;56:47–54. doi: 10.1016/s0920-9964(01)00217-1. [DOI] [PubMed] [Google Scholar]
- Drgon T, Montoya I, Johnson C, Liu QR, Walther D, Hamer D, Uhl GR. Genome-wide association for nicotine dependence and smoking cessation success in NIH research volunteers. Mol Med. 2009;15:21–27. doi: 10.2119/molmed.2008.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Straub RE, Goldberg TE, Yakub I, Callicott JH, Hariri AR, Mattay VS, Bertolino A, Hyde TM, Shannon-Weickert C, Akil M, Crook J, Vakkalanka RK, Balkissoon R, Gibbs RA, Kleinman JE, Weinberger DR. Variation in GRM3 affects cognition, prefrontal glutamate, and risk for schizophrenia. Proc Natl Acad Sci U S A. 2004;101:12604–12609. doi: 10.1073/pnas.0405077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing JA. Detecting alcoholism. The CAGE questionnaire. Jama. 1984;252:1905–1907. doi: 10.1001/jama.252.14.1905. [DOI] [PubMed] [Google Scholar]
- Fallin MD, Lasseter VK, Avramopoulos D, Nicodemus KK, Wolyniec PS, McGrath JA, Steel G, Nestadt G, Liang KY, Huganir RL, Valle D, Pulver AE. Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet. 2005;77:918–936. doi: 10.1086/497703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JI, Vrijenhoek T, Markx S, Janssen IM, van der Vliet WA, Faas BH, Knoers NV, Cahn W, Kahn RS, Edelmann L, Davis KL, Silverman JM, Brunner HG, van Kessel AG, Wijmenga C, Ophoff RA, Veltman JA. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol Psychiatry. 2008;13:261–266. doi: 10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- Fujii Y, Shibata H, Kikuta R, Makino C, Tani A, Hirata N, Shibata A, Ninomiya H, Tashiro N, Fukumaki Y. Positive associations of polymorphisms in the metabotropic glutamate receptor type 3 gene (GRM3) with schizophrenia. Psychiatr Genet. 2003;13:71–76. doi: 10.1097/01.ypg.0000056682.82896.b0. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR. Genetics of alcohol dependence. Hum Genet. 2009;126:91–99. doi: 10.1007/s00439-009-0701-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Weiss R, Brady K, Poling J, Krauthammer M, Farrer L, Kranzler HR. Genomewide linkage scan for nicotine dependence: identification of a chromosome 5 risk locus. Biol Psychiatry. 2007;61:119–126. doi: 10.1016/j.biopsych.2006.08.023. [DOI] [PubMed] [Google Scholar]
- Grant BF, Hasin DS, Chou SP, Stinson FS, Dawson DA. Nicotine dependence and psychiatric disorders in the United States: results from the national epidemiologic survey on alcohol and related conditions. Arch Gen Psychiatry. 2004;61:1107–1115. doi: 10.1001/archpsyc.61.11.1107. [DOI] [PubMed] [Google Scholar]
- Hansell NK, Agrawal A, Whitfield JB, Morley KI, Zhu G, Lind PA, Pergadia ML, Madden PA, Todd RD, Heath AC, Martin NG. Long-term stability and heritability of telephone interview measures of alcohol consumption and dependence. Twin Res Hum Genet. 2008;11:287–305. doi: 10.1375/twin.11.3.287. [DOI] [PubMed] [Google Scholar]
- Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- Hettema JM, Corey LA, Kendler KS. A multivariate genetic analysis of the use of tobacco, alcohol, and caffeine in a population based sample of male and female twins. Drug Alcohol Depend. 1999;57:69–78. doi: 10.1016/s0376-8716(99)00053-8. [DOI] [PubMed] [Google Scholar]
- Hishimoto A, Liu QR, Drgon T, Pletnikova O, Walther D, Zhu XG, Troncoso JC, Uhl GR. Neurexin 3 polymorphisms are associated with alcohol dependence and altered expression of specific isoforms. Hum Mol Genet. 2007;16:2880–2891. doi: 10.1093/hmg/ddm247. [DOI] [PubMed] [Google Scholar]
- Hu G, Agarwal P. Human disease-drug network based on genomic expression profiles. PLoS ONE. 2009;4:e6536. doi: 10.1371/journal.pone.0006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt RD, Offord KP, Croghan IT, Gomez-Dahl L, Kottke TE, Morse RM, Melton LJ., 3rd Mortality following inpatient addictions treatment. Role of tobacco use in a community-based cohort. Jama. 1996;275:1097–1103. doi: 10.1001/jama.275.14.1097. [DOI] [PubMed] [Google Scholar]
- Johnson C, Drgon T, Liu QR, Walther D, Edenberg H, Rice J, Foroud T, Uhl GR. Pooled association genome scanning for alcohol dependence using 104,268 SNPs: validation and use to identify alcoholism vulnerability loci in unrelated individuals from the collaborative study on the genetics of alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2006;141:844–853. doi: 10.1002/ajmg.b.30346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny PJ, Gasparini F, Markou A. Group II metabotropic and alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate (AMPA)/kainate glutamate receptors regulate the deficit in brain reward function associated with nicotine withdrawal in rats. J Pharmacol Exp Ther. 2003;306:1068–1076. doi: 10.1124/jpet.103.052027. [DOI] [PubMed] [Google Scholar]
- Knopik VS, Heath AC, Madden PA, Bucholz KK, Slutske WS, Nelson EC, Statham D, Whitfield JB, Martin NG. Genetic effects on alcohol dependence risk: re-evaluating the importance of psychiatric and other heritable risk factors. Psychol Med. 2004;34:1519–1530. doi: 10.1017/s0033291704002922. [DOI] [PubMed] [Google Scholar]
- Koopmans JR, van Doornen LJ, Boomsma DI. Association between alcohol use and smoking in adolescent and young adult twins: a bivariate genetic analysis. Alcohol Clin Exp Res. 1997;21:537–546. [PubMed] [Google Scholar]
- Kuper H, Tzonou A, Kaklamani E, Hsieh CC, Lagiou P, Adami HO, Trichopoulos D, Stuver SO. Tobacco smoking, alcohol consumption and their interaction in the causation of hepatocellular carcinoma. Int J Cancer. 2000;85:498–502. [PubMed] [Google Scholar]
- Lachman HM, Fann CS, Bartzis M, Evgrafov OV, Rosenthal RN, Nunes EV, Miner C, Santana M, Gaffney J, Riddick A, Hsu CL, Knowles JA. Genomewide suggestive linkage of opioid dependence to chromosome 14q. Hum Mol Genet. 2007;16:1327–1334. doi: 10.1093/hmg/ddm081. [DOI] [PubMed] [Google Scholar]
- Lessov CN, Martin NG, Statham DJ, Todorov AA, Slutske WS, Bucholz KK, Heath AC, Madden PA. Defining nicotine dependence for genetic research: evidence from Australian twins. Psychol Med. 2004;34:865–879. doi: 10.1017/s0033291703001582. [DOI] [PubMed] [Google Scholar]
- Li CY, Mao X, Wei L. Genes and (common) pathways underlying drug addiction. PLoS Comput Biol. 2008;4:e2. doi: 10.1371/journal.pcbi.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti ME, Lhuillier L, Kaupmann K, Markou A. Metabotropic glutamate 2/3 receptors in the ventral tegmental area and the nucleus accumbens shell are involved in behaviors relating to nicotine dependence. J Neurosci. 2007;27:9077–9085. doi: 10.1523/JNEUROSCI.1766-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QR, Drgon T, Johnson C, Walther D, Hess J, Uhl GR. Addiction molecular genetics: 639,401 SNP whole genome association identifies many “cell adhesion” genes. Am J Med Genet B Neuropsychiatr Genet. 2006;141:918–925. doi: 10.1002/ajmg.b.30436. [DOI] [PubMed] [Google Scholar]
- Liu YZ, Pei YF, Guo YF, Wang L, Liu XG, Yan H, Xiong DH, Zhang YP, Levy S, Li J, Haddock CK, Papasian CJ, Xu Q, Ma JZ, Payne TJ, Recker RR, Li MD, Deng HW. Genome-wide association analyses suggested a novel mechanism for smoking behavior regulated by IL15. Mol Psychiatry. 2009 doi: 10.1038/mp.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukola A, Broms U, Maunu H, Widen E, Heikkila K, Siivola M, Salo A, Pergadia ML, Nyman E, Sammalisto S, Perola M, Agrawal A, Heath AC, Martin NG, Madden PA, Peltonen L, Kaprio J. Linkage of nicotine dependence and smoking behavior on 10q, 7q and 11p in twins with homogeneous genetic background. Pharmacogenomics J. 2008;8:209–219. doi: 10.1038/sj.tpj.6500464. [DOI] [PubMed] [Google Scholar]
- Macgregor S, Visscher PM, Montgomery G. Analysis of pooled DNA samples on high density arrays without prior knowledge of differential hybridization rates. Nucleic Acids Res. 2006;34:e55. doi: 10.1093/nar/gkl136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor S, Zhao ZZ, Henders A, Nicholas MG, Montgomery GW, Visscher PM. Highly cost-efficient genome-wide association studies using DNA pools and dense SNP arrays. Nucleic Acids Res. 2008;36:e35. doi: 10.1093/nar/gkm1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden PA, Bucholz KK, Martin NG, Heath AC. Smoking and the genetic contribution to alcohol-dependence risk. Alcohol Res Health. 2000;24:209–214. [PMC free article] [PubMed] [Google Scholar]
- Madden PA, Heath AC. Shared genetic vulnerability in alcohol and cigarette use and dependence. Alcohol Clin Exp Res. 2002;26:1919–1921. doi: 10.1097/01.ALC.0000040960.15151.30. [DOI] [PubMed] [Google Scholar]
- Madden PA, Heath AC, Martin NG. Smoking and intoxication after alcohol challenge in women and men: genetic influences. Alcohol Clin Exp Res. 1997;21:1732–1741. [PubMed] [Google Scholar]
- Madden PA, Heath AC, Pedersen NL, Kaprio J, Koskenvuo MJ, Martin NG. The genetics of smoking persistence in men and women: a multicultural study. Behav Genet. 1999;29:423–431. doi: 10.1023/a:1021674804714. [DOI] [PubMed] [Google Scholar]
- Madden PA, Pedersen NL, Kaprio J, Koskenvuo MJ, Martin NG. The epidemiology and genetics of smoking initiation and persistence: crosscultural comparisons of twin study results. Twin Res. 2004;7:82–97. doi: 10.1375/13690520460741471. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Marti SB, Cichon S, Propping P, Nothen M. Metabotropic glutamate receptor 3 (GRM3) gene variation is not associated with schizophrenia or bipolar affective disorder in the German population. Am J Med Genet. 2002;114:46–50. doi: 10.1002/ajmg.1624. [DOI] [PubMed] [Google Scholar]
- Maussion G, Carayol J, Lepagnol-Bestel AM, Tores F, Loe-Mie Y, Milbreta U, Rousseau F, Fontaine K, Renaud J, Moalic JM, Philippi A, Chedotal A, Gorwood P, Ramoz N, Hager J, Simonneau M. Convergent evidence identifying MAP/microtubule affinity-regulating kinase 1 (MARK1) as a susceptibility gene for autism. Hum Mol Genet. 2008;17:2541–2551. doi: 10.1093/hmg/ddn154. [DOI] [PubMed] [Google Scholar]
- Melquist S, Craig DW, Huentelman MJ, Crook R, Pearson JV, Baker M, Zismann VL, Gass J, Adamson J, Szelinger S, Corneveaux J, Cannon A, Coon KD, Lincoln S, Adler C, Tuite P, Calne DB, Bigio EH, Uitti RJ, Wszolek ZK, Golbe LI, Caselli RJ, Graff-Radford N, Litvan I, Farrer MJ, Dickson DW, Hutton M, Stephan DA. Identification of a novel risk locus for progressive supranuclear palsy by a pooled genomewide scan of 500,288 single-nucleotide polymorphisms. Am J Hum Genet. 2007;80:769–778. doi: 10.1086/513320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossner R, Schuhmacher A, Schulze-Rauschenbach S, Kuhn KU, Rujescu D, Rietschel M, Zobel A, Franke P, Wolwer W, Gaebel W, Hafner H, Wagner M, Maier W. Further evidence for a functional role of the glutamate receptor gene GRM3 in schizophrenia. Eur Neuropsychopharmacol. 2008;18:768–772. doi: 10.1016/j.euroneuro.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Norton N, Williams HJ, Dwyer S, Ivanov D, Preece AC, Gerrish A, Williams NM, Yerassimou P, Zammit S, O’Donovan MC, Owen MJ. No evidence for association between polymorphisms in GRM3 and schizophrenia. BMC Psychiatry. 2005;5:23. doi: 10.1186/1471-244X-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak G, Boukhadra J, Shaikh SA, Kennedy JL, Le Foll B. Association of a polymorphism in the NRXN3 gene with the degree of smoking in schizophrenia: A preliminary study. World J Biol Psychiatry. 2009:1–7. doi: 10.1080/15622970903079499. [DOI] [PubMed] [Google Scholar]
- Owens JC, Balogh SA, McClure-Begley TD, Butt CM, Labarca C, Lester HA, Picciotto MR, Wehner JM, Collins AC. Alpha4beta2* nicotinic acetylcholine receptors modulate the effects of ethanol and nicotine on the acoustic startle response. Alcohol Clin Exp Res. 2003;27:1867–1875. doi: 10.1097/01.ALC.0000102700.72447.0F. [DOI] [PubMed] [Google Scholar]
- Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, Huynh KD, Brunner F, Corneveaux J, Osborne D, Wollmer MA, Aerni A, Coluccia D, Hanggi J, Mondadori CR, Buchmann A, Reiman EM, Caselli RJ, Henke K, de Quervain DJ. Common Kibra alleles are associated with human memory performance. Science. 2006;314:475–478. doi: 10.1126/science.1129837. [DOI] [PubMed] [Google Scholar]
- Penninx BW, Beekman AT, Smit JH, Zitman FG, Nolen WA, Spinhoven P, Cuijpers P, De Jong PJ, Van Marwijk HW, Assendelft WJ, Van Der Meer K, Verhaak P, Wensing M, De Graaf R, Hoogendijk WJ, Ormel J, Van Dyck R. The Netherlands Study of Depression and Anxiety (NESDA): rationale, objectives and methods. Int J Methods Psychiatr Res. 2008;17:121–140. doi: 10.1002/mpr.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergadia ML, Agrawal A, Loukola A, Montgomery GW, Broms U, Saccone SF, Wang JC, Todorov AA, Heikkila K, Statham DJ, Henders AK, Campbell MJ, Rice JP, Todd RD, Heath AC, Goate AM, Peltonen L, Kaprio J, Martin NG, Madden PA. Genetic linkage findings for DSM-IV nicotine withdrawal in two populations. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:950–959. doi: 10.1002/ajmg.b.30924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Rosenberger A, Janicke N, Kohler K, Korb K, Kulle B, Bickeboller H. Surrogate phenotype definition for alcohol use disorders: a genome-wide search for linkage and association. BMC Genet. 2005;6(Suppl 1):S55. doi: 10.1186/1471-2156-6-S1-S55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Pergadia ML, Loukola A, Broms U, Montgomery GW, Wang JC, Agrawal A, Dick DM, Heath AC, Todorov AA, Maunu H, Heikkila K, Morley KI, Rice JP, Todd RD, Kaprio J, Peltonen L, Martin NG, Goate AM, Madden PA. Genetic linkage to chromosome 22q12 for a heavy-smoking quantitative trait in two independent samples. Am J Hum Genet. 2007;80:856–866. doi: 10.1086/513703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Saccone NL, Swan GE, Madden PA, Goate AM, Rice JP, Bierut LJ. Systematic biological prioritization after a genome-wide association study: an application to nicotine dependence. Bioinformatics. 2008;24:1805–1811. doi: 10.1093/bioinformatics/btn315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Wilhelmsen K, Smith TL, Feiler HS, Lind P, Lange LA, Kalmijn J. Autosomal linkage analysis for the level of response to alcohol. Alcohol Clin Exp Res. 2005;29:1976–1982. doi: 10.1097/01.alc.0000187598.82921.27. [DOI] [PubMed] [Google Scholar]
- Sham P, Bader JS, Craig I, O’Donovan M, Owen M. DNA Pooling: a tool for large-scale association studies. Nat Rev Genet. 2002;3:862–871. doi: 10.1038/nrg930. [DOI] [PubMed] [Google Scholar]
- Spinola M, Leoni VP, Galvan A, Korsching E, Conti B, Pastorino U, Ravagnani F, Columbano A, Skaug V, Haugen A, Dragani TA. Genome-wide single nucleotide polymorphism analysis of lung cancer risk detects the KLF6 gene. Cancer Lett. 2007;251:311–316. doi: 10.1016/j.canlet.2006.11.029. [DOI] [PubMed] [Google Scholar]
- Steer S, Abkevich V, Gutin A, Cordell HJ, Gendall KL, Merriman ME, Rodger RA, Rowley KA, Chapman P, Gow P, Harrison AA, Highton J, Jones PB, O’Donnell J, Stamp L, Fitzgerald L, Iliev D, Kouzmine A, Tran T, Skolnick MH, Timms KM, Lanchbury JS, Merriman TR. Genomic DNA pooling for whole-genome association scans in complex disease: empirical demonstration of efficacy in rheumatoid arthritis. Genes Immun. 2007;8:57–68. doi: 10.1038/sj.gene.6364359. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, de Geus EJ, Willemsen G, James MR, Smit JH, Zandbelt T, Arolt V, Baune BT, Blackwood D, Cichon S, Coventry WL, Domschke K, Farmer A, Fava M, Gordon SD, He Q, Heath AC, Heutink P, Holsboer F, Hoogendijk WJ, Hottenga JJ, Hu Y, Kohli M, Lin D, Lucae S, Macintyre DJ, Maier W, McGhee KA, McGuffin P, Montgomery GW, Muir WJ, Nolen WA, Nothen MM, Perlis RH, Pirlo K, Posthuma D, Rietschel M, Rizzu P, Schosser A, Smit AB, Smoller JW, Tzeng JY, van Dyck R, Verhage M, Zitman FG, Martin NG, Wray NR, Boomsma DI, Penninx BW. Genome-wide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Mol Psychiatry. 2009;14:359–375. doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano A, Sanna S, Uda M, Deiana B, Usala G, Busonero F, Maschio A, Scally M, Patriciu N, Chen WM, Distel MA, Slagboom EP, Boomsma DI, Villafuerte S, Sliwerska E, Burmeister M, Amin N, Janssens AC, van Duijn CM, Schlessinger D, Abecasis GR, Costa PT., Jr Genome-wide association scan for five major dimensions of personality. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, Stacey SN, Bergthorsson JT, Thorlacius S, Gudmundsson J, Jonsson T, Jakobsdottir M, Saemundsdottir J, Olafsdottir O, Gudmundsson LJ, Bjornsdottir G, Kristjansson K, Skuladottir H, Isaksson HJ, Gudbjartsson T, Jones GT, Mueller T, Gottsater A, Flex A, Aben KK, de Vegt F, Mulders PF, Isla D, Vidal MJ, Asin L, Saez B, Murillo L, Blondal T, Kolbeinsson H, Stefansson JG, Hansdottir I, Runarsdottir V, Pola R, Lindblad B, van Rij AM, Dieplinger B, Haltmayer M, Mayordomo JI, Kiemeney LA, Matthiasson SE, Oskarsson H, Tyrfingsson T, Gudbjartsson DF, Gulcher JR, Jonsson S, Thorsteinsdottir U, Kong A, Stefansson K. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochigi M, Suga M, Ohashi J, Otowa T, Yamasue H, Kasai K, Kato T, Okazaki Y, Kato N, Sasaki T. No association between the metabotropic glutamate receptor type 3 gene (GRM3) and schizophrenia in a Japanese population. Schizophr Res. 2006;88:260–264. doi: 10.1016/j.schres.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, Fehr C, Scherbaum N, Steffens M, Ludwig KU, Frank J, Wichmann HE, Schreiber S, Dragano N, Sommer WH, Leonardi-Essmann F, Lourdusamy A, Gebicke-Haerter P, Wienker TF, Sullivan PF, Nothen MM, Kiefer F, Spanagel R, Mann K, Rietschel M. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True WR, Xian H, Scherrer JF, Madden PA, Bucholz KK, Heath AC, Eisen SA, Lyons MJ, Goldberg J, Tsuang M. Common genetic vulnerability for nicotine and alcohol dependence in men. Arch Gen Psychiatry. 1999;56:655–661. doi: 10.1001/archpsyc.56.7.655. [DOI] [PubMed] [Google Scholar]
- Uhl GR, Drgon T, Johnson C, Li CY, Contoreggi C, Hess J, Naiman D, Liu QR. Molecular genetics of addiction and related heritable phenotypes: genome-wide association approaches identify “connectivity constellation” and drug target genes with pleiotropic effects. Ann N Y Acad Sci. 2008a;1141:318–381. doi: 10.1196/annals.1441.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE. Molecular genetics of nicotine dependence and abstinence: whole genome association using 520,000 SNPs. BMC Genet. 2007;8:10. doi: 10.1186/1471-2156-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE, David SP, Niaura R, Lerman C. Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Arch Gen Psychiatry. 2008b;65:683–693. doi: 10.1001/archpsyc.65.6.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink JM, Smit AB, de Geus EJ, Sullivan P, Willemsen G, Hottenga JJ, Smit JH, Hoogendijk WJ, Zitman FG, Peltonen L, Kaprio J, Pedersen NL, Magnusson PK, Spector TD, Kyvik KO, Morley KI, Heath AC, Martin NG, Westendorp RG, Slagboom PE, Tiemeier H, Hofman A, Uitterlinden AG, Aulchenko YS, Amin N, van Duijn C, Penninx BW, Boomsma DI. Genome-wide association study of smoking initiation and current smoking. Am J Hum Genet. 2009;84:367–379. doi: 10.1016/j.ajhg.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink JM, Willemsen G, Boomsma DI. Heritability of smoking initiation and nicotine dependence. Behav Genet. 2005;35:397–406. doi: 10.1007/s10519-004-1327-8. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Bucan M. Pathway-Based Approaches for Analysis of Genomewide Association Studies. Am J Hum Genet. 2007:81. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Ma WL, Yang ML, Liu SY, Wang DX. Effect of chronic cigarette smoking on large-conductance calcium-activated potassium channel and Kv1.5 expression in bronchial smooth muscle cells of rats. Sheng Li Xue Bao. 2004;56:573–578. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.