Abstract
Biochemical studies have suggested conflicting roles for the E3 ubiquitin ligase constitutive photomorphogenesis protein 1 (Cop1; also known as Rfwd2) in tumorigenesis, providing evidence for both the oncoprotein c-Jun and the tumor suppressor p53 as its targets. Here we present what we believe to be the first in vivo investigation of the role of Cop1 in cancer etiology. Using an innovative genetic approach to generate an allelic series of Cop1, we found that Cop1 hypomorphic mice spontaneously developed malignancy at a high frequency in the first year of life and were highly susceptible to radiation-induced lymphomagenesis. Further analysis revealed that c-Jun was a key physiological target for Cop1 and that Cop1 constitutively kept c-Jun at low levels in vivo and thereby modulated c-Jun/AP-1 transcriptional activity. Importantly, Cop1 deficiency stimulated cell proliferation in a c-Jun–dependent manner. Focal deletions of COP1 were observed at significant frequency across several cancer types, and COP1 loss was determined to be one of the mechanisms leading to c-Jun upregulation in human cancer. We therefore conclude that Cop1 is a tumor suppressor that functions, at least in part, by antagonizing c-Jun oncogenic activity. In the absence of evidence for a genetic interaction between Cop1 and p53, our data strongly argue against the use of Cop1-inhibitory drugs for cancer therapy.
Introduction
The turnover and the activity of several key oncoproteins and tumor suppressors are controlled by the ubiquitin proteasome system (UPS). The UPS comprises two discrete steps: the covalent attachment of multiple ubiquitin molecules to the protein substrate and degradation of the polyubiquitylated protein by the 26S proteasome complex. The first step is mediated by at least three enzymes: the ubiquitin-activating enzyme E1, the ubiquitin-conjugating enzyme E2, and the ubiquitin ligase E3. Few reports have linked E1 and E2 to cancer development; in contrast, cumulative evidence indicates alterations in the activity of some E3 ligases in the etiology of human malignancies (1). Because E3 ligases are the specific recognition element of the system and are themselves potentially “drugable’” enzymes, they can serve as potential cancer targets as well as cancer biomarkers.
A direct molecular link between deregulation of E3 ligase activity and cancer is illustrated by the well-described oncogenic activity of the RING finger–containing protein Mdm2. In physiological conditions, Mdm2 directly binds the p53 tumor suppressor protein and via its RING finger domain acts as an E3 ligase to promote p53 ubiquitylation and proteasomal degradation (2). In tumors, p53 function is altered either by inactivating mutations of the TP53 gene itself or as the result of aberrant expression and/or functional dysregulation of some of its modulators, including MDM2. Indeed, MDM2 is amplified and/or overexpressed in a significant fraction of human tumors that retain wild-type TP53 (3, 4). Mdm2 is therefore a promising cancer target whose inhibition is expected to unleash latent p53 tumor suppressor activity in tumors harboring wild-type TP53 (5).
Transfection studies in cancer cell lines have suggested that COP1, another RING finger–containing protein, also targets p53 for ubiquitylation and proteasomal degradation (6). Moreover, COP1-mediated p53 degradation was seen to be impaired in response to DNA damage allowing p53 stabilization and activation (7). Consistent with these reports, COP1 overexpression was reported in breast and ovarian adenocarcinomas, and in tumors that retained wild-type p53, overexpression of COP1 correlated with a marked decrease in p53 protein levels and attenuation of its downstream target p21 (8). COP1 would therefore favor tumorigenesis by inactivating the p53 tumor suppressor pathway. Based on these reports, COP1, like MDM2, appears to be an attractive drug target for cancer therapy.
In apparent contradiction to this view, biochemical experiments have also identified the oncoprotein c-Jun as a target of COP1 (9). However, in this context COP1 does not appear to function as an E3 ligase per se but rather permits the recruitment of c-Jun to a multisubunit ubiquitin ligase containing DET1, DDB1, cullin 4A, and ROC1 (10). c-Jun, a component of the transcription factor AP-1, is a positive regulator of cell proliferation and oncogenic transformation (11–13). c-Jun is activated by various stimuli, such as growth factors, cytokines, bacterial and viral infections, and physical and chemical stresses, many of which function as tumor promoters or are known to activate the oncogenic MAP kinase signaling cascades (ERK, p38MAPK, JNK) (12). In response to genotoxic stress, for instance, JNKs are activated and translocate to the nucleus, where they phosphorylate c-Jun at N-terminal serine residues 63 and 73 within its transactivation domain (14, 15). Such phosphorylation events are thought to dramatically potentiate c-Jun transcriptional activity by favoring the recruitment of essential cofactors such as p300 and CBP to target promoters (16–18). JNK-mediated phosphorylation of c-Jun also protects c-Jun from constitutive proteasome-dependent degradation, via a mechanism that is as yet unclear (19).
c-Jun oncogenic activity has been attributed to its ability to promote cell cycle progression and prevent apoptotic cell death (12, 13). Evidence for the relevance of c-Jun for cancer development originally came from mouse genetics (20–22). More recent studies provide substantial evidence for a causative link between c-Jun overexpression and the development of human malignancies (23–25).
Investigations of Cop1 function have thus implicated this E3 ligase in tumor development, but the proposed function of Cop1 in the regulation on the one hand of p53, a tumor suppressor, and on the other of c-Jun, an oncogene, has created a conundrum with respect to its role in tumorigenesis. All experiments to date have been performed in vitro on cancer cell lines and therefore do not take into account the tissue diversity and complexity of an entire living organism. To explore Cop1 physiological function and, in particular, to clarify its role in tumorigenesis in vivo, we generated what we believe to be the first Cop1-mutant mouse line. We used an innovative gene trap approach that enabled us to generate an allelic series of Cop1 in mice from a single insertion event in ES cells. Notably, our study demonstrates that such an approach can be used in a generic way to create allelic series in mice. We show that while complete loss of Cop1 is incompatible with life, mice with hypomorphic Cop1 expression are viable. Importantly, these mice are tumor prone, thus providing strong genetic support of an etiological role for Cop1 in tumor suppression. Moreover, our biochemical and genetic studies highlight a key role for Cop1 in the regulation of c-Jun stability and AP-1 transcriptional activity in vivo. We show that Cop1 negatively regulates cell proliferation in a c-Jun–dependent manner, thus providing molecular mechanistic insights into its tumor suppression function. Importantly, our extensive analyses fail to provide any evidence for a role of Cop1 in the regulation of p53, thus arguing against its use as a target in cancer therapy.
Results
An allelic series of Cop1.
In order to study the physiological function of Cop1 in vivo and in particular its role in tumor development, we set out to generate viable mice with compromised Cop1 function. We anticipated that complete genetic ablation of Cop1 function might be deleterious for embryonic development in light of its putative role as a negative regulator of p53 (6). Germline inactivation of Mdm2 and Mdmx, two key negative regulators of p53, indeed lead to embryonic lethality (26, 27). Thus, we used a more sophisticated strategy to create an allelic series in the Cop1 gene. This strategy is based on the use of a gene trap vector design that contains loxP sites flanking the splice acceptor (SA) of the gene trapping element (Figure 1A). Preliminary studies in ES cells showed that Cre-mediated excision of the SA site can be used to convert the trapped null allele into a hypomorphic allele (W.C. Skarnes, unpublished observations). Removal of the SA site results in splicing around the vector to restore normal wild-type transcript at a reduced level. Based on the analysis of a random insertion in an X-linked gene in male (XY) ES cells, deletion of the SA restored full-length transcripts to approximately 5%–10% of the wild-type expression level (data not shown). This observation prompted us to generate mice with such a gene trap event in the Cop1 gene.
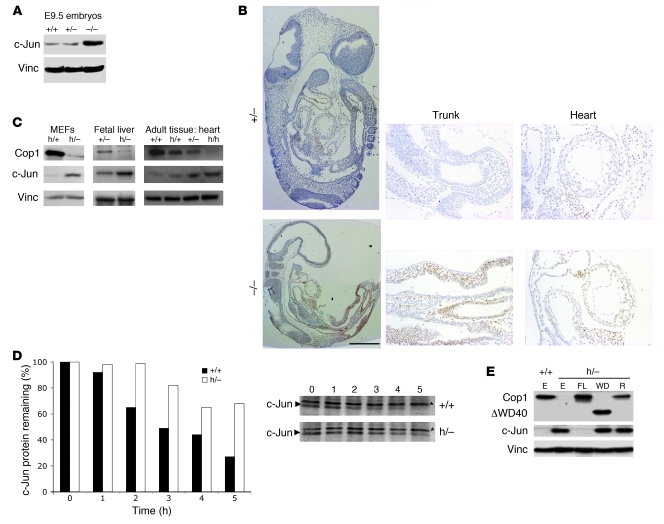
Figure 1. The gene trap approach developed for this study allows the generation of an allelic series of Cop1.
(A) The structure of the pGT01xf gene trap vector along with the structures of the wild-type, trapped (assimilated to a null allele), and hypomorphic alleles. The hypomorphic allele is generated by Cre-mediated conversion of the trapped allele. Exons are indicated by black boxes. En2 intr1, intron 1 of Engrailed2; βgeo, lacz/neomycin reporter/selection cassette. (B) A representative PCR analysis of E9.5 embryos with the indicated genotypes (left panel). The 3 primers used are described in Methods. Discrimination of the wild-type, null, and hypomorphic alleles by PCR using 2 different primer sets (right panel). (C) Wild-type (+/+ or +/–), homozygous trapped Cop1–/– (–/–), and heterozygous embryos harboring one hypomorphic and one trapped allele (h/–) at E10.5 and E14.5, respectively. Control (+/–) mice and mice homozygous for the hypomorphic mutation at P21. Scale bars (left to right): 1 mm, 5 mm, and 3 cm. (D) Total lysates were prepared from E9.5 embryos with the indicated genotypes. Cop1 and vinculin were detected by Western blotting. The relative amount of Cop1 in the various protein lysates is indicated as percentage relative to the amount in wild-type mice (+/+; 100%).
Two different ES clones (XRO653 and XRO177) were used to produce independent Cop1-mutant mouse lines. Since the two lines produced identical phenotypes, we describe herein only one of these lines. In clone XRO653, the pGT01xf vector inserted in intron 6 (Figure 1A). The trapped allele therefore produces a hybrid transcript encoding the N-terminal part of Cop1 fused to the β-galactosidase protein. Since the hybrid protein thus generated would only retain part of the Cop1 functional E3 ligase RING finger domain and lack all C-terminus WD40 repeats that form an essential protein-protein interaction domain, it is expected to be nonfunctional. Nonetheless, the hybrid protein could not be detected in embryos homozygous for the trapped allele (data not shown), suggesting that translation of the fusion transcript does not occur efficiently and/or the hybrid protein is very unstable in vivo. Western blotting analysis failed to detect Cop1–β-galactosidase products using either a monoclonal antibody (28A4) recognizing an epitope located at the N-terminal part of the fusion protein or an antibody recognizing the β-galactosidase protein. Moreover, whole mount lacZ staining of E9.5 and E11.5 embryos harboring at least one trapped allele did not detect β-galactosidase activity, despite elevated and ubiquitous activity of the Cop1 endogenous promoter at these stages of development (data not shown). Finally, Cop1-null embryos, which were generated using a classical gene targeting approach, phenocopied the Cop1 homozygous trapped mutants (see below for a description of the phenotype; V.M. Dixit, personal communication). We therefore concluded that the Cop1 trapped mutation thus generated corresponds to a null mutation. The Cop1 trapped allele is hereafter referred to as Cop1–.
Cop1+/– mice are viable and fertile. However, none of the 215 offspring from heterozygous intercrosses were Cop1– homozygous (Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI45784DS1), suggesting that Cop1-null embryos die during embryogenesis. Although expected Mendelian ratios were observed in embryos isolated at E9.5, the proportion of homozygous embryos declined at later stages, such that no live homozygous embryos were identified among 24 embryos at E12.5 (Supplemental Table 1 and Figure 1B for a representative result of our PCR-based genotyping strategy). At E10.5 all Cop1–/– embryos were smaller than their littermates and developmentally retarded (Figure 1C). Histological and immunohistochemical analyses of E9.5 embryos did not reveal any significant perturbation of cell proliferation or survival. Specifically, we did not observe any significant increase in the number of cells positive for the apoptotic marker cleaved/activated caspase-3 (data not shown). Neither did we detect a significant decrease in the number of cells immunoreactive to the mitosis marker P-H3 (Supplemental Figure 1). The heart malformation seen in most E9.5 Cop1-null embryos (Supplemental Figure 1) suggests that these embryos die from cardiovascular defects. Consistent with such defects, hypoxia-induced genes are among the most significantly upregulated in E9.5 Cop1-null embryos as revealed by genome-wide gene expression profiling (Affymetrix Mouse Genome 430 2.0 Array; GEO GSE18636). Further analyses will be required to establish the specific nature of these cardiovascular defects.
To pursue our goal of generating viable Cop1-deficient mice, we crossed Cop1+/– mice with a Cre-deleter strain in an attempt to generate a hypomorphic allele. We reasoned that low levels, but not complete absence, of Cop1 activity may rescue the recessive lethality associated with the null allele. We chose the Sox2-Cre transgenic line because it promotes high-efficiency recombination in all epiblast-derived tissues (28). A PCR-based genotyping strategy confirmed efficient Cre-mediated excision of the SA site; the non-recombined allele could not be detected by PCR in E9.5 Cre-positive embryos homozygous for the trap allele (Figure 1B, right panel, lane 7). Viable mice heterozygous for the converted, putative hypomorphic allele (Cop1hypo/+) were obtained and either intercrossed or crossed with Cop1+/– mice. No viable Cop1hypo/– mice were obtained (Supplemental Table 2). Timed harvests revealed that Cop1hypo/– embryos were largely indistinguishable, based on appearance and size, from either wild-type or heterozygous (Cop1+/–) littermates prior to E12.5 (Supplemental Table 2), but they all subsequently displayed anemia and hydrops fetalis (Figure 1C). Most Cop1hypo/– embryos died at around E15.5 (Supplemental Table 2). In keeping with the phenotype of the Cop1-null embryos, hydrops fetalis is often the consequence of severe cardiovascular defects.
Importantly, most Cop1hypo/hypo mice were viable, as they were found at a near Mendelian distribution at P4 (Supplemental Table 3). A 15%–20% reduction in body weight was evident in both male and female Cop1hypo/hypo mice compared with wild-type mice at P4, P10, P20, and P40 (Supplemental Figure 2). All organs examined were smaller in Cop1hypo/hypo mice than in wild-type mice. Histological examination of most vital organs and tissues failed to reveal any obvious defects that could account for the reduction in body weight. Of note, most Cop1hypo/hypo mice exhibited a ventral white spotting phenotype (Supplemental Figure 2A, right panel), which could have been a consequence of defects in melanoblast or melanocyte survival, proliferation, or cell migration (29).
In order to confirm hypomorphic expression of Cop1 from the Cre-recombined allele, we harvested E9.5 embryos of various genotypes and performed Western blot analyses (Figure 1D). As predicted, the presence of one Cre-recombined allele (hereafter referred to as the hypomorphic allele; Cop1hypo) led to the expression of low levels of wild-type Cop1. Cop1 protein expression was reduced by 95% in Cop1hypo/– and by 90% in Cop1hypo/hypo embryos.
Together our data show that Cop1 is required for embryonic development. Importantly, our genetic approach allowed the generation of viable Cop1-mutant mice (Cop1hypo/hypo mice) with a 90% decrease in Cop1 expression.
p53 stability is not affected by Cop1 deficiency.
A number of phenotypes associated with Cop1 deficiency are compatible with the previously described role for Cop1 as a negative regulator of p53. Loss of Cop1, similar to loss of Mdmx, leads to an embryonic lethal phenotype at mid-gestation (30, 31). The reduction in body weight observed in Cop1hypo/hypo mice was also reported in mice with hypomorphic expression of Mdm2 (32). To our surprise, however, we found no biochemical evidence for an increase in p53 levels and/or transcriptional activity upon Cop1 inactivation. Western blot analyses failed to detect any increase in basal p53 steady-state levels in E9.5 Cop1-null embryos (Supplemental Figure 3A) and in E14.5 fetal liver from Cop1hypo/+ embryos or any of the tissues analyzed (thymus, spleen, liver, brain, heart, skin) from Cop1hypo/hypo mice (data not shown). Similarly, p53 could not be detected by immunohistochemistry in Cop1-deficient embryonic or adult tissues (data not shown). Steady-state p53 levels were even slightly but reproducibly lower in Cop1hypo/– than in Cop1+/+ early-passage mouse embryonic fibroblasts (MEFs) (Supplemental Figure 3B). Importantly, cycloheximide chase revealed no increase in stability of p53 in Cop1hypo/– MEFs. Consistent with previous studies (33), the estimated p53 half-life was approximately 25 minutes in MEFs of both Cop1 genotypes (Supplemental Figure 3C). Moreover, Cop1hypo/– MEFs proliferated normally in culture and did not undergo p53-mediated growth arrest on a 3T3 schedule (see below and Supplemental Figure 4C). All of these observations are in sharp contrast to what is seen in cells deficient for Mdmx and/or Mdm2 (31, 34).
We also examined the extent and kinetics of p53 stabilization in response to stress. p53 protein levels increase in response to stress signals, such as UV irradiation (35). UV exposure led to accumulation of p53 at comparably high levels in Cop1-proficient and -deficient MEFs (Supplemental Figure 3D). Similar results were obtained when cells were exposed to doxorubicin, a DNA-damaging agent inducing stabilization of p53, or to nutlin-3a, an Mdm2 antagonist (ref. 36 and data not shown). Consistent with these findings, none of the known p53 transcriptional targets were upregulated in E9.5 Cop1-null embryos as assessed by genome-wide gene expression profiling (Affymetrix Mouse Genome 430 2.0 Array; GEO GSE18636). Basal expression levels of key p53 transcriptional targets, such as p21, Mdm2, cyclin G1, Ptprv, Puma, and NoxA, were comparable in Cop1-deficient and -proficient MEFs. p53-dependent induction of the same targets in response to UV was also comparable in these cells (Supplemental Figure 3E). Finally, we did not observe any genetic rescue of the embryonic lethality associated with Cop1 loss on a Tp53-deficient background (Supplemental Figure 3F, upper panel). Moreover, p53 loss did not rescue or delay the embryonic lethal phenotype of Cop1hypo/– mice (Supplemental Figure 3F, lower panel) or the growth defects or white spotting phenotype of Cop1hypo/hypo mice (data not shown). This is again in contrast to the phenotypes associated with Mdmx and Mdm2 deficiencies, all of which are entirely p53 dependent (26). Together these genetic and biochemical data strongly argue against a role for Cop1 in the regulation of p53 stability and activity.
Cop1 regulates c-Jun protein stability in vivo.
Transfection studies had previously identified the oncoprotein c-Jun as a substrate for Cop1-mediated degradation (9, 10). We therefore examined c-Jun protein levels by Western blotting and immunohistochemistry in several Cop1-deficient embryonic and adult tissues (Figure 2). Strikingly, as compared with the relevant controls, a robust increase in c-Jun steady-state levels was observed in E9.5 Cop1-null embryos (Figure 2, A and B), in Cop1hypo/– early-passage MEFs (Figure 2C), and in Cop1hypo/– E14.5 fetal liver extracts (Figure 2C), as well as in several Cop1hypo/hypo tissues in which basal levels of c-Jun were detectable (such as brain, heart, and to a lesser extent thymus). A strict inverse correlation between Cop1 and c-Jun levels was observed in all those tissues (Figure 2C shows heart as a representative example). Immunofluorescence analysis showed that c-Jun accumulates in the nuclei of the Cop1hypo/– MEFs (data not shown).
Figure 2. Cop1 regulates c-Jun protein stability in vivo.
(A–C) Cop1 disruption causes a dramatic increase in c-Jun protein levels in vivo. (A) Total lysates were prepared from E9.5 embryos with the indicated Cop1 genotypes. c-Jun and vinculin (Vinc) were detected by Western blotting. (B) Immunohistochemistry for c-Jun on parasagittal sections from E9.5 Cop1+/– and Cop1–/– embryos. Scale bar (left panels): 1 mm. Original magnification (right panels), ×10. (C) Total lysates were prepared from MEFs, fetal liver cells, and heart of mice with the indicated Cop1 genotypes. c-Jun, Cop1, and vinculin were detected by Western blotting. (D) Determination of c-Jun half-life in early-passage MEFs. MEFs were metabolically pulse labeled using [35S]methionine for 2 hours, and protein extracts were subsequently prepared at the indicated time points. Endogenous c-Jun was immunoprecipitated from these extracts and analyzed by Western blotting. The c-Jun signal was quantified and normalized to a stable and aspecific protein (*). (E) Cells with the indicated Cop1 genotypes were transduced with empty lentivirus (E) or lentivirus encoding full-length (FL) Cop1, a Cop1 deletion mutant lacking its WD40 domain (WD), and a Cop1 RING finger mutant (R). Samples were analyzed by Western blotting for protein expression as indicated. Vinculin served as loading control.
Since Cop1 was also reported to interact with JunB and JunD, two other Jun family bZIP transcription factors (10), we also examined steady-state levels of these proteins in Cop1-deficient cells. We did not observe, however, marked differences in JunB or JunD abundance in these cells as compared with Cop1 wild-type cells (data not shown).
Importantly, the increase in c-Jun protein levels was not accompanied by significant change in c-Jun mRNA levels (data not shown). Metabolic radiolabeling experiments revealed a clear increase in c-Jun stability in Cop1hypo/– early-passage MEFs as compared with control cells. Whereas c-Jun half-life was approximately 3 hours in Cop1 wild-type cells, it was prolonged to more than 6 hours in Cop1hypo/– MEFs (Figure 2D). This observation suggests that Cop1 actively modulates c-Jun turnover. To demonstrate that the increase in c-Jun protein levels was a direct consequence of Cop1 deficiency, we reestablished Cop1 wild-type expression levels in Cop1hypo/– MEFs using a lentivirus-based approach. Expression of full-length Cop1 led to a significant decrease in c-Jun steady-state protein levels. In contrast, expression of a RING finger mutant or a Cop1 WD40 mutant that can no longer interact with c-Jun (10) did not cause any significant decrease in c-Jun levels (Figure 2E).
In conclusion, these data indicate that Cop1 directly modulates c-Jun protein stability in vivo. Cop1-mediated degradation of c-Jun requires an intact WD40 domain, which is essential for direct Cop1–c-Jun interaction. Our data also demonstrate that Cop1-dependent degradation of c-Jun requires an intact Cop1 RING finger domain.
Cop1 modulates AP-1 transcriptional activity in vivo.
To test whether Cop1 modulates c-Jun/AP-1 transcriptional activity in vivo, we intercrossed Cop1+/– and Cop1hypo/+ mice with the AP-1–luciferase (AP1-luc) reporter transgenic mice (37). Surprisingly, basal luciferase activity was comparable and barely detectable in both AP1-luc;Cop1hypo/+ and AP1-luc;Cop1hypo/– MEFs (Figure 3A).
Figure 3. Cop1 modulates AP-1 activity in vivo.
(A–C) MEFs were exposed to 30 J/m2 irradiation. (A) Comparison of AP-1–dependent luciferase activity in Cop1+/+ and Cop1hypo/– MEFs. The data represent the mean ± SD of 3 independent experiments. (B) Samples were analyzed by Western blotting. (C) Transcriptional analysis of c-Jun target genes by Q-RT-PCR analyses. The data were normalized to the level of expression in nontreated control, which is set to 1. The data represent the mean ± SD of 3 independent experiments. (D) Western blot analysis of early-passage MEFs. (E) Transcriptional analysis of c-Jun target genes by Q-RT-PCR analyses in p53-deficient MEFs. The data were normalized to the level of expression in control MEFs, which is set to 1. The data represent the mean ± SD of 2 independent experiments. (F) Comparison of AP-1–dependent luciferase activity in different organs from P6 Cop1hypo/+ and Cop1hypo/hypo mice. The data were first normalized to the total protein levels and then to the luciferase activity observed in Cop1hypo/+ mice, which is set to 1. The data represent the mean ± SD of 4 independent biological replicates. (G) Comparison of AP-1–dependent luciferase activity in skin from P6 Cop1hypo/+ and Cop1hypo/hypo mice exposed to UV-B irradiation (750 mJ/cm2). The data were normalized first to the total protein levels and then to the luciferase activity observed in Cop1hypo/+ mice, which is set to 1. The data represent the mean ± SD of 4 independent biological replicates. (H) Immunohistochemistry for phosphorylated (Ser63) c-Jun on skin sections from P6 Cop1+/+ and Cop1hypo/hypo mice. Mice were either nontreated (NT) or exposed to UV-B irradiation. Original magnification, ×20.
c-Jun transcriptional activity is tightly controlled by posttranslational modification. In response to genotoxic stress, such as UV exposure, JNKs are activated and phosphorylate c-Jun within its transactivation domain to dramatically potentiate c-Jun transcriptional activity (16–18). Accordingly, a rapid and robust increase in c-Jun phosphorylation was readily detectable in UV-treated Cop1+/+ MEFs (Figure 3B). There was a concomitant increase in c-Jun/AP-1 transcriptional activity as evaluated by both the increase in luciferase activity in the AP1-luc;Cop1+/+ cells and induction of selected endogenous c-Jun transcription targets (Figure 3, A and C). In the same experimental conditions, the total pool of phosphorylated c-Jun was more abundant and persistent in Cop1hypo/– MEFs than in control cells. Induction of AP-1 transcriptional activity, as measured by luciferase activity in MEFs positive for the AP1-luc transgene or levels of endogenous expression of c-Jun target genes by quantitative RT-PCR (Q-RT-PCR), was more pronounced in cells compromised for Cop1 function (Figure 3, A and C). Importantly, even though c-Jun basal protein levels were high compared with those in control cells, c-Jun phosphorylation could not be detected in untreated Cop1hypo/– MEFs (Figure 3B).
Inactivation of the p53 pathway also leads to increased basal JNK activity in fibroblasts (38). Accordingly, p53 loss led to a readily detectable increase in c-Jun phosphorylation in Cop1 wild-type MEFs (Figure 3D). In accordance with the above results, the c-Jun phosphorylated pool was more abundant in Cop1hypo/–Tp53–/– double mutant cells than in Cop1+/+Tp53–/– cells. Importantly, the increase in c-Jun phosphorylation correlated with a significant increase in AP-1 transcriptional activity as measured by Q-RT-PCR on selected downstream targets (Figure 3E). This increase in transcriptional activity was more pronounced in Cop1-deficient cells than in Cop1-proficient cells. Thus, in MEFs, Cop1 deficiency significantly increased AP-1 activity upon exposure to stimulatory signals such as UV radiation or p53 inactivation.
Consistent with the MEF data, we did not detect any significant increase in basal luciferase activity in E9.5 Cop1-null compared with Cop1 wild-type embryos (data not shown). Basal luciferase activity in 6.5-day-old Cop1hypo/hypo mice was also comparable to that of control (Cop1hypo/+) littermates in spleen, kidney, heart, and lung and was elevated by less than 2-fold in thymus and brain (Figure 3F). Moreover, none of the known c-Jun transcriptional targets were found to be upregulated in E9.5 Cop1-null embryos as assessed by genome-wide gene expression profiling analysis (Affymetrix GeneChip Mouse Gene 1.0 ST Array; data not shown). We also examined by Q-RT-PCR the consequences of Cop1 downregulation on the expression levels of selected c-Jun targets including Mmp1, Bim, cyclin D1, Plau, and c-Jun itself. In keeping with the above data, there was no detectable induction of these targets either in Cop1-compromised E9.5 embryos or in Cop1hypo/hypo tissues (data not shown).
MAP kinase signaling and consequently basal AP-1 activity in young mice are considerably higher in the skin than in any other organs (39). Accordingly, luciferase activity was approximately 1,000-fold higher in the skin than in thymus, brain, spleen, lung, heart, and kidney of both Cop1 control (Cop1hypo/+) and mutant AP1-luc mice (data not shown). Importantly, in the skin basal AP-1 activity was markedly (7-fold) higher in Cop1-mutant mice than in Cop1hypo/+ mice (Figure 3G). UV-B irradiation is known to induce a readily detectable increase in AP-1 activity in the skin of young mice (40). As expected, a clear increase in AP-1 activity was observed in Cop1hypo/+ mice upon UV-B exposure. AP-1 activity was again more pronounced in Cop1hypo/hypo mice than in the Cop1hypo/+ mice under these experimental conditions (Figure 3G). Hence, Cop1 modulates both basal and UV-induced AP-1 transcriptional activity in vivo. Crucially, it only modulates basal AP-1 activity in specific cell contexts, such as in tissues experiencing high basal MAP kinase activity. Notably, phosphorylated c-Jun could be detected by immunohistochemistry in the skin of both wild-type and Cop1-mutant mice following UV-B exposure; it could also be detected in the epidermis of Cop1hypo/hypo mice prior to UV-B irradiation (Figure 3H).
Together these data indicate that Cop1 modulates AP-1 transcriptional activity. Cop1 deficiency leads to a significant increase in basal AP-1 activity in specific cell contexts in vivo such as skin and in response to stimulatory signals such as UV exposure or increased genomic instability caused by p53 loss. Importantly, this increase strictly correlates with an increase in c-Jun protein levels and phosphorylation.
Cop1 deficiency stimulates cell proliferation in a c-Jun–dependent manner.
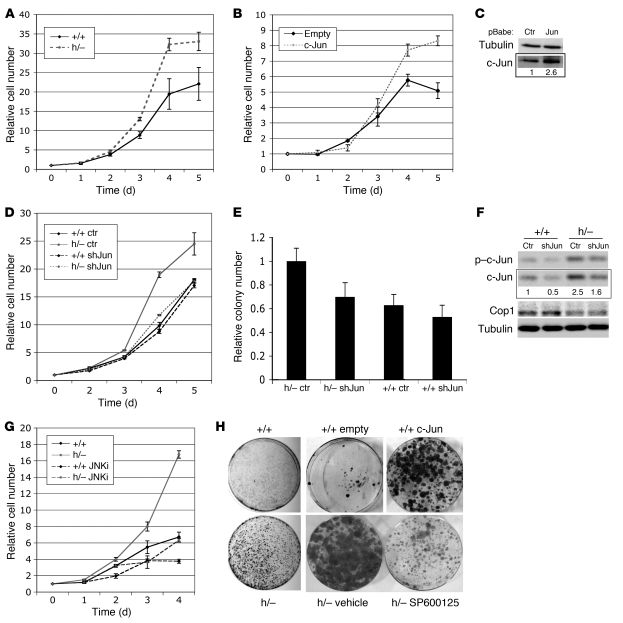
A large body of evidence suggests that increased c-Jun/AP-1 activity positively impacts cell proliferation, cell transformation, and tumor progression (41). We therefore examined the impact of Cop1 deficiency on cell proliferation using the MEFs as a cellular system. Despite high levels of c-Jun expression, MEFs deficient for Cop1 did not show any significant increase in proliferation potential as assessed by direct measurement of cell proliferation or BrdU incorporation assays (Supplemental Figure 4). Wild-type MEFs undergo 10–12 population doublings in culture, after which they cease to divide and develop a so-called senescent phenotype. No significant differences were observed between Cop1-deficient and wild-type cultures on a 3T3 schedule (Supplemental Figure 4). To directly test whether increased c-Jun protein levels are sufficient to enhance MEF proliferation, these cells were transduced with c-Jun–encoding retrovirus, selected for proviral integration, and plated at equal densities. Both cell proliferation and clonogenic assays showed that c-Jun overexpression alone is not sufficient to increase MEF proliferation (data not shown).
We have shown here that Cop1 deficiency only significantly affects AP-1 transcriptional activity in MEFs that are exposed to specific stimulatory signals including loss of p53. We therefore examined cell proliferation of MEFs deficient for both Cop1 and Tp53. Importantly, on a Tp53-null genetic background, early-passage Cop1hypo/– cells grew faster in culture than their wild-type counterparts (Figure 4A). This was further confirmed by clonogenic assays, where both cloning efficiency and colony size were significantly augmented in Cop1-compromised cells (Figure 4H). In addition, overexpression of c-Jun alone also led to a significant increase in the proliferation rate of cells lacking p53 (Figure 4, B, C, and H). Together, these data indicate that Cop1 deficiency promotes proliferation of cells lacking a functional p53 pathway via alterations in c-Jun stability.
Figure 4. Cop1 deficiency stimulates cell proliferation in a c-Jun–dependent manner.
(A–H) All experiments described were performed with cells lacking p53. (A) Growth curve of Cop1+/+ and Cop1hypo/– MEFs; cells were seeded in triplicate at 2 × 105 cells/60-mm dish. The numbers refer to the mean values ± SD of 2 independent MEF cultures. (B) Growth curves of MEFs infected with recombinant retroviruses (pBABE) encoding c-Jun or puromycin resistance only. Cells were seeded in triplicate at 2 × 105 cells/60-mm dish. The numbers refer to the mean values ± SD of 2 independently infected MEF cultures. (C) Samples from B were analyzed by Western blotting. (D) Growth curves of cells expressing shRNAmir–c-Jun (shJun) or shRNAmir-scramble (Ctr). Cells were seeded in triplicate at 2 × 105 cells/60-mm dish. The numbers refer to the mean values ± SD of 2 independently infected MEF cultures. (E) Colony assays were performed with cells expressing shRNAmir–c-Jun or shRNAmir-scramble (Ctr). Data are presented relative to the number of colonies obtained in the Cop1hypo/– cultures infected with the control shRNA, which is set to 1. The numbers refer to the mean values ± SD of 2 independently infected MEF cultures. (F) Samples from D and E were analyzed by Western blotting. (G) Growth curve of Cop1+/+ and Cop1hypo/– MEFs exposed to 10 μM SP600125; cells were seeded in triplicate at 105 cells/100-mm dish. The numbers refer to the mean values ± SD of 2 independent MEF cultures. (H) Colony assays using cells from A, B, and G.
To directly test whether the increase in c-Jun protein levels is causally involved in the increased proliferation potential of Cop1-depleted cells, we generated MEFs expressing shRNAs targeting c-Jun (using GIPZ Lentiviral ShRNAmiR construct 2), from which we subsequently derived polyclonal cell lines expressing distinct c-Jun levels. Cell proliferation and clonogenic assays showed that c-Jun depletion caused cells to significantly reduce their proliferation activity (Figure 4, D and E). Cop1hypo/– cultures expressing c-Jun levels that were comparable to those of their wild-type counterparts (Figure 4F) no longer exhibited increased proliferative capacity. A close correlation between the extent of c-Jun knockdown and the biological effect was observed, arguing against RNAi off-target effects. Moreover, similar results were obtained with cells infected with another GIPZ Lentiviral ShRNAmiR construct (construct 1; data not shown).
c-Jun has to be fully activated by JNKs to promote cell cycle progression (13, 21, 42). Consistent with this view, we observed a strict correlation between the phosphorylation status of c-Jun and proliferative capacity. Indeed, we show that Cop1 deficiency leads to accumulation of a non-phosphorylated pool of c-Jun in Tp53-wild-type MEFs, most likely due to very low basal JNK activities in these cells. These cells did not exhibit any significantly increased proliferative capacity compared with control cells. In contrast, while p53 loss led only to a slight increase in phospho–c-Jun in Cop1 wild-type cells, a dramatic increase in phospho–c-Jun was observed in cells that were also deficient for Cop1 (Figure 3D). These cells proliferated much faster in culture than did the Cop1 control cells. To establish a causative link between the phosphorylation status of c-Jun and the proliferative capacity, we examined cell proliferation of MEFs of different genotypes exposed to different concentrations of the JNK inhibitor SP600125 (hereafter referred to as JNKi) (43). This drug inhibits cell proliferation of immortalized c-Jun wild-type cells, but not c-Jun–/– cells, indicating that it does so through specific effects on the JNK/c-Jun pathway (44). JNKi had little effect on viable cell mass of the immortalized Tp53–/– fibroblasts up to a concentration of 10 μM (data not shown). At such concentrations, JNKi dramatically inhibited proliferation of Cop1hypo/hypoTp53–/– fibroblasts as revealed by both cell proliferation and clonogenic assays (Figure 4, G and H). The inhibitory effect on cell proliferation was also evident in the Cop1+/+Tp53–/– cultures (Figure 4F). Notably, a 10-μM concentration of the JNKi was previously shown to efficiently inhibit JNK activity in comparable experimental conditions (44). Based on these results, we conclude that Cop1 controls cell proliferation by modulating the oncogenic activity of the JNK/c-Jun pathway. Notably, as all these experiments were conducted on a Tp53-null background, Cop1-dependent control of cell proliferation was independent of p53.
Cop1-hypomorphic mice are tumor-prone.
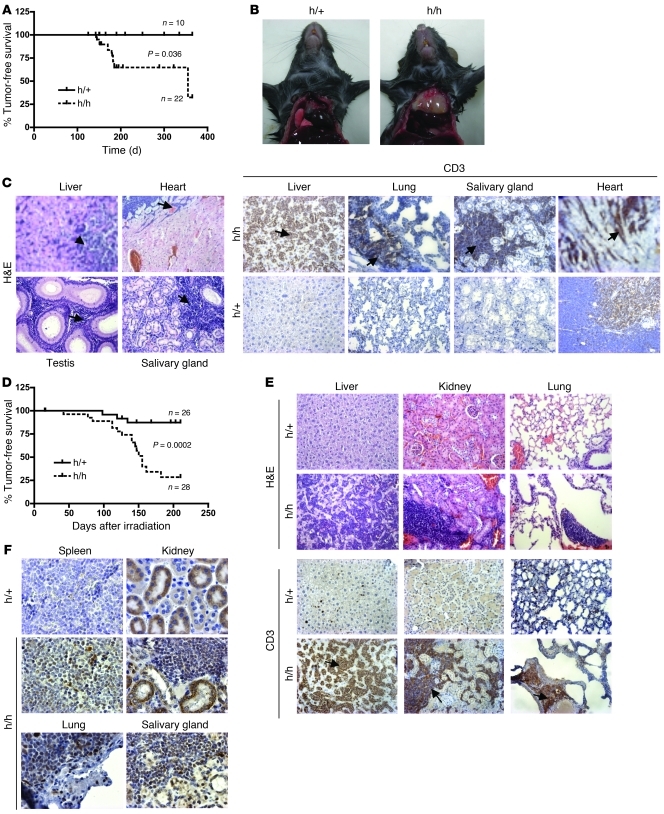
The viable Cop1hypo/hypo mice represent a valuable genetic tool to study the role of Cop1 in tumorigenesis in vivo. The observation that Cop1 counteracts c-Jun–mediated stimulation of cell proliferation predicts that Cop1 may function as a tumor suppressor in vivo. In keeping with this, histopathological analysis revealed that half of the Cop1hypo/hypo mice spontaneously developed tumors in their first year of life (Figure 5A). Most of these mice developed T cell lymphoma with consistent involvement of the cranial mediastinum (thymus) and variable neoplastic infiltrates in multiple other organs including lymph nodes, spleen, lung, liver, salivary gland, kidney, heart, and testis (Figure 5, B and C). Cytomorphology and CD3+B220– immunophenotype of the lymphomas were consistent with a precursor T cell lymphoblastic origin (Figure 5C and data not shown). One mouse developed concurrent histiocytic sarcoma and follicular B cell lymphoma with primary hepatosplenic and nodal involvement. Testicular teratoma was observed in two of the Cop1hypo/hypo males, whereas one female developed an undifferentiated, highly anaplastic uterine tumor. Multifocal melanocytic hyperplasia was also observed in the skin of 4 Cop1hypo/hypo mice. In contrast, no signs of hyperplasia or tumor development were observed in any of the control littermates examined (Cop1hypo/+; 10 of 10). These data indicate that Cop1 deficiency predisposes mice to tumorigenesis.
Figure 5. Cop1hypo/hypo mice are tumor prone.
(A–C) Spontaneous tumor formation in Cop1hypo/hypo mice. (A) 50% of Cop1hypo/hypo mice analyzed (n = 22) by histopathological examination spontaneously developed tumors in their first year of life. None of the Cop1hypo/+ control littermates analyzed (n = 10) showed hyperplasia or tumor development during the course of the experiment (P = 0.036). (B) Most Cop1hypo/hypo mice developed T cell lymphoma. Thymi from Cop1hypo/+ and Cop1hypo/hypo littermates at 20 weeks; the thymus of the Cop1hypo/hypo mouse is tumorous. (C) Left panels: H&E staining of sections of organs from a Cop1hypo/hypo mouse that developed lymphoma (lymphoid infiltrates are indicated by the arrows). Right panels: Immunohistochemistry for CD3 on selected organs from Cop1hypo/+ and Cop1hypo/hypo mice; a dramatic increase in CD3+ cells in Cop1hypo/hypo tissues supports a T cell origin of the infiltrates (arrows). Original magnification, ×14. (D and E) Cop1 deficiency leads to increased infiltration of CD3+ cells and tumorigenesis following exposure to a single sublethal dose (4 Gy) of ionizing radiation. (D) Survival following 4 Gy of whole body irradiation decreased in the Cop1hypo/hypo group (n = 28) at 14–32 weeks following irradiation in comparison with the group of control Cop1hypo/+ littermates (n = 26) (Kaplan-Meier log-rank analysis, P = 0.0002). (E) H&E staining of sections of several organs from a Cop1hypo/+ control and a Cop1hypo/hypo mouse that had developed lymphoma. Immunohistochemistry on several organs from Cop1hypo/hypo mouse shows a dramatic increase in the number of CD3+ cells, suggesting a T cell origin of the infiltrates (see arrows). Original magnification, ×14. (F) Immunohistochemistry on several organs shows high levels of phosphorylated (Ser63) c-Jun in the infiltrates of Cop1hypo/hypo mouse. Original magnification, ×40.
To test this predisposition further, Cop1hypo/hypo and control mice were exposed to a single dose of 4 Gy whole body γ-radiation at 7–12 weeks of age and observed weekly until moribund, then sacrificed and autopsied. The oncogenic effects of radiation in both humans and experimental animals are well documented (45). Labored breathing, hunched body posture, and lethality were observed in most Cop1hypo/hypo animals at 14–30 weeks following irradiation (Figure 5D). Fewer than 10% of the age-matched Cop1hypo/+ controls showed clinical signs of disease or died during the course of the experiment (35 weeks after irradiation). Pathological examination of moribund Cop1hypo/hypo mice revealed the development of aggressive lymphomas with prominent mediastinal (thymic) involvement and extensive infiltration in multiple organs (Figure 5E). Distribution, pattern of growth, cytomorphology, and CD3+B220– immunophenotype of the lymphomas were consistent with a precursor T cell lymphoblastic origin (Figure 5E and data not shown). None of the age-matched Cop1hypo/+ controls exhibited signs of lymphoma or other malignancies (32 weeks after irradiation). Notably, immunohistochemistry indicated that the lymphoma cells found in irradiated Cop1hypo/hypo mice or in mice that developed tumors spontaneously expressed the phosphorylated form of c-Jun (Figure 5F). This observation indicates that in this model, phosphorylated c-Jun may positively impact lymphoma development in a cell-intrinsic manner.
In summary, we found that Cop1hypo/hypo mice have accelerated spontaneous tumor development compared with control mice (Cop1hypo/+) and have a significantly increased incidence of γ-irradiation–induced T cell lymphomas. These data provide the first genetic evidence to our knowledge for a role of Cop1 in tumor suppression.
Loss of COP1 expression in human cancer.
To explore the relevance of our observations in human tumors, we searched the Tumorscape database ( http://www.broadinstitute.org/tumorscape) for evidence of somatic copy number alterations of COP1 across several human cancer types. Focal COP1 deletions are relatively rare (<8% in all cancer types analyzed) but are found in several tumor types including acute lymphoblastic leukemia, an observation that nicely correlates with our mouse data. Interestingly, focal deletions were also observed in melanoma, a tumor type in which the JNK/c-Jun pathway has recently been implicated, and in prostate cancer. Oncomine microarray data were also meta-analyzed to evaluate COP1 gene expression in various human tumors (Oncomine 4.4 Research Edition; http://www.oncomine.org). In keeping with the comparative genomic hybridization (CGH) data, evidence of a significant decrease in COP1 expression was found in glioblastoma, oligondendroglioma, ductal breast carcinoma, and melanoma as compared with expression in normal matching tissues (data not shown). A significant decrease in COP1 expression was also observed in prostate carcinoma (Figure 6A). A large body of evidence suggests that high c-Jun levels favor both androgen-dependent and hormone-refractory human prostate cancer development and/or progression (46, 47). c-Jun is frequently overexpressed in advanced human prostate cancers, and forced expression of c-Jun in prostate cancer cells promotes tumorigenicity (48). We therefore reasoned that loss of COP1 function could contribute, at least partly, to elevated levels of c-Jun in prostate cancer. We analyzed the expression of COP1 and c-Jun in a panel of human prostate cancer cell lines (data not shown). Strikingly, we found an inverse correlation between COP1 and c-Jun protein levels in all prostate cancer cell lines analyzed (Figure 6B shows expression in a few selected cell lines). For instance, while COP1 was readily detectable in LNCaP cells, basal c-Jun steady-state levels were very low. Treatment of these cells with the proteasome inhibitor MG-132 led to a significant increase in c-Jun protein levels, consistent with the notion that c-Jun undergoes constitutive proteasome-dependent degradation in these cells (Figure 6C). Moreover, COP1 knockdown by siRNA led to a significant increase in basal c-Jun protein levels, indicating that c-Jun constitutive degradation is COP1 dependent in these cells (Figure 6D). Importantly, we found that although the chromosomal region encoding COP1 (1q25.1–q25.2) was duplicated in the prostate cancer cell line PC3, the COP1 gene was either completely or partly (exons 17–19) deleted. These rearrangements led to loss of expression of COP1 at both the mRNA and protein levels (Figure 6B and data not shown). In contrast, c-Jun protein levels were elevated in these cells. Reintroduction of COP1 alone, but not of a RING finger deletion mutant, into these cells was sufficient to significantly decrease c-Jun protein levels and decrease cell proliferation (Figure 6, D and E, and data not shown). Together, these observations support the hypothesis that loss of COP1 expression is one of the mechanisms leading to upregulation of the oncoprotein c-Jun in human cancer. Notably, PC3 and its derivatives are TP53-mutant cell lines, indicating that the ability of COP1 to modulate the proliferation rate of these cells is independent of wild-type TP53.
Figure 6. Inverse correlation between COP1 and c-JUN protein levels in human prostate cancers.
(A) COP1 gene-centric expression analysis as revealed by Oncomine. COP1 is significantly underexpressed in a subset of prostate cancers relative to normal prostate tissue (P = 0.001). The y axis represents normalized expression units. Box-and-whisker plots indicate median values of the data and 25th and 75th percentiles of the data sets. Whiskers indicate minimum and maximum data values that are not outliers. P value was calculated by using the Student’s t test. The number of samples (n) in each class is indicated. (B) Western blot analysis of COP1 and c-JUN in several prostate cancer cell lines. COP1 expression is lost in PC3 cells. γ-Tubulin (γ-TUB) served as loading control. (C) Detection by Western blotting of c-JUN and COP1 in PC3 and LNCaP cells treated with the proteasome inhibitor MG-132 for 2, 4, or 6 hours before lysis. γ-Tubulin served as loading control. (D) PC3 cells were transfected with FLAG-tagged expression vectors for COP1. COP1 expression in LNCaP cells was knocked down by siRNA. Western blot analysis confirmed ectopic and knockdown COP1 expression. An inverse correlation between COP1 and c-JUN protein levels was observed. γ-Tubulin served as loading control. (E) Growth curve of PC3 cells transfected with FLAG-tagged empty and COP1 expression vectors; cells were seeded in triplicate at 1.5 × 105 cells/60-mm dish. The numbers refer to the mean values ± SD of 3 independent transfection experiments.
Discussion
Transfection studies have previously implicated Cop1 in ubiquitin-mediated degradation of proteins with opposing functions, including the tumor suppressor p53 and oncoprotein c-Jun, thus creating a conundrum with respect to its role in tumorigenesis. To resolve this issue, we set out to investigate Cop1 function in the physiological setting of a living organism. We used a gene trap–based approach to generate both Cop1-null and hypomorphic alleles. At the technical level, our approach demonstrates the possibility of using the available libraries of mouse ES cells with insertions of the pGT0lxf and pGT0lxr vectors in a generic way to create an allelic series in mice (for a list of genes trapped with these vectors, see http://www.genetrap.org). Crucially, our approach allowed us to make the following key observations in relation to the physiological role of Cop1. First, Cop1 is required for embryonic development, as Cop1-null embryos die around mid-gestation. Second, Cop1 functions as a tumor suppressor in vivo. Mice expressing hypomorphic Cop1 levels are viable but spontaneously develop malignancies at a high frequency at an early age and are susceptible to radiation-induced lymphomagenesis. Third, Cop1 regulates c-Jun protein stability and transcriptional activity in vivo. In contrast, our extensive analyses did not provide any biochemical or genetic evidence in support of a role for Cop1 in the regulation of p53 stability or activity. Fourth, Cop1 deficiency stimulates cell cycle progression in a c-Jun–dependent manner. This observation may indicate a causative molecular link between Cop1 deficiency and the increased susceptibility to tumorigenesis.
We show that Cop1 has intrinsic vital roles in the embryo proper. As we used the Sox2-Cre transgenic line to generate mice with hypomorphic Cop1 expression, in which Cre expression is restricted to epiblast-derived tissues, lethality in Cop1-null embryos is unlikely to be caused by placental defects, one the most common causes of embryonic lethality in mid-gestation (49). Our preliminary phenotypic analyses indicate that the embryonic lethality might result from cardiovascular defects. Consistent with this finding, the hydrops fetalis phenotype seen in Cop1hypo/– mice is also indicative of a role for Cop1 in cardiovascular development. At the molecular level, this phenotype cannot be solely attributed to deregulated c-Jun levels, since we have not been able to rescue/delay the defects and lethality on a c-Jun–null background (data not shown). Among the putative candidates for the mediators of embryonic phenotype are the regulators of glucose and lipid metabolism ACC, CRTC2 (or TORC2), and FoxO1, all of which have been identified as putative targets of Cop1-mediated ubiquitylation and degradation in transfection studies (50–52). A putative role for Cop1 in metabolism could also explain the white spotting phenotype observed in most Cop1hypo/hypo mice. Such a phenotype has previously been attributed to an overall growth deficiency (53), which could itself be a consequence of metabolic defects. Alternatively, white spotting might result from Cop1-mediated degradation of a key regulator(s) of melanoblast or melanocyte survival, proliferation, or cell migration (29). Overall, the pleiotropic defects observed in Cop1-deficient mice most likely reflect the involvement of Cop1 in the regulation of several key factors involved in cell fate, differentiation, proliferation, and survival.
Our data provide the first genetic evidence to our knowledge for a key role of Cop1 in the regulation of c-Jun protein stability. Two other E3 ubiquitin ligases have previously been implicated in c-Jun degradation: Fbw7 (54, 55) and Itch (56, 57). Fbw7- and Itch-mediated c-Jun degradation requires priming phosphorylation events. Itch-dependent degradation of c-Jun depends on JNK-mediated phosphorylation and activation of Itch itself (58). GSK-mediated phosphorylation of c-Jun on Ser243 creates a high-affinity binding site for Fbw7 and is required for its degradation by Fbw7 (55). In neuronal cells, Fbw7 depletion results in accumulation of JNK-phosphorylated c-Jun, stimulation of AP-1 activity, and neuronal apoptosis (54). Hence, both Itch and Fbw7 preferentially target phosphorylated c-Jun and are thus functionally involved in c-Jun degradation only in specific contexts. In contrast, our data argue that Cop1 targets the non-phosphorylated pool of c-Jun. Indeed, despite very high levels of basal c-Jun, phosphorylation at the JNK phosphorylation sites was not detected in most Cop1-deficient cells. Moreover, Cop1 deficiency caused accumulation of c-Jun in tissues with low/no basal JNK activity (most adult tissues). Thus, we propose that Cop1 is the only E3 ligase identified so far that contributes to constitutive UPS-dependent degradation of c-Jun in these tissues.
Unexpectedly, our genetic and biochemical data fail to support a role for Cop1 in the regulation of p53 stability. Since c-Jun has been shown to repress Tp53 transcription (59), one could argue that the increase in c-Jun levels upon Cop1 inactivation might mask a possible contribution of Cop1 to the regulation of p53. Accordingly, a slight but reproducible decrease in p53 mRNA levels was seen in Cop1-deficient cells. However, we did not observe any increase in p53 protein levels in E9.5 embryos lacking both Cop1 and c-Jun (data not shown). Even more importantly, p53 half-life was comparable in Cop1-deficient and -proficient cells. Together, our data fail to support a model in which physiological levels of Cop1 prevent accumulation of p53 in vivo. Crucially, these data strongly argue against the use of Cop1-inhibitory drugs for cancer therapy.
Consistent with the above, examination of the Cop1-hypomorphic mice revealed an essential role for Cop1 in tumor suppression — rather than oncogenesis — in vivo. Notably, spontaneous tumor development, mainly acute lymphoblastic T cell lymphomas, was only observed in 50% of these mice; in animals that developed tumors, the mean tumor latency was rather long (~6 months). We did not detect any evidence of immature T cell hyperproliferation and/or thymic hyperplasia in young Cop1-hypomorphic mice. These observations indicate that a decrease in Cop1 expression predisposes mice to tumorigenesis; however, additional oncogenic events are likely to be required for full-blown tumor formation. Such oncogenic lesions occur spontaneously at a low frequency or can be induced upon systemic genotoxic injury caused, for instance, by whole body γ-irradiation. Strikingly, nearly all of the irradiated Cop1hypo/hypo mice succumbed to tumors, in contrast to similarly treated Cop1 wild-type mice. Genetic analyses of radiation-induced thymic lymphomas had revealed recurrent allelic loss and mutations at a handful of loci including Tp53 (60). We propose that Cop1 deficiency cooperates with these oncogenic events to accelerate tumor formation in vivo. Interestingly, Cop1 deficiency stimulated MEF proliferation in cells deficient for p53 and not in p53 wild-type cells.
Molecularly, we identified the oncoprotein c-Jun as a key physiological target of Cop1-mediated protein degradation in vivo, thus providing a molecular link between Cop1 deficiency and increased tumor susceptibility. This view is supported by our observation that Cop1 deficiency stimulates fibroblast proliferation in a c-Jun–dependent manner. c-Jun oncogenic activity is indeed often attributed to its ability to promote cell cycle progression. Notably, however, the increased incidence of γ-irradiation–induced lymphomagenesis of the Cop1-hypomorphic mice is not rescued when endogenous c-Jun is replaced by a c-Jun mutant allele (JunA/A) in which the two main JNK phosphorylation sites, Ser63 and Ser73, are mutated to alanines (ref. 42 and D. Migliorini, unpublished observations). This result indicates that c-Jun may not be the only Cop1 targets that positively contribute to cancer formation. Recent biochemical evidence indicates that Cop1 targets the three PEA3 group members (61). This group consists of the three highly conserved Ets transcription factors, ERM, ETV1, and PEA3, which are often overexpressed in different types of cancers and have been shown to promote tumorigenesis and metastasis (62, 63). Further genetic experiments are required to assess the specific contributions of the different putative Cop1 targets to the lymphoma phenotype.
Finally, consistent with an evolutionary conserved role of COP1 in tumor suppression, high-resolution CGH analysis indicated that the COP1 locus is deleted in a substantial fraction of tumors across several cancer types ( http://www.broadinstitute.org/tumorscape). We confirmed that COP1 expression is lost in the PC3 prostate cancer cell line through allelic loss. Importantly, we show that reintroduction of COP1 in these cells significantly inhibited cell proliferation. Consistent with our mouse data, a perfect inverse correlation between COP1 and c-Jun levels was observed in these cells. Collectively, these findings argue that COP1 loss of function contributes to the development of human malignancies, at least partly through upregulation of c-Jun levels and activity.
Methods
Plasmids.
DNAs encoding wild-type mouse c-Jun in pBabe (Puro) were a gift from A. Behrens (London Research Institute, Lincoln’s Inn Fields Laboratories, London, United Kingdom). FLAG-tagged human COP1 construct was described previously (10). Mouse GIPZ Lentiviral ShRNAmiR constructs were used to knock down c-Jun in MEFs (Open Biosystems). The hairpin sequences of these constructs were as follows: construct 1, 5′-TGCTGTTGACAGTGAGCGAGCATGTGCTGTGATCATTTATTAGTGAAGCACAGATTAATAAATGATCACAGCACATGCCTGCCTACTGCCTCGGA-3′ and construct 2, 5′-TGCTGTTGACAGTGAGCGCCCTTTGCTTGTGCATATTTATTAGTGAAGC CACAGATGTAATAAATATGCACAAGCAAAGGTTGCCTACTGCCTCGGA-3′. The pENTR3C-mCop1 plasmid was used to generate all Cop1 lentiviral expression vectors. A DraI/XhoI fragment from pENTR3C-mCOP1 was inserted into the EcoRV/XhoI sites of pLV.CMV.bc.PURO to generate the pLV.CMV.bc.PURO-Cop1FL (full-length) expression vector. pLV.CMV.bc.PURO-Cop1 was digested with PshAI and HpaI and ligated onto itself to generate pLV.CMV.bc.PURO-Cop1ΔWD40. We substituted two of the structural RING finger domain residues of Cop1 into alanine (H155A and C158A) using QuikChange II Site-Directed Mutagenesis (Stratagene) and the pENTR3C-mCop1 as template to generate pENTR3C-Cop1RING. A SalI and EcoRV fragment was extracted from pENTR3C-Cop1RING and inserted into HpaI/SalI-digested pLV.CMV.bc.PURO-Cop1 to obtain the pLV.CMV.bc.PURO-Cop1RING mutant vector.
Generation of Cop1-mutant alleles and genotyping.
An ES clone (XR0653) with a single gene trap insertion of pGT01lxf in the Cop1 locus was obtained from the International Gene Trap Consortium. The pGT01lxf vector contains an SA site flanked by loxP sites and a β-geo cassette for positive selection (Figure 1A). To precisely locate the insertion site of the trapping vector, a PCR fragment was amplified, using primers at the 5′ end of the HindIII-linearized plasmid (5′-GGAAAGGGTAAAGTGGTAGG-3′) and in intron 6 of the Cop1 locus (5′-GTTTTCATTAAAGGGAGTCCTC-3′), cloned, and sequenced. The insertion site was mapped to facilitate the design of PCR-based genotyping strategies (see below). This clone was used to generate mice with the unmodified trapped allele (Cop1gt(XR0653)Byg, abbreviated to Cop1–) that were subsequently bred to Sox2-Cre transgenic mice to remove the SA and generate a Cop1-hypomorphic allele (Cop1hypo).
Genotyping.
Genotyping of the Cop1-trapped allele (Cop1gt(XR0653)Byg, abbreviated to Cop1–) was done by PCR using a 3-primer-based method. Two primers are in intron 6 (one forward: 5′-GTTTTCATTAAAGGGAGTCCTC-3′ and one reverse: 5′-CCTAATATGGCTGGGTTATTC-3′) and one primer is in the trapping vector (5′-GGAAAGGGTAAAGTGGTAGG-3′). Genotyping of the Cop1-hypomorphic allele was done by PCR using the following primers: 5′-GCCTGGGGTACCCTATTGGA-3′ and 5′-GGCCTCAGGAAGATCGCACT-3′. Cre-mediated excision of the SA site results in a shift from a 722-bp for the trapped allele to a 277-bp PCR product for the hypomorphic allele. Genotyping of the p53 and c-Jun alleles (a gift from E.F. Wagner, National Cancer Research Centre [CNIO], Madrid, Spain) were done as previously described (64, 65). 2xTRE-luciferase reporter transgenic mice were established by R.A. Flavell and coworkers; genotyping of these mice was performed as previously described (37).
Animal manipulations and histopathological analyses.
All animal experiments were approved by the University of Gent (UGent) and Leuven (K.U.Leuven) Animal Care and Use Ethical Committee (laboratory license number LA1210573). Mice used for these studies were of mixed genetic backgrounds and consisted of 87%–94% C57BL/6 and 6%–13% 126/SvJ. P5 mice were exposed to 750 mJ/cm2 UV-B irradiation using TL20W/12RS Philips UV lamps (peak emission at 310 nm) and a plastic lid to block UV-C emission. The UV-B dose was monitored with an IL1700 radiometer equipped with a SED005/TLS312/W detector (International Light). Where indicated, groups of 8- to 12-week-old mice received a single 4-Gy dose of whole body γ-irradiation from a 137Cs source. After irradiation, mice were monitored twice weekly, and mice showing clinical signs of disease (e.g., hunched posture, labored breathing, immobility) were euthanized. Mice were sacrificed by CO2 asphyxiation, followed by cervical dislocation, and a complete necropsy was performed. Salivary glands, thymus, thyroids, lungs, heart, liver, spleen, kidneys, pancreas, stomach, small intestine, large intestine, urogenital tract, cervical and mesenteric lymph nodes, brain, sternum, and vertebrae with bone marrow were collected in 10% neutral buffered formalin and routinely processed for histological evaluation with H&E. Classification of hematopoietic neoplasm was achieved following the guidelines established by the Hematopathology Subcommittee of the Mouse Models of Human Cancer Consortium (MMHCC) (66, 67). Mice that did not show clinical signs of disease were euthanized 35 weeks after irradiation.
Cell culture.
PC3 and derivatives, BPH1, BPH1205, and LNCAP cell lines were grown in RPMI-1640, while VCaP and DU145 were propagated in DMEM supplemented with 10% fetal bovine serum, 2 mM GlutaMAX (Invitrogen), and 100 U/ml penicillin and 100 mg/ml streptomycin. PC3 cells were transfected with FUGENE HD (Roche). ON-TARGETplus SMARTpool siRNA oligonucleotides (Dharmacon RNAi Technologies, Thermo Scientific) were transfected with Lipofectamine 2000 (Invitrogen). The SMARTpool contains 4 different siRNA molecules that target the following sequences: CAUAAGAACCUGUUAGCUA, GAAUUGGUAUGAAGGGUUA, GCUAAUGUGUGCUGUGUUA, and CUACAAGGAUGUCUCGUAU. Cell lysates were prepared 48 hours after plasmid DNA transfection and 72 hours after oligonucleotide transfection. MEFs were prepared from E12.5 embryos, genotyped, and cultured as previously described (31). MEFs were used at a passage number 5 or less for all analyses except 3T3, which was performed as described previously (68). MEFs were treated with doxorubicin (0.2 μg/ml), nutlin-3a (5 μM), and SP600125 (10 μM) or exposed to UV-C (30 J/cm2). Proliferation and clonogenic assays were performed exactly as previously described (31, 68). For metabolic labeling, cells were washed with methionine-free medium (GIBCO, Life Technologies) and labeled with 200 μCi [35S]methionine (ICN) for 2 hours in methionine-free medium with dialyzed bovine serum. For a chase, the radioactive medium was removed, and cells were washed several times with normal medium and then grown under standard conditions.
Retro- and lentivirus preparation and infection.
pBABE and pBABE-c-Jun with puromycin resistance were transfected into phoenix Eco packaging cells by using CaPO4-based transfection. pLV.CMV.bc.PURO, pLV.CMV.bc.PURO-Cop1FL, pLV.CMV.bc.PURO-Cop1ΔWD40, pENTR3C-Cop1RING, and the GIPZ-shRNAmir–c-Jun constructs with puromycin resistance were cotransfected with packaging and envelop plasmids into HEK293T cells. The pLV.CMV.bc.GFP or GIPZ-shRNAmirz–scramble (GFP) vectors were used to control both the transfection and transduction efficiencies. We collected virus-containing media on days 2 and 3 after transfection. Log-phase target cells were infected with appropriate viruses in media plus 8 μg/ml Polybrene. Media was changed the following day. Twenty-four hours after changing media, selection was performed with puromycin (2 μg/ml) for 3 days.
Histology and immunohistochemistry.
Embryos or dissected organs were fixed overnight in 4% PFA/1× PBS, paraffin embedded, sectioned (5–7 μm), and stained in H&E. For BrdU experiments, pregnant females were injected intraperitoneally with 100 μg BrdU/g body weight and sacrificed 2 hours later. Immunohistochemistry was performed using the following antisera: anti-BrdU (BU33, Sigma-Aldrich); anti–cleaved caspase-3 (Cell Signaling Technology); anti–phosphorylated histone H3 (pHH3) (Calbiochem); anti-p53 (CM5, Novocastra); anti–c-Jun (H79, Santa Cruz Biotechnology Inc.); anti–phosphorylated (Ser63) c-Jun (Cell Signaling Technology); anti-CD3 (Dako); anti-B220 (RA3-6B2, Santa Cruz Biotechnology Inc.). HRP-based detection was performed with the appropriate secondary antibodies and developed with DAB from Vector Laboratories according to the manufacturer’s instructions. Sections were counterstained with hematoxylin.
AP-1 luciferase assays.
MEFs were exposed to UV-C, cultured for an additional 8 hours, and extracted with luciferase lysis buffer (Promega). Luciferase activity was measured using a luminometer (Monolight 2010, Pegasus Scientific Inc.). The luciferase assay reaction was verified to measure the linear range. For the ex vivo luciferase measurements, multiple organs were isolated from 5.5-day-old mice and the relative AP-1 activity was measured as previously described (39). Results are expressed as relative AP-1 activity (RLU).
Western blotting analysis.
Lysates were prepared as previously described (31). Extracts (75 mg each) were fractionated by SDS-PAGE and transferred to nitrocellulose. The primary antibodies used were as follows: anti-Cop1 (28A4, a gift from V. Dixit, Department of Physiological Chemistry, Genentech Inc., South San Francisco, California, USA); anti–c-Jun (H79, Santa Cruz Biotechnology Inc.); anti–phosphorylated (Ser63) c-Jun (Cell Signaling Technology); anti-p53 (1C12, Cell Signaling Technology), anti-p21 (F5, Santa Cruz Biotechnology Inc.), anti-p19ARF (ab80, Abcam), anti-FLAG (Sigma-Aldrich); and anti–β-tubulin and anti–γ-tubulin (Sigma-Aldrich) or anti-vinculin (clone hVIN-1, Sigma-Aldrich) or β-actin (ab8229, Abcam) for normalization. Secondary antibodies used include peroxidase-conjugated goat anti-rabbit IgG and anti-mouse IgG (Pierce). Proteins were detected using ChemiDoc-It 500 (UVP). VisionWorksLS software (UVP) was used to calculate the signal level of each sample.
TaqMan real-time Q-RT-PCR assays.
Total RNA was prepared from cell pellets using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s protocol. Total RNA (1 μg) was reverse transcribed in a final volume of 20 μl using a SuperScript kit (Invitrogen). These assays were performed following the manufacturer’s specifications (PE Applied Biosystems). Primer pairs and TaqMan probes were designed by Applied Biosystems (Assays On Demand). Each sample was analyzed in triplicate.
Cancer gene expression studies.
For the analysis of COP1 expression data, Oncomine ( http://www.oncomine.org) was searched for studies comparing tumor with normal tissue containing probes for COP1. Gene expression differences were analyzed by 2-sided Student’s t test using GraphPad Prism software version 4.03. Details of standard normalization methods and statistical calculations are provided on the Oncomine website.
Statistics.
Numerical values are reported as mean ± SD. For comparisons between 2 groups, P values were calculated using either paired or unpaired 2-tailed Student’s t tests. A P value of 0.05 or less was accepted as statistically significant.
Supplementary Material
Acknowledgments
We thank Natalie Farla, Natacha Rosseel, and Rose Van Isacker for excellent technical assistance. We thank Torik Ayoubi and Joke Allemeers from the VIB MicroArray Facility (MAF) for help with the gene expression profiling experiments. We thank V. Dixit, A. Vitari, A. Behrens, J. Haigh, and M. Skipper for helpful discussions and comments on the manuscript. We thank E. Wagner for providing the c-Jun–KO mice. A. Zwolinska is supported by the Research Foundation — Flanders (FWO). J-.C. Marine received support from the EMBO Young Investigator program. This work was supported in part by Geconcerteerde Onderzoek Aangelegenheden (GOA, University Ghent, Belgium), Association for International Cancer Research, “Belgian Foundation against Cancer,” and EU (FP6 program, ACTIVEP53, contract 503576). This publication reflects only the authors’ views. The European Commission is not liable for any use that may be made of the information herein.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(4):1329–1343. doi:10.1172/JCI45784.
See the related Commentary beginning on page 1263.
References
- 1.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6(5):369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 2.Marine JC, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2010;17(1):93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- 3.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358(6381):80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 4.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26(15):3453–3459. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wade M, Wahl GM. Targeting Mdm2 and Mdmx in cancer therapy: better living through medicinal chemistry? Mol Cancer Res. 2009;7(1):1–11. doi: 10.1158/1541-7786.MCR-08-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dornan D, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429(6987):86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 7.Dornan D, et al. ATM engages autodegradation of the E3 ubiquitin ligase COP1 after DNA damage. Science. 2006;313(5790):1122–1126. doi: 10.1126/science.1127335. [DOI] [PubMed] [Google Scholar]
- 8.Dornan D, et al. COP1, the negative regulator of p53, is overexpressed in breast and ovarian adenocarcinomas. Cancer Res. 2004;64(20):7226–7230. doi: 10.1158/0008-5472.CAN-04-2601. [DOI] [PubMed] [Google Scholar]
- 9.Bianchi E, et al. Characterization of human constitutive photomorphogenesis protein 1, a RING finger ubiquitin ligase that interacts with Jun transcription factors and modulates their transcriptional activity. J Biol Chem. 2003;278(22):19682–19690. doi: 10.1074/jbc.M212681200. [DOI] [PubMed] [Google Scholar]
- 10.Wertz IE, et al. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science. 2004;303(5662):1371–1374. doi: 10.1126/science.1093549. [DOI] [PubMed] [Google Scholar]
- 11.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072(2–3):129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 12.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4(5):E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 13.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3(11):859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 14.Derijard B, et al. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76(6):1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 15.Kyriakis JM, et al. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369(6476):156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 16.Arias J, et al. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370(6486):226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 17.Bannister AJ, Oehler T, Wilhelm D, Angel P, Kouzarides T. Stimulation of c-Jun activity by CBP: c-Jun residues Ser63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene. 1995;11(12):2509–2514. [PubMed] [Google Scholar]
- 18.Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J Biol Chem. 1995;270(28):16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- 19.Fuchs SY, Dolan L, Davis RJ, Ronai Z. Phosphorylation-dependent targeting of c-Jun ubiquitination by Jun N-kinase. Oncogene. 1996;13(7):1531–1535. [PubMed] [Google Scholar]
- 20.Wang ZQ, Liang J, Schellander K, Wagner EF, Grigoriadis AE. c-fos-induced osteosarcoma formation in transgenic mice: cooperativity with c-jun and the role of endogenous c-fos. Cancer Res. 1995;55(24):6244–6251. [PubMed] [Google Scholar]
- 21.Behrens A, Jochum W, Sibilia M, Wagner EF. Oncogenic transformation by ras and fos is mediated by c-Jun N-terminal phosphorylation. Oncogene. 2000;19(22):2657–2663. doi: 10.1038/sj.onc.1203603. [DOI] [PubMed] [Google Scholar]
- 22.Eferl R, et al. Liver tumor development. c-Jun antagonizes the proapoptotic activity of p53. Cell. 2003;112(2):181–192. doi: 10.1016/s0092-8674(03)00042-4. [DOI] [PubMed] [Google Scholar]
- 23.Mathas S, et al. Aberrantly expressed c-Jun and JunB are a hallmark of Hodgkin lymphoma cells, stimulate proliferation and synergize with NF-kappa B. EMBO J. 2002;21(15):4104–4113. doi: 10.1093/emboj/cdf389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez-Bergami P, et al. Rewired ERK-JNK signaling pathways in melanoma. Cancer Cell. 2007;11(5):447–460. doi: 10.1016/j.ccr.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mariani O, et al. JUN oncogene amplification and overexpression block adipocytic differentiation in highly aggressive sarcomas. Cancer Cell. 2007;11(4):361–374. doi: 10.1016/j.ccr.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 26.Marine JC, Francoz S, Maetens M, Wahl G, Toledo F, Lozano G. Keeping p53 in check: essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 2006;13(6):927–934. doi: 10.1038/sj.cdd.4401912. [DOI] [PubMed] [Google Scholar]
- 27.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6(12):909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 28.Hayashi S, Lewis P, Pevny L, McMahon AP. Efficient gene modulation in mouse epiblast using Sox2Cre transgenic mouse strain. Gene Expr Patterns. 2002;2(1–2):93–97. doi: 10.1016/S0925-4773(02)00292-7. [DOI] [PubMed] [Google Scholar]
- 29.Baxter LL, Hou L, Loftus SK, Pavan WJ. Spotlight on spotted mice: a review of white spotting mouse mutants and associated human pigmentation disorders. Pigment Cell Res. 2004;17(3):215–224. doi: 10.1111/j.1600-0749.2004.00147.x. [DOI] [PubMed] [Google Scholar]
- 30.Parant J, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29(1):92–95. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- 31.Migliorini D, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol. 2002;22(15):5527–5538. doi: 10.1128/MCB.22.15.5527-5538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendrysa SM, McElwee MK, Michalowski J, O’Leary KA, Young KM, Perry ME. mdm2 Is critical for inhibition of p53 during lymphopoiesis and the response to ionizing irradiation. Mol Cell Biol. 2003;23(2):462–472. doi: 10.1128/MCB.23.2.462-473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krummel KA, Lee CJ, Toledo F, Wahl GM. The C-terminal lysines fine-tune P53 stress responses in a mouse model but are not required for stability control or transactivation. Proc Natl Acad Sci U S A. 2005;102(29):10188–10193. doi: 10.1073/pnas.0503068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Francoz S, et al. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci U S A. 2006;103(9):3232–3237. doi: 10.1073/pnas.0508476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26(9):1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- 36.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 37.Rincon M, Flavell RA. AP-1 transcriptional activity requires both T-cell receptor-mediated and co-stimulatory signals in primary T lymphocytes. EMBO J. 1994;13(18):4370–4381. doi: 10.1002/j.1460-2075.1994.tb06757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramaswamy NT, Pelling JC. Mutational status of the p53 gene modulates the basal level of jun N-terminal kinase and its inducibility by ultraviolet irradiation in A1-5 rat fibroblasts. Mol Carcinog. 1999;25(4):262–272. doi: 10.1002/(sici)1098-2744(199908)25:4<262::aid-mc5>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 39.Zhong SP, Ma WY, Quealy JA, Zhang Y, Dong Z. Organ-specific distribution of AP-1 in AP-1 luciferase transgenic mice during the maturation process. Am J Physiol Regul Integr Comp Physiol. 2001;280(2):R376–R381. doi: 10.1152/ajpregu.2001.280.2.R376. [DOI] [PubMed] [Google Scholar]
- 40.Huang C, Ma WY, Hanenberger D, Cleary MP, Bowden GT, Dong Z. Inhibition of ultraviolet B-induced activator protein-1 (AP-1) activity by aspirin in AP-1-luciferase transgenic mice. J Biol Chem. 1997;272(42):26325–26331. doi: 10.1074/jbc.272.42.26325. [DOI] [PubMed] [Google Scholar]
- 41.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103(2):239–252. doi: 10.1016/S0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 42.Behrens A, Sibilia M, Wagner EF. Amino-terminal phosphorylation of c-Jun regulates stress-induced apoptosis and cellular proliferation. Nat Genet. 1999;21(3):326–329. doi: 10.1038/6854. [DOI] [PubMed] [Google Scholar]
- 43.Bennett BL, et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc Natl Acad Sci U S A. 2001;98(24):13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du L, et al. Inhibition of cell proliferation and cell cycle progression by specific inhibition of basal JNK activity: evidence that mitotic Bcl-2 phosphorylation is JNK-independent. J Biol Chem. 2004;279(12):11957–11966. doi: 10.1074/jbc.M304935200. [DOI] [PubMed] [Google Scholar]
- 45.Barcellos-Hoff MH, Park C, Wright EG. Radiation and the microenvironment — tumorigenesis and therapy. Nat Rev Cancer. 2005;5(11):867–875. doi: 10.1038/nrc1735. [DOI] [PubMed] [Google Scholar]
- 46.Edwards J, Krishna NS, Mukherjee R, Bartlett JM. The role of c-Jun and c-Fos expression in androgen-independent prostate cancer. J Pathol. 2004;204(2):153–158. doi: 10.1002/path.1605. [DOI] [PubMed] [Google Scholar]
- 47.Chen SY, et al. c-Jun enhancement of androgen receptor transactivation is associated with prostate cancer cell proliferation. Oncogene. 2006;25(54):7212–7223. doi: 10.1038/sj.onc.1209705. [DOI] [PubMed] [Google Scholar]
- 48.Ouyang X, et al. Activator protein-1 transcription factors are associated with progression and recurrence of prostate cancer. Cancer Res. 2008;68(7):2132–2144. doi: 10.1158/0008-5472.CAN-07-6055. [DOI] [PubMed] [Google Scholar]
- 49.Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet. 2001;2(7):538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- 50.Qi L, et al. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science. 2006;312(5781):1763–1766. doi: 10.1126/science.1123374. [DOI] [PubMed] [Google Scholar]
- 51.Dentin R, et al. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature. 2007;449(7160):366–369. doi: 10.1038/nature06128. [DOI] [PubMed] [Google Scholar]
- 52.Kato S, Ding J, Pisck E, Jhala US, Du K. COP1 functions as a FoxO1 ubiquitin E3 ligase to regulate FoxO1-mediated gene expression. J Biol Chem. 2008;283(51):35464–35473. doi: 10.1074/jbc.M801011200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oliver ER, Saunders TL, Tarle SA, Glaser T. Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development. 2004;131(16):3907–3920. doi: 10.1242/dev.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nateri AS, Riera-Sans L, Da Costa C, Behrens A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science. 2004;303(5662):1374–1378. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- 55.Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8(1):25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 56.Gao M, et al. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science. 2004;306(5694):271–275. doi: 10.1126/science.1099414. [DOI] [PubMed] [Google Scholar]
- 57.Fang D, Kerppola TK. Ubiquitin-mediated fluorescence complementation reveals that Jun ubiquitinated by Itch/AIP4 is localized to lysosomes. Proc Natl Acad Sci U S A. 2004;101(41):14782–14787. doi: 10.1073/pnas.0404445101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang L, et al. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell. 2006;124(3):601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 59.Schreiber M, et al. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13(5):607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kominami R, Niwa O. Radiation carcinogenesis in mouse thymic lymphomas. Cancer Sci. 2006;97(7):575–581. doi: 10.1111/j.1349-7006.2006.00218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baert JL, et al. The E3 ubiquitin ligase complex component COP1 regulates PEA3 group member stability and transcriptional activity. Oncogene. 2010;29(12):1810–1820. doi: 10.1038/onc.2009.471. [DOI] [PubMed] [Google Scholar]
- 62.de Launoit Y, et al. PEA3 family of transcription factors and the regulation of oncogenesis. Bull Cancer. 1996;93(10):985–989. [PubMed] [Google Scholar]
- 63.Jané-Valbuena J, et al. An oncogenic role for ETV1 in melanoma. Cancer Res. 2010;70(5):2075–2084. doi: 10.1158/0008-5472.CAN-09-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jacks T, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4(1):1–7. doi: 10.1016/S0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 65.Hilberg F, Aguzzi A, Howells N, Wagner EF. c-jun is essential for normal mouse development and hepatogenesis. Nature. 1993;365(6442):179–181. doi: 10.1038/365179a0. [DOI] [PubMed] [Google Scholar]
- 66.Kogan SC, et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100(1):238–245. doi: 10.1182/blood.V100.1.238. [DOI] [PubMed] [Google Scholar]
- 67.Morse HC 3rd, et al. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood. 2002;100(1):246–258. doi: 10.1182/blood.v100.1.246. [DOI] [PubMed] [Google Scholar]
- 68.Danovi D, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol. 2004;24(13):5835–5843. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.