Abstract
Background
Plasmodium falciparum is responsible for the most acute form of human malaria. Most recent studies demonstrate that it belongs to a monophyletic lineage specialized in the infection of great ape hosts. Several other Plasmodium species cause human malaria. They all belong to another distinct lineage of parasites which infect a wider range of primate species. All known mammalian malaria parasites appear to be monophyletic. Their clade includes the two previous distinct lineages of parasites of primates and great apes, one lineage of rodent parasites, and presumably Hepatocystis species. Plasmodium falciparum and great ape parasites are commonly thought to be the sister-group of all other mammal-infecting malaria parasites. However, some studies supported contradictory origins and found parasites of great apes to be closer to those of rodents, or to those of other primates.
Results
To distinguish between these mutually exclusive hypotheses on the origin of Plasmodium falciparum and its great ape infecting relatives, we performed a comprehensive phylogenetic analysis based on a data set of three mitochondrial genes from 33 to 84 malaria parasites. We showed that malarial mitochondrial genes have evolved slowly and are compositionally homogeneous. We estimated their phylogenetic relationships using Bayesian and maximum-likelihood methods. Inferred trees were checked for their robustness to the (i) site selection, (ii) assumptions of various probabilistic models, and (iii) taxon sampling. Our results robustly support a common ancestry of rodent parasites and Plasmodium falciparum's relatives infecting great apes.
Conclusions
Our results refute the most common view of the origin of great ape malaria parasites, and instead demonstrate the robustness of a less well-established phylogenetic hypothesis, under which Plasmodium falciparum and its relatives infecting great apes are closely related to rodent parasites. This study sheds light on the evolutionary history of Plasmodium falciparum, a major issue for human health.
Background
Malaria is an overwhelming public health problem all over the world. It kills one to three million people annually and infects 200 to 500 million others [1]. Human malaria is induced by infections caused by a range of eukaryotic protists belonging to the phylum Apicomplexa. These organisms possess an endosymbiont of red algal origin [2] derived into an apical organelle, the apicoplast. This organelle is specialized in host cell invasion [3]. Within Apicomplexa, malaria parasites, also called Haemosporidia, are characterized by their infection of vertebrate hosts, haemoglobin digestion, and a complex life cycle involving dipteran vectors feeding from their vertebrate hosts' blood [4].
Haemosporidia include the genera Leucocytozoon (bird parasites), Haemoproteus and Parahaemoproteus (Sauria, i.e. bird and reptile parasites), Plasmodium (saurian and mammalian parasites) and Hepatocystis (mammalian parasites) [4-6]. These five genera have long been defined by their morphological differences (e.g. storage of products of haemoglobin degradation in the case of Haemoproteus, Parahaemoproteus, Plasmodium and Hepatocystis), variations in their life cycle (e.g. asexual replication stage in erythrocytes for Haemoproteus, Parahaemoproteus and Plasmodium) and host and vector specificity [4,5,7,8]. However, studies which attempted to link these phenotypic and life history traits with molecular data concluded that the latter provide deeper insight into the evolutionary history of Haemosporidia, and allow the identification of cryptic species which cannot be distinguished using only microscopy observations [9,10]. The increasing availability of molecular data has enabled numerous studies of Haemosporidia phylogeny, improving our understanding of the evolutionary history of malaria parasites.
To date, five Plasmodium species have been shown to cause human malaria: P. falciparum, P. vivax, P. malariae, P. ovale and P. knowlesi [11-14]. P. falciparum has stimulated the interest of the scientific community, largely because it is the most virulent, but also because of the very high A+T contents of its genome. Indeed, it has an average content of 80% A+T over its nuclear genome, with intergenic regions frequently displaying more than 90% A+T [15]. Furthermore, the question of the origin of P. falciparum is intriguing: its 18 S rRNA gene diverges strikingly from that of the other Plasmodium species which infect primates, of which four cause human malaria [16-19]. Another parasite of interest is P. reichenowi, which infects chimpanzees. P. reichenowi has long been the only known close relative of P. falciparum [6,18-24]. However, since 2009, it has been recognized that both P. falciparum and P. reichenowi belong to a wider monophyletic lineage of parasites specialized in the infection of great ape hosts: gorilla, chimpanzee, bonobo and human [25-28].
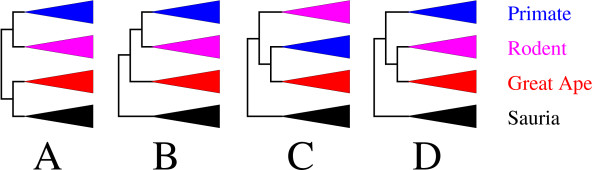
Despite intense efforts to obtain accurate molecular phylogenies, the position of P. falciparum and of great ape malaria parasites within Haemosporidia remains unclear. In recent decades, many molecular phylogenies have been produced which clustered P. falciparum (alone, or with P. reichenowi) closer to avian than to mammalian parasites (Figure 1A). This led to the hypothesis of a recent switch from avian to human (and great ape) hosts, rather than a shared ancestry of P. falciparum and other mammalian malaria parasites [16-19,21,29-35]. However, it has recently been recognized that, in these early works, the clustering of P. falciparum with avian malaria parasites may have resulted mainly from stochastic noise due to the insufficient number of sequences analyzed [35]. More recent studies relied on larger data sets and demonstrated that all malaria parasites known to exclusively infect mammals, including P. falciparum, are monophyletic [6,20-28,36]. Hence, this monophyletic clade of mammalian malaria parasites includes (i) a lineage infecting great apes [25-28], (ii) a distantly related and less specialized lineage infecting primates [11], (iii) a third lineage of rodent parasites [6,37], and (iv) species of the Hepatocystis genus infecting bats and primates [5,6,21]. Most studies inferred great ape parasites to be a sister-group to all other mammal parasites [6,20-22,24,25,27,28,36] (Figure 1B). However, a few studies yielded contradictory results. Some suggested that great ape parasites could be closer to other primate parasites [14] (Figure 1C), while others suggested that great ape parasites could be related to rodent parasites [23,27] (Figure 1D).
Figure 1.
Competing hypotheses for the origin of Plasmodium falciparum and great ape malaria parasites. A: The avian origin hypothesis [16-19,21,29-35]. 1B: Great ape parasites sister-group to all other mammal parasites [6,20-22,24,25,27,28,36]. 1C: Close relationship with primate malaria parasites [14]. 1D: Close relationship with rodent malaria parasites [23,27].
In the present study, we distinguished between these mutually exclusive hypotheses to identify the origin of Plasmodium falciparum and other great ape parasites. We analyzed three genes from 33 publicly available complete Haemosporidian mitochondrial genomes, as well as 51 additional sets of one to three mitochondrial genes from Hepatocystis and Plasmodium species infecting various mammalian and saurian hosts. Concatenated nucleotide and amino-acid alignments were analyzed using various probabilistic models of sequence evolution, applying both maximum-likelihood (ML) and Bayesian inference (BI) methods. Statistical measurements of fit and posterior predictive experiments enabled the adequacy of model assumptions to the data to be checked. We also evaluated the robustness of the obtained phylogeny to the taxonomic sampling by removing and adding taxa. Our phylogenetic analyses provide robust support for a close relationship between great ape and rodent parasites (Figure 1D).
Results and Discussion
Preliminary measures
In this study, we analyzed coding genes from the 33 complete Haemosporidian mitochondrial genomes available in 2009 (see Methods and Additional file 1, Table S1). Each genome includes only three coding genes. In this section, we describe the general features of the resulting alignments.
In the three concatenated genes and 33 taxa data set, 54% of the 1099 amino-acid sites, and 51% of the 3308 nucleotide sites were constant. The nucleotide and amino-acid alignments are thus highly conserved. Genes displayed the typical pattern in which substitution rates are higher at the third and first codon positions, and lower at the second codon positions. The first and second codon positions are highly conserved (55% and 77% of constant positions, respectively) and the saturation plot indicates that they are slightly saturated (slopes of 0.19 and 0.89 respectively, Additional file 2, Figure S1). Although third codon positions evolved more rapidly and are thus more saturated (20% of constant sites, slope of 0.09, Additional file 2, Figure S1), another standard test for saturation (PAUP 4.0 [38], partition-homogeneity test by codon position) indicated that they are not significantly more saturated than the first and second codon positions (p = 1).
The nucleotide data set had a 74% A+T content and was compositionally homogeneous (p = 0.83, PAUP χ2 test of compositional homogeneity across taxa). However, the amount of A+T and the compositional homogeneity differed strikingly depending on the codon position. First and second codon positions displayed relatively low A+T contents (68% and 64% respectively) and were homogeneous (p = 1, PAUP χ2 test of compositional homogeneity across taxa). In contrast, third codon positions had a high A+T content (89%) and were compositionally heterogeneous (p = 0, PAUP χ2 test of compositional homogeneity across taxa). This difference in composition between codon positions suggests that the typical drift of Haemosporidian species toward A+T richness was negatively selected at first and second codon positions and less constrained at third positions, most likely due to constraints at the protein level.
Standard phylogenetic analyses
Phylogenetic reconstructions in this section were obtained with the most widely used models, which have both maximum-likelihood (ML) and Bayesian implementations. MrAIC [39] estimated that, among 56 models of nucleotide substitution, the best AIC score [40] was achieved by the most general homogeneous and reversible model (GTR, "general time reversible" [41]), combined with four discrete gamma categories of "rates across sites" [42], plus an additional rate category for invariant sites (model GTRnt + Γ4 + I, where subscript nt stands for nucleotides). This model was also selected by MrAIC with individual codon positions. We used the PhyloBayes 3.0 software [43] for Bayesian analyses, which does not implement the invariant rate category. Bayesian analyses of nucleotide alignments were thus performed under the GTRnt + Γ4 model.
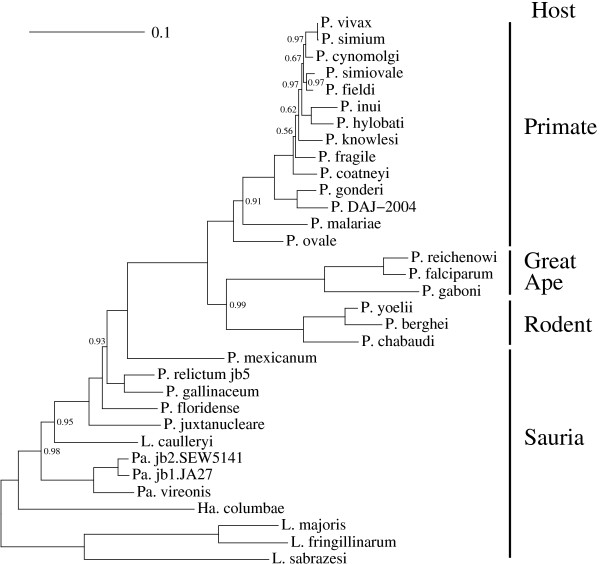
ML and Bayesian analyses of the nucleotide data set under GTRnt + Γ4 + I and GTRnt + Γ4 models, respectively, strongly supported a clade containing the 20 mammal infecting Plasmodium species (Figure 2) [6,20-25,27,28,36]. The mammal malaria parasite clade comprises three strongly supported main lineages. One lineage is specialized in infecting great ape hosts and includes P. falciparum, P. reichenowi and P. gaboni [25-28]. The second lineage is characterized by African and Asian primate hosts and comprises 14 Plasmodium species [11]. The third lineage includes P. berghei, P. yoelii and P. chabaudi, the African rodent malaria parasites [37]. Most interestingly, the two lineages of great ape and rodent parasites clustered together with strong posterior probability (PP = 0.99, Figure 2). ML analysis also yielded significant support for this clade (Shimodaira-Hasegawa-like support SH = 0.93, bootstrap support BS = 0.82, Table 1).
Figure 2.
Phylogeny of mitochondrial genes of 33 Haemosporidian species. Bayesian phylogenetic reconstruction under the GTRnt + Γ4 model, using PhyloBayes 3.0 [43]. P. falciparum and two of its relatives infecting great ape hosts, P. reichenowi and P. gaboni, form a monophyletic clade with three rodent parasites, P. yoelii, P. berghei and P. chabaudi (posterior probability PP = 0.99). Posterior probabilities equal to 1 were removed. Abbreviations "P.": Plasmodium species, "He.": Hepatocystis species, "Ha.": Haemoproteus species, "Pa.": Parahaemoproteus species, "L.": Leucocytozoon species.
Table 1.
Support dependency on the model assumptions
| Model | Type of Data | Statistical Support | ||
|---|---|---|---|---|
| PP | SH | BS | ||
| GTRnt + Γ4 (+I) | ||||
| CAT +GTRnt + Γ4 | Nucleotide | 0.995 | * | * |
| GTRnt +BP + Γ 4 | 0.949 | * | * | |
| JTT + Γ4 (+I) | 0.919 | 0.46 | 0.416 | |
| GTRaa + Γ4 | 0.978 | * | * | |
| CAT + Γ4 | Amino Acid | 0.629 | * | * |
| CAT +JTT + Γ4 | 0.802 | * | * | |
| CAT +BP + Γ4 | 0.747 | * | * | |
Support dependency of the great ape and rodent parasites monophyly on assumptions of various probabilistic models of substitution (GTRnt, GTRaa and JTT: single matrix model, CAT: site heterogeneous mixture model, BP : time heterogeneous model). Rates across site model components are defined as + Γ4 + I under maximum-likelihood (ML) and as + Γ4 under Bayesian inference (BI) methods. PP: Posterior probability (BI), SH: Shimodaira-Hasegawa-like support (ML), and BS: bootstrap support (ML). "*": not applicable.
First and second codon positions, and third codon positions were analyzed separately, using the same models of nucleotide evolution as previously. Both data sets supported, although weakly, a clade containing great ape and rodent malaria parasites, hereafter denoted "monophyly of great ape and rodent parasites" (Additional file 3, Table S2, column "Rodent").
Finally, ProtTest [44] determined that the best fit to the amino-acid alignment was provided by the JTT + Γ4 + I + F model [45]. We used the JTT + Γ4 model for the Bayesian analysis of the amino-acid alignment. The monophyly of great ape and rodent parasites received weak SH and bootstrap support (SH = 0.46, BS = 0.41), but relatively high posterior probability (PP = 0.92, Table 1).
SH and BS supports are more conservative than posterior probabilities, which are generally expected to be higher [46,47]. Moreover, given the high level of conservation of protein and individual codon position alignments (see above), the complete nucleotide alignment is expected to provide more phylogenetic signal and higher supports. Thus, all previous results are congruent, with differing but explainable levels of confidence, and support a monophyly of great ape and rodent parasites.
Assessment of model violation and robustness to the model choice
To evaluate the influence of potential model violations, we used improved Bayesian models implemented in PhyloBayes 3.0 software [43]. The fits of these additional models to the data were measured using cross-validated likelihood (see Methods and Additional file 4, Table S3). Moreover, we applied posterior predictive tests which measure the model ability to accurately reproduce observed features of the data (see Methods and Additional file 5, Table S4). In this section, we only comment on experiments where posterior predictive tests were not rejected (i.e. there was significant violation of model assumptions). In this case, we conclude that a specific feature of the data is correctly anticipated by the model assumptions.
We wanted to evaluate the potential effects of site saturation on our estimates. Site-heterogeneous mixture models such as "CAT" [48] (see Methods) efficiently deal with violations caused by high saturation levels [48,49]. However, this model might lack resolution power, especially in the case of small data sets [48]. In addition, we wanted to measure the potential effect of compositional biases on our estimates. Consequently, we analyzed data sets under the time-heterogeneous model "BP" [50,51], which is designed to deal with compositional heterogeneity across taxa (see Methods).
Saturation in the nucleotide alignment
Among the three additional models considered, CAT + GTRnt + Γ4 [43] (see Methods) yielded the best fit to the nucleotide data set, and it outperformed GTRnt + Γ4 by 103 points of cross-validated likelihood (Additional file 4, Table S3). Moreover, posterior predictive tests showed that this model correctly anticipated the level of saturation of the nucleotide data set (p > 0.28, Additional file 5, Table S4). This suggests that the CAT + GTRnt + Γ4 model is not misled by site saturation. CAT + GTRnt + Γ4 strongly supported the monophyly of great ape and rodent parasites, considering either all codon positions (PP = 0.99, Table 1), first and second (see Additional file 6, Figure S2), or third codon positions (PP = 0.92 and PP = 0.65 respectively, Additional file 3, Table S2).
Compositional heterogeneity in the nucleotide alignment
Third codon positions were compositionally heterogeneous (see above χ2 tests), and they carried 54% of variable sites. Hence, potential convergence of sequence compositions could have misled the previously used time-homogeneous models [50-52]. Interestingly, posterior predictive tests showed that the compositional heterogeneity across taxa of first and second codon positions was correctly anticipated under the time-homogeneous models GTRnt + Γ4 and CAT + GTRnt + Γ4 (p > 0.44, Additional file 5, Table S4), suggesting that these models are relatively robust to compositional changes in this data set.
The time-heterogeneous model GTRnt + BP + Γ4 [50] (see Methods) explicitly accounts for variations in composition across taxa. It correctly anticipated the observed compositional heterogeneity, considering either all, first and second, and third codon positions (p > 0.11, Additional file 5, Table S4), suggesting that this model is unlikely to be misled by compositional heterogeneity across taxa. The monophyly of great ape and rodent parasites was strongly supported under the GTRnt + BP + Γ4 model, considering either all codon positions (PP = 0.95, Table 1), first and second, or third codon positions (PP = 0.99 and PP = 0.72 respectively, Additional file 3, Table S2).
Model violations in the amino-acid alignment
According to the analysis of the amino-acid data set, methodological bias may arise from the use of the universal replacement model JTT. The peculiar A+T rich composition of Haemosporidian genes could lead to slightly different estimations for the exchange rate parameters and hence alter the probability of clustering great ape and rodent parasites together. Accordingly, we used the GTRaa model (where subscript aa stands for amino acids) which does not rely on a pre-estimated replacement matrix like JTT. The GTRaa + Γ4 model strongly supported the monophyly of great ape and rodent parasites (PP = 0.98, Table 1).
Cross-validations indicated that, among the 12 alternative models, the site-heterogeneous model CAT + JTT + Γ4 [43] (see Methods) provided the best fit to the amino-acid data set (Additional file 4, Table S3). According to posterior predictive tests, this model correctly anticipated the saturation level observed in the data (p > 0.07, Additional file 5, Table S4). Moreover, the site- and time- heterogeneous model CAT + BP + Γ4 [51] (see Methods) correctly anticipated the level of saturation of the amino-acid data set (p = 0.41). However, the posterior predictive test for compositional heterogeneity across taxa was rejected under CAT + BP + Γ4 (p = 0.02), although as expected, this model anticipated compositional heterogeneity better than do time-homogeneous models (p = 0.001, Additional file 5, Table S4). Both the two last models moderately supported the monophyly of rodent and great ape parasites (PP = 0.80 and PP = 0.75 under CAT + JTT + Γ4 and CAT + BP + Γ4 respectively, Table 1). In both cases, few variable amino-acid positions are interpreted under highly parameter rich models, and moderate support is therefore to be expected.
Most importantly, all results in this section are congruent with our initial estimate. This suggests that measured model violations do not significantly bias the relationship of great ape parasites among mammal parasites. In other words, the monophyly of great ape and rodent parasites appears to be robust to the choice of the model, as well as to its assumptions and dimensionality.
Robustness to taxon sampling
To avoid possible biases resulting from an ad hoc set of sequences, it is important to assess the robustness of this phylogenetic association with respect to taxon selection, which was achieved by analyses of 30 different taxonomic samples. Among previously considered data sets, the complete nucleotide data set had the highest number of variable positions to interpret (1627), and our experiments showed that it is unlikely to induce strong model violation, whatever the phylogenetic model considered. Hence, in the following sections, we present phylogenetic analyses of complete nucleotide alignments performed under models GTRnt + Γ4 + I (ML) and GTRnt + Γ4 (Bayesian inference).
Robustness to taxon removal
First, we checked the influence on phylogenetic reconstructions of the selected great ape parasites. According to the initial taxon selection, this lineage includes P. falciparum, P. reichenowi and P. gaboni. Six additional combinations of these three taxa were devised (see Methods). The weakest (but still relatively high) support for the monophyly of great ape and rodent parasites was obtained when P. reichenowi was considered as the only representative of its lineage (PP = 0.93, SH = 0.78, BS = 0.68, Additional file 7, Table S5). Second, we devised six combinations of the three rodent parasites, P. berghei, P. yoelii and P. chabaudi. The data set with P. berghei as the only representative of its lineage yielded the weakest support for the monophyly of great ape and rodent parasites (PP = 0.56, BS = 0.52, Additional file 7, Table S5), and the ML tree weakly supported the alternative hypothesis of a monophyly of primate and rodent parasites (i.e. Figure 1B, SH = 0.05). Third, six combinations of primate parasites were considered. Only the combination with African primate parasites (P. gonderi and P. DAJ-2004) as the only representatives of their lineage supported the alternative hypothesis of a monophyly of primate and rodent parasites (i.e. Figure 1B, PP = 0.85, SH = 0.26, BS = 0.48, Additional file 7, Table S5). The five other combinations of primate parasites supported the monophyly of great ape and rodent parasites (PP > 0.96, SH > 0.60, BS > 0.71, Additional file 7, Table S5). Finally, we investigated the robustness to the taxon composition of the outgroup (i.e. mammal and saurian parasites were considered as ingroup and outgroup, respectively). All six devised outgroups yielded high support for the monophyly of great ape and rodent parasites (PP > 0.98, SH > 0.64, BS > 0.74, Additional file 7, Table S5).
Hence, with the exception of three taxonomic samples, in which (i) P. reichenowi, (ii) P. berghei and (iii) P. gonderi and P. DAJ-2004 were considered as the only representatives of their respective lineages, all other 21 combinations of taxa provided good support for the association of great ape and rodent parasites (PP > 0.96, SH > 0.60, BS > 0.71, Additional file 7, Table S5).
Robustness to taxon addition
First, (i) 8 CytB genes from great ape parasites, (ii) 10 pairs of CytB and Cox1 genes from rodent parasites and, (iii) 27 pairs of CytB and Cox1 genes from Plasmodium species infecting a wide range of sauria hosts (Additional file 8, Table S6) were added in turn to the initial 33-taxon data set. The association of great ape and rodent parasites was still strongly supported (PP > 0.99, SH > 0.77, BS > 0.78, Additional file 7, Table S5), and the lineages of great ape, rodent, and mammal parasites were each still shown to be monophyletic (PP = 1).
Second, all previous taxa were analyzed together (33 taxa + 8 great ape + 10 rodent + 27 saurian parasites), yielding a 78-taxon tree. The monophyly of great ape and rodent parasites was still strongly supported (PP = 0.99, SH = 0.86, BS = 0.76, Additional file 7, Table S5).
Third, we added six Hepatocystis species to the initial 33-taxon nucleotide data set. These six parasites were monophyletic (PP = 1). They clustered within the clade of mammal parasites, which was then composed of four monophyletic main lineages. The monophyly of great ape and rodent parasites was weakly supported (PP = 0.40, SH = 0.05, BS = 0.38), but this low support was entirely due to high uncertainty with respect to the position of Hepatocystis within mammalian malaria parasites. Indeed, Hepatocystis were located, with weak support, in five positions on trees in which great ape and rodent parasites were located close together (e.g. Hepatocystis as a sister-group to great ape parasites: PP = 0.28, BS = 0.28, or to rodent parasites: PP = 0.31, BS = 0.29). We evaluated posterior and bootstrap support for the great ape being close to, but not necessarily monophyletic with, rodent parasites (i.e. the great ape plus rodent parasite lineage could also include Hepatocystis). When support was summed over the three possible positions of Hepatocystis relative to the association of great ape and rodent parasites, then parasites of great apes and of rodents were located close together with strong support (PP = 0.99 and BS = 0.87, Additional file 7, Table S5).
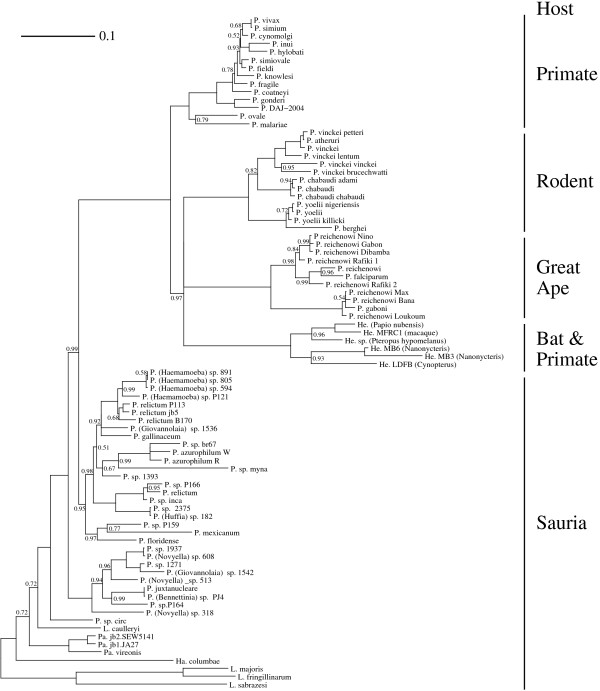
Fourth, the six Hepatocystis species were analyzed simultaneously with all the previous 78 taxa, yielding an 84-taxon tree (Figure 3). A strict monophyly of great ape and rodent parasites was weakly supported (PP = 0.28, SH = 0, BS = 0.31), due to high uncertainty with respect to the position of Hepatocystis (e.g. Hepatocystis as a sister-group to great ape parasites: PP = 0.36, BS = 0.25, or to rodent parasites: PP = 0.35, BS = 0.25). However, disregarding the exact position of Hepatocystis, great ape and rodent parasites were located close together with high support (PP = 0.99, BS = 0.75, Additional file 7, Table S5).
Figure 3.
Phylogeny of 84 Haemosporidian species. Bayesian phylogenetic reconstruction under the GTRnt + Γ4 model, using PhyloBayes 3.0 [43]. Posterior probabilities equal to 1 were removed, edges with posterior probability PP < 0.5 were collapsed. Abbreviations "P.": Plasmodium species, "He.": Hepatocystis species, "Ha.": Haemoproteus species, "Pa.": Parahaemoproteus species, "L.": Leucocytozoon species.
Thus, all six previous additions of mitochondrial genes did not alter the result indicating a likely close phylogenetic relationship of great ape and rodent parasites. Overall, statistical supports averaged over the 30 taxonomic samples considered in this section showed a robust relationship of great ape and rodent parasites 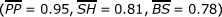 . This suggests that this relationship does not depend solely on the selection of the taxa we considered here. However, the uncertainty concerning the exact position of Hepatocystis species within mammal parasites challenges the above mentioned monophyly of a clade only comprising parasites of great apes and of rodents, because it would be possible for Hepatocystis to cluster within that clade. Nonetheless, whatever the true position of Hepatocystis may be, it does not contradict our main result indicating that great ape parasites are unlikely to be a sister-group to all other mammal parasites [6,20-22,24,25,27,28,36] (Figure 1B), but instead, probably share a more recent common ancestor with rodent parasites [23,27] (Figure 1D).
. This suggests that this relationship does not depend solely on the selection of the taxa we considered here. However, the uncertainty concerning the exact position of Hepatocystis species within mammal parasites challenges the above mentioned monophyly of a clade only comprising parasites of great apes and of rodents, because it would be possible for Hepatocystis to cluster within that clade. Nonetheless, whatever the true position of Hepatocystis may be, it does not contradict our main result indicating that great ape parasites are unlikely to be a sister-group to all other mammal parasites [6,20-22,24,25,27,28,36] (Figure 1B), but instead, probably share a more recent common ancestor with rodent parasites [23,27] (Figure 1D).
Comparison with previous studies
In previous studies, the hypothesis of great ape parasites being a sister-group to all other mammal parasites was defended by analyses of mitochondrial genes [6,21,22,24,27,35] or complete mitochondrial genomes [24,25,28], of nuclear coding genes [53] and ribosomal RNA [20], or by combining genes from nuclear and mitochondrial genomes with genes from the apicoplast genome [36]. Moreover, considering different rooting assumptions breaking the monophyly of mammal parasites, this result was also obtained through analyses of nuclear 18 S rRNA [11,18], nuclear genes [29,32,34] and mitochondrial cytochrome b genes [21].
Most of these studies of the Haemosporidia phylogeny relied on a single gene, and only a few taxa data sets, which might lack phylogenetic signal [35]. In contrast, two recent studies analyzed larger data sets. The first study analyzed a large number of taxa (40), but few concatenated genes (4) [36], whereas the second focused on a large number of genes (104), but considered very few taxa (8) [53]. Next, we suggest possible reasons for the disagreement between the results of these two studies and ours.
Comparison with a taxa-wide phylogenetic analysis
As a general guideline, wider taxon sampling usually helps to resolve phylogenies more accurately, provided enough genes are available to overcome stochastic noise, and are also sufficiently conserved to avoid systematic errors [54]. In line with this idea, Martinsen et al. [36] analyzed four concatenated genes for a relatively wide sample of 40 taxa. Among the previous works, their experimental conditions are thus the closest to ours. But, intriguingly, our results do not confirm theirs. We suggest that the disagreement between the two studies is due to several factors, the first being the differences in the phylogenetic markers analyzed.
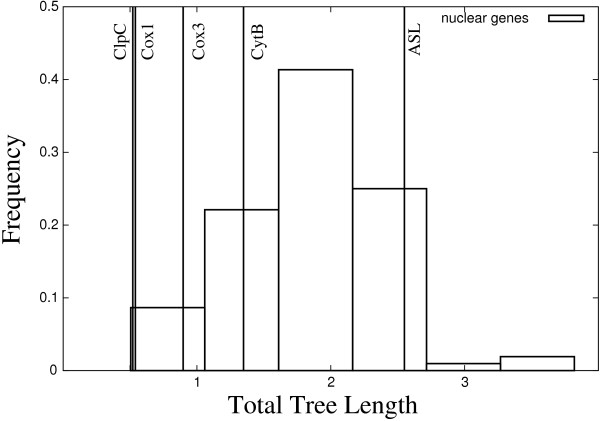
Both Martinsen et al. [36] and our study considered CytB and Cox1 mitochondrial genes. However, Martinsen et al. [36] additionally analyzed adenylosuccinate lyase (ASL) and caseinolytic protease C (ClpC) genes, whereas we analyzed the third mitochondrial gene Cox3. In order to compare global rates of evolution between these genes, we measured the total lengths of gene trees [55], for a common sub set of eight taxa (P. falciparum, P. reichenowi, P. vivax, P. knowlesi, P. berghei, P. chabaudi, P yoelii and P. gallinaceum). Our values indicate that the ASL genes evolved 3 to 5 times faster than the slowest evolving genes: ClpC, Cox1 and Cox3 (Figure 4). The signal to noise ratio is expected to be higher for slowly evolving phylogenetic markers, and fast rates of evolution generally reduce the accuracy of inferred phylogenetic trees [49,56]. In addition, we considered ASL and ClpC genes of 18 and 27 taxa respectively, for taxonomic samples as close as possible to our originally selected 33 taxa (Additional file 9, Table S7). The rapidly evolving gene, ASL, did not support a monophyly of mammal parasites, suggesting strong systematic errors (Additional file 10, Figure S3). In contrast, the slow evolving gene, ClpC, supported this monophyly (PP = 0.99, SH = 0.94, BS = 0.51), but did not support any particular position of P. falciparum within mammal parasites (Additional file 11, Figure S4). However, a recent study of 14 ClpC genes supported a common ancestry of great ape and rodent parasites [57].
Figure 4.
Estimation of length of 8-taxa trees, for 109 genes. Maximum-likelihood estimation of total lengths of 8- and 7- taxa gene trees, under GTRnt + Γ4 + I using PhyML 3.0 [47]. Taxa: P. falciparum, P. reichenowi, P. vivax, P. knowlesi, P. berghei, P. chabaudi, P yoelii and P. gallinaceum. Vertical lines: lengths of 8-taxa trees for CytB, Cox3 and Cox1 mitochondrial genes, and of 7-taxa trees, for ClpC and ASL (no sequence available for P. reichenowi, see accessions in Additional file 9, Table S7). Square boxes: distribution of lengths of 8-taxa trees over the 104 orthologous nuclear genes of Dávalos and Perkins [53].
Moreover, most CytB, ClpC and ASL sequences analyzed by Martinsen et al. [36] were partial CDS. Thus, even considering four genes, their alignment covered 2334 nucleotide sites, representing 70% of the 3308 nucleotide sites considered in the present study. Finally, they considered fewer mammalian malaria parasites than we did (11 taxa in the study of Martinsen et al. [36], versus 20 to 44 taxa in our work), and they considered P. falciparum as the only representative of the lineage of great ape parasites, together with P. vivax and P. knowlesi as the only representatives of primate parasites. All these differences in experimental conditions (i.e. saturation of ASL gene, fewer sites, fewer mammal parasites close to P. falciparum) could together contribute to the difference between the results of Martinsen et al. [36] and ours.
Comparison with a genome-wide phylogenetic analysis
Dávalos and Perkins [53] extracted a set of 104 putative orthologous nuclear genes from the eight complete genomes of Plasmodium species sequenced to date (P. falciparum, P. reichenowi, P. vivax, P. knowlesi, P. berghei, P. chabaudi, P yoelii and P. gallinaceum). Their phylogenetic analyses of individual genes displayed discrepancies with respect to the inferred trees. Approximately half the 104 genes supported a monophyly of primate and rodent parasites (Figure 1B) [6,20-22,24,25,27,28,36]. Alternative hypotheses of a monophyly of great ape and primate parasites (Figure 1C) [14], and of great ape and rodent parasites (Figure 1D) [23,27], were each supported by nearly a quarter of these 104 genes. Interestingly, for the same reduced sample of eight taxa, our phylogenetic analyses of mitochondrial genes, taken separately, displayed comparable discrepancies (Table 2). Moreover, the supports obtained showed a near-random resolution of the internal branch of the 8-taxon individual gene trees. This suggests that, rather than showing a global preference of individual genes for the monophyly of primate and rodent parasites, the analysis of individual genes by Dávalos and Perkins [53] might have been strongly influenced by stochastic noise.
Table 2.
Effects of reduced taxon and site sampling
| Supports for Great Ape parasites sister-group of: | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Number of Taxa | Genes | Rodent | Primate+Rodent | Primate | ||||||
| PP, | SH, | BS | PP, | SH, | BS | PP, | SH, | BS | ||
| CytB | 0.221, | *, | 0.306 | 0.612, | 0.29, | 0.444 | 0.166, | *, | 0.250 | |
| 8 Taxa | Cox1 | 0.152, | *, | 0.374 | 0.848, | 0.45, | 0.610 | 0.000, | *, | 0.016 |
| Cox3 | 0.922, | 0.63, | 0.659 | 0.002, | *, | 0.013 | 0.070, | *, | 0.313 | |
| Conc. | 0.971, | 0.58, | 0.655 | 0.029, | *, | 0.316 | 0.000, | *, | 0.029 | |
| CytB | 0.911, | 0.77, | 0.548 | 0.074, | *, | 0.225 | 0.013, | *, | 0.009 | |
| 33 Taxa | Cox1 | 0.022, | *, | 0.336 | 0.976, | 0.61, | 0.606 | 0.001, | *, | 0.043 |
| Cox3 | 0.985, | 0.90, | 0.801 | 0.003, | *, | 0.011 | 0.012, | *, | 0.092 | |
| Conc. | 0.999, | 0.93, | 0.821 | 0.001, | *, | 0.174 | 0.000, | *, | 0.004 | |
Dependency of clade support on a reduced (8 Taxa) or extended (33 Taxa) taxon sampling, and on individual (CytB, Cox1 and Cox3 genes) or concatenated (Conc.) gene analyses. Models GTRnt + Γ4 + I and GTRnt + Γ4 were applied to nucleotide alignments, under maximum-likelihood (ML) and Bayesian (BI) methods, respectively. Cells display support as [PP, SH, BS] with PP: posterior probability (BI), SH: Shimodaira-Hasegawa-like support (ML), and BS: bootstrap support (ML). "*": not applicable. Main lineages of mammalian parasites are defined according to their host preference: "Rodent", "Primate" and "Great Ape" (see Additional file 1, Table 1).
Increasing the amount of signal by concatenating genes helps to alleviate the effects of stochastic noise. The analysis of the three concatenated mitochondrial genes, for the eight taxa, supported the monophyly of great ape and rodent parasites (PP = 0.97, SH = 0.58, BS = 0.65; Table 2). In contrast, the concatenation of the 104 nuclear genes yielded strong support for the monophyly of primate and rodent parasites [53] (Figure 1B). We estimated the total tree lengths of each of these 104 genes, as well as of their concatenation. First, mitochondrial genes evolved as slowly as the  fraction of the 104 nuclear genes which displayed the slowest rate of evolution (Figure 4). Second, total tree length estimated for the 104 gene concatenation indicated a fast average rate of evolution, about two times faster than that of the three mitochondrial genes. These observations confirm the fact that mitochondrial genes are well conserved, and corroborate the conclusion of Dávalos and Perkins [53] indicating that most of their 104 genes are highly saturated and evolved relatively fast. Thus, the monophyly of primate and rodent parasites (Figure 1B), obtained by the latter authors from a large concatenation of 104 genes, most likely results from systematic errors that may be due to the high saturation level of most genes [53,58], but also presumably to the small sample of only eight taxa [54].
fraction of the 104 nuclear genes which displayed the slowest rate of evolution (Figure 4). Second, total tree length estimated for the 104 gene concatenation indicated a fast average rate of evolution, about two times faster than that of the three mitochondrial genes. These observations confirm the fact that mitochondrial genes are well conserved, and corroborate the conclusion of Dávalos and Perkins [53] indicating that most of their 104 genes are highly saturated and evolved relatively fast. Thus, the monophyly of primate and rodent parasites (Figure 1B), obtained by the latter authors from a large concatenation of 104 genes, most likely results from systematic errors that may be due to the high saturation level of most genes [53,58], but also presumably to the small sample of only eight taxa [54].
Comparison with two corroborated studies
In contrast, our results support a common origin of great ape and rodent parasites (Figure 1D). This corroborates results of a recent study published by Perkins [23] who, to the best of our knowledge, was the first to mention this hypothesis. This author sequenced seven new mitochondrial genomes and reconstructed the phylogeny of a sample of 24 taxa. A similar phylogeny was also obtained from 38 partial CytB sequences [27]. However, the two previous studies did not discuss the robustness of this result, but instead suggested it should be considered with caution [23,27], as most previous studies of mitochondrial genes supported a monophyly of primate and rodent parasites (Figure 1B) [6,21,22,24,24,25,27,28,35].
However, in our results, this hypothesis never obtained significant statistical support: at most, it reached PP = 0.35 and BS = 0.57 with the 33-taxon amino-acid alignment (Additional file 3, Table S2, column "Primate+Rodent"), and it obtained averaged supports of 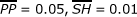 and
and 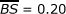 over the 30 additional taxon samples considered (Additional file 7, Table S5, column "Primate+Rodent").
over the 30 additional taxon samples considered (Additional file 7, Table S5, column "Primate+Rodent").
Interestingly, each time the monophyly of great ape and rodent parasites (Figure 1D) was not significantly supported, the only alternative hypothesis which could not be statistically rejected was the monophyly of primate and rodent parasites (i.e. support for "Primate+Rodent" in Additional files 3 and 7, Tables S2 and S5, where PP > 0.05). Moreover, nine additional samples of 19 taxa were drawn so that trees would display long branches, and we obtained five trees in which great ape parasites were shown to be a sister-group to primate and rodent parasites (Additional file 12, Table S8). This suggests that the monophyly of primate and rodent parasites (Figure 1B), rather than the monophyly of great ape and rodent parasites (Figure 1D), might result from the effect of long branch attraction. Thus, the slight tendency of mitochondrial genes to weakly support a monophyly of primate and rodent parasites, along with differences in taxon sampling and gene selection, could explain the disagreements between our results and most previous studies.
Conclusions
With special focus on the still unclear phylogenetic position of great ape parasites, which include Plasmodium falciparum [25-28], in this study, we explored the phylogeny of Haemosporidian species by analyzing their mitochondrial genes. We showed that these genes have evolved relatively slowly and are mostly compositionally homogeneous, which characterizes them as potentially accurate phylogenetic markers. Corroborating many results obtained over the past few years, we obtained a monophyly of mammalian malaria parasites. Within that clade, we observed four main host-specialized lineages of parasites: Plasmodium species infecting (i) primate, (ii) rodent and (iii) great ape hosts, and (iv) Hepatocystis species infecting bats and primates. The inferred relationships within host-specialized lineages of Plasmodium parasites are congruent with the literature [11,26,37]. Hepatocystis species have received little attention to date. According to our results, these parasites may have diverged within mammalian malaria parasites, but their exact origin remains unclear. Our results support a common ancestry of great ape and rodent parasites (Figure 1D). We showed that this phylogenetic relationship is robust to various experimental conditions, demonstrating that it is unlikely to arise from an artefact of tree reconstruction.
Our study focused on mitochondrial genes. Nevertheless, it is still unclear if mitochondrial genomes match the Haemosporidian species tree or not [23]. This question could be answered by comparing the respective phylogenies of the three genomes hosted by Haemosporidian species (i.e. phylogenies of the mitochondrial, nuclear and apicoplast genomes). However, appropriate data sets for phylogenetic reconstruction of nuclear and apicoplast genomes are not yet available. A successful strategy for resolving a gene- and taxa- wide phylogeny would involve targeted sequencing of identified slowly evolving genes from the apicoplast and nuclear genomes. Careful phylogenetic analyses of such new and accurate phylogenetic markers will likely help to definitely resolve the phylogenetic origins of Plasmodium falciparum and other great ape parasites.
Methods
Mitochondrial gene and protein alignments
Complete mitochondrial genomes of 33 Haemosporidian parasites [22,23,25,59-66] were downloaded from the NCBI website (Additional file 1, Table S1). Available taxa included four Leucocytozoon species infecting birds [23,66]. These parasites are the closest relatives of other already identified Haemosporidia [6], and were used as outgroups. We collected mitochondrial genomes of four Haemoproteus and Parahaemoproteus species [23,62], and of five Plasmodium species [22,23,62,65] infecting saurian hosts (birds and reptiles). We collected a set of 20 mitochondrial genomes of mammal malaria parasites, including those of three rodent [59], three great ape [25,60,61] and 14 primate parasites [22,63,64].
Mitochondrial genomes of Haemosporidian species are vestigial and have a typical length of 6, 000 base pairs [67,68]. They form linear concatemers, each repeated unit encoding fragments of ribosomal genes together with three coding genes involved in the electron transport chain: cytochrome b and cytochrome oxidase subunits I and III (denoted as CytB, Cox1 and Cox3, respectively). Both nucleotide coding sequences and their translations into amino acids were retrieved according to annotated CDS. No annotation of the mitochondrial genes was available for Plasmodium relictum jb5, Parahaemoproteus jb1.JA27 and Parahaemoproteus jb2.SEW5141. In this case, gene sequences were extracted manually and unambiguously, given the high conservation level of start and end positions shared by all 30 other annotated genes.
Nucleotide sequences were aligned using MACSE (Ranwez V, Harispe S, Delsuc F, and Douzery EJP, "MACSE: Multiple Alignment of Coding Sequences accounting for frameshifts and stop codons", manuscript in preparation). This method computes the alignment of coding nucleotide sequences with respect to their possible translations. It attempts to minimize the occurrence of frameshifts and stop codons. We applied the relevant codon table, as indicated on the NCBI website [69]. MACSE identified and corrected three long frameshifts in the Cox3 gene of P. berghei, and in CytB genes of Ha. columbae and L. caulleryi. These frameshifts resulted in erroneous translations of the corresponding publically available amino-acid sequences. Consequently, we used the translated alignment computed by MACSE, rather than the alignment of the official NCBI translations. Individual gene and protein alignments were filtered with Gblocks 0.91 [70], and allowing a maximum of half gap states per site (option -b5 = h). Filtered alignments of nucleotide and amino-acid sequences were finally concatenated. This yielded two concatenations, one of 3308 nucleotide sites (number of nucleotide sites for CytB: 1125, Cox1: 1434, Cox3: 749), and one of 1099 amino-acid positions. The assembled data sets are available at the following URL: http://www.lirmm.fr/mab/blanquart
Phylogenetic inferences
Description of phylogenetic models
Data sets were analyzed under various probabilistic models of molecular evolution. We applied the JTT [45] replacement model to the amino-acid data set. We applied the most general time reversible model GTRnt, where subscript nt denotes nucleotides, to the nucleotide data sets [41]. These substitution models were run using both maximum-likelihood (ML) and Bayesian inference (BI) methods. This allowed for the use of different statistical supports with different meanings, and comparison of the phylogenies estimated with the two approaches.
ML phylogenetic reconstructions were performed using PhyML 3.0 [47]. Irrespective of the substitution model (GTRnt, JTT) used for the analysis, the phylogenetic model additionally involved four discrete categories of gamma distributed rates across sites (denoted + Γ4, [42]), plus an invariant site category (denoted +I). The proportion of invariant sites and the shape parameter of the gamma distribution were estimated from the data. When the nucleotide data sets were analyzed, all eight free parameters of the GTRnt substitution models were estimated from the data (GTRnt + Γ4 + I, 10 degrees of freedom). In the case of the amino-acid alignment analyzed under JTT, stationary probabilities were set to empirical frequencies of amino acids measured over the whole data set (JTT + Γ4 + I + F, 21 degrees of freedom). Note that these models were identified as the available ML models that best fit the sequence alignments, according to the AIC criterion [40].
Bayesian phylogenetic reconstructions were performed using PhyloBayes 3.0 [43]. For all Bayesian experiments performed in this study, two independent MCMC chains - each starting from a random point - were run for up to 100, 000 cycles. One MCMC sample was saved every 10 cycles, and the first 500 samples were discarded as "burnin". The eight free parameters of GTRnt and the amino-acid frequencies of JTT (19 free parameters), were estimated from the data.
We also applied more general and parameter rich models of evolution, implemented in a Bayesian framework. We used GTRaa, where subscript aa indicates a GTR model dedicated to amino-acid sequences. This model directly estimates the exchange rate parameters from the data (208 degrees of freedom). Models JTT and GTRaa homogeneously apply a single substitution model to the whole data set. However, in some cases, this parameterization is prone to violations by the data, resulting in wrong phylogenetic inferences [51]. Consequently, we applied the site-heterogeneous mixture model CAT to the amino-acid alignment, which implements a mixture of stationary probability vectors across sites [48]. The CAT model was combined with free (+GTRnt) or empirical (+JTT) relative exchange rates, applied to the nucleotide and amino-acid alignments, respectively. Both CAT + GTRnt and CAT + JTT models were combined with discretized gamma rates across sites (+ Γ4). Finally, we analyzed both nucleotide and amino-acid alignments under time-heterogeneous models of evolution. The BP model component allows for changes over time of the substitution model stationary probabilities and hence, estimates the compositional drift of the sequences [50]. We applied the GTRnt + BP + Γ4 [50] and the CAT + BP + Γ4 [51] models to the nucleotide and the amino-acid alignments, respectively.
Additional models with Bayesian implementations were compared using cross-validation (see below). In addition to the + Γ4 model of rate variation across sites, we applied a covarion model (+COV) which enabled us to estimate site specific rate variations (i.e. heterotachy) [71]. In addition to JTT and GTRaa, we considered the MtREV [72] empirical rate matrix. Finally, in addition to the mixture model CAT, we considered the empirical mixture models UL2 and UL3 [73]. These components allowed 13 and 3 models of evolution to be derived and applied to amino-acid and nucleotide alignments, respectively.
Cross-validation experiments
The fit of the models implemented in a Bayesian framework was estimated by cross-validation, as implemented in PhyloBayes 3.0 [43]. Ten replicate data sets were randomly drawn. The learning part of each replicate data set comprised 90% of the sites of the whole alignment. The 10% of remaining sites were used to compute the cross-validated likelihood. The tree topology was considered as a free parameter. Note that, for computational reasons, fits of time-heterogeneous models GTRnt + BP + Γ4 and CAT + BP + Γ4 were not evaluated.
Posterior predictive experiments
Model violations were measured by posterior predictive experiments, as implemented in PhyloBayes 3.0 [43]. We applied a test statistic measuring the compositional heterogeneity across taxa. The test statistic "composition" was defined as the maximum of the χ2 distances separating each sequence composition from the composition of the whole data set [52]. We applied two test statistics to measure site saturation. The test statistic "site diversity" measures the mean state diversity across sites [48] (e.g. a constant site has a diversity of 1). The test statistic "homoplasy" considers the averaged number of convergence and reversion events per site, as displayed by inferred stochastic mapping [49]. A posterior predictive test compares the value VO of a test statistic measured given the observed data, to the distribution of that test statistic measured over simulated replicate data sets. Each replicate data set was simulated given an a posteriori drawn sample of parameters. The p-value indicates the probability of observing a test statistic as extreme as VO, under the null hypothesis stating that the model assumptions are true. Failure to reject a posterior predictive test indicates that the model assumptions allow to realistically reproduce the observation VO based on real data.
Saturation plot
Saturation of the phylogenetic signal of each codon position was illustrated by a saturation plot [74]. For each pair of taxa in an alignment, we plotted their "pairwise similarity distance" (i.e. y-coordinates: number of sites displaying different states, normalized by the alignment length), versus the distance separating these two taxa along the tree branches (i.e. x-coordinates: the sum of branch lengths from the two taxa to their common ancestor). We used a fixed tree topology estimated from all codon positions (Figure 2). Branch lengths and other model parameters were evaluated according to separate codon positions.
Estimation of tree node supports
Under ML analysis, statistical support of tree branches was estimated from 1000 bootstrap replicates and, in addition, using the Shimodaira-Hasegawa-like test (SH) implemented in PhyML 3.0 [47,75]. Bayesian analysis classically provides a collection of samples drawn from the a posteriori distribution. The posterior probability of observing a given phylogenetic association between two lineages is then approximated by its frequency among sampled trees. Given a monophyletic target lineage A (e.g. great ape parasites), we extracted from a tree collection the list of all its N different sister-groups Bn (e.g. rodent parasites). We then computed the frequency 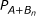 : the posterior support of clade A + Bn (e.g. great ape plus rodent parasites). The same approach was used for the bootstrap support, but not for the SH support which applies only to clades that belong to the ML tree.
: the posterior support of clade A + Bn (e.g. great ape plus rodent parasites). The same approach was used for the bootstrap support, but not for the SH support which applies only to clades that belong to the ML tree.
Taxon sampling
Let A and B each be a monophyletic lineage within a phylogenetic tree, and let them form a well-supported monophyletic clade A + B according to an initial sample of taxa. To ensure that the target relationship between A and B does not result from stochastic noise (lack of signal) or systematic error (model violation), we checked its robustness to taxon sampling (e.g. [49]). Given a lineage C (possibly equal to A or B) including k taxa, we checked that every combination of 1 to k - 1 taxa of C yielded a congruent phylogeny with respect to the phylogenetic relationship A + B. If lineage C was composed of too many taxa, we selected only a few relevant taxon combinations among all those available.
We focused on the robustness of the association of lineage A: great ape parasites, with lineage B: rodent parasites. Each of these two lineages was considered in turn as a sampled lineage C. According to our initial selection of 33 taxa, both these lineages were composed of 3 taxa and, 6 combinations of single or pairs of representatives were considered. We also considered in turn the group of primate parasites (14 taxa) and the saurian parasite outgroup (13 taxa) as sampled lineages C. For each of these sampled lineages, only 6 combinations of single or pairs of sub-groups were considered. For the 14 primate parasites, the 3 sub-groups were: (a) P. malariae and P. ovale (infecting humans), (b) P. gonderi and P. DAJ-2004 (African primate parasites), and (c) 10 Plasmodium species infecting Asian primates (Additional file 1, Table S1). For the 13 saurian parasites, the 3 sub-groups were (a) the 5 Plasmodium species, (b) the 4 Haemoproteus and Parahaemoproteus species, and (c) the 4 Leucocytozoon species (Additional file 1, Table S1). All these 24 sub data sets were obtained from the nucleotide alignment by simply discarding the relevant sequences without renewed aligning.
Finally, up to 51 malaria parasites were added to the initial selection of 33 taxa. We aligned all 84 taxa following the alignment procedure described above. Note that for CytB, 38 of the 51 additional sequences were partial CDS (see Additional file 8, Table S6). In order to retain more sites, complete and partial CytB genes were filtered separately with Gblocks, and then manually reassembled into an 84-gene alignment. All 6 additions of taxa to the 33-taxon data set were obtained from the whole concatenation of genes of the 84 taxa by discarding the relevant sequences.
Authors' contributions
SB designed and conducted all the experiments. OG provided guidance throughout the study. Both authors contributed to the writing of the paper.
Supplementary Material
Supplementary Table S1, Accession numbers of 33 mitochondrial genomes, species and host names. Accession numbers of 33 complete mitochondrial genomes of Haemosporidian parasites, parasite names, and host names retrieved from NCBI annotations (b: host names complemented from Leclerc et al. 2004 [11]). "P.": Plasmodium species, "Ha.": Haemoproteus species, "Pa.": Parahaemoproteus species, "L.": Leucocytozoon species.
Supplementary Figure S1, Saturation plot of codon positions. Saturation plot of codon positions of the 33 taxa and 3 concatenated genes data set, computed with a Bio++ script [80]. Each dot represents the comparison of the similarity distance (y coordinate) versus the tree distance (x coordinate), for a pair of taxa. Tree branch lengths were estimated under the GTRnt + Γ4 model (PhyloBayes 3.0, [43]), for 3 data sets corresponding to each codon position, and using the tree topology estimated from the whole nucleotide data set (Figure 2).
Supplementary Table S2, Analyses of the 33 taxa and 3 mitochondrial gene data sets. Dependency of clade support on codon positions ("Cod. pos."), amino-acid translation and on assumptions of various probabilistic models of substitution (GTRnt, GTRaa and JTT: single matrix model, CAT: site heterogeneous mixture model, BP: time heterogeneous model). Rates across site model components are defined as + Γ4 + I under maximum-likelihood (ML) and as + Γ4 under Bayesian (BI) methods. Cells display support as [PP, SH, BS], with PP: posterior probability (BI), SH: Shimodaira-Hasegawa-like support (ML), and BS: bootstrap support (ML). "*": not applicable. Main lineages of mammal parasites are defined according to their host preference: "Rodent", "Primate" and "Great Ape" (see Additional file 1, Table S1).
Supplementary Table S3, Fit of Bayesian models. Cross-validation estimations of the fit of Bayesian models to the 33 taxa and the three concatenated gene data sets. Models applied to the nucleotide and amino-acid data set are compared to the best fitting ML models, GTRnt + Γ4 and JTT + Γ4, respectively. Models are defined according to their components. Substitution model: GTRnt, GTRaa, MtREV and JTT, exchange rate parameters; CAT, UL2 and UL3, site heterogeneous mixture models. Rates across sites models: + Γ4, discretized gamma rates (Yang 1994, [42]); +COV, covarion model (Tuffley and Steel 1998, [71]).
Supplementary Table S4, p-values of posterior predictive tests performed on the 33 taxa and 3 mitochondrial gene data sets. Data sets were analyzed under various probabilistic models of substitution (GTRnt, GTRaa and JTT: single matrix model, CAT: site heterogeneous mixture model, BP: time heterogeneous model, + Γ4: Rates across site model component). Posterior predictive test "Composition" measures compositional heterogeneity across taxa, "Site Diversity" and "Homoplasy" measure the level of saturation of the phylogenetic signal. "Cod. pos.": codon positions. "*": not applicable.
Supplementary Figure S2, Phylogenetic tree of first and second codon positions analyzed under CAT + GTR + Γ4. Bayesian phylogenetic reconstruction using PhyloBayes 3.0 [43]. The CAT + GTRnt + Γ4 substitution model was applied to first and second codon positions of the 33 taxa data set. P. falciparum and 2 of its relatives infecting great ape hosts, P. reichenowi and P. gaboni, formed a monophyletic clade with 3 rodent parasites, P. yoelii, P. berghei and P. chabaudi (posterior probability PP = 0.92). Posterior probabilities equal to 1 were removed.
Supplementary Table S5, Robustness of the support to the removal and addition of taxa. All codon positions were analyzed under GTR + Γ4 and GTR + Γ4 + I models, for Bayesian and ML methods, respectively. Addition or removal of taxa to the complete nucleotide data set comprising 33 taxa and 3 concatenated genes, 3308 sites. Phylogenetic analyses were performed under models GTRnt + Γ4 + I and GTRnt + Γ4, for maximum-likelihood (ML) and Bayesian (BI) methods, respectively. Cells display support as [PP, SH, BS], with PP: posterior probability (BI), SH: Shimodaira-Hasegawa-like support (ML), and BS: bootstrap support (ML). "*": not applicable. (a): PP and BS are summed over various positions of Hepatocystis species. Main lineages of mammal parasites are defined according to their host preference: "Rodent", "Primate" and "Great Ape" (see Additional file 1 and 8, Tables S1 and S6). "-" removal of species. "+" addition of species. "P. fal.": P. falciparum; "P. rei.": P. reichenowi; "P. gab.": P. gaboni; "P. yoe.": P. yoelii; "P. ber.": P. berghei; "P. cha.": P. chabaudi; "Hum": human primate parasites P. malariae and P. ovale; "Afr.": African primate parasites P. gonderi and P. DAJ-2004; "Asi": 10 Asian primate parasites; "Pla.": Plasmodium species infecting saurian hosts; "Hae.": Haemoproteus and Parahaemoproteus species; "Leu.": Leucocytozoon species; "Haemo." Haemosporidian species.
Supplementary Table S6, Additional mitochondrial genes. Accession numbers of 51, 41 and 1 additional CytB, Cox1 and Cox3 genes, respectively, and parasite and host names (1 partial CytB genes). References: (a) Perkins and Schall (2002) [6]; (b) Perkins et al. (2007) [37]; (c) Cheesman et al. (2009) [76]; (d) Hall et al. (2005) [77]; (e) Escalante et al. (1998) [21]; (f) Seethamchai et al. (2008) [78]; (g) Martinsen et al. (2008) [36]; (h) Martinsen et al. (2007) [79]; (i) CytB + Cox1 + Cox3, Perkins (2008) [23]; (j) Rich et al. (2009) [26]. Abbreviation: "P.": Plasmodium species, "He.": Hepatocystis species.
Supplementary Table S7, Accession numbers of ClpC and ASL genes. Accession numbers of 27 ClpC and 18 ASL genes. (a): Taxa used for the estimation of the length of the 7-taxa tree.
Supplementary Figure S3, Phylogenetic tree of 18 ASL genes. Bayesian reconstruction under the GTRnt + Γ4 model. Edges with PP < 0.9 were collapsed, and PP = 1 are not shown. The ASL phylogeny is not congruent with a monophyly of mammal malaria parasites.
Supplementary Figure S4, Phylogenetic tree of 27 ClpC genes. Bayesian reconstruction under the GTRnt + Γ4 model. Edges with PP < 0.9 were collapsed, and PP = 1 are not shown. The ClpC phylogeny supports the monophyly of mammalian malaria parasites (PP = 0.99).
Supplementary Table S8, Robustness of clade support in trees designed to display long branches. Each of the 9 taxon samples comprised the three most distantly related Leucocytozoon species, the 14 primate parasites, and two single representatives of parasites of great apes and of rodents, respectively. In each case, 14 taxa were removed from the complete nucleotide data-set comprising 33 taxa and 3 concatenated genes, 3308 sites. Phylogenetic analyses were performed under models GTRnt + Γ4 + I and GTRnt + Γ4, for maximum likelihood (ML) and Bayesian (BI) methods, respectively. Cells display support as follows: [PP, SH, BS], with PP: posterior probability (BI), SH: Shimodaira-Hasegawa-like support ("*": not applicable, ML), and BS: bootstrap support (ML). Main lineages of mammal parasites are defined according to their host preference: "Rodent", "Primate" and "Great Ape" (see Additional file 1, Table S1). "P. fal.": P. falciparum; "P. rei.": P. reichenowi; "P. gab.": P. gaboni; "P. yoe.": P. yoelii; "P. ber.": P. berghei; "P. cha.": P. chabaudi.
Contributor Information
Samuel Blanquart, Email: samuel.blanquart@inria.fr.
Olivier Gascuel, Email: olivier.gascuel@lirmm.fr.
Acknowledgements
We wish to thank Emeric Sevin, Martin Taylor, Frédéric Delsuc and Nicolas Lartillot for helpful comments on this manuscript, Julien Dutheil and Vincent Ranwez for sharing code, Liliana Dávalos for sharing data, and François Renaud for helpful discussions about Haemosporidia evolution. We wish to thank the Associate Editor, Hervé Philippe, who requested the correction of sequence frameshifts during the processing of this manuscript. We thank Nick Goldman and the European Molecular Biology Laboratory (EMBL), as well as Nicolas Galtier and the European Research Council grant 232971 (ERC PopPhyl), who supported the continuation of this work during the post-doctoral positions they offered to SB. We thank the EMBL-European Bioinformatics Institute (EBI) for the computational facilities we used in this study. This work was supported by the French "Agence Nationale de la Recherche", ANR PlasmoExplore.
References
- Volkman SK, Barry AE, Lyons EJ, Nielsen KM, Thomas SM, Choi M, Thakore SS, Day KP, Wirth DF, Hartl DL. Recent origin of Plasmodium falciparum from a single progenitor. Science. 2001;293(5529):482–484. doi: 10.1126/science.1059878. [DOI] [PubMed] [Google Scholar]
- Foth BJ, McFadden GI. The apicoplast: a plastid in Plasmodium falciparum and other Apicomplexan parasites. Int Rev Cytol. 2003;224:57–110. doi: 10.1016/s0074-7696(05)24003-2. full_text. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Williamson DH, Wilson RJ. A circular DNA in malaria parasites encodes an RNA polymerase like that of prokaryotes and chloroplasts. Mol Biochem Parasitol. 1991;44:115–123. doi: 10.1016/0166-6851(91)90227-W. [DOI] [PubMed] [Google Scholar]
- Valkiunas G. Avian malaria parasites and other haemosporidia. New York: CRC Press, Boca Raton; 2005. [Google Scholar]
- Levine ND. The protozoan phylum Apicomplexa. Florida: CRC Press, Boca Raton; 1988. [Google Scholar]
- Perkins SL, Schall JJ. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J Parasitol. 2002;88(5):972–978. doi: 10.1645/0022-3395(2002)088[0972:AMPOMP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Manwell RD. The problem of species, with special reference to the malaria parasites. Ann Trop Med. 1936;30:435–439. [Google Scholar]
- Garnham PCC. Malaria parasites and other haemosporidia. Oxford: Blackwell Scientific; 1966. [Google Scholar]
- Perkins SL. Species concepts and malaria parasites: detecting a cryptic species of Plasmodium. Proc Biol Sci. 2000;267(1459):2345–2350. doi: 10.1098/rspb.2000.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinsen ES, Paperna I, Schall JJ. Morphological versus molecular identification of avian Haemosporidia: an exploration of three species concepts. Parasitology. 2006;133(3):279–288. doi: 10.1017/S0031182006000424. [DOI] [PubMed] [Google Scholar]
- Leclerc MC, Hugot JP, Durand P, Renaud F. Evolutionary relationships between 15 Plasmodium species from new and old world primates (including humans): an 18 S rDNA cladistic analysis. Parasitology. 2004;129(6):677–684. doi: 10.1017/S0031182004006146. [DOI] [PubMed] [Google Scholar]
- Singh B, Kim-Sung L, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, Thomas A, Conway DJ. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363(9414):1017–1024. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- Cox-Singh J, Singh B. Knowlesi malaria: newly emergent and of public health importance? Trends Parasitol. 2008;24(9):406–410. doi: 10.1016/j.pt.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garamszegi LZ. Patterns of co-speciation and host switching in primate malaria parasites. Malar J. 2009;8(110):1–15. doi: 10.1186/1475-2875-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Iyer LM, Wellems TE, Miller LH. Plasmodium biology: genomic gleanings. Cell. 2003;115(7):771–785. doi: 10.1016/S0092-8674(03)01023-7. [DOI] [PubMed] [Google Scholar]
- Waters AP, Higgins DG, McCutchan TF. Plasmodium falciparum appears to have arisen as a result of lateral transfer between avian and human hosts. Proc Natl Acad Sci USA. 1991;88(8):3140–3144. doi: 10.1073/pnas.88.8.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AP, Higgins DG, McCutchan TF. Evolutionary relatedness of some primate models of Plasmodium. Mol Biol Evol. 1993;10(4):914–923. doi: 10.1093/oxfordjournals.molbev.a040038. [DOI] [PubMed] [Google Scholar]
- Escalante AA, Ayala FJ. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proc Natl Acad Sci USA. 1994;91(24):11373–11377. doi: 10.1073/pnas.91.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Ayala FJ. Evolutionary origin of Plasmodium and other Apicomplexa based on rRNA genes. Proc Natl Acad Sci USA. 1995;92(13):5793–5797. doi: 10.1073/pnas.92.13.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qari SH, Shi YP, Pieniazek NJ, Collins WE, Lal AA. Phylogenetic relationship among the malaria parasites based on small subunit rRNA gene sequences: monophyletic nature of the human malaria parasite, Plasmodium falciparum. Mol Phylogenet Evol. 1996;6:157–165. doi: 10.1006/mpev.1996.0068. [DOI] [PubMed] [Google Scholar]
- Escalante AA, Freeland DE, Collins WE, Lal AA. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc Natl Acad Sci USA. 1998;95(14):8124–8129. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T, Culleton R, Otani H, Horii T, Tanabe K. Big bang in the evolution of extant malaria parasites. Mol Biol Evol. 2008;25(10):2233–2239. doi: 10.1093/molbev/msn171. [DOI] [PubMed] [Google Scholar]
- Perkins SL. Molecular systematics of the three mitochondrial protein-coding genes of malaria parasites: Corroborative and new evidence for the origins of human malaria. Mitochondrial DNA. 2008;19(6):471–478. doi: 10.1080/19401730802570926. [DOI] [PubMed] [Google Scholar]
- Roy S, Irimia M. Origins of human malaria: rare genomic changes and full mitochondrial genomes confirm the relationship of Plasmodium falciparum to other mammalian parasites but complicate the origins of Plasmodium vivax. Mol Biol Evol. 2008;25(6):1192–1198. doi: 10.1093/molbev/msn069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollomo B, Durand P, Prugnolle F, Douzery EJP, Arnathau C, Nkoghe D, Leroy E, Renaud F. A new malaria agent in African hominids. PLoS Pathog. 2009;5(5):e1000446. doi: 10.1371/journal.ppat.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich SM, Leendertz FH, Xu G, Lebreton M, Djoko CF, Aminake MN, Takang EE, Diffo JL, Pike BL, Rosenthal BM, Formenty P, Boesch C, Ayala FJ, Wolfe ND. The origin of malignant malaria. Proc Natl Acad Sci USA. 2009;106(35):14902–14907. doi: 10.1073/pnas.0907740106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prugnolle F, Durand P, Neel C, Ollomo B, Ayala FJ, Arnathau C, Etienne L, Mpoudi-Ngole E, Nkoghe D, Leroy E, Delaporte E, Peeters M, Renaud F. African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. Proc Natl Acad Sci USA. 2010;107(4):1458–1463. doi: 10.1073/pnas.0914440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krief S, Escalante AA, Pacheco MA, Mugisha L, André C, Halbwax M, Fischer A, Krief JM, Kasenene JM, Crandfield M, Cornejo OE, Chavatte JM, Lin C, Letourneur F, Grüner AC, McCutchan TF, Rénia L, Snounou G. On the diversity of malaria parasites in African apes and the origin of Plasmodium falciparum from Bonobos. PLoS Pathog. 2010;6(2):e1000765. doi: 10.1371/journal.ppat.1000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutchan TF, Kissinger JC, Touray MG, Rogers MJ, Li J, Sullivan M, Braga EM, Krettli AU, Miller LH. Comparison of circumsporozoite proteins from avian and mammalian malarias: biological and phylogenetic implications. Proc Natl Acad Sci USA. 1996;93(21):11889–11894. doi: 10.1073/pnas.93.21.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Goldman IF, De-Rijk P, De-Wachter R, Collins WE, Qari SH, Lal AA. Phylogenetic study of the genus Plasmodium based on the secondary structure-based alignment of the small subunit ribosomal RNA. Mol Biochem Parasitol. 1997;90:317–321. doi: 10.1016/S0166-6851(97)00121-7. [DOI] [PubMed] [Google Scholar]
- Rathore D, Wahl AM, Sullivan M, McCutchan TF. A phylogenetic comparison of gene trees constructed from plastid, mitochondrial and genomic DNA of Plasmodium species. Mol Biochem Parasitol. 2001;114:89–94. doi: 10.1016/S0166-6851(01)00241-9. [DOI] [PubMed] [Google Scholar]
- Kedzierski L, Escalante AA, Isea R, Black CG, Barnwell JW, Coppel RL. Phylogenetic analysis of the genus Plasmodium based on the gene encoding adenylosuccinate lyase. Infect Genet Evol. 2002;1(4):297–301. doi: 10.1016/S1567-1348(02)00031-X. [DOI] [PubMed] [Google Scholar]
- Kissinger JC, Souza PC, Soares CO, Paul R, Wahl AM, Rathore D, McCutchan TF, Krettli AU. Molecular phylogenetic analysis of the avian malarial parasite Plasmodium Novyella juxtanucleare. J Parasitol. 2002;88(4):769–773. doi: 10.1645/0022-3395(2002)088[0769:MPAOTA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Vargas-Serrato E, Corredor V, Galinski MR. Phylogenetic analysis of CSP and MSP-9 gene sequences demonstrates the close relationship of Plasmodium coatneyi and Plasmodium knowlesi. Infect Genet Evol. 2003;3:67–73. doi: 10.1016/S1567-1348(03)00007-8. [DOI] [PubMed] [Google Scholar]
- Hagner SC, Misof B, Maier WA, Kampen H. Bayesian analysis of new and old malaria parasite DNA sequence data demonstrates the need for more phylogenetic signal to clarify the descent of Plasmodium falciparum. Parasitol Res. 2007;101(3):493–503. doi: 10.1007/s00436-007-0499-6. [DOI] [PubMed] [Google Scholar]
- Martinsen ES, Perkins SL, Schall JJ. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): Evolution of life-history traits and host switches. Mol Phylogenet Evol. 2008;47:261–273. doi: 10.1016/j.ympev.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Perkins SL, Sarkar IN, Carter R. The phylogeny of rodent malaria parasites: simultaneous analysis across three genomes. Infect Genet Evol. 2007;7:74–83. doi: 10.1016/j.meegid.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP* 4.0 beta 5: Phylogenetic Analysis Using Parsimony and Other Methods. Sinauer Associates Sunderland, MA; 2001. [Google Scholar]
- MrAIC v. 1.4.4. http://www.abc.se/~nylander/
- Akaike H. In: Proceedings 2nd International Symposium on Information Theory. Petrov C, editor. Budapest, Akademia Kiado; 1973. Information theory and an extension of the maximum likelihood principle; pp. 267–281. [Google Scholar]
- Lanave C, Preparata G, Saccone C, Serio G. A new method for calculating evolutionary substitution rates. J Mol Evol. 1984;20:86–93. doi: 10.1007/BF02101990. [DOI] [PubMed] [Google Scholar]
- Yang Z. Maximum-likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 1994;39(3):306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. PhyloBayes 3. A Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25(17):2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- Abascal F, Zardoya R, Posada D. ProtTest: Selection of bestfit models of protein evolution. Bioinformatics. 2005;21(9):2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thomton JM. The rapid generation of mutation data matrices from protein sequences. Computational Applied Bioscience. 1992;8(3):275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Douady CJ, Delsuc F, Boucher Y, Doolittle WF, Douzery EJP. Comparison of Bayesian and maximum likelihood bootstrap measures of phylogenetic reliability. Mol Biol Evol. 2003;20(2):248–254. doi: 10.1093/molbev/msg042. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate Maximum-Likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21(6):1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, Brinkmann H, Roure B, Lartillot N, Lang BF, Philippe H. Detecting and overcoming systematic errors in genome-scale phylogenies. Syst Biol. 2007;56(3):389–399. doi: 10.1080/10635150701397643. [DOI] [PubMed] [Google Scholar]
- Blanquart S, Lartillot N. A Bayesian compound stochastic process for modeling nonstationary and nonhomogeneous sequence evolution. Mol Biol Evol. 2006;23(11):2058–2071. doi: 10.1093/molbev/msl091. [DOI] [PubMed] [Google Scholar]
- Blanquart S, Lartillot N. A site- and time-heterogeneous model of amino-acid replacement. Mol Biol Evol. 2008;25(5):842–858. doi: 10.1093/molbev/msn018. [DOI] [PubMed] [Google Scholar]
- Foster PG. Modeling Compositional Heterogeneity. Systematic Biology. 2004;53(3):485–495. doi: 10.1080/10635150490445779. [DOI] [PubMed] [Google Scholar]
- Dávalos LM, Perkins SL. Saturation and base composition bias explain phylogenomic conflict in Plasmodium. Genomics. 2008;91(5):433–442. doi: 10.1016/j.ygeno.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Heath TA, Hedtke SM, Hillis DM. Taxon sampling and the accuracy of phylogenetic analyses. J Syst Evol. 2008;46(3):239–257. [Google Scholar]
- Anisimova M, Bielawski JP, Yang Z. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol. 2001;18(8):1585–1592. doi: 10.1093/oxfordjournals.molbev.a003945. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Cases in which parsimony or compatibility method will be positively misleading. Syst Zool. 1978;27(4):401–410. doi: 10.2307/2412923. [DOI] [Google Scholar]
- Hughes AL, Verra F. Malaria Parasite Sequences from Chimpanzee Support the Co-Speciation Hypothesis for the Origin of Virulent Human Malaria (Plasmodium falciparum) Mol Phylogenet Evol. 2010;57:135–143. doi: 10.1016/j.ympev.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6(5):361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- Vaidya AB, Akella R, Suplick K. Sequences similar to genes for two mitochondrial proteins and portions of ribosomal RNA in tandemly arrayed 6-kilobase-pair DNA of a malarial parasite. Mol Biochem Parasitol. 1989;35(2):97–107. doi: 10.1016/0166-6851(89)90112-6. [DOI] [PubMed] [Google Scholar]
- Conway DJ, Fanello C, Lloyd JM, Al-Joubori BM, Baloch AH, Somanath SD, Roper C, Oduola AM, Mulder B, Povoa MM, Singh B, Thomas AW. Origin of Plasmodium falciparum malaria is traced by mitochondrial DNA. Mol Biochem Parasitol. 2000;111:163–171. doi: 10.1016/S0166-6851(00)00313-3. [DOI] [PubMed] [Google Scholar]
- Joy DA, Feng X, Mu J, Furuya T, Chotivanich K, Krettli AU, Ho M, Wang A, White NJ, Suh E, Beerli P, Su XZ. Early origin and recent expansion of Plasmodium falciparum. Science. 2003;300(5617):318–321. doi: 10.1126/science.1081449. [DOI] [PubMed] [Google Scholar]
- Beadell JS, Fleischer R. A restriction enzyme-based assay to distinguish between avian Haemosporidians. J Parasitol. 2005;91(3):683–685. doi: 10.1645/GE-3412RN. [DOI] [PubMed] [Google Scholar]
- Jongwutiwes S, Putaporntip C, Iwasaki T, Ferreira MU, Kanbara H, Hughes AL. Mitochondrial genome sequences support ancient population expansion in Plasmodium vivax. Mol Biol Evol. 2005;22(8):1733–1739. doi: 10.1093/molbev/msi168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Joy DA, Duan J, Huang Y, Carlton J, Walker J, Barnwell J, Beerli P, Charleston MA, Pybus OG, Su XZ. Host switch leads to emergence of Plasmodium vivax malaria in humans. Mol Biol Evol. 2005;22(8):1686–1693. doi: 10.1093/molbev/msi160. [DOI] [PubMed] [Google Scholar]
- Omori S, Sato Y, Isobe T, Yukawa M, Murata K. Complete nucleotide sequences of the mitochondrial genomes of two avian malaria protozoa, Plasmodium gallinaceum and Plasmodium juxtanucleare. Parasitol Res. 2007;100(3):661–664. doi: 10.1007/s00436-006-0333-6. [DOI] [PubMed] [Google Scholar]
- Omori S, Sato Y, Hirakawa S, Isobe T, Yukawa M, Murata K. Two extra chromosomal genomes of Leucocytozoon caulleryi; complete nucleotide sequences of the mitochondrial genome and existence of the apicoplast genome. Parasitol Res. 2008;103(4):953–957. doi: 10.1007/s00436-008-1083-4. [DOI] [PubMed] [Google Scholar]
- Feagin JE. The extrachromosomal DNAs of apicomplexan parasites. Annu Rev Microbiol. 1994;48:81–104. doi: 10.1146/annurev.mi.48.100194.000501. [DOI] [PubMed] [Google Scholar]
- Wilson RJ, Williamson DH. Extrachromosomal DNA in the Apicomplexa. Microbiol Mol Biol Rev. 1997;61:1–16. doi: 10.1128/mmbr.61.1.1-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldritt SM, Joseph JT, Wirth DF. Sequence identification of cytochrome b in Plasmodium gallinaceum. Mol Cell Biol. 1989;9(9):3614–3620. doi: 10.1128/mcb.9.9.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Tuffley C, Steel M. Modeling the covarion hypothesis of nucleotide substitution. Mathematical Biosciences. 1998;147:63–91. doi: 10.1016/S0025-5564(97)00081-3. [DOI] [PubMed] [Google Scholar]
- Adachi J, Hasegawa M. Model of amino acid substitution in proteins encoded by mitochondrial DNA. J Mol Evol. 1996;42(4):459–468. doi: 10.1007/BF02498640. [DOI] [PubMed] [Google Scholar]
- Le SQ, Lartillot N, Gascuel O. Phylogenetic mixture models for proteins. Philos Trans R Soc Lond B Biol Sci. 2008;363(1512):3965–76. doi: 10.1098/rstb.2008.0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffroy O, Brinkmann H, Delsuc F, Philippe H. Phylogenomics: the beginning of incongruence? Trends Genet. 2006;22(4):225–231. doi: 10.1016/j.tig.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55(4):539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Cheesman S, Tanabe K, Sawai H, O'Mahony E, Carter R. Strain-specific immunity may drive adaptive polymorphism in the merozoite surface protein 1 of the rodent malaria parasite Plasmodium chabaudi. Infect Genet Evol. 2009;9(2):248–255. doi: 10.1016/j.meegid.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Hall N, Karras M, Raine JD, Carlton JM, Kooij TW, Berriman M, Florens L, Janssen CS, Pain A, Christophides GK, James K, Rutherford K, Harris B, Harris D, Churcher C, Quail MA, Ormond D, Doggett J, Trueman HE, Mendoza J, Bidwell SL, Rajandream MA, Carucci DJ, Yates JR, Kafatos FC, Janse CJ, Barrell B, Turner CM, Waters AP, Sinden RE. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307(5706):82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- Seethamchai S, Putaporntip C, Malaivijitnond S, Cui L, Jongwutiwes S. Malaria and Hepatocystis species in wild macaques, southern Thailand. Am J Trop Med Hyg. 2008;78(4):646–653. [PubMed] [Google Scholar]
- Martinsen ES, Waite JL, Schall JJ. Morphologically defined subgenera of Plasmodium from avian hosts: test of monophyly by phylogenetic analysis of two mitochondrial genes. Parasitology. 2007;134(4):483–490. doi: 10.1017/S0031182006001922. [DOI] [PubMed] [Google Scholar]
- Dutheil J, Gaillard S, Bazin E, Glffémin S, Ranwez V, Galtier N, Belkhir K. Bio++: a set of C++ libraries for sequence analysis, phylogenetics, molecular evolution and population genetics. BMC Bioinformatics. 2006;7(188):1–6. doi: 10.1186/1471-2105-7-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1, Accession numbers of 33 mitochondrial genomes, species and host names. Accession numbers of 33 complete mitochondrial genomes of Haemosporidian parasites, parasite names, and host names retrieved from NCBI annotations (b: host names complemented from Leclerc et al. 2004 [11]). "P.": Plasmodium species, "Ha.": Haemoproteus species, "Pa.": Parahaemoproteus species, "L.": Leucocytozoon species.
Supplementary Figure S1, Saturation plot of codon positions. Saturation plot of codon positions of the 33 taxa and 3 concatenated genes data set, computed with a Bio++ script [80]. Each dot represents the comparison of the similarity distance (y coordinate) versus the tree distance (x coordinate), for a pair of taxa. Tree branch lengths were estimated under the GTRnt + Γ4 model (PhyloBayes 3.0, [43]), for 3 data sets corresponding to each codon position, and using the tree topology estimated from the whole nucleotide data set (Figure 2).
Supplementary Table S2, Analyses of the 33 taxa and 3 mitochondrial gene data sets. Dependency of clade support on codon positions ("Cod. pos."), amino-acid translation and on assumptions of various probabilistic models of substitution (GTRnt, GTRaa and JTT: single matrix model, CAT: site heterogeneous mixture model, BP: time heterogeneous model). Rates across site model components are defined as + Γ4 + I under maximum-likelihood (ML) and as + Γ4 under Bayesian (BI) methods. Cells display support as [PP, SH, BS], with PP: posterior probability (BI), SH: Shimodaira-Hasegawa-like support (ML), and BS: bootstrap support (ML). "*": not applicable. Main lineages of mammal parasites are defined according to their host preference: "Rodent", "Primate" and "Great Ape" (see Additional file 1, Table S1).
Supplementary Table S3, Fit of Bayesian models. Cross-validation estimations of the fit of Bayesian models to the 33 taxa and the three concatenated gene data sets. Models applied to the nucleotide and amino-acid data set are compared to the best fitting ML models, GTRnt + Γ4 and JTT + Γ4, respectively. Models are defined according to their components. Substitution model: GTRnt, GTRaa, MtREV and JTT, exchange rate parameters; CAT, UL2 and UL3, site heterogeneous mixture models. Rates across sites models: + Γ4, discretized gamma rates (Yang 1994, [42]); +COV, covarion model (Tuffley and Steel 1998, [71]).
Supplementary Table S4, p-values of posterior predictive tests performed on the 33 taxa and 3 mitochondrial gene data sets. Data sets were analyzed under various probabilistic models of substitution (GTRnt, GTRaa and JTT: single matrix model, CAT: site heterogeneous mixture model, BP: time heterogeneous model, + Γ4: Rates across site model component). Posterior predictive test "Composition" measures compositional heterogeneity across taxa, "Site Diversity" and "Homoplasy" measure the level of saturation of the phylogenetic signal. "Cod. pos.": codon positions. "*": not applicable.
Supplementary Figure S2, Phylogenetic tree of first and second codon positions analyzed under CAT + GTR + Γ4. Bayesian phylogenetic reconstruction using PhyloBayes 3.0 [43]. The CAT + GTRnt + Γ4 substitution model was applied to first and second codon positions of the 33 taxa data set. P. falciparum and 2 of its relatives infecting great ape hosts, P. reichenowi and P. gaboni, formed a monophyletic clade with 3 rodent parasites, P. yoelii, P. berghei and P. chabaudi (posterior probability PP = 0.92). Posterior probabilities equal to 1 were removed.
Supplementary Table S5, Robustness of the support to the removal and addition of taxa. All codon positions were analyzed under GTR + Γ4 and GTR + Γ4 + I models, for Bayesian and ML methods, respectively. Addition or removal of taxa to the complete nucleotide data set comprising 33 taxa and 3 concatenated genes, 3308 sites. Phylogenetic analyses were performed under models GTRnt + Γ4 + I and GTRnt + Γ4, for maximum-likelihood (ML) and Bayesian (BI) methods, respectively. Cells display support as [PP, SH, BS], with PP: posterior probability (BI), SH: Shimodaira-Hasegawa-like support (ML), and BS: bootstrap support (ML). "*": not applicable. (a): PP and BS are summed over various positions of Hepatocystis species. Main lineages of mammal parasites are defined according to their host preference: "Rodent", "Primate" and "Great Ape" (see Additional file 1 and 8, Tables S1 and S6). "-" removal of species. "+" addition of species. "P. fal.": P. falciparum; "P. rei.": P. reichenowi; "P. gab.": P. gaboni; "P. yoe.": P. yoelii; "P. ber.": P. berghei; "P. cha.": P. chabaudi; "Hum": human primate parasites P. malariae and P. ovale; "Afr.": African primate parasites P. gonderi and P. DAJ-2004; "Asi": 10 Asian primate parasites; "Pla.": Plasmodium species infecting saurian hosts; "Hae.": Haemoproteus and Parahaemoproteus species; "Leu.": Leucocytozoon species; "Haemo." Haemosporidian species.
Supplementary Table S6, Additional mitochondrial genes. Accession numbers of 51, 41 and 1 additional CytB, Cox1 and Cox3 genes, respectively, and parasite and host names (1 partial CytB genes). References: (a) Perkins and Schall (2002) [6]; (b) Perkins et al. (2007) [37]; (c) Cheesman et al. (2009) [76]; (d) Hall et al. (2005) [77]; (e) Escalante et al. (1998) [21]; (f) Seethamchai et al. (2008) [78]; (g) Martinsen et al. (2008) [36]; (h) Martinsen et al. (2007) [79]; (i) CytB + Cox1 + Cox3, Perkins (2008) [23]; (j) Rich et al. (2009) [26]. Abbreviation: "P.": Plasmodium species, "He.": Hepatocystis species.
Supplementary Table S7, Accession numbers of ClpC and ASL genes. Accession numbers of 27 ClpC and 18 ASL genes. (a): Taxa used for the estimation of the length of the 7-taxa tree.
Supplementary Figure S3, Phylogenetic tree of 18 ASL genes. Bayesian reconstruction under the GTRnt + Γ4 model. Edges with PP < 0.9 were collapsed, and PP = 1 are not shown. The ASL phylogeny is not congruent with a monophyly of mammal malaria parasites.
Supplementary Figure S4, Phylogenetic tree of 27 ClpC genes. Bayesian reconstruction under the GTRnt + Γ4 model. Edges with PP < 0.9 were collapsed, and PP = 1 are not shown. The ClpC phylogeny supports the monophyly of mammalian malaria parasites (PP = 0.99).
Supplementary Table S8, Robustness of clade support in trees designed to display long branches. Each of the 9 taxon samples comprised the three most distantly related Leucocytozoon species, the 14 primate parasites, and two single representatives of parasites of great apes and of rodents, respectively. In each case, 14 taxa were removed from the complete nucleotide data-set comprising 33 taxa and 3 concatenated genes, 3308 sites. Phylogenetic analyses were performed under models GTRnt + Γ4 + I and GTRnt + Γ4, for maximum likelihood (ML) and Bayesian (BI) methods, respectively. Cells display support as follows: [PP, SH, BS], with PP: posterior probability (BI), SH: Shimodaira-Hasegawa-like support ("*": not applicable, ML), and BS: bootstrap support (ML). Main lineages of mammal parasites are defined according to their host preference: "Rodent", "Primate" and "Great Ape" (see Additional file 1, Table S1). "P. fal.": P. falciparum; "P. rei.": P. reichenowi; "P. gab.": P. gaboni; "P. yoe.": P. yoelii; "P. ber.": P. berghei; "P. cha.": P. chabaudi.