SUMMARY
Plants have evolved sophisticated surveillance systems to recognize pathogen effectors delivered into host cells. RPM1 is an NB-LRR immune receptor that recognizes the Pseudomonas syringae effectors AvrB and AvrRpm1. Both effectors associate with and affect the phosphorylation of RIN4, an immune regulator. Although the kinase and the specific mechanisms involved are unclear, it has been hypothesized that RPM1 recognizes phosphorylated RIN4. Here we identify RIPK as a RIN4-interacting receptor-like protein kinase that phosphorylates RIN4. In response to bacterial effectors, RIPK phosphorylates RIN4 at amino acid residues T21, S160, and T166. RIN4 phosphomimetic mutants display constitutive activation of RPM1-mediated defense responses and RIN4 phosphorylation is induced by AvrB and AvrRpm1 during P. syringae infection. RIPK knockout lines exhibit reduced RIN4 phosphorylation and blunted RPM1-mediated defense responses. Taken together, our results demonstrate that the RIPK kinase associates with and modifies an effector-targeted protein complex to initiate host immunity.
INTRODUCTION
Plants are continuously exposed to pathogenic microorganisms. Plants lack circulating immune cells and do not possess adaptive immunity. As a result, each plant cell has the capability to recognize non-self molecules through a large number of plant innate immune receptors. Plant pattern recognition receptors displayed on the plasma membrane recognize conserved pathogen associated molecular patterns (PAMPs), such as bacterial flagellin (Zipfel et al., 2004), resulting in PAMP-triggered immunity (PTI) (Chisholm et al., 2006). These PTI receptors are structurally similar to animal toll-like receptors (Vance et al., 2009).
PTI is thought to limit initial pathogen invasion and multiplication. In order to cause disease, virulent microorganisms must interfere with plant immune perception. Many plant pathogenic bacteria use their type three secretion system (T3SS) to deliver a large number of effector proteins (20–40) into host cells to promote pathogenesis (Cui et al., 2009). Effectors can suppress immune responses by targeting immune receptors as well as downstream signaling components (Cui et al., 2009).
As a second line of defense, plants possess intracellular nucleotide-binding domain, leucine-rich repeat containing (NB-LRR) immune receptors that specifically recognize pathogen effectors delivered into plant cells during infection. Plant NB-LRR immune receptors are structurally analogous to animal NLR (nucleotide-binding domain, leucine-rich repeat containing) receptors (Ting et al., 2008). NB-LRR activation results in effector-triggered immunity (ETI). Plant NB-LRRs can be further subdivided into two classes: proteins that contain a Toll/Interleukin-1 receptor-like region and those that contain a coiled-coil region near their N-termini. The distinct N-terminal domains of plant NB-LRR proteins influence the requirement for downstream signaling components (Feys and Parker, 2000). Individual NB-LRRs can recognize effectors from all pathogen classes. There are ∼150 NB-LRRs in Arabidopsis, which would seem to be too few to efficiently recognize the multitude of pathogen effectors encountered. This has led to the guard hypothesis which states that plant NB-LRRs recognize pathogens by monitoring (or guarding) for effector-mediated perturbations of a conserved host targets (Jones and Dangl, 2006). Investigating the plant protein RIN4 has provided evidence supporting this hypothesis. RIN4 is an Arabidopsis protein that can act to regulate both ETI and PTI (Widjaja et al., 2009; Kim et al., 2005a; Liu et al., 2009; Mackey et al., 2002). Not surprisingly, multiple Pseudomonas syringae effectors with the ability to suppress PTI target RIN4 (Axtell et al., 2003; Coaker et al., 2005; Luo et al., 2009; Mackey et al., 2002; Wilton et al., 2010). RIN4 overexpression and knockout lines exhibit decreased and increased PTI-based responses, respectively, indicating that genetically RIN4 is a negative regulator of immune responses (Kim et al., 2005b). RIN4 is also monitored by two Arabidopsis NB-LRR immune receptors, RPM1 and RPS2 (Bent et al., 1994; Grant et al., 1995). The P. syringae effector AvrRpt2 is a protease that directly targets RIN4 (Axtell et al., 2003; Coaker et al., 2005). RIN4 cleavage results in RPS2-mediated ETI (Axtell and Staskawicz, 2003; Mackey et al., 2003). RPM1 recognizes the unrelated P. syringae effectors AvrB and AvrRpm1. Both effectors associate with RIN4 and induce RIN4 phosphorylation (Mackey et al., 2002).
Pathogen perception by plant immune receptors leads to massive transcriptional reprogramming of the host cell, directing expression towards defense. A hallmark of ETI-based responses is the hypersensitive response (HR), a form of programmed cell death at the site of infection (Chisholm et al., 2006). Prior to transcriptional reprogramming, protein kinases act to relay the signals of non-self perception. MAP kinase cascades downstream of receptor activation can act as both positive and negative regulators of plant defense (Rodriguez et al., 2010). Recent studies also highlight the important of plant receptor-like cytoplasmic kinases (RLCKs) in mediating both PTI and ETI. The RLCK BIK1 has been found to play an important role in PTI signaling through its interaction with and phosphorylation of the flagellin PAMP receptor FLS2 (Lu et al., 2010; Zhang et al., 2010). In tomato, the RLCK Pto can directly interact with the P. syringae effectors AvrPto and AvrPtoB and this interaction is necessary for ETI (Kim et al., 2002). In Arabidopsis, the PBS1 RLCK is targeted by the P. syringae AvrPphB effector protease (Shao et al., 2003). AvrPphB-mediated cleavage of PBS1 is recognized by the RPS5 NB-LRR immune receptor (Shao et al., 2003). Taken together, these results highlight the importance of plant RLCKs as central players in mediating both PAMP and effector-triggered immune responses.
In this study, we investigated RPM1-induced ETI by purifying and identifying the phosphorylated RIN4 complex in Arabidopsis. One of the identified proteins, RPM1 induced protein kinase (RIPK, At2g05940), belongs to the receptor-like cytoplasmic kinase (RLCK) family. RIPK can directly phosphorylate RIN4 in vitro and RIN4 phosphorylation is induced by the bacterial effectors AvrB and AvrRpm1 in planta. Phosphorylation of RIN4 on amino acid residue T166 is sufficient to induce RPM1 activation. RIPK can also phosphorylate and interact with AvrB. We hypothesize that AvrB enhances RIPK phosphorylation activity or substrate specificity, resulting in increased RIN4 phosphorylation. These results demonstrate that the NB-LRR immune receptor RPM1 monitors for RIN4’s phosphorylation status and RIN4 phosphorylation is indirectly triggered by the AvrB and AvrRpm1 effectors through RIPK and related kinases.
RESULTS
Purification and Identification of RIPK
Previously, we purified members of the RIN4 protein complex in the absence of pathogen stimulus (Liu et al., 2009). In order to investigate dynamic changes in RIN4’s interaction with other proteins during defense signaling, we immunoprecipitated and identified proteins that can interact with RIN4 in transgenic plants expressing Dexamethasone (Dex) inducible avrRpm1. After Dex application, RIN4 is rapidly phosphorylated within 2–3h. We used whole leaf protein extracts to purify RIN4 and associated proteins after Dex treatment across three biological replicates. The rpm1/rps2/rin4 genetic background was used as a negative control. We were able to identify RIN4 across all three biological replications (Table S1). In order to be characterized as a RIN4 associated protein, the proteins identified by mass spectrometry needed to be identified by at least two unique peptides, localized to the plasma membrane or cytosol, present in at least two biological replicates for the positive control, and absent or highly reduced in the negative control. Two proteins met these criteria: the RLCK RIPK (At2g05940) and clathrin (At3g08530), a protein that plays a major role in coated vesicles for endo- and exo-cytosis. RIPK was previously identified in a cDNA screen as upregulated during RPM1 defense responses, but was never genetically or biochemically characterized (de Torres et al., 2003). A large number of contaminating chloroplast proteins were identified by mass spectrometry, which may be due to the onset of the hypersensitive response in Dex∷avrRpm1 lines and subsequent organelle disruption. This dataset is substantially different than the proteins associated with RIN4 in the absence of pathogen stimulus (Liu et al., 2009), indicating that RIN4 interacts with different proteins during ETI.
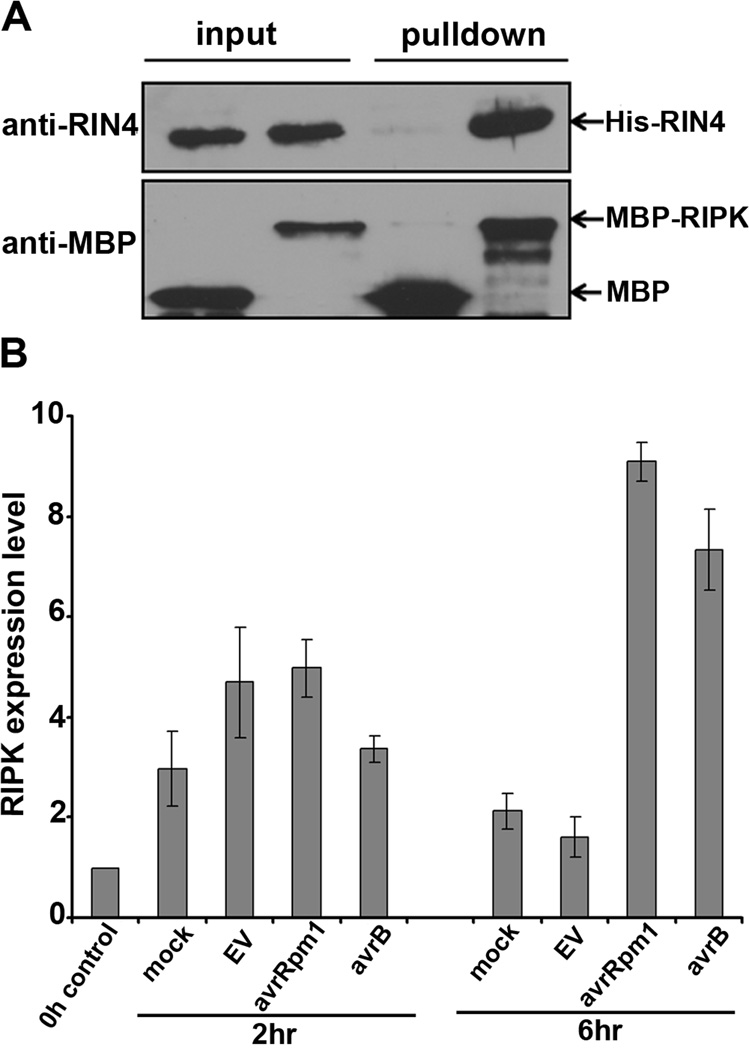
Since RIPK is a putative protein kinase and RIN4 is known to be phosphorylated during RPM1-induced ETI, their association was investigated in greater detail. Purified recombinant His-RIN4 and RIPK fused to maltose binding protein (MBP-RIPK) can directly interact in vitro by MPB pulldowns (Fig 1A). Agrobacterium-mediated transient expression in Nicotiana benthamiana was used to determine RIPK localization in planta. RIPK and related RLCKs contain an N-terminal palmitoylation/myristoylation motif, which should serve as a targeting signal for plasma membrane localization. Consistent with this prediction RIPK-GFP localizes to the plasma membrane (Fig S1). RIPK-GFP also co-localized with a known plasma membrane localized receptor like kinase fused to CFP (At4g23740) (Caplan et al., 2009). Quantitative RT-PCR reveals that RIPK transcription is induced at a high level in response to AvrB and AvrRpm1 (Fig 1B). Therefore, the Dex∷avrRpm1 plants used for immunoaffinity chromatography likely expressed high levels of RIPK, enabling our ability to purify RIPK in RIN4-associated fractions.
Figure 1. RIPK interacts with RIN4 and RIPK expression is induced by avrB and avrRpm1.
(A) Maltose Binding Protein (MBP) pulldown between purified recombinant His-RIN4 and MBP-RIPK in vitro. MBP alone is used as a negative control. Proteins were subjected to immunoblot analyses with antibodies recognizing RIN4 and MBP. (B) Quantitative Real Time PCR (qRT-PCR) illustrating RIPK induction upon bacterial inoculation. Four-week-old Arabidopsis leaves were syringe infiltrated with 2.5×107 cfu/ml of Pst DC3000 with the broad host range vector pVSP61carrying empty vector (EV), avrRpm1, or avrB. Y-axis indicates fold change. Error bars represent means ± standard deviation for qRT-PCR, n = 3. See also Figure S1 and Table S1.
RIPK knockout and overexpression lines exhibit enhanced disease resistance and susceptibility, respectively
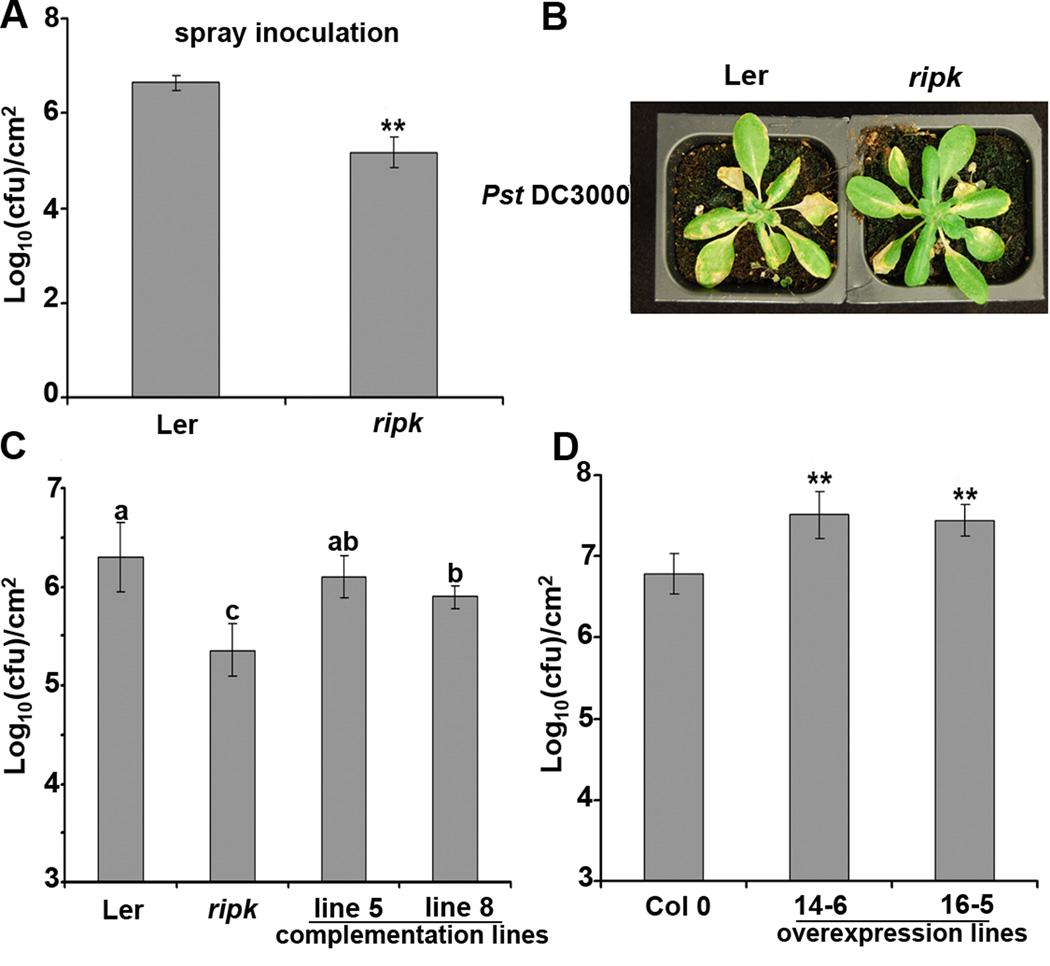
To determine whether RIPK plays a role in plant innate immunity, a RIPK T-DNA knockout (GT22343) was isolated in the Landsberg erecta ecotype (Ler, Fig S2A–B). The ripk knockout (KO) did not exhibit severe morphological defects, but did exhibit significantly narrower leaf width than wild-type Ler (Fig S2C–D). Ler and the ripk KO were inoculated with P. syringae pv tomato (Pst) DC3000 by both spray and syringe infiltration. No obvious differences were detected between ripk and wild-type plants after syringe infiltration with virulent bacteria. However, ripk conferred significant enhanced disease resistance after spray inoculation (Fig 2A–B), implying resistance occurs at an early stage of bacterial infection. This enhanced disease resistance phenotype in the ripk KO can be complemented in native promoter RIPK transgenic lines (Fig 2C, Fig S2G). RIPK overexpression lines exhibit enhanced disease susceptibility (Fig 2D, Fig S2F). ripk was introgressed into the Columbia 0 (Col 0) ecotype and ripk in a Col 0 background also exhibited enhanced resistance to Pst DC3000 (Fig S2E). Taken together, these results indicate that RIPK plays an important role in plant innate immunity and genetically acts as a negative regulator of plant basal defense responses.
Figure 2. The ripk knockout line is more resistant, while RIPK overexpression lines are more susceptible to Pst DC3000.
(A) and (B) Four-week-old Ler and ripk plants were spray-inoculated with 1×109 cfu/ml of Pst DC3000. Four days post-inoculation, plants were subjected to growth curve analysis and photographed. (C) Complementation analyses of the ripk knockout with npro∷RIPK-myc. Two homozygous T3 complementation lines and controls were subjected to spray inoculation as described above. (D) 35S∷RIPK-HA overexpression lines are more susceptible to Pst DC3000. Two homozygous T3 overexpression lines were subjected to spray inoculation as described above. Statistical differences were detected by a two-tailed t-test for (A) and (D), alpha = 0.01, and by Fisher’s LSD for (C), alpha = 0.05. Error bars represent means ± standard deviation, n = 6. The data shown are representative of three independent experiments with similar results. See also Figure S2.
RIPK can phosphorylate RIN4 in vitro and RIN4 phosphorylation induces RPM1 activation
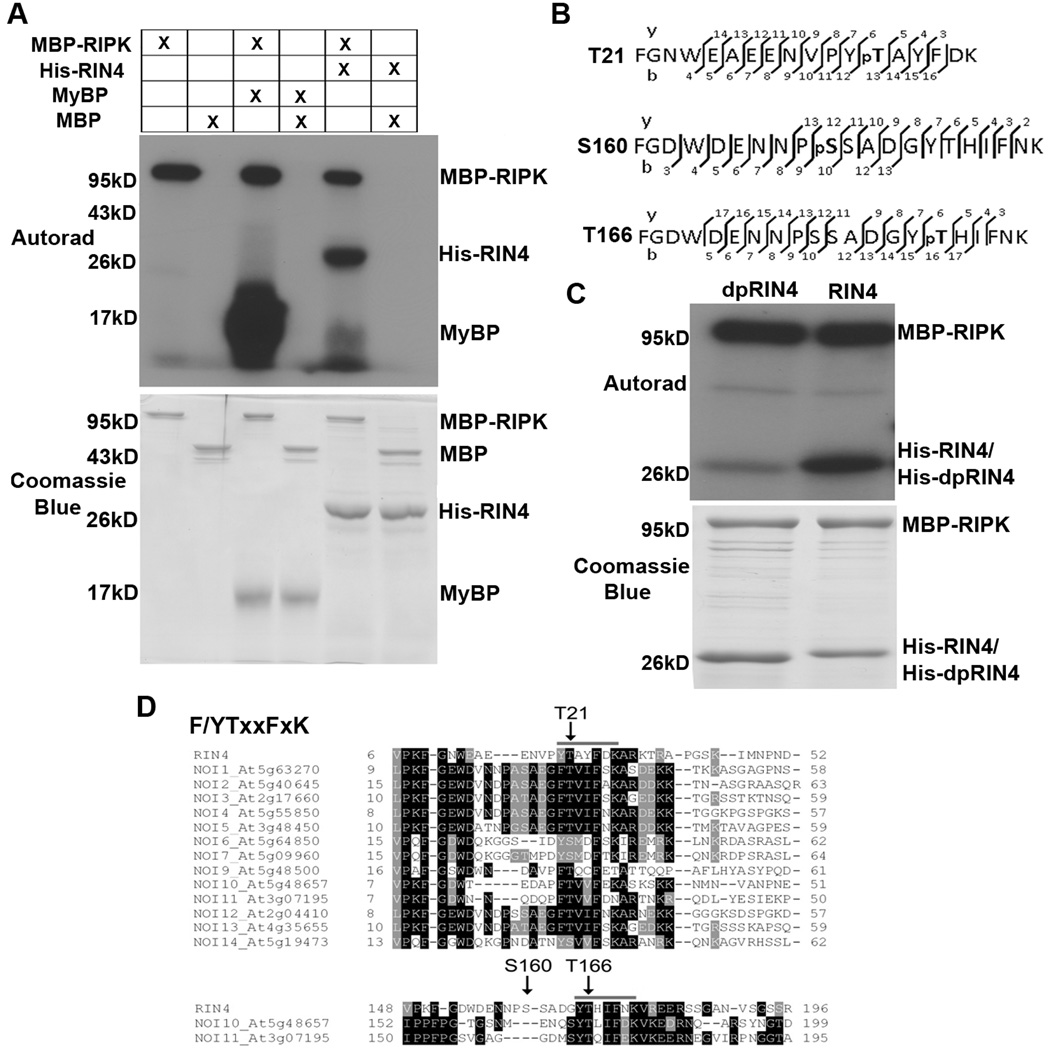
RIPK belongs to the RLCK subfamily and contains a predicted STYKc kinase domain, implying that it is likely a functional kinase. To test this possibility, myelin basic protein (MyBP) was used as a substrate for in vitro kinase activity assays using recombinant proteins purified from E. coli. RIPK can strongly phosphorylate MyBP, indicating that it is a functional kinase (Fig 3A). Moreover, RIPK is an autophosphorylating kinase. RIPK can also strongly phosphorylate RIN4 in vitro (Fig 3A). We tested RIPK’s substrate specificity using recombinant bovine serum albumin, the plant folding catalyst and cyclophilin ROC1, as well as the P. syringae effector AvrRpt2 (Fig 6D, Fig S3A). RIPK was unable to efficiently phosphorylate any of these substrates. RIN4 phosphorylation sites were mapped by mass spectrometry, and three residues were identified from high quality fragmentation spectra: Threonine 21, Serine 160 and Threonine 166 (Fig 3B, Fig S3B). All sites were mutated to alanine in concert to mimic dephosphorylation (dpRIN4). Recombinant dpRIN4 protein could no longer be efficiently phosphorylated by RIPK, indicating that these residues are the major RIPK-mediated phosphorylation sites (Fig 3C). Sequence analysis of residues adjacent to these phosphorylation sites in RIN4 and related proteins revealed a conserved F/YTxxFxK motif, surrounding T21 and T166 in RIN4 (Fig 3D). There are ∼15 Arabidopsis proteins with homology to RIN4; these proteins all share a common NOI domain (nitrate-induced domain, Pfam: PF05627) and can be cleaved by AvrRpt2 (Chisholm et al., 2005; Kim et al., 2005a). Thirteen of the proteins are smaller than RIN4 and span the first RIN4 NOI domain, whereas NOI10 and NOI11 are larger and possess two NOI domains. The F/YTxxFxK motif is conserved between RIN4 and other Arabidopsis NOI proteins (Fig 3D) as well as RIN4 orthologs in other plant species.
Figure 3. RIPK phosphorylates RIN4 in vitro.
(A) Kinase assays using recombinant MBP-RIPK, His-RIN4, MyBP (myelin basic protein), and MBP (maltose binding protein). The kinase assay was initiated by adding γ-32P-ATP to the reaction mixture and phosphorylated proteins were visualized by autoradiography (upper panel). SDS-PAGE gel stained with coomassie blue (lower panel). (B) RIN4 phosphorylation sites detected by LC-MS/MS. The vertical bars represent the observed fragmentation sites of the precursor ion in the MS2 spectrum. The observed y and b ions are numbered. (C) RIPK cannot efficiently phosphorylate a RIN4 dephosphorylation mimic (dpRIN4, RIN4(T21A/S160A/T166A)). Recombinant His-dpRIN4, His-RIN4, and MBP-RIPK were incubated in a radiolabeled kinase assay as described in (A). (D) The F/YTxxFxK motif surrounding RIN4 T21 and T166. ClustalW alignment of RIN4 and other NOI proteins in Arabidopsis. RIN4 phosphorylated residues are indicated. See also Figure S3.
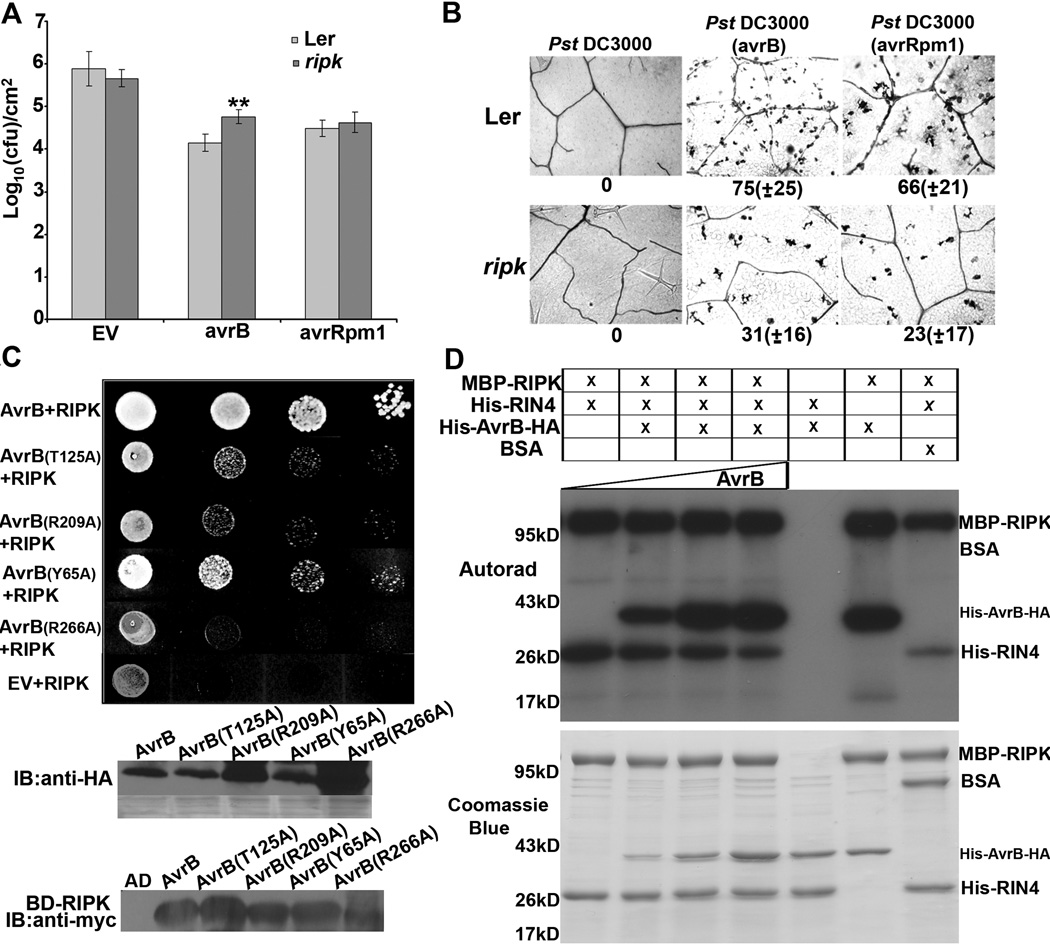
Figure 6. RIPK interacts with and phosphorylates AvrB in vitro.
(A) The ripk knockout is more susceptible to Pst DC3000(avrB). Four-week-old Ler and ripk plants were syringe infiltrated with 0.5×105 cfu/ml, and growth curve analyses were performed 4 days post inoculation. Statistical differences were detected by a two-tailed t-test, alpha = 0.01. Error bars represent means ± standard deviation, n = 6. . The experiments were repeated three times with similar results. (B) The ripk knockout exhibits decreased single-cell HR compared to Ler after infiltration with Pst DC3000(avrB) or (avrRpm1). Leaves were infiltrated with 2.5×105 cfu/ml of bacteria, stained with trypan blue 16h post-inoculation, and photographed to visualize cell death. Lower panel = number of dead cells detected on 12 leaf images. Error bars represent means ± standard deviation, n = 12 (leaves/genotype). Experiments were repeated two times with similar results. (C) AvrB interacts with RIPK by yeast two-hybrid. Mutations in AvrB’s RIN4 (T125, R209) and ADP binding sites (Y65, R266) impaired AvrB and RIPK interactions in yeast. Lower panels: immunoblot analyses demonstrating HA-AvrB and myc-RIPK expression in yeast. (D) Purified His-AvrB-HA and His-RIN4 recombinant proteins were incubated with MBP-RIPK and the kinase reaction was initiated by adding γ-32P-ATP. Phosphorylated proteins were visualized by autoradiography. Increasing the amount of AvrB in the reaction, denoted by the triangle, increased the amount of AvrB phosphorylation by RIPK. SDS-PAGE gel stained with coomassie blue demonstrates recombinant protein purity (lower panel). BSA (bovine serum albumin) served as a negative control. See also Figure S5.
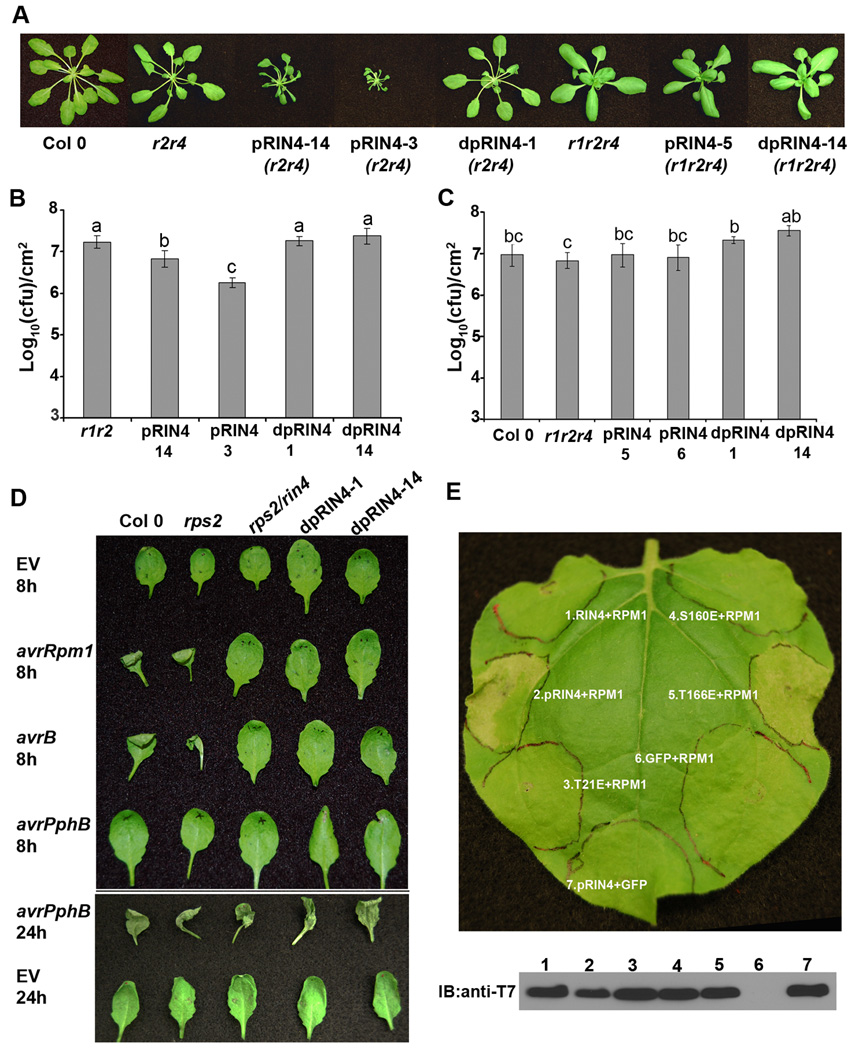
In order to investigate whether the RIN4 phosphorylation sites we identified can induce RPM1-mediated ETI, we generated a phosphomimetic mutant of RIN4 (pRIN4) by mutating T21, S160, and T166 in concert to glutamic acid. Both pRIN4 and dpRIN4 were transformed into rps2/rin4 under control of the RIN4 native promoter. In the rps2/rin4 background, pRIN4 transgenic plants exhibited a severe dwarf phenotype and were lesion mimics under long day conditions, which is characteristic of autoactive defense responses (Fig 4A). However, rps2/rin4 transformed with dpRIN4 appears phenotypically normal. We then transformed the same constructs into rpm1/rps2/rin4. We examined over 200 individual T1 transformants and all pRIN4 and dpRIN4 transgenic plants appeared phenotypically normal in the absence of RPM1. RIN4 mRNA expression in Col 0, npro∷pRIN4 and npro∷dpRIN4 were similar. However, the pRIN4 and dpRIN4 proteins were detectable, but at a lower level than wild-type RIN4 by immunoblot analysis, suggesting that mutant RIN4 proteins are not as stable as wild-type. Taken together, these results indicate that RPM1 recognizes RIN4 phosphorylation in planta.
Figure 4. RPM1 recognizes phosphorylated RIN4.
(A) RIN4 phosphorylation mimics induce RPM1-dependent dwarfism. npro∷pRIN4 (T21E/S160E/T166E) and npro∷dpRIN4 (T21A/S160A/T166A) were transformed into rpm1/rps2 (r1/r2), rps2/rin4 (r2/r4) and rpm1/rps2/rin4 (r1/r2/r4). Representative pictures were taken from 4 week old T1 plants. (B) and (C) Four-week-old Arabidopsis plants were spray inoculated with 1×109 cfu/ml of Pst DC3000. Growth curve analysis was performed 4 days post inoculation. Statistical differences were detected by Fisher’s LSD, alpha = 0.05. Experiments were performed on homozygous T3 lines in the rps2/rin4 background and T2 lines in the rpm1/rps2/rin4 background. Transgenic lines originate from the T1 plants as indicated in (A). Error bars represent means ± standard deviation, n = 6. (D) Homozygous T3 transgenic lines expressing npro∷dpRIN4 in an rps2/rin4 background are no longer able to elicit HR in response to avrB or avrRpm1. Plants were syringe infiltrated with 2.5×107 cfu/ml of Pst DC3000 carrying empty vector, avrRpm1, avrB or avrPphB. Plants were photographed at 8h and 24h post-inoculation. (E) RPM1 recognizes RIN4 phosphorylation at T166. RPM1, T7-RIN4, T7-pRIN4(T21E/S160E/T166E), T7-RIN4(T21E), T7-RIN4(S160E), and T7-RIN4(T166E) were co-expressed in N. benthamiana using Agrobacterium-mediated transient expression. Leaf disks were sampled 40h post-infiltration for anti-T7 immunoblotting. Leaves were photographed for RPM1-induced HR 72h post-infiltration. The data shown are representative of three independent experiments with similar results.
To determine if transgenic lines expressing npro∷dpRIN4 can still activate RPM1-mediated ETI in the presence of AvrB and AvrRpm1, high density bacterial infiltrations were performed. Transgenic lines expressing npro∷dpRIN4 in the rps2/rin4 background were syringe infiltrated with Pst DC3000 expressing avrB, avrRpm1, or avrPphB. High density inoculations of these effectors in plants possessing the NB-LRR immune receptors RPM1 and RPS5 (recognizes AvrPphB) results in a macroscopic HR 6–24 hours post inoculation. We were unable to detect an RPM1-mediated HR on npro∷dpRIN4 lines in the rps2/rin4 background, indicating that RIN4 phosphorylation is required for recognition of both AvrB and AvrRpm1 (Fig 4D). Transgenic npro∷dpRIN4 lines were able to induce an HR in response to avrPphB, indicating that they do not exhibit defects in ETI mediated by other NB-LRRs (Fig 4D).
We were able to recapitulate RPM1-induced ETI in N. benthamiana using transient expression. Co-expression of RPM1 with T7-pRIN4, but not wild type T7-RIN4 or T7-dpRIN4, resulted in an RPM1-specific HR (Fig 4E). Using this assay, we assessed the importance of T21, S160, and T166 residues for activating RPM1. Only co-expression of T7-RIN4(T166E) with RPM1 resulted in a HR, indicating that T166 is the major residue recognized by RPM1 (Fig 4E). Interestingly, T7-pRIN4 induced a faster HR after co-expression with RPM1 than T7-RIN4(T166E) (40h versus 72h), suggesting that phosphorylation of T21 and S160 enhance RPM1-mediated ETI.
Growth curve analyses were performed with pRIN4 lines, dpRIN4 lines, and their corresponding genetic controls to examine whether pRIN4 and dpRIN4 alter plant defense responses to virulent Pst DC3000. In the rps2/rin4 background, npro∷pRIN4 lines displayed enhanced disease resistance compared to rps2/rin4, likely due to the constitutive activation of RPM1 (Fig 4B). npro∷pRIN4 lines in the rpm1/rps2/rin4 background exhibited similar bacteria growth as rpm1/rps2/rin4 (Fig 4C). These data support the hypothesis that npro∷pRIN4 transgenic lines exhibit enhanced disease resistance due to constitutive activation of defense responses mediated by RPM1. Notably, dpRIN4 slightly compromised disease resistance in the absence of RPM1 (Fig 4C). Nonphosphorylated derivatives of RIN4 could misregulate PTI responses by interacting with a different set of client proteins than wild-type or phosphorylated RIN4 during defense signaling.
Agrobacterium-mediated transient expression in N. benthamiana was used to determine pRIN4 localization in planta. Previously RIN4 was also shown to be plasma membrane localized in the absence of phosphorylation (Kim et al., 2005a; Mackey et al., 2002). Consistent with these findings, both RIN4-GFP and pRIN4-GFP localize to the plasma membrane in N. benthamiana (Fig S1B).
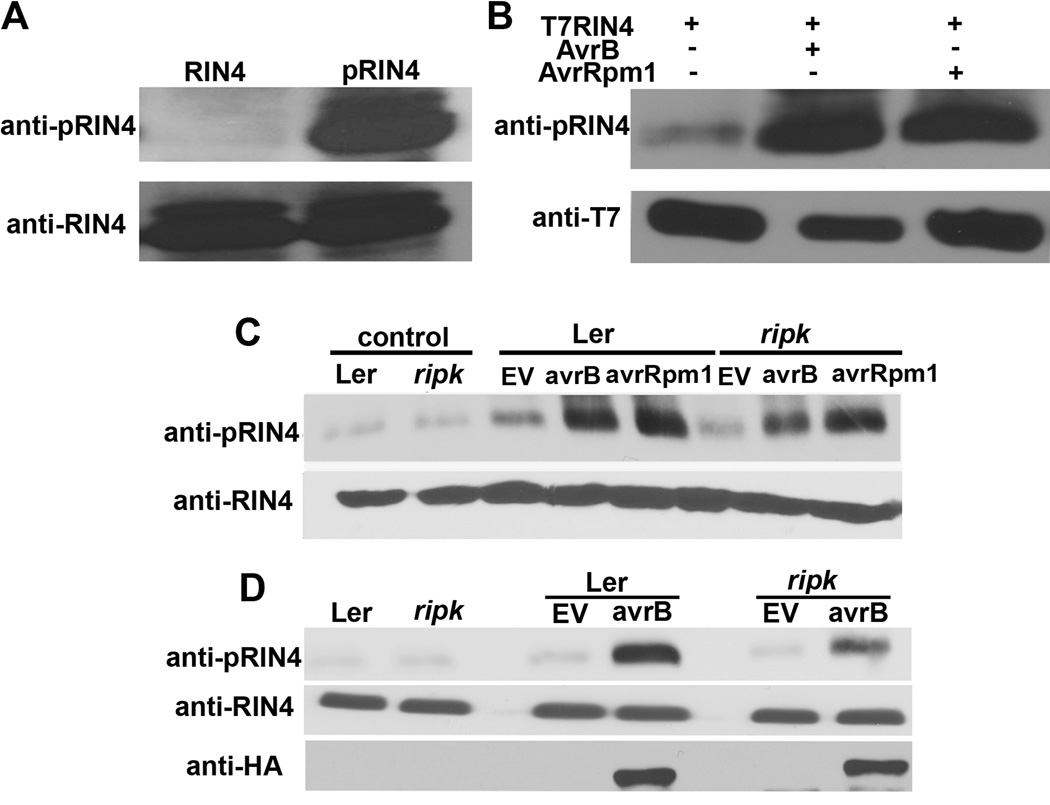
RIN4 is phosphorylated in planta in the presence of AvrB or AvrRpm1 and RIN4 phosphorylation is reduced in the ripk knockout
In order to further investigate the importance of RIN4 phosphorylation for RPM1-induced ETI in planta, a peptide antibody was generated that recognizes phosphorylated RIN4 T166. In vitro kinase assays using recombinant RIN4 and RIPK demonstrated anti-pRIN4(T166) specificity (Fig 5A). We transiently expressed AvrB and AvrRpm1 along with T7-RIN4 in N. benthamiana. In the absence of either effector, T7-RIN4 phosphorylation is very weak. RIN4 phosphorylation can be strongly induced by either AvrB or AvrRpm1 in N. benthamiana (Fig 5B). Infiltration of Pst DC3000(avrB) or (avrRpm1) also induces RIN4 phosphorylation in Arabidopsis (Fig 5C). We were able to observe a reproducible decrease in RIN4 phosphorylation in the ripk KO after infiltration with Pst DC3000(avrB), but the results with avrRpm1 were more variable (Fig 5C, Fig S4). The decrease in phosphorylated RIN4 is not due to differences in effector expression between Ler and ripk as bacterial infiltrations with Pst DC3000(avrB-HA) verified that this effector is expressed at similar levels in the ripk KO compared to wild-type Arabidopsis (Fig 5D).
Figure 5. RIN4 phosphorylation is induced by AvrB and AvrRpm1 in vivo.
(A) An antibody raised against a phosphorylated T166 RIN4 peptide (CGADGYpTHIFNK) specifically recognizes phosphorylated RIN4. Lane 1 (RIN4) = recombinant His-RIN4 and MBP-RIPK proteins incubated in the absence of ATP, Lane 2 (pRIN4) = recombinant His-RIN4 and MBP-RIPK proteins incubated in the presence of ATP. Immunoblot analyses with antibodies recognizing phosphorylated RIN4 (anti-pRIN4, upper panel) and RIN4 (anti-RIN4, lower panel). (B) T7-RIN4 phosphorylation in N. benthamiana after co-expression with AvrB or AvrRpm1 using Agrobacterium-mediated transient expression. Protein was extracted from leaf disks 40h post-infiltration and subjected to immunoblot analyses. (C) RIN4 phosphorylation in Arabidopsis Ler and ripk after delivery of AvrB and AvrRpm1. Arabidopsis leaves were infiltrated with 5×107cfu/ml of Pst DC3000 and Pst DC3000(avrB) or (avrRpm1). Immunoblot analysis was performed 6h post-inoculation. Upper panel = phosphorylated RIN4 immunoblot, lower panel = anti-RIN4 immunoblot, control = 0h time point, EV = Pst DC3000 control. (D) AvrB-HA is expressed at similar levels in Ler and ripk plants. Ler and ripk plants were syringe infiltrated with 5×107cfu/ml of Pst DC3000(avrB-HA). Immunoblot analysis was performed 6h post-inoculation. The data shown are representative of three independent experiments with similar results. See also Figure S4.
We tested the ripk knockout for alterations in RPM1-mediated ETI by growth curves, macroscopic HR, and single-cell HR assays, but were not able to detect complete elimination of RPM1 defense responses. However, the ripk knockout conferred enhanced susceptibility to Pst DC3000 carrying AvrB in the Ler background (Fig 6A). We were not able to detect a significant difference between Ler and ripk after infiltration with Pst DC3000(avrRpm1) (Fig 6A). This may be because RPM1-mediated ETI responses are much stronger and more rapid in response to AvrRpm1, masking an intermediate effect. Using low density bacterial infiltrations to assess single-cell HR development, the ripk knockout exhibited reduced single-cell HR in response to Pst DC3000 carrying avrB or avrRpm1 (Fig 6B). RIPK is a member of a large family of RLCKs in Arabidopsis, and it is likely that related RLCKs can phosphorylate RIN4 when RIPK is no longer present. Taken together, these experiments as well as those described above indicate that RIN4 phosphorylation mediated by RIPK in the presence of AvrB and AvrRpm1 results in the activation of RPM1.
RIPK can phosphorylate the AvrB effector
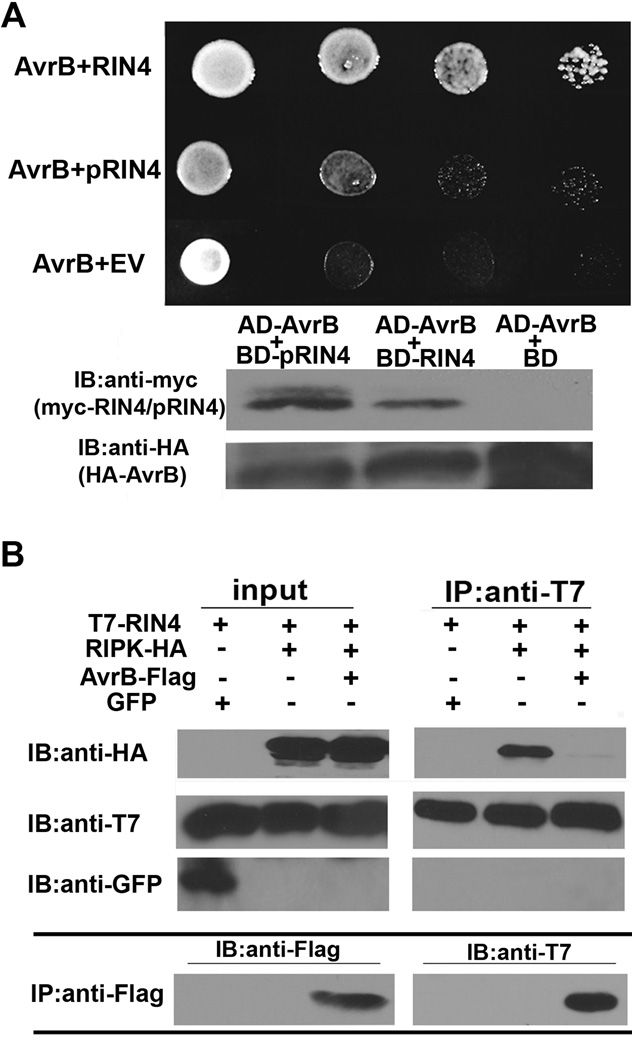
Because the ripk knockout is partially compromised in RPM1-mediated recognition of AvrB, we investigated the interaction of RIPK with AvrB. RIPK and AvrB interact by yeast two-hybrid (Fig 6C). Several other P. syringae effectors were screened for interactions with RIPK. AvrRpt2, AvrPto, AvrPtoB, and AvrRps4 could not interact with RIPK by yeast two-hybrid, indicating that the RIPK-AvrB interaction is specific (Fig S5). AvrRpm1 also could not interact with RIPK by yeast two-hybrid, but we have not been able to detect an interaction between AvrRpm1 and any other proteins using this assay and the effector may not be properly folded in yeast (Fig S5). It is well known that AvrB can also interact with RIN4 in yeast, in vitro and by co-immunoprecipitations in planta (Desveaux et al., 2007; Mackey et al., 2002). In vitro kinase assays were performed with AvrB. AvrB’s crystal structure possesses limited structural similarity to protein kinases (Desveaux et al., 2007) and to a fic domain core conserved in proteins with AMPylation activity (Kinch et al., 2009). However, we were unable to detect any kinase activity with recombinant AvrB protein (Fig 6D) or AvrB-mediated AMPylation activity by mass spectrometry. Surprisingly, RIPK can also phosphorylate AvrB in vitro (Fig 6D). Mutations in the RIN4 or ADP binding sites of AvrB dramatically impaired AvrB-RIPK interactions by yeast two-hybrid (Fig 6C).
Protein interaction dynamics
To investigate AvrB and RIPK protein associations, a combination of yeast two-hybrid and co-immunoprecipitation experiments were performed in N. benthamiana. We were able to detect a robust interaction between AvrB and RIN4, but not pRIN4 by yeast-two hybrid (Fig 7A). We were also able to detect an interaction with AvrB-FLAG and T7-RIN4 by co-immunoprecipitation (Fig 7B). Given the inability of pRIN4 to interact with AvrB in yeast, it is likely that the pool of RIN4 associated with AvrB in plants is not phosphorylated. Co-immunoprecipitations demonstrate that T7-RIN4 and RIPK-HA can interact in the absence of AvrB, and AvrB-FLAG disrupts the RIPK-RIN4 complex (Fig 7B).
Figure 7. RIN4 phosphorylation status alters protein associations.
(A) RIN4 and RIN4 phosphorylation mimic (pRIN4, RIN4(T21E/S160E/T166E)) interaction with AvrB by yeast-two hybrid. Lower panel: immunoblot analyses demonstrating myc-RIN4 and HA-AvrB expression in yeast. (B) AvrB disrupts the RIN4 and RIPK complex in vivo. GFP, T7-RIN4, RIPK-HA, and AvrB-FLAG were transiently expressed in N. benthamiana by Agrobacterium-mediated protein expression. T7-RIN4 was immunoprecipitated with T7 antisera and associated proteins were detected by immunoblot analyses. Bottom panel: Anti-FLAG immunoprecipitation. AvrB-FLAG expression could only be detected by immunoprecipitation in the input due to low-level expression.
DISCUSSION
In this paper, we report the identification of RIPK, a member of the large RLCK family present in plants that can phosphorylate RIN4. RPM1 recognizes RIN4 phosphorylation, leading to activation of ETI. Transient expression of RIN4 and RIN4 phosphomimics in N. benthamiana revealed that RIN4(T166E) induces an RPM1-dependent HR after 72h in the absence of AvrB and AvrRpm1, demonstrating that this residue is crucial for RPM1-mediated recognition (Fig 4). However, RIN4(T21E/S160E/T166E) induced a faster RPM1-dependent HR by 40h, suggesting that phosphorylation of T21 and S160 may alter RIN4 structure, functioning cooperatively with T166 in triggering ETI. S160 and T166 lie within the AvrB binding site of RIN4 (Desveaux et al., 2007; Kim et al., 2005a). Here, we show that AvrB is no longer able to interact with pRIN4 (Fig 7).
The data presented in this paper and others provide models for RPM1 activation by AvrB. AvrB is delivered into the plant cell by the T3SS, where it is acylated and targeted to the plasma membrane (Nimchuk et al., 2000). AvrB has been previously hypothesized to be activated in the host by nucleotide binding and subsequent phosphorylation via a host kinase (Desveaux et al., 2007). As RIPK can phosphorylate AvrB in vitro (Fig 6D), it is possible that this is also the kinase that is responsible for AvrB activation in planta. We hypothesize that AvrB can then promote RIPK-mediated phosphorylation of RIN4 leading to the activation of RPM1. Delivery of AvrB during infection results in the upregulation of RIPK, implicating an amplification of this response occurs. AvrB possesses some limited structural homology to protein kinases (Desveaux et al., 2007). However, we were unable to detect any kinase activity using recombinant AvrB protein (Fig 6). It is still possible that AvrB can act as a bona fide kinase in planta and may phosphorylate RIN4 directly. In this scenario, AvrB could work in concert with RIPK or mimic RIPK activity to phosphorylate RIN4 and activate RPM1.
Models of NB-LRR activation hypothesize that these immune receptors exist in an auto-repressed state where they are bound to ADP. Recognition of cognate effector proteins is hypothesized to induce NB-LRR conformational changes, displacing the LRR from the NB domain and enabling nucleotide exchange and hydrolysis leading to the activation of downstream signaling (Takken and Tameling, 2009). RPM1-mediated recognition of phosphorylated RIN4 likely coincides or comes before RPM1 nucleotide exchange and hydrolysis, as mutations in the P-loop of multiple NB-LRRs abolishes their activity. Other examples of NB-LRR activation in plants by monitoring host kinases exist for the NB-LRR receptors Prf and RPS5. In Arabidopsis RPS5 is activated upon PBS1 cleavage, mediated by the effector AvrPphB (Shao et al., 2003). In the absence of RPS5, AvrPphB promotes bacterial virulence by cleaving PBS1 and related protein kinases (Zhang et al., 2010). Similarly, the tomato kinase Pto is targeted by the effectors AvrPto and AvrPtoB, resulting in the activation of the NB-LRR receptor Prf (Mucyn et al., 2006). Recognition of cleaved PBS1 and potential post-translational modification of Pto by RPS5 and Prf is probably analogous to the recognition of phosphorylated RIN4 by RPM1.
Although saturating genetic screens have been conducted for loci involved in RPM1-mediated ETI, and no protein kinases were identified, suggesting that functional redundancy is likely to exist with respect to the kinase(s) that phosphorylate RIN4 (Tornero et al., 2002). Consistent with these findings, ripk KO lines exhibit reduced RIN4 phosphorylation, single cell HR, as well as enhanced bacterial growth compared to wild-type after inoculation with Pst DC3000(avrB) (Fig 6, Fig 5, Fig S4). These results validate the importance of RIPK in RPM1-mediated ETI, but indicate that there are other related RLCKs that can also phosphorylate RIN4 and/or activate AvrB in the absence of RIPK. ripk KO lines exhibit reduced single cell HR, but are not impaired in bacterial growth and exhibit a slight decrease in RIN4 phosphorylation after inoculation with DC3000(avrRpm1) (Fig 5, Fig 6, Fig S4). AvrRpm1 induces a much stronger ETI response than AvrB and also induces a more significant size shift in RIN4 phosphorylation, indicating that RIN4 may be phosphorylated at additional sites in the presence of AvrRpm1 (Kim et al., 2005b). Alternatively, AvrRpm1 may also induce other post-translational modifications of RIN4 that are perceived by RPM1 and enhance RPM1-mediated ETI.
AvrB was recently shown to interact with the Arabidopsis mitogen activated protein kinase MPK4 (Cui et al., 2010). AvrB’s interaction with MPK4 enables this effector to modulate plant hormone signaling and enhance pathogen virulence in an RPM1-independent manner (Cui et al., 2010). MPK4 is also able to interact with and phosphorylate RIN4, and RIN4 overexpression and KO plants possess reduced and enhanced expression of the jasmonic acid marker gene PDF1.2 (Cui et al., 2010). The AvrB-MPK4 and MPK4-RIN4 interactions are likely distinct from that of RIPK as they are RPM1-independent. However, these data support the hypothesis that AvrB targets host protein kinases, such as RIPK and MPK4, to modulate their activity and/or substrate specificity.
RIN4 is a negative regulator of PTI, while both AvrB and AvrRpm1 suppress PTI (Kim et al., 2005b; Shang et al., 2006). Like RIN4, the ripk KO exhibits enhanced disease resistance and RIPK overexpression lines exhibit enhanced disease susceptibility to virulent Pst DC3000 (Fig 2, Fig S2). These results indicate that phosphorylation of RIN4 and/or other NOI proteins may block PTI-based responses in genotypes lacking RPM1. Phosphorylation of RIN4 could inhibit the negative regulation of basal defense by altering RIN4 structure and enabling RIN4 to interact with a different set of plant proteins. Consistent with this hypothesis, MS revealed that phosphorylated RIN4 co-immunoprecipitates a different set of plant proteins than RIN4 in the absence of pathogen stimulus (Table S1) (Liu et al., 2009). Furthermore, AvrB can no longer interact with phosphorylated RIN4. The observation that RIPK overexpression does not result in activation of RPM1 (Fig 2D), indicates that the presence of AvrB or AvrRpm1 is necessary to induce a high-level of RIN4 hyperphosphorylation. Thus, other targets of RIPK may also be important regulators of PTI and explain the enhanced disease susceptibility phenotype observed in RIPK overexpression lines. RIPK phosphorylates RIN4’s F/YTxxFxK motif, which is conserved between RIN4, other NOI proteins, and other plant RIN4 orthologs (Fig 3). This would impose a selective pressure on both effectors to maintain their ability to induce phosphorylation of RIN4 and related proteins.
Interestingly, RIPK mRNA expression is upregulated (∼2 fold) after inoculation with virulent DC3000. As RIPK is a negative regulator of basal defense, enhancing and/or altering RIPK expression may be a virulence strategy used by multiple bacterial effectors. RIN4 is a known target of multiple bacterial effectors, including AvrB, AvrRpm1, AvrRpt2, HopF2, and AvrPto (Axtell and Staskawicz, 2003; Luo et al., 2009; Mackey et al., 2003; Wang et al., 2010; Wilton et al., 2010). It is possible that effectors could directly target RIPK, as in the case of AvrB, or targeting of RIN4 could lead to altered RIPK protein associations and enhanced phosphorylation activity. Future experiments addressing the importance of RIPK for bacterial virulence in rin4 knockout lines as well as the identification of additional RIPK targets will help address this hypothesis. Future investigations focusing on the function of RIPK during PTI, elucidating RIPK substrates before and after infection, as well as the function of proteins phosphorylated by RIPK will significantly advance our understanding of plant innate immune signaling.
EXPERIMENTAL PROCEDURES
Plant materials and growth conditions
A homozygous gene trap Ler mutant (GT22343), in RIPK (At2g05940), was ordered from Cold Spring Harbor. The homozygous line was crossed to Columbia 0 (Col 0) three times and selfed in order to make a KO line in the Col 0 background. In the text, the rps2, rpm1, and rin4 mutants refer to rps2–101c, rpm1–3, and the rin4 T-DNA knockout (Boyes et al., 1998; Mackey et al., 2002; Mindrinos et al., 1994). Plant growth conditions, bacterial inoculations and growth assays were performed as described previously (Liu et al., 2009). Single-cell HR assays were performed as described (Coll et al., 2010).
Transgenic Arabidopsis were generated by following the floral dip transformation procedure (Bent, 2006). The RIPK open reading frame (ORF) was amplified and cloned into the BamH I/Xho I site of binary vector pMD-1 (Tai et al., 1999), driven by the 35S promoter and fused with a C terminal HA tag. The transgenic plants were selected using 25µg/mL kanamycin. The RIN4 phosphorylation mimics (pRIN4) and dephosphorylation mimics (dpRIN4) were generated by PCR-based mutagenesis. T21, S160, and T166 were mutated to alanine in concert to generate dpRIN4. T21, S160, and T166 were mutated to glutamic acid to generate pRIN4 and individual phosphorylation mimics. RIN4 ORFs were then cloned into the pENTR/D-TOPO’s Asc I site, fused with the 2.1kb RIN4 promoter, and recombined into the PGWB 16 binary vector (Nakagawa et al., 2007) by gateway cloning. In order to make the ripk complementation lines, the RIPK gene and its promoter (1.5kb) were cloned into PGWB16 (Nakagawa et al., 2007) by gateway cloning. Two independent T3 lines were used for all bacterial inoculations, with the exception of pRIN4 and dpRIN4 transformed into rpm1/rps2/rin4, where two independent T2 lines were used.
Yeast two-hybrid
The yeast two hybrid experiments were performed as previously described (Liu et al., 2009), with the exception that the Matchmaker pGADT7 and pGBKT7 vectors (Clonetech) were modified to be gateway compatible.
Protein complex purification and identification
Dex∷avrRpm1 (Bennett et al., 2005) and wild type Col 0 plants were sprayed with 20µM Dexamethasone containing 0.025% silwett L-77. Leaf tissue was harvested for protein complex purification 2 hours later. RIN4 protein complex purification was performed as previously described (Liu et al., 2009).
RT-PCR and qRT-PCR
Total RNA was extracted by using the Trizol method and subjected to DNase I digestion (Invitrogen). First strand cDNA was synthesized by using 5µg of total RNA with a cDNA synthesis Kit (Promega) in a 20µl reaction. The expression level of RIPK (At2g05940) was normalized to the expression of Actin2 (At3g18780). RT-PCR was run for 28 cycles for Actin2 and 35 cycles for RIPK. qRT-PCR was carried out with a Biorad SsoFast™ EvaGreen Supermix according to manufacturer’s directions, with Actin2 as reference gene. The primers for all experiments are listed in supplementary data (Table S2).
In vitro kinase activity assays
RIN4 ORFs were cloned in the E. coli pRSET A (Invitrogen) expression vector’s BamH I/Xho I sites. avrB was cloned into the modified Gateway compatible pETDuet vector (Novagen) with an N terminal 6xHis tag and a C terminal HA tag. RIPK was cloned in the pMAL-C4X vector (New England Biolabs) using BamH I and Xho I restriction sites. E. coli Rosetta cells were used for recombinant protein expression. Cloning and purification of GST-ROC1, His-AvrRpt2-HA, and His-RIN4 have been previously described (Coaker et al., 2005). MBP-RIPK expression was induced with 0.3mM IPTG for 3hrs at 28°C. MBP-RIPK was purified by amylose affinity chromatography. AvrB protein expression was induced at 16°C for 7hrs as previously described (Desveaux et al., 2007) and protein was purified by Ni-NTA affinity chromatography. Recombinant protein purity was assessed by SDS-PAGE. Kinase activity assays were performed in 20mM Tris-HCl (pH7.5), 10mM MgCl2, 1mM CaCl2, 10µM ATP, 1mM DTT, 1µg kinase and 1∼2 µg recombinant protein as substrates, in a total volume of 30µl. The assay was initiated by adding 1µl (10uCi) 32P-ATP and incubated for 40min at 30°C. The reaction was terminated by the addition of 3x laemmli loading buffer (Laemmli, 1970) and subsequent incubation at 95°C for 5min. The proteins were separated on a 12% SDS-PAGE gel and signals were visualized by X-ray film exposure.
Recombinant protein pull-down assays
MBP-RIPK and His-RIN4 (3µg each) were incubated with amylose beads (10µl) in TEN100 buffer (20mM Tris-HCl pH7.4, 100mM NaCl, 0.1mM EDTA and 0.2% Triton X-100) on an earthquake shaker for 1hr, followed by two in-tube washes. The mixture was transferred to a micro spin column (Bio-Rad) and washed twice with 500µl of NETN300 (20mM Tris-HCl, pH7.4, 300mM NaCl, 0.1mM EDTA and 0.5% NP-40). The proteins were eluted from the beads with 10mM maltose and the eluate boiled in 1x laemmli buffer for 5min. Proteins were then separated on a 12% SDS-PAGE gel and immunoblotted with anti-RIN4 and anti-MBP (Affinity BioReagents) antisera, respectively. The secondary goat anti-rabbit or anti-mouse IgG-HRP conjugates (Bio-rad) were used for detection via enhanced chemiluminescence (Pierce).
RIN4 phosphorylation in planta
An antibody (anti-pRIN4) was generated in rabbit against a phosphorylated T166 RIN4 peptide (CGADGYpTHIFNK). Crude antisera were affinity purified and tested for specificity by ELISA (Open Biosystems). AvrB and AvrRpm1 cloned into pVSP61 (Innes et al., 1993) or AvrB-HA cloned into pBBR1 MCS-2 (Desveaux et al., 2007) were delivered by Pst DC3000 to Arabidopsis at 5×107 cfu/ml and leaf samples were taken at 6hrs after inoculation. For immunoblotting, tissue samples were ground in liquid N2 and protein was extracted in buffer containing 50 mM HEPES (pH7.5), 50mM NaCl, 10 mM EDTA, 0.2% Triton X-100, complete protease inhibitors (Roche) and phosphatase inhibitors (Fisher). The resulting samples were then analyzed by western blotting and probed with primary anti-pRIN4 (1:3000) antibody.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by a grant from the NIH (1R01GM092772-01) to GC. JME is supported by an NSF IGERT graduate research training grant (DGE-0653984). Mass spectrometry was performed at the UC Davis Proteomics Facility.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES AND NOTES
- Axtell MJ, Chisholm ST, Dahlbeck D, Staskawicz BJ. Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol. Microbiol. 2003;49:1537–1546. doi: 10.1046/j.1365-2958.2003.03666.x. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- Bennett M, Mehta M, Grant M. Biophoton imaging: a nondestructive method for assaying R gene responses. Mol. Plant Microbe Interact. 2005;18:95–102. doi: 10.1094/MPMI-18-0095. [DOI] [PubMed] [Google Scholar]
- Bent A. Arabidopsis thaliana floral dip transformation method. Meth. Mol. Biol. 2006;343:87–103. doi: 10.1385/1-59745-130-4:87. [DOI] [PubMed] [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- Boyes DC, Nam J, Dangl JL. The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl. Acad. Sci. USA. 1998;95:15849–15854. doi: 10.1073/pnas.95.26.15849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan JL, Zhu X, Mamillapalli P, Marathe R, Anandalakshmi R, Dinesh-Kumar SP. Induced ER chaperones regulate a receptor-like kinase to mediate antiviral innate immune response in plants. Cell Host Microbe. 2009;6:457–469. doi: 10.1016/j.chom.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Dahlbeck D, Krishnamurthy N, Day B, Sjolander K, Staskawicz BJ. Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2. Proc. Natl. Acad. Sci. USA. 2005;102:2087–2092. doi: 10.1073/pnas.0409468102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coaker G, Falick A, Staskawicz B. Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science. 2005;308:548–550. doi: 10.1126/science.1108633. [DOI] [PubMed] [Google Scholar]
- Coll NS, Vercammen D, Smidler A, Clover C, Van Breusegem F, Dangl JL, Epple P. Arabidopsis type I metacaspases control cell death. Science. 2010:1393–1397. doi: 10.1126/science.1194980. [DOI] [PubMed] [Google Scholar]
- Cui H, Wang Y, Xue L, Chu J, Yan C, Fu J, Chen M, Innes RW, Zhou JM. Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe. 2010;7:164–175. doi: 10.1016/j.chom.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Xiang T, Zhou JM. Plant immunity: a lesson from pathogenic bacterial effector proteins. Cell. Microbiol. 2009;11:1453–1461. doi: 10.1111/j.1462-5822.2009.01359.x. [DOI] [PubMed] [Google Scholar]
- de Torres M, Sanchez P, Fernandez-Delmond I, Grant M. Expression profiling of the host response to bacterial infection: the transition from basal to induced defence responses in RPM1-mediated resistance. Plant J. 2003;33:665–676. doi: 10.1046/j.1365-313x.2003.01653.x. [DOI] [PubMed] [Google Scholar]
- Desveaux D, Singer AU, Wu AJ, McNulty BC, Musselwhite L, Nimchuk Z, Sondek J, Dangl JL. Type III effector activation via nucleotide binding, phosphorylation, and host target interaction. PLoS Pathogens. 2007;3:e48. doi: 10.1371/journal.ppat.0030048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Parker JE. Interplay of signaling pathways in plant disease resistance. Trends Genet. 2000;16:449–455. doi: 10.1016/s0168-9525(00)02107-7. [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- Innes RW, Bisgrove SR, Smith NM, Bent AF, Staskawicz BJ, Liu YC. Identification of a disease resistance locus in Arabidopsis that is functionally homologous to the RPG1 locus of soybean. Plant J. 1993;4:813–820. doi: 10.1046/j.1365-313x.1993.04050813.x. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kim HS, Desveaux D, Singer AU, Patel P, Sondek J, Dangl JL. The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc. Natl. Acad. Sci. USA. 2005a;102:6496–6501. doi: 10.1073/pnas.0500792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MG, da Cunha L, McFall AJ, Belkhadir Y, DebRoy S, Dangl JL, Mackey D. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell. 2005b;121:749–759. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Lin NC, Martin GB. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell. 2002;109:589–598. doi: 10.1016/s0092-8674(02)00743-2. [DOI] [PubMed] [Google Scholar]
- Kinch LN, Yarbrough ML, Orth K, Grishin NV. Fido, a novel AMPylation domain common to fic, doc, and AvrB. PLoS ONE. 2009;4:e5818. doi: 10.1371/journal.pone.0005818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK.Cleavage of structural proteins during the assembly of the head of bacteriophage T4 Nature 1970. p227 [DOI] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Fuglsang AT, Palmgren M, Staskawicz B, Coaker G. RIN4 functions with the plasma membrane H+-ATPase to regulate stomatal apertures during pathogen attack. PLoS Biol. 2009;7:e1000139. doi: 10.1371/journal.pbio.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA. 2010;107:496–501. doi: 10.1073/pnas.0909705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Caldwell KS, Wroblewski T, Wright ME, Michelmore RW. Proteolysis of a negative regulator of innate immunity is dependent on resistance genes in tomato and Nicotiana benthamiana and induced by multiple bacterial effectors. Plant Cell. 2009;21:2458–2472. doi: 10.1105/tpc.107.056044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF, 3rd, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- Mindrinos M, Katagiri F, Yu GL, Ausubel FM. The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell. 1994;78:1089–1099. doi: 10.1016/0092-8674(94)90282-8. [DOI] [PubMed] [Google Scholar]
- Mucyn TS, Clemente A, Andriotis VM, Balmuth AL, Oldroyd GE, Staskawicz BJ, Rathjen JP. The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell. 2006;18:2792–2806. doi: 10.1105/tpc.106.044016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. and Bioengineering. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- Nimchuk Z, Marois E, Kjemtrup S, Leister RT, Katagiri F, Dangl JL. Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell. 2000;101:353–363. doi: 10.1016/s0092-8674(00)80846-6. [DOI] [PubMed] [Google Scholar]
- Rodriguez MC, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 2010;61:621–649. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- Shang Y, Li X, Cui H, He P, Thilmony R, Chintamanani S, Zwiesler-Vollick J, Gopalan S, Tang X, Zhou JM. RAR1, a central player in plant immunity, is targeted by Pseudomonas syringae effector AvrB. Proc. Natl. Acad. Sci. USA. 2006;103:19200–19205. doi: 10.1073/pnas.0607279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- Tai TH, Dahlbeck D, Clark ET, Gajiwala P, Pasion R, Whalen MC, Stall RE, Staskawicz BJ. Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl. Acad. Sci. USA. 1999;96:14153–14158. doi: 10.1073/pnas.96.24.14153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken FL, Tameling WI. To nibble at plant resistance proteins. Science. 2009;324:744–746. doi: 10.1126/science.1171666. [DOI] [PubMed] [Google Scholar]
- Ting JP, Willingham SB, Bergstralh DT. NLRs at the intersection of cell death and immunity. Nat. Rev. Immunol. 2008;8:372–379. doi: 10.1038/nri2296. [DOI] [PubMed] [Google Scholar]
- Tornero P, Chao RA, Luthin WN, Goff SA, Dangl JL. Large-scale structure-function analysis of the Arabidopsis RPM1 disease resistance protein. Plant Cell. 2002;14:435–450. doi: 10.1105/tpc.010393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: Discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li J, Hou S, Wang X, Li Y, Ren D, Chen S, Tang X, Zhou J-M. A Pseudomonas syringae ADP-ribosyltransferase inhibits Arabidopsis mitogen-activated protein kinase kinases. Plant Cell. 2010;22:2033–2044. doi: 10.1105/tpc.110.075697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widjaja I, Naumann K, Roth U, Wolf N, Mackey D, Dangl JL, Scheel D, Lee J. Combining subproteome enrichment and Rubisco depletion enables identification of low abundance proteins differentially regulated during plant defense. Proteomics. 2009;9:138–147. doi: 10.1002/pmic.200800293. [DOI] [PubMed] [Google Scholar]
- Wilton M, Subramaniam R, Elmore J, Felsensteiner C, Coaker G, Desveaux D. The type III effector HopF2Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc. Natl. Acad. Sci. USA. 2010;107:2349–2354. doi: 10.1073/pnas.0904739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010;7:290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.